Abstract
Heavy meromyosin (HMM) of myosin II and cofilin each binds to actin filaments cooperatively and forms clusters along the filaments, but it is unknown whether the two cooperative bindings are correlated and what physiological roles they have. Fluorescence microscopy demonstrated that HMM-GFP and cofilin-mCherry each bound cooperatively to different parts of actin filaments when they were added simultaneously in 0.2 μM ATP, indicating that the two cooperative bindings are mutually exclusive. In 0.1 mM ATP, the motor domain of myosin (S1) strongly inhibited the formation of cofilin clusters along actin filaments. Under this condition, most actin protomers were unoccupied by S1 at any given moment, suggesting that transiently bound S1 alters the structure of actin filaments cooperatively and/or persistently to inhibit cofilin binding. Consistently, cosedimentation experiments using copolymers of actin and actin-S1 fusion protein demonstrated that the fusion protein affects the neighboring actin protomers, reducing their affinity for cofilin. In reciprocal experiments, cofilin-actin fusion protein reduced the affinity of neighboring actin protomers for S1. Thus, allosteric regulation by cooperative conformational changes of actin filaments contributes to mutually exclusive cooperative binding of myosin II and cofilin to actin filaments, and presumably to the differential localization of both proteins in cells.
Actin filaments perform a variety of important functions in eukaryotic cells, ranging from cytokinesis, lamellipodial extension, adhesion, intracellular transport and nuclear functions, in a manner dependent on the interaction with specific actin binding proteins (ABPs). In a number of cases, researchers have successfully explained the regulation of ABPs by local biochemical signaling, such as phosphorylation and changes in the concentration of signaling molecules. However, not all localized activities of ABPs are fully explained by specific biochemical signaling. Thus, the spatial and temporal regulation of the activity of each ABP in a cell remain major unresolved issues in cell biology.
Meanwhile, a number of biochemical and biophysical studies have established that binding of ABPs induces specific conformational changes in actin filaments, which are in certain cases known to propagate along the filament, i.e., cooperative conformation changes. The pioneering work of Oosawa and colleagues demonstrated that the addition of two-headed soluble fragment of skeletal muscle myosin (heavy meromyosin or HMM) increases the fluorescence intensity of labeled actin, and that this effect became saturated in the presence of a 1/20 molar ratio of HMM to actin1. Subsequently, a number of studies reported similar, myosin-induced cooperative conformational changes in actin filaments detected using different methods (e.g., refs 2,3). Furthermore, a large body of evidence has accumulated that demonstrates that cofilin binding also changes the structure of actin protomers in a cooperative manner. For instance, cofilin binding super-twists the helix of actin filaments by ~25%, involving changes in the atomic structure of each actin protomer4,5,6,7, and this conformational change is propagated to a neighboring bare zone4,8 on the pointed end side7. A single bound cofilin molecule affects the structure of ~100 actin protomers in the filament9,10,11, and increases the affinity for a second cofilin molecule within the ~65 nm vicinity12. Furthermore, binding of certain ABPs to actin filaments is cooperative. Cofilin forms tight clusters along the filament, leaving other parts of the filament mostly bare4,5,7,13. HMM also binds to actin filaments cooperatively under certain conditions14,15. In the presence of low concentrations of ATP, HMM is sparsely bound along the entire length of a fraction of actin filaments, while leaving other filaments bare15. In both cases, HMM and cofilin molecules in clusters along actin filaments do not directly contact with each other6,15. Thus, the cooperative binding of HMM and that of cofilin to actin filaments most likely involve cooperative conformational changes of actin protomers within the filaments.
Stretching actin filaments untwists actin filaments16,17. This mechanosensitivity of actin filaments might provide an additional mechanism to regulate ABP binding18,19. For instance, stretched actin filaments are resistant to the severing activity of cofilin both in vivo and in vitro20, whereas an in vivo study suggested that stretching actin filaments increases the affinity for myosin II19.
This raises an interesting possibility that cooperative conformational changes of actin filaments may contribute to the determination of the function of actin filaments by recruiting specific ABPs15,18,19,21,22. Furthermore, if two ABPs induce different cooperative structural changes in actin filaments, cooperative binding of those two ABPs should be mutually exclusive, leading to functional differentiation of the filaments. Here, we tested this latter prediction experimentally using two different approaches. Myosin II and cofilin were chosen as the candidates of mutually incompatible ABPs, not only because they are major ABPs, but also because these two proteins cause opposite changes in the helical twist of actin filaments, show opposite responses to the stretching of actin filaments, and have different intracellular localizations.
