Abstract
Semiconductor nanoparticles particularly quantum dots (QDs) are interesting alternatives to organic fluorophores for a range of applications such as biosensing, imaging and therapeutics. Addition of a programmable scaffold such as DNA to QDs further expands the scope and applicability of these hybrid nanomaterials in biology. In this review, the most important stages of preparation of QD–DNA conjugates for specific applications in biology are discussed. Special emphasis is laid on (i) the most successful strategies to disperse QDs in aqueous media, (ii) the range of different conjugation with detailed discussion about specific merits and demerits in each case, and (iii) typical applications of these conjugates in the context of biology.
Keywords: conjugation, purification, biosensing and therapeutics
1. Introduction
Inherent material properties of semiconductor nanoparticles (NPs) or quantum dots (QDs) lend in tremendous potential for applications in biology [1–4]. Properties such as size tunable emission, high fluorescent quantum yield and resistance to photobleaching make QDs highly desirable candidates for several biosensing and tracking applications. Additionally, high surface to volume ratio, multiple chemically reactive ligands on the surface and high biomolecule loading capacity have augmented the use of QDs in targeted delivery and therapeutics. For most of these specific applications, QDs are first conjugated with biomolecules such as nucleic acids, proteins or signalling molecules. The last couple of decades have seen a lot of development in the routes of functionalization of QDs with these biomolecules. In this review, different approaches to transfer QD from organic media to aqueous media and subsequent biofunctionalization with nucleic acids are discussed. Quantum dot–DNA conjugates bring together the material and biological properties of both QD and DNA. As discussed in subsequent sections, these conjugates can be tailored for multiple physical, chemical and biological applications.
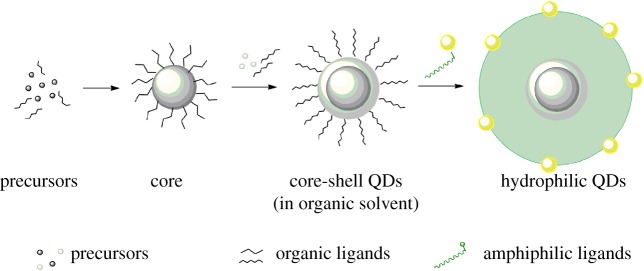
QDs are semiconductor nanomaterials synthesized by systematic formation of a core and a capping shell. The major photophysical and optical properties of QDs arise from the core and shell. The shell additionally assists in passivation of the surface defects of the core, enabling to thus preserve the properties of the core and prevent environmental damage [5]. Organic ligands are bound to the shell of QDs, which need to be systematically over-coated by amphiphilic moieties or displaced by hydrophilic ligands in order to make the nanocrystals water soluble. A step-wise scheme to generate QDs dispersed in water is shown in figure 1. Synthesis of core/shell type QDs can be also carried out directly in water, albeit with limited control. For a comprehensive overview on mechanistic details of QD synthesis, readers are referred to the review by Rodriguez et al. [6].
Figure 1.
Steps to synthesize quantum dots for biological applications. A schematic for synthesis of quantum dots from precursors to core/shell structures to dispersion in water is shown. QDs dispersed in water are ideal candidates for bioconjugation and eventual biological applications. (Online version in colour.)
In the next section, different strategies to make QDs water soluble are described in detail.
2. Methods to disperse quantum dots in aqueous media
Several methods commonly employed to transition QDs from organic to aqueous media are discussed. These methods are chosen such that the nanoparticle diameters are less than 30 nm, which is desirable for a range of biological applications including targeting and therapeutic delivery.
2.1. Encapsulation
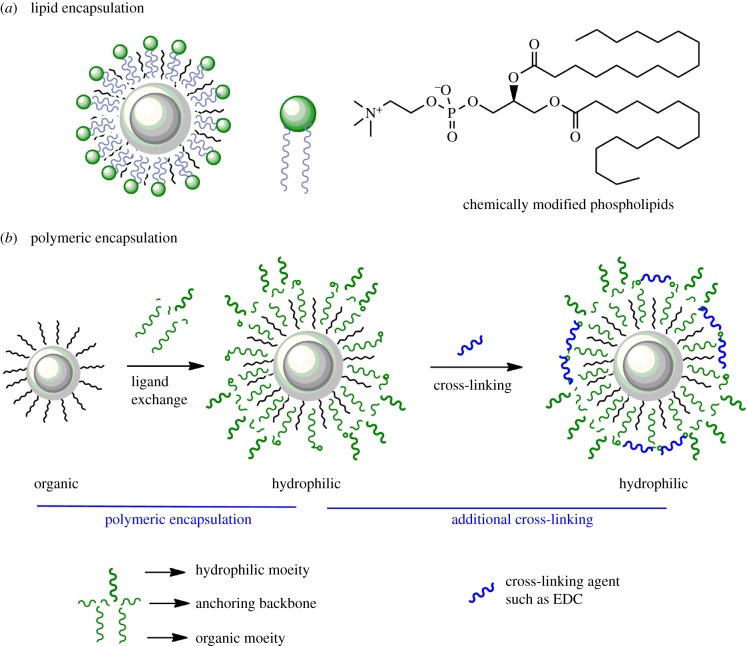
One of the fundamental methods to solubilize QDs in aqueous media involves encapsulation with phospholipids [7,8]. Pioneered by Dubertret et al., this strategy involves over-coating the existing organic ligands with amphiphilic phospholipids (figure 2a). The aliphatic chains of phospholipid can organize within the organic ligands on the surface of QDs by virtue of the hydrophobic interactions. The composition of the phospholipid formulations can be tailored for specific applications to yield fairly monodisperse QDs. In this study, these formulations remained stable for up to a month at a broad range of salt conditions. Though upon inception it was one of the most radical approaches to transfer QDs in aqueous solutions, the strategy has some inherent disadvantages. As the strategy is based on over-coating the existing ligands, the inherent size of dispersed particles is large. Additionally, the phospholipids can easily desorb from the surface of QDs (high koff) in a concentration-dependent manner. Thus, these formulations lose stability at higher dilutions. Phospholipids-coated QDs can also be transfected into cells, which undergo rapid aggregation and reduced cytoplasmic mobility [9,10]. In order to overcome several of these issues, one of the first reports by Wu et al. demonstrated that octylamine-modified poly-acrylic acid based polymer can be used to coat QDs, and further used to selectively detect multiple biological ligands in cells [11]. Another strategy of encapsulation by Pellegrino et al. used polymeric ligands based on poly(maleic anhydride alt-1-tetradecene) [12]. Herein, the QDs were first coated with these oligomeric ligands which are then cross-linked on the surface of QDs (by specific linker molecules) followed by hydrolyzation of the unreacted anhydride groups (figure 2b). This step makes the polymeric shell amphiphilic. This strategy compactly encages the QDs with average diameter between 11 and 13 nm. In this work, however, no bioconjugation or targeting was shown. In another work soon after, Kairdolf et al. demonstrated that QDs encapsulated with poly(acrylic acid)-octylamine polymer can be cross-linked and hydroxylated by reaction with 1,3-diamino-2-propanol (DAP) in the presence of EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride) and NHS (N-hydroxysuccinimide) [13]. This strategy further neutralized the surface charge on QDs, maintained small hydrodynamic diameter (Dh, 13–14 nm) and conserved high quantum yield (QY) up to 60–65%. Additionally, due to near neutral surface potential, the QDs displayed minimal non-specific adsorption on HeLa cells. Encapsulation via the polymeric shell in general has been acknowledged as a fairly robust method to generate QDs with high QY and preserved solution stability [7,14–16]. Similarly QDs encapsulated with PEG-based polymers are the current state of the art for several commercial preparations from manufactures such as ThermoFisher Scientific (formerly Invitrogen).
Figure 2.
Two approaches for encapsulation of quantum dots in hydrophilic shells. Quantum dots can be dispersed in water by encapsulation within (a) layer of amphiphilic phospholipids, (b) hydrophobic polymers that are subsequently made hydrophilic by derivatization with specific functional groups. (Online version in colour.)
2.2. Ligand exchange
In order to disperse QDs in aqueous solutions, systematic change of surface ligands is carried out. Type of exchanging ligands determines overall properties of QDs including hydrodynamic radius, fluorescent quantum yield and colloidal stability. A typical procedure to exchange ligands on the surface of QDs involves precipitation of the nanocrystals out of the organic medium and then re-dispersion in the presence of excess of the amphiphilic ligands. The resuspended solution is usually kept at high temperature (60–90°C) for 5–18 hours to facilitate exchange and enable sufficient surface coverage. After ligand exchange, the QDs display polar functional groups that impart solubility in aqueous media. These ligands are bound to the QD surface by non-covalent bonds. Several designs of these ligands have been tuned to increase bond strength, stability and solution properties of QDs. Similarly the procedure for ligand exchange may additionally influence the polydispersity of the water-solubilized QDs [17,18].
Typical ligands employed for phase transfer of nanocrystals are based on the affinity of specific functional groups for the QD surface. The chemical groups, which have been shown to have high affinity for the QD surfaces include thiols (discussed below), carboxylic acids [19], phosphonic acids [20] and amines. For the scope of this review, selected thiol-based ligands are discussed in detail. The ligands that are commonly used to coat the surface of QDs can be broadly classified into two groups—small molecule ligands and higher molecular weight polymeric ligands.
2.2.1. Small molecular weight ligands
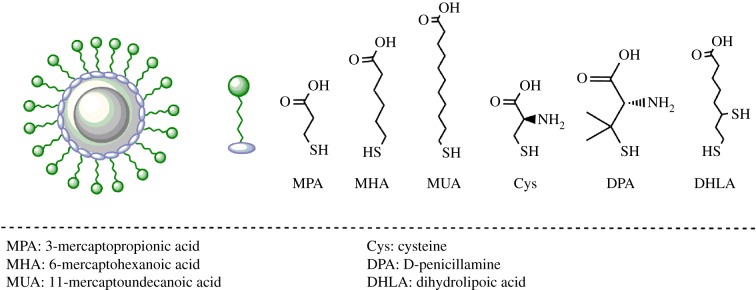
These ligands are generally derivatives of mercaptoacetic acids or mercaptoethyl amines (figure 3). First demonstrated by Chan & Nie close to a couple of decades back, the most popular mercapto alkyl acid based ligand is mercaptopropionic acid (MPA) [2,17]. The others include mercaptohexanoic acid (MHA) [21], mercaptoundecanoic acid (MUA) [22], etc. These ligands have a thiol functional group (–SH) on one end and carboxylic acid (–COOH) on other end of the molecule. The carboxylic acid group not only imparts water solubility, but can also be used for conjugation of biomolecules. Initially demonstrated by Liu et al., thioalkyl amine based ligands including cysteamine [4,23] and thiol-containing amino acids such as cysteine [23] and D-penicillamine [24,25] have also been used for ligand exchange. These ligands bind to the QD surface via the thiol functionality and terminate in a hydrophilic amine group, which can be further used for conjugation. It has also been observed in the literature that thioalkyl amine derivatives often result in less stable dispersions of QDs in hydrophilic media; and thus are used in moderation [23,26].
Figure 3.
Small molecule ligands with affinity towards the surface of quantum dots. Several small molecular weight thiolated ligands have been used to impart hydrophilicity. Thiols on these ligands bind to the surface of quantum dots and the terminal functional groups (carboxylic acid or amine) impart water solubility. (Online version in colour.)
Monothiol-based ligands are widely used for ligand exchange of QDs. However, they also suffer from inherent disadvantages. The anchoring of monothiol ligands to the QD surface is mitigated by a single non-covalent bond, which is prone to oxidation. This causes reduced stability of QD in aqueous solutions. Owing to desorption, surface coverage of these passivating ligands on the QD are reduced, that results in aggregation of the dots. Also, protonation of thiolate (at lower pH) or by UV irradiation causes leaching of the ligands from the QD surface. In order to circumvent these issues of instability, dithiol-bearing ligands are being increasingly used. Pioneered by the groups of Mattoussi & Bawendi, the most popular dithiol containing ligand for capping the surface of QDs is dihydrolipoic acid (DHLA) [27–33]. Dihydrolipoic acid is a small molecule ligand obtained by the reduction of α-lipoic acid. The binding and long-term stability of DHLA-coated QDs is higher than monothiol ligands. Owing to high negative charge density at physiological pH, DHLA-coated QDs often show high non-specific adsorption of proteins that in turn affect their biological applicability. In order to specifically address this issue, variants of DHLA have been developed. These variants often include a passivating layer of polyethylene glycol (PEG) along with the dithiol [34,35]. Another variant of DHLA-based small molecule ligand includes zwitterions instead of PEG that impart hydrophilicity and antifouling properties to the QDs [32].