Results
Fluorescence microscopic examination in the presence of low concentrations of ATP
We previously found that when Dictyostelium discoideum (Dd) HMM was fused with GFP (HMM-GFP), and was mixed with excess rabbit skeletal (sk) actin filaments labeled with 1/20 (mol/mol) rhodamine phalloidin in the presence of 0.1~1 μM ATP, certain filaments were diffusely labeled with GFP along the entire lengths, whereas other filaments were left unbound15. This was termed weak cooperativity in the sense that several unbound actin protomers were present between bound HMM molecules. Use of low concentration of ATP was essential to obtain the weak cooperative binding of HMM to actin filaments; in the absence of ATP, HMM-GFP was uniformly bound along all actin filaments because the affinity was too high, and in the presence of higher concentration of ATP, no binding was detected during the observation period. We speculated that the low concentration of ATP allows dissociation-association cycles to facilitate relocation of HMM-GFP to sections of actin filaments with a high affinity for HMM, and eventually stabilizes the binding when depleted by the ATPase activity15. In a similar experiment, Suarez et al. reported that when fluorescently labeled yeast cofilin was mixed with actin filaments, it formed tight clusters along the filaments while apparently leaving other sections of the filaments bare13.
We thus queried what would happen if fluorescent-cofilin and HMM-GFP coexisted in actin solutions in the presence of low concentrations of ATP. We first repeated our previous experiments of weak cooperative binding of HMM-GFP to excess actin filaments, except that in the present study we used Dd actin filaments that were loosely immobilized on a positively charged surface in advance. Furthermore, because phalloidin interferes with cofilin binding23, actin was visualized by chemical labeling with Cy3-succinimide and used without stabilization with phalloidin. The pH of the solution was reduced from 7.4 to 6.5, because slightly acidic condition suppresses the severing activity of cofilin23,24,25 and allows the examination of cofilin binding in the same buffer. Despite these changes in experimental conditions, 7 nM Dd HMM-GFP was bound along limited sections of the actin filaments while apparently leaving other sections of the filament bare (Fig. 1A). We noticed that, however, the binding of HMM-GFP along the filaments was more punctate than in our earlier observation15. We suspect that this difference was primarily due to the use of Dd actin instead of sk actin.
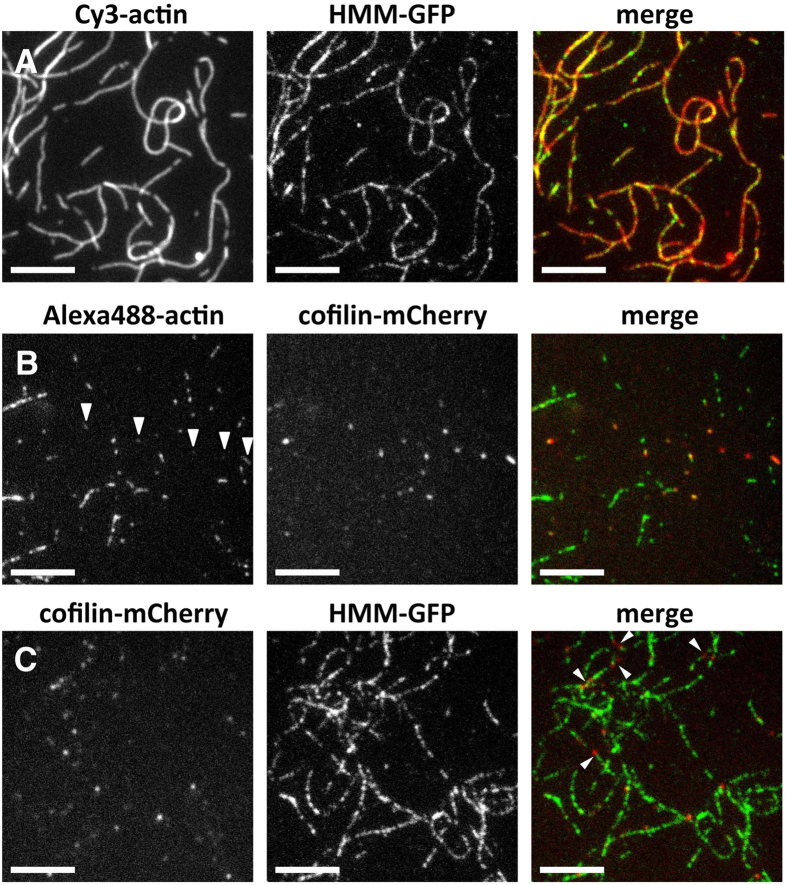
Figure 1. Fluorescence microscopic observation of binding of Dd HMM-GFP and Hs cofilin-mCherry to Dd actin filaments.