2.2.2. Polymeric ligands
Using the design principles from small molecule ligands, several polymeric ligands have been synthesized. These polymeric ligands comprise several monomers with affinity for QD surface, several charged or hydrophilic groups that impart water solubility and some reactive groups that can be used for bioconjugation. Polymers can thus be considered a unified scaffold that comprises several desirable modalities. The synthesis is often carried out by copolymerization of stoichiometrically predefined monomers. Another alternative includes synthesis of a polymer backbone and then conjugation of water solubilizing and functionalizing entities on this scaffold [36]. After the synthesis of polymers, these preparations are added either directly to the QDs (in the case of ligands based on PEG, imidazole, etc.) or to QDs pre-exchanged with MPA or other small ligands (particularly for thiol-based ligands including DHLA).
Based on the nature of synthesis of QDs, types of ligands and nature of reducing agents, variable protocols are optimized [37]. For example, for disulfide containing ligands, prior reduction of the ligands assists in generation of thiol and favours dynamic interactions with the QD surface [38,39]. In the case of some imidazole- or pyridine-based polymers, this reduction step is not required [40,41]. There have been multiple designs of such polymers with various compositions of surface anchoring and water solubilizing monomers. Interested readers are referred to a review by Sperling & Parak for a comprehensive description of various polymeric scaffolds used to disperse QDs in aqueous media [42].
Surface exchange via polymeric ligands has profound advantages over mono/bidentate small molecule based ligands. Owing to the multidentate nature of interactions between the surface and the polymers, QDs display higher stability in dilute conditions for prolonged periods of time. A very interesting comparison by Giovanelli et al. discussed the time-dependent desorption of dithiol versus poly-thiol ligands from the surface of QDs via ligand competition experiments. It was shown that the rate of desorption (koff) of polymeric ligands was more than 300× lower than that of dithiol-based ligands [38]. Secondly, based on the type of polymers, the stability is not affected by variation in pH. The polymers derived from thiol-based linkers have higher tolerance to a range of pH (4.5–9 or higher), whereas polyhistidine-based ligands are often unstable below pH 6. Despite the nature of ligands, QDs coated with hydrophilic ligands have both high stability and QY in neutral to basic pH. Additionally, QDs coated with polymeric ligands also have additional functional groups incorporated for conjugation where specific covalent attachment of biomolecules could be obtained. Polymer-based ligands have also expanded the intracellular stability of the conjugates. An inherent issue with the use of polymeric ligands is the concomitant increase in the size of QDs. The amphiphilic coating can easily increase the hydrodynamic radius 2–3-fold. Therefore, current efforts are attempting to devise polymeric scaffolds that not only provide the advantages discussed above, but also retain smaller size of nanocrystals.
Another type of ligand exchange strategy not discussed in detail here includes coating of QDs with silane derivatives. Silica shells can be grown around QDs either by silanization (smaller Dh, less than 10 nm) or by encapsulation (larger Dh, 20–30 nm). Often growth of silica shells around QDs is a multistep process. However, the hydrophilic QDs thus obtained have both moderately high QY and stability, particularly in alkaline media. Interested readers are referred to a review by Selvan for additional details and current state of the art [43].
The strategies mentioned above discuss the types of ligands and their assembly onto the surface of QDs via different exchange methods. The overall goal of different ligand exchange strategies is to achieve bright and compact QDs in biological buffers. Some of the desirable properties include (i) homogeneous monodisperse population of QDs, (ii) retaining high fluorescence quantum yield, (iii) long-term colloidal stability, (iv) small size, (v) minimal non-specific interactions with biological media, and (vi) synthetic reactivity to chemical conjugation. Previous and current research aims to achieve water-solubilized QDs with several of these properties. In general, transition from organic solvent to aqueous media is accompanied by reduction in quantum yield. Type of ligands and the exchange procedure tends to affect the state of aggregation of the capped QDs. The ligand exchange procedures generate major population of single nanocrystals homogeneously coated with the polymeric ligands. However, along with these QDs, there are sub-populations of dimers or oligomeric aggregates of QDs that form, and increase the heterogeneity of samples. Bigger aggregates can be removed by centrifugation, but smaller clusters (dimers, etc.) are often impossible to get rid of. Also, smaller ligands maintain compactness of the QDs, but display poor stability in dilute conditions and long durations. Contrarily, QDs coated with polymeric ligands have higher colloidal stability, but the QDs are both heterogeneous and substantially bigger. Current efforts are dedicated to optimize several of these parameters based on the specific target applications. After dispersing QDs in aqueous solutions, the next step is to functionalize them with specific biomolecules, such as nucleic acids.
3. Strategies to conjugate DNA to quantum dots dispersed in aqueous media
Nucleic acids, particularly DNA, are one of the most stable classes of biomolecules. The natural selection of DNA as the hereditary material in the course of evolution is attributed to this remarkable tolerance to chemical reactivity. There are several very interesting characteristics of DNA. Three of the most relevant in the context of this discussion include (i) polyanionic nature, (ii) hybridizability, and (iii) persistence length. In subsequent sections, the use of these properties for conjugation, detection and material design using QD–DNA conjugates will be discussed.
Specific conjugation of DNA to QDs (and other NPs) is brought about by reaction of particular functional groups on DNA with those on QDs. These reactive functional groups are incorporated during the solid phase synthesis of DNA. These modifications produce specifically end-labelled DNA, where single functional groups (such as –NH2, –SH, –N3) can be chemically inserted along with hydrocarbon linker chain of desired length. This linker chain projects the functional group away from the oligonucleotide chain, and hence increases the solvent accessibility and reactivity of the functional group. Additional functional chemical groups such as biotin, PEG, etc. can also be incorporated during the synthesis of DNA. These groups often improve the water solubility, reactivity and/or the intracellular uptake of DNA.
The conjugation strategies discussed herein are grouped in two categories: first are the strategies where functional groups on DNA have affinity directly towards the inorganic shells of QDs, and second, where the DNA interacts with specific organic or biomolecular ligands on the QDs.
3.1. Conjugation of DNA to shells of quantum dots
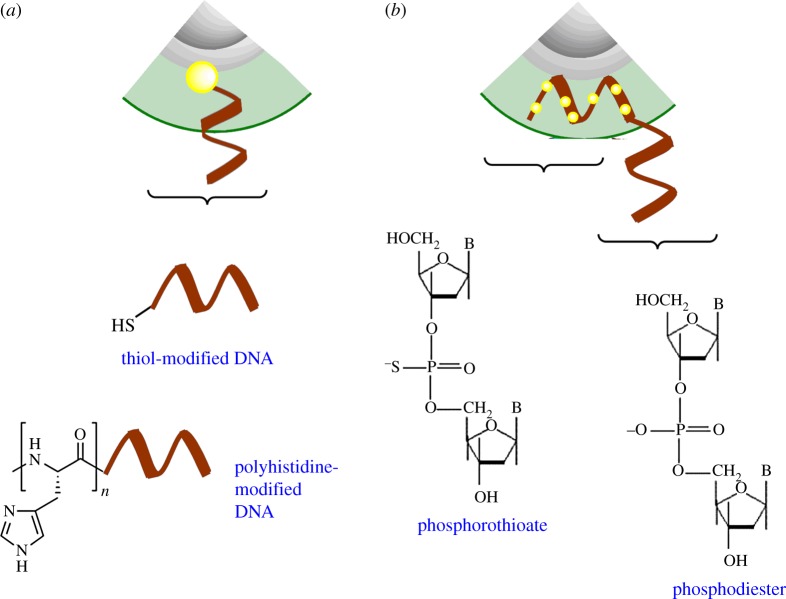
Ligands such as thiols and imidazoles have affinity towards the inorganic shells of QDs (figure 4). These interactions have been used to attach DNA to QDs as described in the subsequent section.
Figure 4.
Conjugation of DNA to the shell of quantum dots. Several variants of chemically modified DNA have affinity towards the metal ions on the surface of the quantum dots. These include (a) thiols (top) and polyhistidine (bottom) and (b) via phosphorothioate modifications in DNA backbone. (Online version in colour.)
3.1.1. Thiolated DNA
This is the most popular category of ligands for the non-covalent attachment to the surface of QDs. Thiols have an intrinsic property to self-assemble on the surface of nanoparticles, including QDs. QDs are first dispersed in water using small molecule ligands such as MPA. Excess MPA is removed by several rounds of centrifugation and resuspension in basic buffers (pH > 8), wherein deprotonation of MPA makes the nanocrystals hydrophilic. Then thiolated DNA is added in excess, such that dynamic equilibrium can replace the original ligands with thiolated DNA. First shown by Mitchell et al., alkylthiol-labelled DNA of various lengths have been self-assembled on a range of colloidal nanoparticles including QDs in high yield and reproducibility [44–46]. In the first report, the exchange was carried out for 1–2 days and several length and sequences of DNA were assembled. These conjugates displayed infinite stability at high concentrations, but were sensitive to pH, photo-oxidation and dilutions. Since then several reports have conjugated thiolated DNA on QDs within shorter times [47,48]. However, general issues with photo-oxidative ligand loss and pH sensitivity are a fundamental limitation of this approach.
3.1.2. Peptide-tagged DNA
Imidazoles are another class of functional groups that have high affinity toward shells of QDs. This affinity has been harvested in the use of polyhistidine peptide tags of various lengths to conjugate DNA to QDs. Sapsford et al. demonstrated that polyhistidine tags could self-assemble over the QD surfaces with almost instantaneous kinetics [49]. Several reports from Mattoussi's group have also demonstrated that DNA conjugation can be carried out on peptide tags with terminal functional groups such as lysine (–NH2), aspartate (–COOH) or cysteine (–SH) using specific reaction conditions [50]. These conjugates could be then purified using affinity or reverse phase liquid chromatography and simply mixed with ligand-exchanged QDs. These conjugates are state of the art and easy to assemble. However, there are certain limitations. Affinity of the imidazole group towards the QD surface is highly pH dependent. Protonation of the N-hydrogen of imidazole functional group (at pH < 6) destabilizes the conjugates. Additionally, because polyhistidine-tagged peptides are highly cationic and DNA is highly polyanionic, the conjugation in solution is complex and pH sensitive, and often results in low yield and aggregation of the biomolecules when synthesized [51]. This issue could, however, be circumvented by using DNA with a neutral polypeptide backbone (PNA) which retains the properties of hybridization of DNA without the high negative charge. Additionally, PNA–DNA duplexes have higher thermodynamic stability than DNA–DNA [52] and are far more expensive than DNA. These two factors limit the use of the PNA-based conjugates for rapid and efficient biosensing applications.
3.1.3. Phosphorothioate-modified DNA
The DNA backbone comprises polyphosphates. The polyphosphates can be synthetically modified to phosphorothioates wherein one of the non-bridging atoms of oxygen is replaced with sulfur. Sulfur has much higher affinity (approx. 3000×) for Cd2+ than oxygen and hence forms stronger complexes with the inorganic shells, including QDs. First demonstrated by Kumar et al. on colloidal gold, this affinity has been utilized to conjugate phosphorothioate-modified DNA to gold nanoparticles (GNPs) [53,54] and later to QDs [55–57] either post or during synthesis. This method is advantageous to link DNA on NPs, but suffers from the limitation that the ‘attached’ DNA is conformationally distorted and often loses its ability to hybridize complementary DNA [56]. However, this is circumvented by introducing a DNA overhang with phosphodiester backbone, displaying the target sequence [58]. Using a combination of phosphorothioate backbone (for binding) and phosphodiester backbone for molecular assembly, these pre-functionalized QDs can be further used for hybridization, biosensing, cellular targeting and other applications.
3.2. Conjugation of DNA to ligands on quantum dots
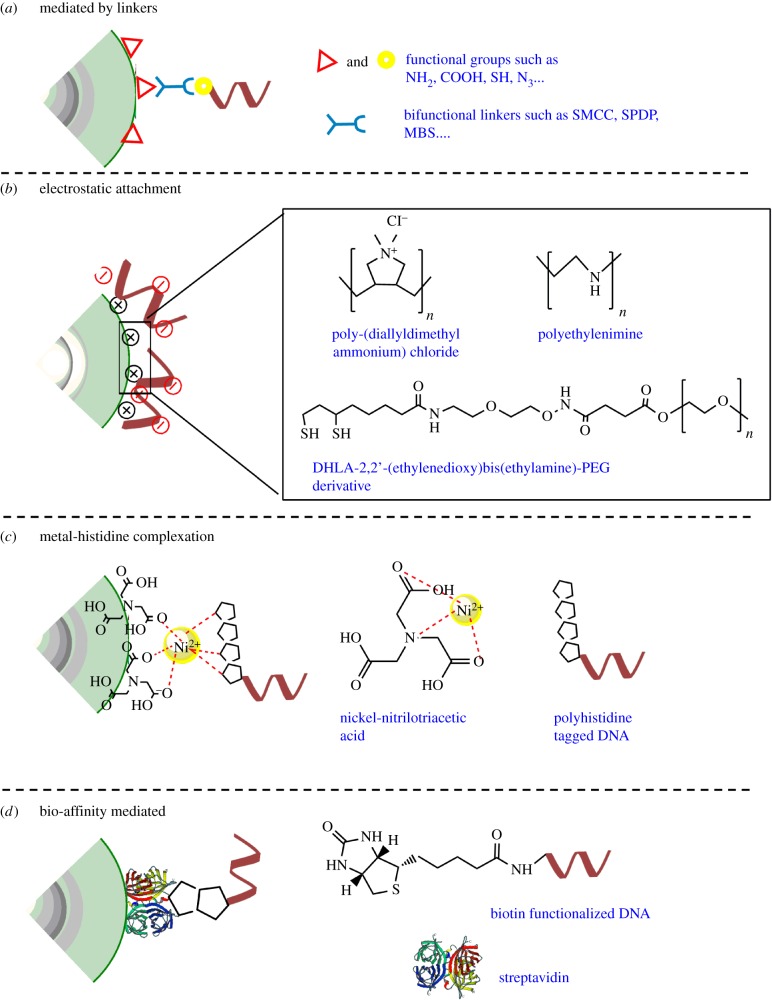
In the previous section, the conjugation strategies were based on the affinity of the functional groups on DNA towards the metal ions on the shells of the QDs. Another popular category of conjugation can be based on affinity of DNA for specific ligands coating the surface of the QDs (figure 5). Some of the examples are discussed below and summarized in figure 5.
Figure 5.
Conjugation of DNA to specific ligands on quantum dots. (a) Covalent conjugation of DNA to QDs can be carried out by commercial bifunctional linkers. (b) Polyanionic phosphodiester backbone of DNA can be adsorbed by cationic surface coatings surface coatings on the QDs via electrostatic interactions. (c) Ni-NTA-modified QDs can be complexed with polyhistidine-modified DNA. Another variation of this strategy includes affinity of polyhistidine tagged DNA directly for the surface of QDs. (d) Biotin-modified DNA can be easily coupled to streptavidin-modified QDs. (Online version in colour.)
3.2.1. Lipid encapsulated conjugates
As discussed in the previous section, amphiphilic phospholipids self-assemble as micelles on the surface of QDs and impart water solubility to QDs. The polar head groups of the phospholipids impart water solubility. As shown by Dubertret et al., the composition of encapsulating phospholipids can be tailored such that a few display functional groups that can be used for conjugation of DNA [7,8]. More recently, Aimé et al. have shown that lipid oligonucleotide conjugates (LONs) can embed within the amphipathic capping layer on QDs by hydrophobic interactions [59]. In this method of conjugation of DNA, the oligonucleotides are first conjugated to the amphiphilic lipids and then added in the overall encapsulating formulation to directly display conjugated DNA on QDs. These methods, however, lack control over the conjugation efficiency. These conjugates also have less applicability in the context of cellular processes, because phospholipid preparations often result in unambiguous cytoplasmic delivery or translocation through lipid bilayer [60–62].
3.2.2. Electrostatic interactions with ligands on quantum dots
The phosphate backbone of DNA is highly negatively charged. This enables attachment of DNA to QDs with positively charged surface coatings by virtue of the electrostatic attraction. There are several examples of QD–DNA conjugates synthesized via this method in the literature. This approach is very simple, instantaneous and results in high DNA loading. Mirkin et al. demonstrated for the first time that DNA loading capacity on GNP can be further enhanced by step-wise increase in the osmolarity of the reaction [63].
Phosphate backbone-mediated electrostatic attachment of DNA on polymer-coated QDs has been shown [45]. In a report by Peng et al., QDs were ligand exchanged with cationic polymers such as poly(diallyldimethylammonium chloride) and associated with DNA by electrostatic interactions [64]. In another report by Lee et al., QDs were first ligand exchanged with amine-modified DHLA and passivated with a layer of high molecular weight PEG. This DHLA–PEG hybrid layer has moderate cationic nature along with antifouling properties and can be used to quantitatively load DNA mediated by electrostatic interactions [65]. Other strategy involves using small molecule ligands such as MPA (negative charge) followed by PEI (polyethylenimine) [66–68]. These high to moderately cationic QDs have been extensively used to electrostatically assemble excess of DNA for biodetection and delivery of therapeutically active agents.
However, electrostatics-mediated attachment of DNA to QD and other nanoparticles has several limitations. Affinity of phosphate towards the positive surface of QDs may lead to conformational distortion of DNA [69]. The interactions between the phosphate backbone of DNA and the QD surface can vary with the pH, and the conjugates tend to aggregate upon long storage. Also, the preparations are often heterogeneous with variations in conjugation yield.
3.2.3. Surface modified quantum dots
In a recent report, Kwon et al. have demonstrated that DNA can be conjugated on QD based on affinity of polyimidazoles for Ni-NTA [70]. In this approach, QDs displaying carboxylic acid functional groups have been conjugated with polyglycine–polyhistidine peptide tag. In parallel, DNA is conjugated to thiolated NTA. Post conjugation, Ni2+chelation of the NTA is carried out, which makes the DNA reactive towards the polyhistidine tag. These two reactants have been shown to self-assemble instantaneously and the complexation can be reversed in presence of excess imidazole. However, this study does not discuss the natural propensity of polyhistidine tags to self-assemble on the inorganic surface of QDs (and other NPs). The specificity of conjugation of the polyglycine–polyhistidine peptide to the carboxylic acid ligands is thus debatable. This approach is conceptually similar to the design of using NTA-modified QDs to associate polyhistidine-tagged proteins [49,71] where tighter control over the number of DNA per QD is imposed.
3.2.4. Affinity for existing conjugated ligands
Biotin-streptavidin binding is one of the strongest non-covalent interactions known in biology. This high affinity has been exploited in multiple scenarios to attach biomolecules to one another. This is also one of the widely used strategies to conjugate DNA to QDs. Herein, QDs are first functionalized with streptavidin by either affinity (peptide) or covalent conjugation methods [3,45,72,73]. Then these QDs are purified from unconjugated proteins and simply mixed with commercial biotinylated DNA. Biotinylated DNA can simply attach to the QD mediated by the streptavidin linker. Each streptavidin has four biotin binding sites, thereby enabling association of more than one DNA per protein. In order to further control the stoichiometry of biotin on streptavidin, molecularly engineered variants of the protein have been used, which have affinity for only one biotin [74,75] (and hence biotinylated DNA) that permit monolabelling (per streptavidin molecule) with biotinylated DNA [76]. Molecular cloning has also realized facile production of higher affinity avidin variants such as dimeric rhizavidin [77,78] or electrostatically neutral variants (at physiological pH) such as neutravidin which further expands the tool-kit of this type of conjugation.
Streptavidin-Bt–DNA conjugates of QD have been easily generated and could be used for various applications. Zhang et al. showed that different lengths of DNA could be easily assembled on the QD surface by simple one-step mixing protocol [79]. However, the only limitations of this approach is that the additional layer of protein often results in an increase in the size of the particle, that eventually limits its applications in contexts of particle tracking at neuronal synapses or ion channels.
Conjugation of DNA to specific ligands on QDs has been demonstrated by different strategies. Each strategy has its own merits and demerits, as described above and listed in table 1. Most reactions are tailored based on the need of specific types of conjugates, such as QY, size, poly-functionalization, ease of preparation, etc. The rapidly expanding library of ligands is paving way for the development of newer generation of conjugates with highly desirable properties for applications in biology.
Table 1.
Comparison of different methods for conjugation of DNA to quantum dots.
| s.no | approach | modification on QD | modification on DNA | advantages | disadvantages | references |
|---|---|---|---|---|---|---|
| 1 | phospholipid encapsulation | organic ligands | amine/thiol | — one step procedure — compatible with range of synthesis — DNA retains hybridizability |
— stability issues — large size of particles — random fusion with membranes — low QY |
[7,8] |
| 2 | electrostatic attraction with polymer on QDs | positively charged hydrophilic ligands | unmodified; interaction with phosphate backbone | — rapid — high DNA loading efficiency — reversible |
— difficult to control stoichiometry of DNA — issues with stability and aggregation |
[65,68] |
| 3 | affinity for cationic shell of QDs | positively charged or neutral | thiol/polyhistidine/phosphorothioate | — rapid assembly — high yield — compact |
— unstable in high dilutions — loss of DNA by photo-oxidation — pH sensitive — interactions with DNA backbone can cause conformational change — low QY due to charge transfer |
[44–46,49,56] |
| 4 | affinity for specific ligand | streptavidin conjugation | biotinylated DNA | — fast and easy — conjugation of DNA independent of length — stoichiometric control — High QY |
— increase in conjugate size due to presence of protein | [3,35,45,72,73,79] |
| Ni-NTA modified | polyhistidine modified | — one step — efficient — high QY |
— pH sensitive stability | [70] | ||
| 5 | covalent conjugation with ligands on QDs | QDs with carboxylic acids | amine modified | — highly stable — pH insensitive — DNA hybridization preserved — moderate-high QY |
— multistep — low yield — nanoparticle size increases |
[80–82] |
| QDs with amines | thiol modified | [83] | ||||
| QDs with thiols | amine modified cyclooctyne | [84] | ||||
| QDs with azide | — reactants are not prone to hydrolysis | — loss of QY | [85] |
4. Types of covalent conjugation reactions
Commercially available linkers are most commonly employed to form covalent bonds between the ligands on QDs and DNA. These linkers (i) either assist in the ‘activation’ of the reacting molecules by forming transient intermediates (using carbodiimide chemistry) or (ii) have two functionally reactive groups on the same linker, one for each reacting molecule (bifunctional linkers). The following section describes the use of these linkers in several examples of covalent conjugation of DNA to QDs based on the reacting functional groups.