In all experiments, actin filaments were loosely immobilized on a positively-charged lipid bilayer and the binding reaction was performed in F-buffer 1 containing 0.2 μM ATP. (A) Cooperative binding of 7 nM HMM-GFP along Cy3-labeled actin filaments, forming short clusters or puncta along the actin filaments. (B) Cooperative binding of 80 nM cofilin-mCherry to Alexa488-labeled actin filaments. Cofilin-mCherry formed tight clusters along filaments. Alexa488 fluorescence of filaments bound with cofilin-mCherry was weaker (arrowheads), presumably due to quenching by FRET with mCherry. (C) Simultaneous addition of 7 nM HMM-GFP and 80 nM cofilin-mCherry to unlabeled actin filaments. Because the concentrations of actin filaments and HMM-GFP were identical to those in (A), most of the filaments were labeled by HMM-GFP in a punctate manner. Other shorter filaments were labeled only by cofilin-mCherry. Additionally, some filaments were labeled by both fluorescent proteins in an alternating punctate manner (arrowheads). Scale bars: 5 μm.
We next added 80 nM human (Hs) cofilin fused with mCherry to Dd actin filaments labeled by Alexa488-succinimide and loosely adsorbed on a positively charged surface. Consistent with an earlier report13, sections of filaments were bound with Hs cofilin-mCherry, while other sections were apparently bare (Fig. 1B). Filaments were shorter than those incubated with HMM-GFP, due to residual severing activity of cofilin.
Finally, a mixture of 80 nM Hs cofilin-mCherry and 7 nM Dd HMM-GFP was allowed to interact with Dd actin filaments loosely adsorbed on a positively charged surface (Fig. 1C). Under this experimental condition, the majority of filaments were labeled by HMM-GFP in a punctate manner, and cofilin-mCherry formed separate puncta. Importantly, punctate staining by HMM-GFP and cofilin-mCherry never overlapped. These results suggest that cooperative bindings of cofilin and HMM to actin filaments are mutually exclusive.
TIRFM observation of the inhibitory effect of S1 on cofilin binding in the presence of 0.1 mM ATP
The mutually exclusive binding of cofilin and HMM to actin filaments could be due either to direct competition of binding sites on actin protomers or to allosteric cooperative conformational changes of actin filaments. To distinguish these two possibilities, binding of cofilin to actin filaments was observed in the presence of the motor domain or subfragment 1 (S1) of sk myosin II and ATP by total internal reflection fluorescence microscopy (TIRFM). The concentration of ATP in this experiment was 0.1 mM, 500-fold higher than that used in the experiment shown above, so that a saturating concentration of ATP was maintained during the observation. Two extra Cys residues at the N-terminus of chicken cofilin 2 were labeled with iodoacetamide fluorescein, and the previous biochemical characterization confirmed that the resultant fluorescein-cofilin binds to actin filaments normally and severs them in a pH-dependent manner26. Sk actin filaments were labeled with Alexa555-succinimide, and were purified by a cycle of depolymerization-polymerization. Cosedimentation experiment confirmed that fluorescein-cofilin interacts normally with Alexa555-actin filaments (Supplementary Fig. S1). Alexa555-actin filaments were loosely immobilized on positively charged inner surfaces of flow cells. Following introduction of 0.3 μM fluorescein-cofilin in 0.1 mM ATP, 40 mM KCl and 20 mM Pipes pH 6.8, the fluorescence of Alexa555 and fluorescein were observed by TIRFM. Spots of fluorescein were visible along actin filaments at 50 s after the addition of fluorescein-cofilin, and the number and size of the fluorescein spots increased over the time course of 10 min (Figs 2A and 3A). In contrast, very few fluorescein-cofilin clusters formed, even after 10 min when 1 μM S1 was added 5 min prior to the addition of fluorescein-cofilin (Figs 2C and 3C). A weaker inhibitory effect on cofilin cluster formation was observed in the presence of 0.3 μM S1 (Figs 2B and 3B). A semi-quantitative evaluation of the inhibitory effects by S1 is summarized in Fig. 3D–F and Supplementary Table S1. Under the present condition (0.1 mM ATP, 40 mM KCl, pH 6.8), binding of S1 to actin filaments is transient, and the majority of actin protomers in the filaments should be free of bound S1 at any given moment, as confirmed by live imaging by high speed atomic force microscopy (Supplementary Fig. S2 and Supplementary Videos S1 and S2). It is therefore obvious that inhibition of cofilin cluster formation by S1 in the presence of ATP does not depend on direct competition of binding sites on actin protomers, leading us to conclude that the inhibition of cofilin cluster formation is primarily allosteric and depends on cooperative conformational changes of actin filaments.
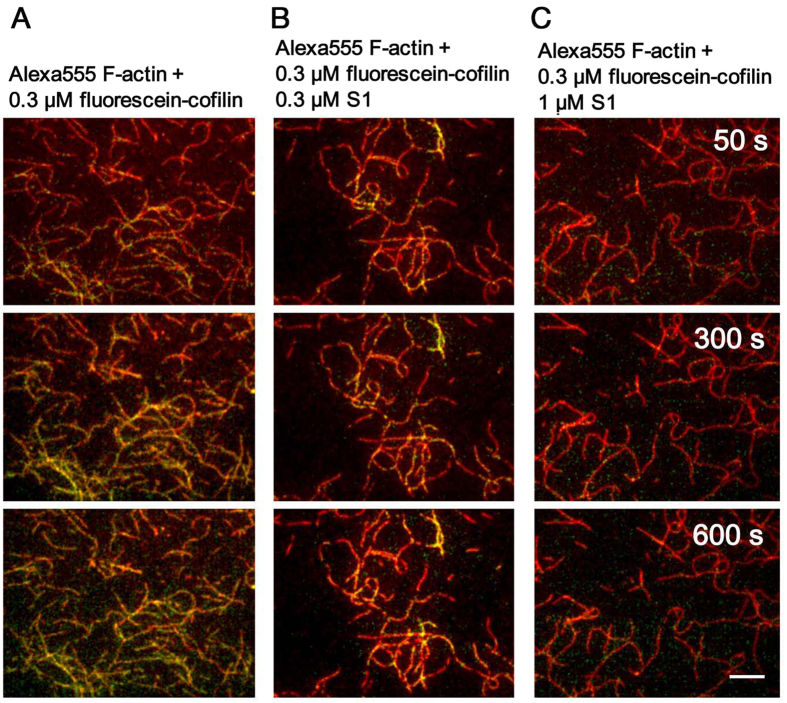
Figure 2. TIRF microscopic observation of inhibitory effects of sk S1 on binding of chicken cofilin to sk actin filament in the presence of 0.1 mM ATP.
Merged TIRF images of Alexa555-actin filaments (red) and fluorescein-cofilin (green) of the same field, captured at different indicated times, shown in s, after the addition of 0.3 μM fluorescein-cofilin without (A) or with 0.3 μM (B) or 1 μM S1 (C). Bar: 5 μm.
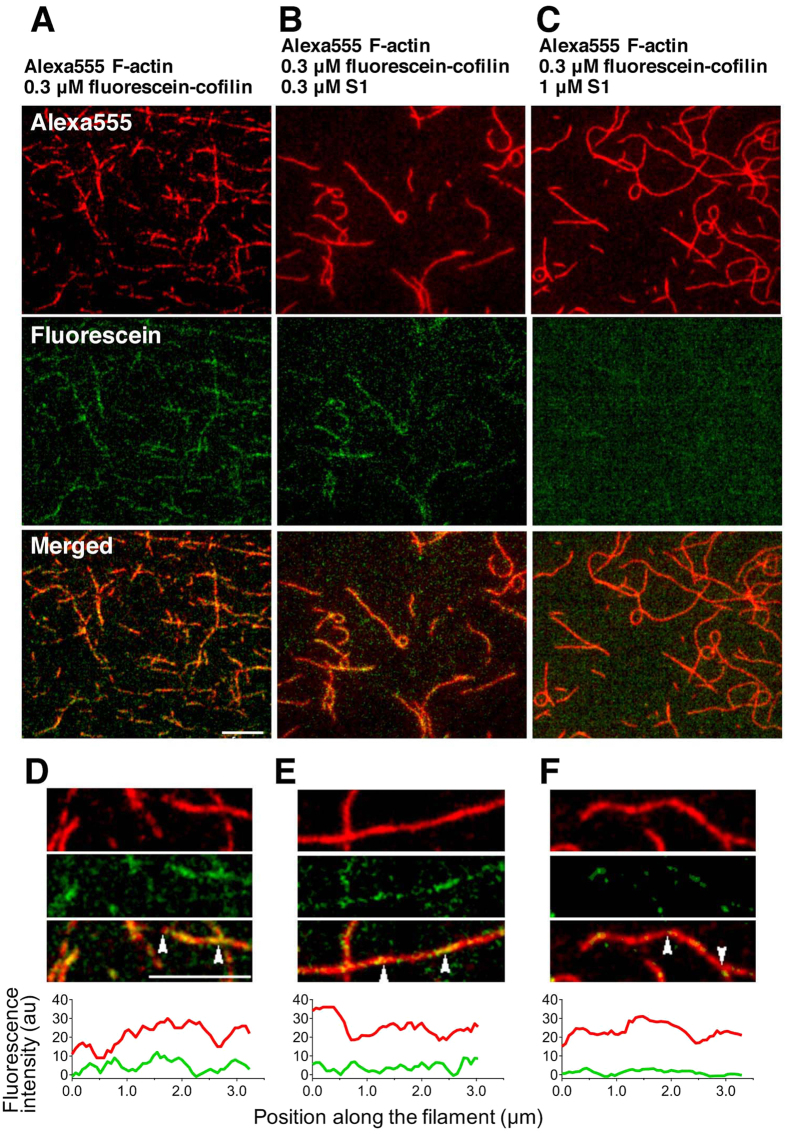
Figure 3. TIRF microscopic observation of inhibitory effects of sk S1 on binding of chicken cofilin to sk actin filament in presence of 0.1 mM ATP.
(A–C) are fluorescence images of Alexa555-sk actin filaments and fluorescein-cofilin and the merge image, captured 10 min after the addition of 0.3 μM fluorescein-cofilin without (A) or with 0.3 μM (B) or 1 μM S1 (C). Bar: 5 μm. (D–F) are enlarged views of a different field of the same sample as (A–C), respectively. The top and middle rows show Alexa555 and fluorescein images, and the bottom row shows the merged image, respectively. Bar: 5 μm. The graphs at the bottom show fluorescence intensity profiles of Alexa555 (red) and fluorescein (green) after background subtraction along the filament segments flanked by the two arrowheads in the merged images immediately above.
Cosedimentation assays using fusion proteins
The mechanism of mutually exclusive cooperative binding of cofilin and HMM to actin filaments was further explored by cosedimentation assays. Initially, we carried out a series of standard cosedimentation experiments using sk actin filaments, sk S1 and human cofilin. Unexpectedly, however, we were unable to find a condition under which S1 in the presence of ATP significantly reduced the amount of cofilin co-pelleting with actin filaments (e.g., Supplementary Fig. S3). One major difference between these cosedimentation experiments and the TIRFM (Fig. 2) and AFM (Supplementary Fig. S2) experiments is the molar