4.1. Amine to carboxylic acid conjugation
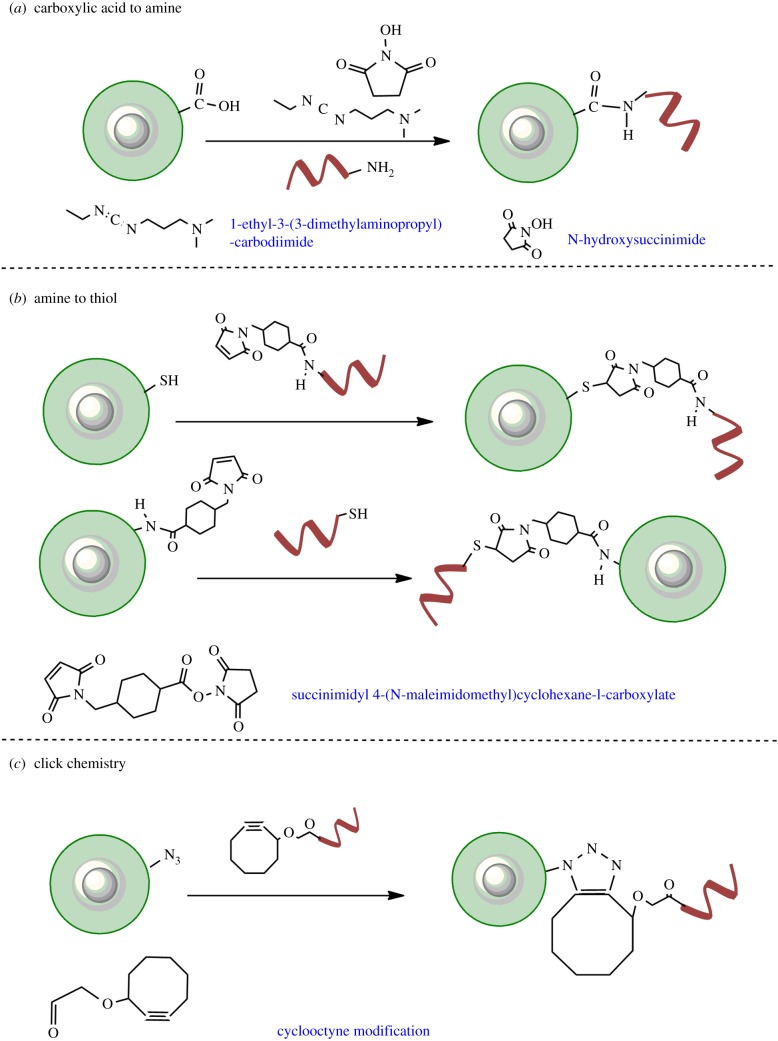
This is one of the extensively used strategies for conjugation of DNA to QDs (figure 6a). The QDs are first coated with ligands that bear carboxylic acid functional groups. These ligands are either small molecules such as MAA (mercapto acetic acid), MPA, MHA and DHLA [35,42,80,81] or derived polymeric scaffolds terminating in carboxylic acid [82,86]. To all of these QDs ‘terminating’ in solvent-exposed carboxylic acid functional groups, conjugation of amine-functionalized DNA is carried out in the presence of EDC and NHS. First, the carboxylic acid group of QDs is activated by reaction with EDC. This step produces a highly reactive (and labile) O-acylisourea intermediate. This intermediate can be immediately mixed with NHS, which results in QD-NHS. These NHS-derived QDs are then reacted immediately with amine-DNA to form a stable amide bond between the capping ligand and DNA. In the case of failure of reaction with NHS, the intermediate hydrolyses and the carboxylic acid on QDs can be regenerated. This method retains the small size of QDs and is often used to conjugate DNA on QDs coated with mercapto alkyl acid based ligands or polymers with terminal acids. However, this reaction is very sensitive to rapid hydrolysis of esters of EDC-NHS. In addition, particularly observed for QDs coated with mercapto alkyl and DHLA ligands, the nanoparticles tend to lose colloidal stability [87] when the terminal carboxylic acids undergo esterification with EDC-NHS. These two factors often result in reduced yields.
Figure 6.
Covalent conjugation reactions used to synthesize QD–DNA conjugates. (a) Carboxylic acid containing ligands on QDs can be conjugated to amine-labelled DNA using EDC-NHS chemistry. (b) Coupling of maleimide-functionalized DNA to thiols on QD (top) and maleimide-functionalized QD to thiolated DNA (bottom) assisted by heterobifunctional linker sSMCC. (c) Copper-free click chemistry employed to conjugate azide-modified QDs to cyclooctyne-modified DNA. (Online version in colour.)
4.2. Amine to thiol reactive linkers
There are two popular types of amine to thiol reactive cross-linkers used for bioconjugation on QDs. The first is sulfosuccinimidyl-4-(N-maleimido-methyl) cyclohexane-1-carboxylate (sSMCC) which has a terminal NHS and a terminal maleimide (figure 6b). In this reaction, amine-labelled DNA is first reacted with NHS to give DNA maleimide. The DNA maleimide is then conjugated with polymer-capped QDs, which have been reduced to display thiols [84]. The same design could also be used to conjugate proteins and antibodies to QDs [88]. Alternatively, a more classical utilization of this approach involves functionalizing QDs displaying primary amines to thiol-modified DNA [8].
Another bifunctional linker called succinimidyl 3-(2-pyridyldithio)propionate (SPDP) can also be used to functionalize amine terminating DNA to QDs [83]. This linker has an NHS group on one end and a cleavable pyridyl disulfide on the other. The general template to use this linker is to first functionalize amine-DNA with the linker. In the second step, the disulfide is cleaved in the presence of a reducing agent to generate reactive thiols. The reducing agent and the pyridine-2 thione group (by-product) thus generated are removed and the DNA functionalized with a reactive thiol is mixed with maleimide-functionalized QD. The thiol-maleimide reaction results in conjugation of DNA.
Apart from conjugation of thiol to maleimide by the reaction described above, SPDP can be also used to couple thiol to thiol by simple disulfide exchange. The pyridyl disulfide moiety in SPDP can transiently react with reactive thiol containing ligands resulting in successful and reversible conjugation. Another specific advantage of using SPDP over SMCC is the control over utilization of the pyridyl disulfide. After reaction of the linker with the primary amine containing biomolecule, the modified biomolecule can be easily stored for long periods without loss of function of the linker. However, in the case of SMCC, the maleimide derivative should be reacted within a day due to hydrolysis of the maleimide. A comparative disadvantage of SPDP over SMCC is the formation of disulfide cross-linking in the case of the former. In the case of reactions carried out in pH 7–8, reformation of disulfide can lead to cross-linking of biomolecules instead of reaction of the maleimide of QD to the thiols of the biomolecule. Hence, tight regulation of pH in the case of this reaction is highly sought after.
4.3. Click chemistry
Click chemistry is the new age biorthogonal labelling technique employed to conjugate a range of biomolecules onto one another. The reaction comprises covalent conjugation of a molecule with functional azide to another molecule with alkyne via cycloaddition (figure 6c). The reaction is often catalysed by Cu+. However, Cu+ cations are known to quench the photoluminescence of QDs [16]. In order to circumvent this issue, Cu-free click chemistry has been employed to conjugate DNA to QD while maintaining high fluorescent quantum yield [85]. Briefly, QDs coated with DHLA-PEG-N3 were mixed with cyclooctyne-modified DNA (thrombin binding aptamer) in a 1 : 30 ratio and left overnight at 4°C. With this approach, approximately 67% of total DNA was conjugated. This is one of the highest yields reported so far.
Covalent conjugation of biomolecules on the surface of QDs is by far the most popular method to prepare biofunctionalized QDs. The linkers are cheap and easily available and most of the reactions can be carried out without the need of highly specialized conditions. These methods also ensure that the DNA (or other biomolecule) remains tightly bound to the QDs and the stability is derived from the innate stability of the encompassing ligands. After preparation of QD–DNA conjugates by various methods discussed above, the conjugates need to be purified from the excess uncoupled reactants. A few of the most popularly employed methods are discussed below.
5. Purification of conjugates of quantum dot–DNA
The search for an ideal method of purification of functionalized QDs from the excess reactants is as old as the hunt for best strategy of conjugation. Almost no biological or chemical application of these specific conjugates can be carried out without purification of excess (uncoupled) DNA. There are three broad methods of separation discussed herein.
5.1. Electrophoresis
Electrophoretic migration of QDs depends on both the type of QD structure and the type of surface chemistry. Ligands such as MPA, DHLA and anionic polymers impart negative charge on QDs, which facilitates migration towards positive terminals [35,38].
Conjugation of proteins with near neutral isoelectric points (pIs) often reduces electrophoretic migration of QDs [89]. Contrarily, conjugation of DNA generally increases anodic migration, which is also affected by the length and number of DNA conjugated [76,79,90].
Trends in altered electrophoretic migration have been used to separate QDs conjugated with DNA from the unconjugated ones [79,91]. Electrophoresis is indeed one of the most simple and routinely used methods of characterization and purification of QD–DNA conjugates. QDs conjugated to DNA have even been purified by the extent of labelling stoichiometry [58]. However, in order to extract conjugated QDs from the gel, tedious extraction processes need to be carried out. The relevant bands of interest should be first excised out of the agarose gel followed by either melting of the gel or prolonged incubation of the gel fragments in relevant buffers. QDs are then re-collected back by centrifugation and concentration. Passing QD–DNA conjugates from several of these methods often result in poor yields and issues with long-term stability.
5.2. Ultracentifugation
Owing to the presence of multiple metallic atoms in a confined volume, QDs (and other NPs) have high density in the core and at the shell. Additional coating of organic polymeric layer does not increase the density of particles. However, general ligand exchange procedures can change the state of aggregation of the samples. Further conjugation of biomolecules on this layer does not contribute towards increase in the density of the biomolecules. This allows for separation of QDs from fluorescent or colourless biomolecule layer [7,83].
5.3. Chromatographic separation
Several examples of purification of QD–DNA conjugates using a variety of chromatographic techniques have been reported in the past. For example, in reactions with high yield of conjugation, the nanoparticle size increases due to the addition of a layer of biomolecules. This facilitates purification of QD–DNA conjugates from unreacted DNA using size exclusion chromatography (SEC) [84,92]. Another approach to purify QD–DNA conjugates from unreacted DNA is by the use of anion exchange chromatography (AEC) [93]. The affinity of diethyl-aminoethyl (DEAE) based matrices for anionic ligands such as DNA is well known [94]. This affinity is used as a method to purify QDs conjugated with DNA from the unconjugated ones. First, a column prepacked with DEAE cellulose beads is equilibrated with buffers of low ionic strength. This enables interaction of the DNA-functionalized QDs with the matrix whereas the unconjugated QDs elute out at this time. Then the entrapped QD–DNA conjugates can be eluted by simply passing a buffer of higher ionic strength through the column. A further modification of this strategy is the use of DEAE-modified magnetic beads [95]. Herein, Uddayasankar et al. tuned the purification strategies to particularly separate QD–DNA conjugates from unreacted QDs by binding to DEAE on the magnetic beads. In this particular study, QDs conjugated to different lengths of DNA were purified by tuning the ionic strength of the interacting and eluting media. The rapid purification times (within minutes) and reduced number of interacting interfaces in the case of magnetic beads were a far more efficient method of purification than cellulose beads.
6. Applications of quantum dot–DNA conjugates
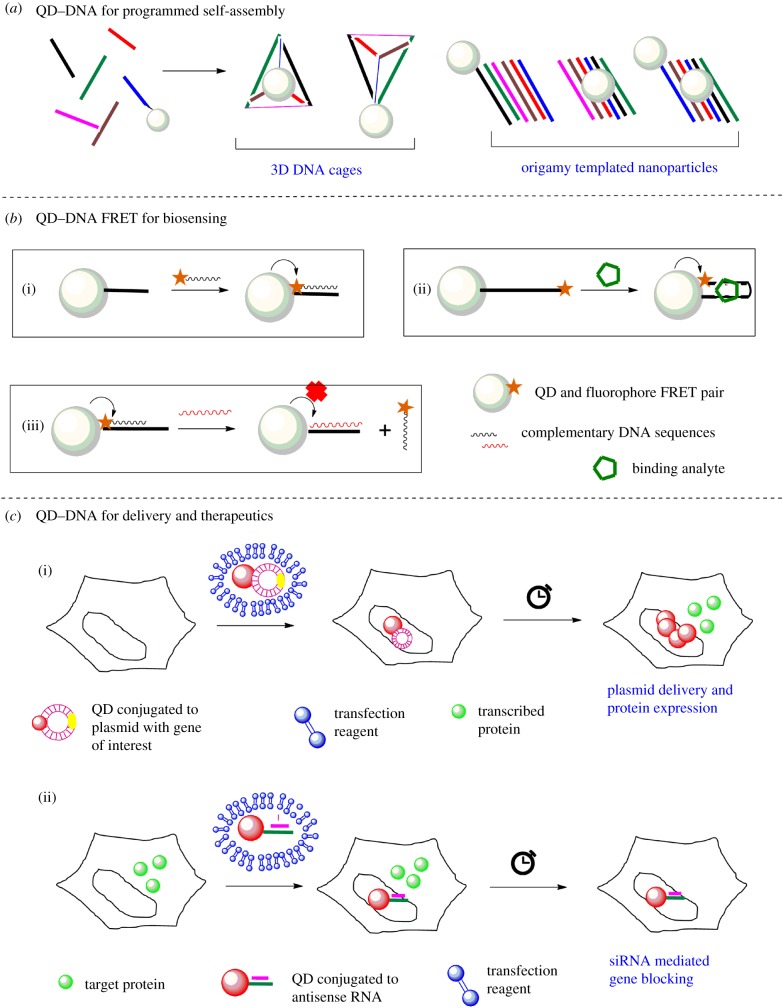
The predictable base pairing properties and persistence length of DNA make it an ideal template for nanoconstruction. Conjugation of electron dense nanoparticles such as gold or QDs to DNA opens up routes for novel designs and nano-assemblies [73,96]. These GNP/QD-functionalized DNA templates can then be used to synthesize novel materials for engineering, charge transport and photo physics. In parallel, such synthetic scaffolds could also be used for biological applications. In this section, three broad categories of applications of QD–DNA conjugates related to biosensing and therapeutics are briefly discussed (figure 7).