ratio between S1 and actin in the reaction mixtures. In the two microscopic experiments, concentrations of actin were very low, in the order of 10 nM, such that concentration of S1, 0.3~1 μM, was 30~100-fold higher than that of actin, whereas in the cosedimentation experiments, the concentrations of S1 were less than that of actin. We speculated that this may be the reason why S1 in the presence of ATP inhibited cofilin binding in the microscopic experiments but not in the cosedimentation experiments, considering the kinetics of the actin-activated ATPase cycle of S1 (Supplementary Fig. S4). If so, this potential problem can be circumvented by using S1 that is physically tethered to actin protomers, as tethering would increase the effective concentration of S1 around actin filaments.
Actin-S1 fusion protein has the whole Dd actin polypeptide inserted in loop 2 of S1 of Dd myosin II using two Gly-based 18 amino acid-residue linkers (Fig. 4A)27. Loop 2 is an actin binding site of S1, and this fusion was intended to mimic the natural actin-S1 complex. Our previous characterization27 showed that the actin-S1 fusion protein was unable to polymerize on its own, which was contrary to the initial expectation. Nonetheless, it was able to copolymerize with sk actin, and the copolymerization significantly enhanced the MgATPase activity of the S1 moiety, albeit not to the level of Vmax. Furthermore, copolymer of actin-S1 fusion protein with actin was indistinguishable from actin filaments sparsely bound with sk S1 by electron microscopy. We therefore concluded that the actin-S1 fusion protein in copolymer with actin mimics certain aspects of frequently interacting actin and S1 with very high local concentration of S1. Based on those two rationales, we attempted to reproduce inhibition of cofilin binding to actin filaments by cooperative conformational changes induced by S1 in the presence of ATP using the actin-S1 fusion protein in cosedimentation experiments.