Figure 7.
General application of QD–DNA conjugates. Three broad categories of applications of QD–DNA conjugates are covered herein. (a) Use of QD–DNA conjugates for design of origami and templated nanoconstructs. (b) Three modes of FRET-based detection and biosensing of target nucleic acids and analytes using QD–DNA conjugates. (c) Use of QD–DNA conjugates for therapeutic applications such as controlled protein expression (top) and siRNA (bottom). (Online version in colour.)
6.1. Synthetic programmable scaffolds
With the increasing popularity and ease of generation of QDs (and other NPs) functionalized by DNA, these synthetic scaffolds have been recently used to develop highly complex, tunable and reconfigurable macromolecular scaffolds (figure 7a). Conjugates of QD–DNA have been assembled into various geometries for about 15 years now. In one of the first reports by Mitchell et al., QD and GNP assemblies were augmented by DNA hybridization [44]. In rather recent examples, QDs emitting at different wavelengths were conjugated to DNA of different lengths and assembled into reversibly programmable structures with controlled valency and orientation by Tikhomirov et al. [97]. These structures could be tuned to mitigate electron transfer thereby allowing control on photophysical properties. In another report by Ko et al., QD–DNA conjugates have been precisely positioned on DNA origami templates using hybridization and used to tune the fluorescence lifetime of QDs via this synthetic scaffold [98]. In yet another approach, electrostatic interactions have been used to encapsulate QDs within a DNA nanocage by Wang et al., to conserve the single molecule aggregation-free state of QDs in solutions [99]. In a similar approach by Wang et al., QD–DNA conjugates have been used for the detection of a correctly ‘folded’ 3D DNA origami cage [100].
These conjugates are not only novel materials for design and assembly, but also display interesting electronic and chemical properties and therefore are valuable for understanding the macromolecular behaviour of complex nanomaterials.
6.2. Biosensing application
The conjugates of QD–DNA have been widely used for detection of nucleic acids including DNA, RNA, mRNA and miRNA, and other molecular ligands. The biosensing modality is introduced to QD–DNA conjugates by using fluorescently labelled complementary probes that can hybridize and ensure a bimolecular process like fluorescence resonance energy transfer (FRET) (figure 7b) [101–103]. The FRET signal can be induced by simple mixing of target polynucleotide sequences [104]. In one of the classic works by Zhang et al., a QD–DNA to Cy5 DNA-based single molecule FRET nano-sensor was developed which was then used to detect single point mutation in clinical samples [105]. This FRET-based biosensing design showed more than 100× superior performance over vastly popular molecular beacons. It was also minimalistic design with no additional requirement for purification. A classic work by Patolsky et al. demonstrated the use of QD–DNA conjugates to monitor the DNA replication [106]. The ‘donor’ (QD–DNA) conjugates were incubated in a soup of dNTPs (wherein dUTP was labelled with acceptor fluorophore) and telomerase. Successful incorporation of dNTPs (i.e DNA synthesis) by the action of telomerase positioned the fluorophore (‘acceptor’) close to the QDs, resulting in a FRET ‘on’ system. Upon successful DNA elongation, both loss of donor intensity and concomitant increase in acceptor intensity were used as a time-dependent read-out for enzymatic activity of telomerase.
A similar work by Suzuki et al. used QD–DNA FRET-based biosensing to characterize the polymerase activity during elongation of the DNA strand (PCR) [107]. In this work, QD–DNA conjugates were used as a template for elongation of DNA by systematic incorporation of fluorophore-labelled nucleotides. Successful elongation of DNA by addition of nucleotides by the activity of DNA polymerase converted the QD–DNA conjugate into a FRET system; where energy transfer could occur between QD–DNA and fluorophore on the newly elongated strand of complementary DNA. This design led to rapid assessment of activity of DNA polymerases.
In a rather recent study, Shahmuradyan & Krull employed an interesting FRET-ON design. They conjugated a pair of thiazole-orange dye-derivative at predefined locations on ssDNA, and conjugated this strand to QD [108]. This thiazole-orange dye can have excitonic interactions with the adjacent dye molecule, resulting in a fluorescent quenched state. These molecules can additionally intercalate in dsDNA efficiently. Therefore, upon hybridization of this DNA strand with its complementary sequence, the thiazole-orange derivatives intercalate within the DNA, thereby losing the electronic overlap resulting in a ‘quenched’ state. This QD–dye–dye assembly could then act as a FRET-ON system. This assembly has been employed to detect single nucleotide polymorphism in complex serum containing media with better signal to noise than conventional QD–dye FRET system.
In the above few examples, QDs have been used as the energy ‘donor’ for the FRET pairs. There are several examples where QDs perform as FRET acceptors, by coupling them with luminescent lanthanide derivatives, which act as energy donors. These lanthanide derivatives have a larger Stoke's shift (approx. 150 nm) and longer fluorescence decay times (millisecond range) along with sharper emission of about 10 nm FWHM. These properties enable multiplexed detection with minimal cross-talk along with expanded FRET distances compared with organic dyes [109]. As demonstrated by several works from the Hildebrandt group, lanthanide complexes employing terbium (Tb) are particularly promising for FRET-based bio-applications, including cell biology and clinical diagnostics [110–112]. In other classic works by the same group, Tb-to-QD FRET has been employed for multiplexed detection of up to three circulating miRNAs in samples in the presence of serum [113]. These works used time-gated FRET detection that was rapid with minimal sample processing (preparation and purification) and high sensitivity with detection limits close to 30 fmol for triplex detection. Needless to say, lanthanide complexes-to-QD FRET are the current state of the art for rapid, amplification-free biosensing.
A variation of this design is when loss of FRET is coupled to the biosensing. An interesting example of this ‘FRET-loss’ method is the work by Zhang et al. where a thrombin binding aptamer was conjugated to QD and blocked by partially complementary sequence. Terminal fluorophore on this complementary sequence formed a FRET pair with QD. Binding of a longer complementary ssDNA or conformational change induced by thrombin to the aptamer, displaced the fluorophore-labelled DNA, thereby causing loss of FRET [85]. In another classic example by Kay et al., the use of conformational change of DNA induced FRET to quantify pH in endosomes of live cells [114].
Another biosensing application of QD–DNA conjugates is in fluorescence in situ hybridization (FISH) [115,116]. The purpose of FISH is to locate specific gene targets in metaphase chromosomes of model organisms for karyotyping, biomarker detection, etc. The fundamental principle involves searching for ‘target’ sequences mediated by hybridization of fluorescent complementary sequence. The probe DNA conjugated to QD is incubated with cells (in metaphase), denatured and fixed on glass slides. The probe DNA is allowed to hybridize with the ‘target’ gene. The slides are thoroughly washed and stained with DAPI (to mark the chromosomes) and imaged using total internal microscopy. Almost at the same time, Bentolila & Weiss and Xiao & Barker demonstrated that the FISH assay using QD–DNA probes have much higher sensitivity and signal to noise ratio in comparison with organic fluorophores.
Another example where QD–DNA conjugates are used for biosensing is in the detection of mRNAs [117]. A high throughput microarray of ‘catch’ DNA sequences was prepared, and biotinylated target sequences of mRNA (unknown) were allowed to hybridize. To this assembly, QDs labelled with streptavidin were passed at a controlled flow rate so as to enable streptavidin-biotin binding. Then the array was washed to get rid of excess background, and QD fluorescence was used as a detection signal. Presence and intensity of fluorescence indicated presence and quantification of target miRNA sequence. Using QDs for this detection, Liang et al. demonstrated the detection limit of less than 1 fmol and dynamic range of sensitivity up to two orders of magnitude, demonstrating the power of QD-enabled molecular diagnostics.
Another category of biosensing and theranostics application of QD–DNA conjugates is the use of QD–aptamers [118]. Aptamers are synthetic sequences of DNA that can be selected to have specific recognition motifs binding to particular targets. These aptamers are designed and selected by SELEX (systematic evolution of ligands by exponential enrichment) where DNA sequences with specific affinity for targets of interest are screened and mutated to improve the affinity by several iterations of synthetic evolution. Functionalization of QDs with these DNA aptamers paves the way for using QDs for molecular diagnostics. The thrombin binding aptamer and the pH sensing sequence discussed in the previous sections are a few of the classic examples of QD–DNA aptamer mediated detection of analytes. Several QD–aptamer conjugates sensitive to different analytes are being rapidly developed. Interested readers are referred to a recent review by Jo et al. for a detailed overview [118].
Using QDs as alternatives to fluorophores for biosensing assays has definitely improved the reliability of detection due to high signal to noise and also substantially increased the limit of detection of analytes, thereby improving the current state of the art. These QD–DNA–fluorophore FRET-based detection methods are robust, sensitive, ratiometric and highly quantitative. The existing challenge is to carefully control the number of DNA and generate homogeneously labelled populations of conjugates. This area is currently actively being investigated.
6.3. Delivery and therapeutics
The conjugates of QDs find applications in delivery and therapeutics primarily because of two reasons: (i) high surface to volume ratio allows loading of multiple copies of biomolecules on single scaffold and (ii) enhanced photostability enables long duration tracking inside cells. There are a few examples where QD–DNA conjugates have been used for intracellular delivery of biologically active materials for therapeutic applications. These examples can be broadly divided into nanoparticle-assisted gene regulation and gene therapy (figure 7c). One of the earliest examples by Srinivasan et al. showed intracellular gene regulation by tracking and delivery of plasmid using QD–DNA conjugates [83]. A plasmid encoding for enhanced green fluorescent protein (EFGP) is labelled with several QDs via PNA–DNA hybridization and transfected into cells. The intracellular and subsequent nuclear delivery of the labelled plasmid is tracked by time-lapse imaging of the QD signal. Also, time-dependent analysis of intracellular GFP signal could be used as a read-out for plasmid-induced protein expression. Similar design of QD–DNA conjugates have been used for by Wu et al. for transfection of plasmid and over expression of a number of proteins in cellulo [119]. An alternative approach by Li et al. involved transcriptional activation of the ‘conjugated’ plasmid in response to spatial stimulus [120]. This design has been implemented in loading plasmid encoding EGFP onto QD surfaces and transfecting this complex into cells. The complexes localize in the nucleus and remain transcriptionally inactive. However, addition of glutathione in the media triggers plasmid release from the QD surface, which in turn makes the plasmid transcriptionally more active. This design provides additional control over the transcription and is a classic example of biosensing and therapeutic application of QD–DNA conjugates.
Another class of therapeutic applications is the use of QD–DNA conjugates for siRNA-mediated gene silencing [121–123]. First demonstrated by Derfus et al., QDs were labelled with siRNA against EGFP and transfected in stable cell lines. The concomitant silencing of EGFP gene was observed by comparison of EGFP signal in trypsinized cells. This design has been further tuned by others to show QD–DNA mediated gene silencing in other cellular contexts. However, several reports also suggest that different nanoparticles can often get trapped in the endo-lysosomal pathway and get degraded. This impedes their full potential in therapeutics.
Therefore, for more widespread applications of QD–DNA conjugates in therapeutics, a rigorous and careful evaluation of nanoparticle surface parameters is absolutely essential.
7. Perspectives
Since the first report of DNA conjugation onto QDs about 15 years back, there has been a lot of progress in the strategies of ligand exchange, coupling and the eventual applications of QD–DNA conjugates. This interesting nanomaterial–biology interface is increasingly developing and the scope and applications of such tailored materials are also expanding. However, there are still some persistent challenges. For example, solution stability along with compact structure of nanobioconjugates can be improved further. Additionally, purification methods involving dilution and centrifugation cause loss of several ligands from the surface of QDs that reduce both the QY and solution stability of QDs. The third and bigger challenge is the reduction of non-specific adsorption on the surfaces of the cells. For any specific intracellular application, the interaction of biofunctionalized QD with cells should be highly specific. However, owing to the presence of DNA, there is often an increased non-specific uptake of QD–DNA conjugates within cells. This is attributed to the interactions of nucleic acids to anionic ligand binding scavenger receptors. This type of uptake of QD–DNA conjugates eventually traps these nanomaterials in the endo-lysosomal system that causes loss of specific functionality. Therefore, careful material design strategies need to be devised that can bypass or overcome this degradation pathway and efficiently deliver nanomaterials to the cytosol. Another challenge is to carefully control the stoichiometry of DNA molecules on the QDs. Current solution approaches have limited success in predicting and realizing specific conjugates with controlled number of DNA molecules, which is essential for several biosensing applications.
Rapidly developing infrastructure, ligand chemistry and understanding of interactions between material and biological interface have the set stage for challenging applications of QD–DNA conjugates such as multiplex biosensing, simultaneous tracking of cellular pathways, and rapid and efficient delivery of therapeutic payload. These advances continue to demonstrate and inspire superior performance of QDs for diversified applications ranging from material design to molecular therapy.
We are unable to accommodate several innovative works of our co-workers from this rapidly expanding community, for the sake of brevity of this review.
Acknowledgements
A.B. expresses her gratitude towards Prof. Yamuna Krishnan and Dr Chloé Grazon for continued support.
Competing interests
We have no competing interests.
Funding
This research was funded by CEFIPRA for PhD fellowship.
References
- 1.Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. 1998. Semiconductor nanocrystals as fluorescent biological labels. Science 281, 2013–2016. ( 10.1126/science.281.5385.2013) [DOI] [PubMed] [Google Scholar]
- 2.Chan WCW, Nie S. 1998. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science 281, 2016–2018. ( 10.1126/science.281.5385.2016) [DOI] [PubMed] [Google Scholar]
- 3.Michalet X, et al. 2005. Quantum dots for live cells, in vivo imaging, and diagnostics. Science 307, 538–544. ( 10.1126/science.1104274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. 2008. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 5, 763–775. ( 10.1038/NMETH.1248) [DOI] [PubMed] [Google Scholar]
- 5.Yin Y, Alivisatos AP. 2005. Colloidal nanocrystal synthesis and the organic–inorganic interface. Nature 437, 664–670. ( 10.1038/nature04165) [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez GR, Hendricks MP, Cossairt BM, Liu H, Owen JS. 2013. Conversion reactions of cadmium chalcogenide nanocrystal precursors. Chem. Mater. 25, 1233–1249. ( 10.1021/cm3035642) [DOI] [Google Scholar]
- 7.Carion O, Malher B, Pons T, Dubertret B. 2007. Synthesis, encapsulation, purification and coupling of single quantum dots in phospholipid micelles for their use in cellular and in vivo imaging. Nat. Protoc. 2, 2383–2390. ( 10.1038/nprot.2007.351) [DOI] [PubMed] [Google Scholar]
- 8.Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A. 2002. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science 298, 1759–1762. ( 10.1126/science.1077194) [DOI] [PubMed] [Google Scholar]
- 9.Voura EB, Jaiswal JK, Mattoussi H, Simon SM. 2004. Tracking metastatic tumor cell extravasation with quantum dot nanocrystals and fluorescence emission-scanning microscopy. Nat. Med. 10, 993–998. ( 10.1038/nm1096) [DOI] [PubMed] [Google Scholar]
- 10.Muro E, Fragola A, Pons T, Lequeux N, Ioannou A, Skourides P, Dubertret B. 2012. Comparing intracellular stability and targeting of sulfobetaine quantum dots with other surface chemistries in live cells. Small 8, 1029–1037. ( 10.1002/smll.201101787) [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Liu H, Liu J, Haley KN, Treadway JA, Larson JP, Ge N, Peale F, Bruchez MP. 2002. Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat. Biotechnol. 21, 41–46. ( 10.1038/nbt764) [DOI] [PubMed] [Google Scholar]
- 12.Pellegrino T, et al. 2004. Hydrophobic nanocrystals coated with an amphiphilic polymer shell: a general route to water soluble nanocrystals. Nano Lett. 4, 703–707. ( 10.1021/nl035172j) [DOI] [Google Scholar]
- 13.Kairdolf BA, Mancini MC, Smith AM, Nie S, Qd Q. 2008. Minimizing nonspecific cellular binding of quantum dots with hydroxyl-derivatized surface coatings. Anal. Chem. 80, 3029–3034. ( 10.1021/ac800068q) [DOI] [PubMed] [Google Scholar]
- 14.Zhang F, Lees E, Amin F, Rivera Gil P, Yang F, Mulvaney P, Parak WJ. 2011. Polymer-coated nanoparticles: a universal tool for biolabelling experiments. Small 7, 3113–3127. ( 10.1002/smll.201100608) [DOI] [PubMed] [Google Scholar]
- 15.Zhou J, Yang Y, Zhang C. 2015. Toward biocompatible semiconductor quantum dots: from biosynthesis and bioconjugation to biomedical application. Chem. Rev. 115, 11 669–11 717. ( 10.1021/acs.chemrev.5b00049) [DOI] [PubMed] [Google Scholar]
- 16.Beaune G, Tamang S, Bernardin A, Bayle-Guillemaud P. 2011. Luminescence of polyethylene glycol coated CdSeTe/ZnS and InP/ZnS nanoparticles in the presence of copper cations. ChemPhysChem 12, 2247–2254. ( 10.1002/cphc.201100266) [DOI] [PubMed] [Google Scholar]
- 17.Clarke SJ, Hollmann CA, Aldaye FA, Nadeau JL. 2008. Effect of ligand density on the spectral, physical, and biological characteristics of CdSe/ZnS quantum dots. Bioconjug. Chem. 19, 562–568. ( 10.1021/bc700404v) [DOI] [PubMed] [Google Scholar]
- 18.Liang M, et al. 2016. Multidentate polymer coatings for compact and homogeneous quantum dots with efficient bioconjugation. J. Am. Chem. Soc. 138, 3382–3394. ( 10.1021/jacs.5b12378) [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Kim M, Wang Y, Wang YA. 2006. Highly luminescent, stable, and water-soluble CdSe/CdS core–shell dendron nanocrystals with carboxylate anchoring groups. Langmuir 22, 6341–6345. ( 10.1021/la052747e) [DOI] [PubMed] [Google Scholar]
- 20.Gomes R, Hassinen A, Szczygiel A, Zhao Q, Hens Z. 2011. Binding of phosphonic acids to CdSe quantum dots: a solution NMR study. J. Phys. Chem. Lett. 2, 145–152. ( 10.1021/jz1016729) [DOI] [Google Scholar]
- 21.Chen C, Yet C, Wang H, Chao C. 1999. Self-assembly of monolayers of cadmium selenide nanocrystals with dual color emission. Langmuir 15, 6845–6850. ( 10.1021/la990165p) [DOI] [Google Scholar]
- 22.Hanaki K, Momo A, Oku T, Komoto A, Maenosono S, Yamaguchi Y, Yamamoto K. 2003. Semiconductor quantum dot/albumin complex is a long-life and highly photostable endosome marker. Biochem. Biophys. Res. Commun. 302, 496–501. ( 10.1016/S0006-291X(03)00211-0) [DOI] [PubMed] [Google Scholar]
- 23.Liu W, Choi HS, Zimmer JP, Tanaka E, Frangioni JV, Bawendi M. 2007. Compact cysteine-coated CdSe(ZnCdS) quantum dots for in vivo applications. J. Am. Chem. Soc. 129, 14 530–14 531. ( 10.1021/ja073790m) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamang S, Beaune G, Texier I, Reiss P. 2011. Aqueous phase transfer of InP/ZnS nanocrystals conserving fluorescence and high colloidal stability. ACS Nano 5, 9392–9402. ( 10.1021/nn203598c) [DOI] [PubMed] [Google Scholar]
- 25.Jiang X, Ro C, Hafner M, Brandholt S, Do M, Nienhaus GU. 2010. Endo- and exocytosis of zwitterionic quantum dot nanoparticles by live HeLa cells. ACS Nano 4, 6787–6797. ( 10.1021/nn101277w) [DOI] [PubMed] [Google Scholar]
- 26.Zheng H, Mortensen LJ, Delouise LA. 2013. Thiol antioxidant-functionalized CdSe/ZnS quantum dots: synthesis, characterization, cytotoxicity. J. Biomed. Nanotechnol. 9, 382–392. ( 10.1166/jbn.2013.1561) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uyeda HT, Medintz IL, Jaiswal JK, Simon SM, Mattoussi H. 2005. Synthesis of compact multidentate ligands to prepare stable hydrophilic quantum dot fluorophores. J. Am. Chem. Soc. 127, 3870–3878. ( 10.1021/ja044031w) [DOI] [PubMed] [Google Scholar]
- 28.Jaiswal JK, Goldman ER, Mattoussi H, Simon SM. 2004. Use of quantum dots for live cell imaging. Nat. Methods 1, 73–78. ( 10.1038/nmeth1004-73) [DOI] [PubMed] [Google Scholar]
- 29.Clapp AR, Goldman ER, Mattoussi H. 2006. Capping of CdSe-ZnS quantum dots with DHLA and subsequent conjugation with proteins. Nat. Protoc. 1, 1258–1266. ( 10.1038/nprot.2006.184) [DOI] [PubMed] [Google Scholar]
- 30.Goldman ER, Mattoussi H, Anderson GP, Medintz IL, Mauro JM. 2005. Fluoroimmunoassays using antibody-conjugated quantum dots. Nanobiotechnol. Protocols 303, 19–34. ( 10.1385/1-59259-901-X:019) [DOI] [PubMed] [Google Scholar]
- 31.Susumu K, Uyeda HT, Medintz IL, Pons T, Delehanty JB, Mattoussi H. 2007. Enhancing the stability and biological functionalities of quantum dots via compact multifunctional ligands. J. Am. Chem. Soc. 129, 13 987–13 996. ( 10.1021/ja0749744) [DOI] [PubMed] [Google Scholar]
- 32.Muro E, Pons T, Lequeux N, Fragola A, Sanson N. 2010. Small and stable sulfobetaine zwitterionic quantum dots for functional live-cell imaging. J. Am. Chem. Soc. 132, 4556–4557. ( 10.1021/ja1005493) [DOI] [PubMed] [Google Scholar]
- 33.Mattoussi H, Mauro JM, Goldman ER, Anderson GP, Sundar VC, Mikulec FV, Bawendi MG. 2000. Self-assembly of CdSe–ZnS quantum dot bioconjugates using an engineered recombinant protein. J. Am. Chem. Soc. 122, 12 142–12 150. ( 10.1021/ja002535y) [DOI] [Google Scholar]
- 34.Susumu K, Mei BC, Mattoussi H. 2009. Multifunctional ligands based on dihydrolipoic acid and polyethylene glycol to promote biocompatibility of quantum dots. Nat. Protoc. 4, 424–436. ( 10.1038/nprot.2008.247) [DOI] [PubMed] [Google Scholar]
- 35.Liu W, Howarth M, Greytak AB, Zheng Y, Nocera DG, Ting AY, Bawendi MG. 2008. Compact biocompatible quantum dots functionalized for cellular imaging. J. Am. Chem. Soc. 130, 1274–1284. ( 10.1021/ja076069p) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin C-AJ, Sperling RA, Li JK, Yang T-Y, Li P-Y, Zanella M, Chang WH, Parak WJ. 2008. Design of an amphiphilic polymer for nanoparticle coating and functionalization. Small 4, 334–341. ( 10.1002/smll.200700654) [DOI] [PubMed] [Google Scholar]
- 37.Zhang F, Lees E, Amin F, Gil PR, Yang F, Mulvaney P, Parak WJ. 2011. Polymer-coated nanoparticles: a universal tool for biolabelling experiments. Small 7, 3113–3127. ( 10.1002/smll.201100608) [DOI] [PubMed] [Google Scholar]
- 38.Giovanelli E, Muro E, Sitbon G, Hanafi M, Pons T, Dubertret B, Lequeux N. 2012. Highly enhanced affinity of multidentate versus bidentate zwitterionic ligands for long-term quantum dot bioimaging. Langmuir 28, 15 177–15 184. ( 10.1021/la302896x) [DOI] [PubMed] [Google Scholar]
- 39.Zhan N, Palui G, Safi M, Ji X, Mattoussi H. 2013. Multidentate zwitterionic ligands provide compact and highly biocompatible quantum dots. J. Am. Chem. Soc. 135, 13 786–13 795. ( 10.1021/ja405010v) [DOI] [PubMed] [Google Scholar]
- 40.Tasso M, Giovanelli E, Zala D, Bouccara S, Fragola A, Hanafi M, Lenkei Z, Pons T, Lequeux N. 2015. Sulfobetaine à vinylimidazole block copolymers: a robust quantum dot surface chemistry expanding bioimaging's horizons. ACS Nano 9, 11 479–11 489. ( 10.1021/acsnano.5b05705) [DOI] [PubMed] [Google Scholar]
- 41.Susumu K, et al. 2014. A new family of pyridine-appended multidentate polymers as hydrophilic surface ligands for preparing stable biocompatible quantum dots. Chem. Mater. 26, 5327–5344. ( 10.1021/cm502386f) [DOI] [Google Scholar]
- 42.Sperling RA, Parak WJ. 2010. Surface modification, functionalization and bioconjugation of colloidal inorganic nanoparticles. Philos. Trans. A. Math. Phys. Eng. Sci. 368, 1333–1383. ( 10.1098/rsta.2009.0273) [DOI] [PubMed] [Google Scholar]
- 43.Selvan ST. 2010. Silica-coated quantum dots and magnetic nanoparticles for bioimaging applications. Biointerphases 5, FA110–FA115. ( 10.1116/1.3516492) [DOI] [PubMed] [Google Scholar]
- 44.Mitchell GP, Mirkin CA, Letsinger RL. 1999. Programmed assembly of DNA functionalized quantum dots. J. Am. Chem. Soc. 121, 8122–8123. ( 10.1021/ja991662v) [DOI] [Google Scholar]
- 45.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. 2005. Quantum dot bioconjugates for imaging, labelling and sensing. Nat. Mater. 4, 435–446. ( 10.1038/nmat1390) [DOI] [PubMed] [Google Scholar]
- 46.Gill R, Willner I, Shweky I, Banin U. 2005. Fluorescence resonance energy transfer in CdSe/ZnS-DNA conjugates: probing hybridization and DNA cleavage. J. Phys. Chem. B 109, 23 715–23 719. ( 10.1021/jp054874p) [DOI] [PubMed] [Google Scholar]
- 47.Zhou D, Piper JD, Abell C, Klenerman D, Kang D-J, Ying L. 2005. Fluorescence resonance energy transfer between a quantum dot donor and a dye acceptor attached to DNA. Chem. Commun. 38, 4807–4809. ( 10.1039/b508911e) [DOI] [PubMed] [Google Scholar]
- 48.Wu S-M, Zhao X, Zhang Z-L, Xie H-Y, Tian Z-Q, Peng J, Lu Z-X, Pang D-W, Xie Z-X. 2006. Quantum-dot-labeled DNA probes for fluorescence in situ hybridization (FISH) in the microorganism Escherichia coli. ChemPhysChem 7, 1062–1067. ( 10.1002/cphc.200500608) [DOI] [PubMed] [Google Scholar]
- 49.Sapsford KE, Pons T, Medintz IL, Higashiya S, Brunel FM, Dawson PE, Mattoussi H. 2007. Kinetics of metal-affinity driven self-assembly between proteins or peptides and CdSe–ZnS quantum dots. J. Phys. Chem. C 111, 11 528–11 538. ( 10.1021/jp073550t) [DOI] [Google Scholar]
- 50.Medintz IL, Berti L, Pons T, Grimes AF, English DS, Alessandrini A, Facci P, Mattoussi H. 2007. A reactive peptidic linker for self-assembling hybrid quantum dot-DNA bioconjugates. Nano Lett. 7, 1741–1748. ( 10.1021/nl070782v) [DOI] [PubMed] [Google Scholar]
- 51.Harrison JG, Balasubramanian S. 1998. Synthesis and hybridization analysis of a small library of peptide–oligonucleotide conjugates. Nucleic Acid Res. 26, 3136–3145. ( 10.1093/nar/26.13.3136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwarz FP, Robinson S, Butler JM. 1999. Thermodynamic comparison of PNA/DNA and DNA/DNA hybridization reactions at ambient temperature. Nucleic Acid Res. 27, 4792–4800. ( 10.1093/nar/27.24.4792) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar A, Phadtare S. 2003. Assembling gold nanoparticles in solution using phosphorothioate DNA as structural interconnects. Curr. Sci. 84, 71–74. [Google Scholar]
- 54.Lee JH, Wernette DP, Yigit MV, Liu J, Wang Z, Lu Y. 2007. Site-specific control of distances between gold nanoparticles using phosphorothioate anchors on DNA and a short bifunctional molecular fastener. Angew. Chem. 46, 9006–9010. ( 10.1002/anie.200702569) [DOI] [PubMed] [Google Scholar]
- 55.Jiang G, Susha AS, Lutich AA, Stefani FD, Feldmann J, Rogach AL. 2009. Cascaded FRET in conjugated polymer/quantum dot/dye-labeled DNA complexes for DNA hybridization detection. ACS Nano 3, 4127–4131. ( 10.1021/nn901324y) [DOI] [PubMed] [Google Scholar]
- 56.Zhou W, Wang F, Ding J, Liu J. 2014. Tandem phosphorothioate modifications for DNA adsorption strength and polarity control on gold nanoparticles. ACS Appl. Mater. Interfaces 6, 140822145406001 ( 10.1021/am504791b) [DOI] [PubMed] [Google Scholar]
- 57.Liu C, Mao G, Su C, Ji X, He Z. 2015. Aptamer functionalised CdTe:Zn2+ quantum dots for the detection of tomato systemin. Anal. Methods 7, 7748–7752. ( 10.1039/C5AY01728A) [DOI] [Google Scholar]
- 58.Farlow J, Seo D, Broaders KE, Taylor MJ, Gartner ZJ, Jun Y. 2013. Formation of targeted monovalent quantum dots by steric exclusion. Nat. Methods 10, 1203–1205. ( 10.1038/nmeth.2682) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aimé A, Beztsinna N, Patwa A, Pokolenko A, Bestel I, Barthélémy P. 2013. quantum dot lipid oligonucleotide bioconjugates: toward a new anti-microRNA nanoplatform. Bioconjug. Chem. 24, 1345–1355. ( 10.1021/bc400157z) [DOI] [PubMed] [Google Scholar]
- 60.Lehn RC, Van Ricci M, Silva PHJ, Andreozzi P, Reguera J, Voı K, Stellacci F, Alexander-katz A. 2014. Lipid tail protrusions mediate the insertion of nanoparticles into model cell membranes. Nat. Commun. 5, 1–11. ( 10.1038/ncomms5482) [DOI] [PubMed] [Google Scholar]
- 61.Chen AA, Derfus AM, Khetani SR, Bhatia SN. 2005. Quantum dots to monitor RNAi delivery and improve gene silencing. Nucleic Acid Res. 33, e190 ( 10.1093/nar/gni188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nel AE, Mädler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. 2009. Understanding biophysicochemical interactions at the nano–bio interface. Nat. Mater. 8, 543–557. ( 10.1038/nmat2442) [DOI] [PubMed] [Google Scholar]
- 63.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. 1996. A DNA based method for rationally assembling nanoparticles into macroscopic materials. Nature 382, 607–609. ( 10.1038/382607a0) [DOI] [PubMed] [Google Scholar]
- 64.Peng H, Zhang L, Kja THM, Soeller C, Travas-sejdic J. 2007. DNA hybridization detection with blue luminescent quantum dots and dye labelled single stranded DNA. J. Am. Chem. Soc. 129, 3048–3049. ( 10.1021/ja0685452) [DOI] [PubMed] [Google Scholar]
- 65.Lee J, Choi Y, Kim J, Park E, Song R. 2009. Positively charged compact quantum dot–DNA complexes for detection of nucleic acids. Chemphyschem 10, 806–811. ( 10.1002/cphc.200800504) [DOI] [PubMed] [Google Scholar]
- 66.Zintchenko A, Susha AS, Concia M, Feldmann J, Wagner E, Rogach AL, Ogris M. 2009. Drug nanocarriers labeled with near-infrared-emitting quantum dots (quantoplexes): imaging fast dynamics of distribution in living animals. Mol. Ther. 17, 1849–1856. ( 10.1038/mt.2009.201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bagalkot V, Gao X. 2011. siRNA-aptamer chimeras on nanoparticles: preserving targeting functionality for effective gene silencing. ACS Nano 5, 8131–8139. ( 10.1021/nn202772p) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park J, et al. 2015. Quantum dots in an amphiphilic polyethyleneimine derivative platform for cellular labeling, targeting, gene delivery, and ratiometric oxygen sensing. ACS Nano 9, 6511–6521. ( 10.1021/acsnano.5b02357) [DOI] [PubMed] [Google Scholar]
- 69.Algar WR, Krull UJ. 2006. Adsorption and hybridization of oligonucleotides on mercaptoacetic acid-capped CdSe/ZnS quantum dots and quantum dot–oligonucleotide conjugates. Langmuir 22, 11 346–11 352. ( 10.1021/la062217y) [DOI] [PubMed] [Google Scholar]
- 70.Kwon H, Hong S, Kim H, Choi Y, Kim J, Song R. 2010. Controlled stoichiometric synthesis of DNA–quantum dot conjugates using Ni-mediated coordination chemistry. Chem. Commun. 46, 8959–8961. ( 10.1039/c0cc03462b) [DOI] [PubMed] [Google Scholar]
- 71.Susumu K, Medintz IL, Delehanty JB, Boeneman K, Mattoussi H. 2010. Modification of poly(ethylene glycol)-capped quantum dots with nickel nitrilotriacetic acid and self-assembly with histidine-tagged proteins. J. Phys. Chem. C 114, 13 526–13 531. ( 10.1021/jp103872j) [DOI] [Google Scholar]
- 72.Han M, Gao X, Su JZ, Nie S. 2001. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat. Biotechnol. 19, 631–635. ( 10.1038/90228) [DOI] [PubMed] [Google Scholar]
- 73.Fu A, Micheel CM, Cha J, Chang H, Yang H, Alivisatos AP. 2004. Discrete nanostructures of quantum dots/Au with DNA. J. Am. Chem. Soc. 126, 10 832–10 833. ( 10.1021/ja046747x) [DOI] [PubMed] [Google Scholar]
- 74.Howarth M, Chinnapen DJ, Gerrow K, Dorrestein PC, Grandy R, Kelleher NL, El-husseini A, Ting AY. 2006. A monovalent streptavidin with a single femtomolar biotin binding site. Nat. Methods 3, 267–273. ( 10.1038/NMETHXXX.A) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu S. 2005. Engineering soluble monomeric streptavidin with reversible biotin binding capability. J. Biol. Chem. 280, 23 225–23 231. ( 10.1074/jbc.M501733200) [DOI] [PubMed] [Google Scholar]
- 76.Howarth M, Liu W, Puthenveetil S, Zheng Y, Marshall LF, Schmidt M, Wittrup KD, Bawendi MG, Ting AY. 2008. Monovalent, reduced-size quantum dots for imaging receptors on living cells. Nat. Methods 5, 397–399. ( 10.1038/nmeth.1206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lim KH, Huang H, Pralle A, Park S. 2013. Stable, high-affinity streptavidin monomer for protein labeling and monovalent biotin detection. Biotechnol. Bioeng. 110, 57–67. ( 10.1002/bit.24605) [DOI] [PubMed] [Google Scholar]
- 78.Helppolainen SH, et al. 2007. Rhizavidin from Rhizobium etli: the first natural dimer in the avidin. Biochem. J. 405, 397–405. ( 10.1042/BJ20070076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y, Liu KJ, Wang T, Shih I, Wang T. 2012. Mapping DNA quantity into electrophoretic mobility through quantum dot nanotethers for high-resolution genetic and epigenetic analysis. ACS Nano 6, 858–864. ( 10.1021/nn204377k) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xing Y, et al. 2007. Bioconjugated quantum dots for multiplexed and quantitative immunohistochemistry. Nat. Protoc. 2, 1152–1165. ( 10.1038/nprot.2007.107) [DOI] [PubMed] [Google Scholar]
- 81.Wu C, Cupps JM, Fan X. 2009. Compact quantum dot probes for rapid and sensitive DNA detection using highly efficient fluorescence resonant energy transfer. Nanotechnology 20, 305502 ( 10.1088/0957-4484/20/30/305502) [DOI] [PubMed] [Google Scholar]
- 82.Sun D, Gang O. 2013. DNA-functionalized quantum dots: fabrication, structural, and physicochemical properties. Langmuir 29, 7038–7046. ( 10.1021/la4000186) [DOI] [PubMed] [Google Scholar]
- 83.Srinivasan C, Lee J, Papadimitrakopoulos F, Silbart LK, Zhao M, Burgess DJ. 2006. Labeling and intracellular tracking of functionally active plasmid DNA with semiconductor quantum dots. Mol. Ther. 14, 192–201. ( 10.1016/j.ymthe.2006.03.010) [DOI] [PubMed] [Google Scholar]
- 84.Banerjee A, Grazon C, Nadal B, Pons T, Krishnan Y, Dubertret B. 2015. Fast, efficient and stable conjugation of multiple DNA strands on colloidal quantum dots. Bioconjug. Chem. 26, 1582–1589. ( 10.1021/acs.bioconjchem.5b00221) [DOI] [PubMed] [Google Scholar]
- 85.Zhang H, Feng G, Guo Y, Zhou D. 2013. Robust and specific ratiometric biosensing using a copper-free clicked quantum dot–DNA aptamer sensor. Nanoscale 5, 10 307–10 315. ( 10.1039/c3nr02897f) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Farokhzad OC, Jon S, Khademhosseini A, Tran TT, Lavan DA, Langer R. 2004. Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells. Cancer Res. 64, 7668–7672. ( 10.1158/0008-5472.CAN-04-2550) [DOI] [PubMed] [Google Scholar]
- 87.Smith AM, Duan H, Rhyner MN, Ruan G. 2006. A systematic examination of surface coatings on the optical and chemical properties of semiconductor quantum dots. Phys. Chem. Chem. Phys. 8, 3895–3903. ( 10.1039/b606572b) [DOI] [PubMed] [Google Scholar]
- 88.Tasso M, Singh MK, Giovanelli E, Fragola A, Loriette V, Regairaz M, Lenkei Z, Lequeux N, Pons T. 2015. Oriented bioconjugation of unmodified antibodies to quantum dots capped with copolymeric ligands as versatile cellular imaging tools. ACS Appl. Mater. Interfaces 7, 26 904–26 913. ( 10.1021/acsami.5b09777) [DOI] [PubMed] [Google Scholar]
- 89.Pons T, Uyeda HT, Medintz IL, Mattoussi H. 2006. Hydrodynamic dimensions, electrophoretic mobility, and stability of hydrophilic quantum dots. J. Phys. Chem. B 110, 20 308–20 316. ( 10.1021/jp065041h) [DOI] [PubMed] [Google Scholar]
- 90.Boeneman K, Deschamps JR, Buckhout-White S, Prasuhn DE, Goldman R, Ancona M, Medintz IL. 2015. Quantum dot DNA bioconjugate: attachment chemistry strongly influences the resulting composite architecture. ACS Nano 4, 7253–7266. ( 10.1021/nn1021346.Quantum) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ackerson CJ, Sykes MT, Kornberg RD. 2005. Defined DNA–nanoparticle conjugates. Proc. Natl Acad. Sci. USA 102, 13 383–13 385. ( 10.1073/pnas.0506290102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Taton TA. 2002. Preparation of gold nanoparticle–DNA. In Current protocols in nucleic acid chemistry, 122.1–12.2.12. Wiley Online Library. ( 10.1002/0471142700.nc1202s09) [DOI] [PubMed] [Google Scholar]
- 93.Carstairs HMJ, Lymperopoulos K, Kapanidis AN, Bath J, Turberfield AJ. 2009. DNA monofunctionalization of quantum dots. ChemBioChem 10, 1781–1783. ( 10.1002/cbic.200900300) [DOI] [PubMed] [Google Scholar]
- 94.Budelier K, Schorr J. 1998. Purification of DNA by anion-exchange chromatography. In Current protocols in molecular biology, pp. 11–18. Wiley Online Library. (doi:0.1002/0471142727.mb0201bs42) [DOI] [PubMed] [Google Scholar]
- 95.Uddayasankar U, Zhang Z, Shergill RT, Gradinaru CC, Krull UJ. 2014. Isolation of monovalent quantum dot-nucleic acid conjugates using magnetic beads. Bioconjug. Chem. 25, 1342–1350. ( 10.1021/bc5002032) [DOI] [PubMed] [Google Scholar]
- 96.Schreiber R, Do J, Roller E-M, Zhang T, Schüller VJ, Nickels PC, Feldmann J, Liedl T. 2013. Hierarchical assembly of metal nanoparticles, quantum dots and organic dyes using DNA origami scaffolds. Nat. Nanotechnol. 9, 74–78. ( 10.1038/nnano.2013.253) [DOI] [PubMed] [Google Scholar]
- 97.Tikhomirov G, Hoogland S, Lee PE, Fischer A, Sargent EH, Kelley SO. 2011. DNA-based programming of quantum dot valency, self-assembly and luminescence. Nat. Nanotechnol. 6, 485–490. ( 10.1038/nnano.2011.100) [DOI] [PubMed] [Google Scholar]
- 98.Ko SH, Du K, Liddle JA. 2013. Quantum-dot fluorescence lifetime engineering with DNA origami constructs. Angew. Chem. 52, 1193–1197. ( 10.1002/anie.201206253) [DOI] [PubMed] [Google Scholar]
- 99.Wang Z, Dick T, Susha AS, Shan M, Kershaw SV, Kwan P, Rogach AL. 2016. Aggregation-free DNA nanocage/quantum dot complexes based on electrostatic adsorption. Colloids Surfaces A Physicochem. Eng. Asp. 495, 62–67. ( 10.1016/j.colsurfa.2016.02.002) [DOI] [Google Scholar]
- 100.Wang Z, Xue Q, Tian W, Jiang W. 2012. Quantitative detection of single DNA molecules on DNA tetrahedron decorated substrates. Chem. Commun. 48, 9661–9663. ( 10.1039/c2cc35208g) [DOI] [PubMed] [Google Scholar]
- 101.Dubertret B. 2005. Quantum dots DNA detectives. Nat. Mater. 4, 797–798. ( 10.1038/nmat1520) [DOI] [PubMed] [Google Scholar]
- 102.Crut A, Géron-Landre B, Bonnet I, Bonneau S, Desbiolles P, Escudé C. 2005. Detection of single DNA molecules by multicolor quantum-dot end-labeling. Nucleic Acids Res. 33, e98 ( 10.1093/nar/gni097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Algar WR, Krull UJ. 2007. Towards multi-colour strategies for the detection of oligonucleotide hybridization using quantum dots as energy donors in fluorescence resonance energy transfer (FRET). Anal. Chim. Acta 581, 193–201. ( 10.1016/j.aca.2006.08.026) [DOI] [PubMed] [Google Scholar]
- 104.Michaelis J, van der Heden van Noort GJ, Seitz O. 2014. DNA-triggered dye Transfer on a quantum dot. Bioconjug. Chem. 25, 18–23. ( 10.1021/bc400494j) [DOI] [PubMed] [Google Scholar]
- 105.Zhang C-Y, Yeh H-C, Kuroki MT, Wang T-H. 2005. Single-quantum-dot-based DNA nanosensor. Nat. Mater. 4, 826–831. ( 10.1038/nmat1508) [DOI] [PubMed] [Google Scholar]
- 106.Patolsky F, Gill R, Weizmann Y, Mokari T, Banin U, Willner I. 2003. Lighting-up the dynamics of telomerization and DNA replication by CdSe–ZnS quantum dots. J. Am. Chem. Soc. 125, 13 918–13 919. ( 10.1021/ja035848c) [DOI] [PubMed] [Google Scholar]
- 107.Suzuki M, Husimi Y, Komatsu H, Suzuki K, Douglas KT. 2008. Quantum dot FRET biosensors that respond to pH, to proteolytic or nucleolytic cleavage, to DNA synthesis, or to a multiplexing combination. J. Am. Chem. Soc. 130, 5720–5725. ( 10.1021/ja710870e) [DOI] [PubMed] [Google Scholar]
- 108.Shahmuradyan A, Krull UJ. 2016. Intrinsically labeled fluorescent oligonucleotide probes on quantum dots for transduction of nucleic acid hybridization. Anal. Chem. 88, 3186–3193. ( 10.1021/acs.analchem.5b04536) [DOI] [PubMed] [Google Scholar]
- 109.Geißler D, Hildebrandt N. 2011. Lanthanide complexes in FRET applications. Curr. Inorg. Chem. 1, 17–35. ( 10.2174/1877944111101010017) [DOI] [Google Scholar]
- 110.Wegner KD, Jin Z, Lindén S, Jennings TL, Hildebrandt N. 2013. Quantum-dot-based Förster resonance energy transfer immunoassay for sensitive clinical diagnostics of low-volume serum samples. ACS Nano 7, 7411–7419. ( 10.1021/nn403253y) [DOI] [PubMed] [Google Scholar]
- 111.Lindén S, Singh MK, Wegner KD, Regairaz M, Dautry F, Treussart F, Hildebrandt N. 2015. Terbium-based time-gated Förster resonance energy transfer imaging for evaluating protein–protein interactions on cell membranes. Dalt. Trans. 44, 4994–5003. ( 10.1039/c4dt02884h) [DOI] [PubMed] [Google Scholar]
- 112.Cardoso M, Santos D, Hildebrandt N. In press. Recent developments in lanthanide-to-quantum dot FRET using time-gated fluorescence detection and photon upconversion. Trends Anal. Chem. ( 10.1016/j.trac.2016.03.005) [DOI] [Google Scholar]
- 113.Jin Z, Geißler D, Qiu X, Wegner KD, Hildebrandt N. 2015. A rapid, amplification-free, and sensitive diagnostic assay for single-step multiplexed fluorescence detection of microRNA. Angew. Chem. 54, 10 024–10 029. ( 10.1002/anie.201504887) [DOI] [PubMed] [Google Scholar]
- 114.Kay ER, Lee J, Nocera DG, Bawendi MG. 2013. Conformational control of energy transfer: a mechanism for biocompatible nanocrystal-based sensors. Angew. Chem. Int. Ed. Engl. 52, 1165–1169. ( 10.1002/anie.201207181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bentolila AL, Weiss S. 2006. Single-step multicolor fluorescence in situ hybridization using semiconductor quantum dot-DNA conjugates. Cell Biochem. Biophys. 45, 59–70. ( 10.1385/CBB) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xiao Y, Barker PE. 2006. Semiconductor nanocrystal probes for human metaphase chromosomes. Nucleic Acid Res. 32, 28–32. ( 10.1093/nar/gnh024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liang R, Li W, Li Y, Tan C, Li J, Jin Y, Ruan K. 2005. An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acid Res. 33, e17 ( 10.1093/nar/gni019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jo H, Ban C. 2016. Aptamer–nanoparticle complexes as powerful diagnostic and therapeutic tools. Exp. Mol. Med. 48, e230 ( 10.1038/emm.2016.44) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu Y, Eisele K, Doroshenko M, Algara-Siller G, Kaiser U. 2012. A quantum dot photoswitch for DNA detection, gene transfection, and live-cell imaging. Small 8, 3465–3475. ( 10.1002/smll.201200409) [DOI] [PubMed] [Google Scholar]
- 120.Li D, Li G, Guo W, Li P, Wang E, Wang J. 2008. Glutathione-mediated release of functional plasmid DNA from positively charged quantum dots. Biomaterials 29, 2776–2782. ( 10.1016/j.biomaterials.2008.03.007) [DOI] [PubMed] [Google Scholar]
- 121.Derfus AM, Chen AA, Min D, Ruoslahti E, Bhatia SN. 2007. Targeted quantum dot conjugates for siRNA delivery. Bioconjug. Chem. 18, 1391–1396. ( 10.1021/bc060367e) [DOI] [PubMed] [Google Scholar]
- 122.Elbakry A, Zaky A, Liebl R, Rachel R, Goepferich A, Breunig M. 2009. Layer-by-layer assembled gold nanoparticles for siRNA delivery 2009. Nano Lett. 9, 2059–2064. ( 10.1021/nl9003865) [DOI] [PubMed] [Google Scholar]
- 123.Qi L, Gao X. 2008. Quantum dot–amphipol nanocomplex time imaging of siRNA. ACS Nano 2, 1403–1410. ( 10.1021/nn800280r) [DOI] [PMC free article] [PubMed] [Google Scholar]