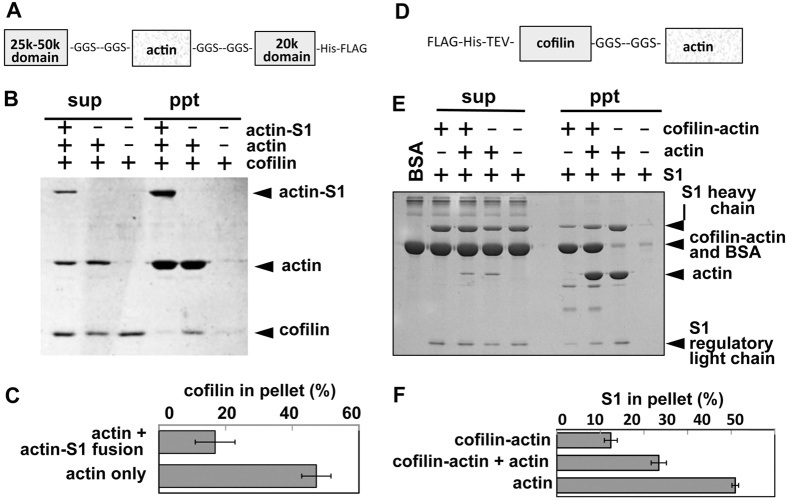
Figure 4. Cosedimentation experiments to assay the affinity of cofilin for copolymer of actin and actin-S1 fusion protein (A–C), and that of S1 for copolymer of actin and cofilin-actin fusion protein (D–F).
(A) Schematic structure of actin-S1 fusion protein. (B) Cosedimentation of 3 μM Dd cofilin with 3 μM Dd actin homopolymer or copolymer (3 μM Dd actin and 1.5 μM actin-S1) in the presence of 70 mM KCl and 2 mM ATP at pH 6.5. Supernatant (sup) and pellet (ppt) fractions after ultracentrifugation were analyzed by SDS-PAGE. (D) Schematic structure of cofilin-actin fusion protein. (E) Cosedimentation of 1.5 μM Dd S1 with 5 μM cofilin-actin fusion protein homopolymer, copolymer (5 μM cofilin-actin, 4 μM Dd actin) or 4 μM Dd, actin homopolymer in the presence of 50 mM KCl, 1 mM ADP and 0.2 mM ATP at pH 6.5. Supernatant (sup) and pellet (ppt) fractions after ultracentrifugation were analyzed by SDS-PAGE. Two-fold larger amounts of pellet fractions were loaded. (C,F) Densitometric quantitation showing mean ± SD from three independent experiments.
The affinity of Dd cofilin for copolymers of actin-S1 fusion protein and Dd actin was compared with that for homopolymers of Dd actin at pH 6.5 and a KCl concentration of 70 mM, the conditions under which cofilin binds actin filaments without rapidly severing them23,24,25. Actin-S1 fusion protein in copolymers strongly inhibited binding of cofilin to neighboring actin protomers (Fig. 4B,C). The molar ratio of actin-S1 fusion protein to actin in this experiment was 1:2, implying that each actin-S1 fusion protein affected more than two neighboring actin protomers.
In the second experiment, we used another fusion protein, in which Dd actin was fused at the C-terminus of Dd cofilin via a Gly-based 14 amino acid residue linker (Fig. 4D). Similar to the actin-S1 fusion protein, this cofilin-actin fusion protein copolymerizes with Dd actin and affects the structure of neighboring actin protomers8. Within the copolymer, the fusion protein did not segregate from actin to form tight clusters, although its distribution within copolymer was not necessarily uniform8. This is a clear advantage over using actin filaments bound with cofilin because cofilin binds to actin filaments highly cooperatively and forms tight clusters, which would hinder quantitative interpretation of the impact of cofilin binding at a given molar ratio to actin.
The affinity of Dd S1 for copolymers of cofilin-actin fusion protein and Dd actin was compared with that for homopolymers of Dd actin in the presence of 0.2 mM ATP, 1 mM ADP, and 50 mM KCl at pH 6.5. Under this condition, S1 binds actin filaments with an intermediate affinity (Fig. 4E,F). Notably, much less S1 cosedimented with copolymers of cofilin-actin fusion protein and actin than with actin homopolymers.
Actin protomers in copolymers with actin-S1 fusion protein retained their ability to bind Dd S1 and skeletal HMM (Supplementary Fig. S5), and actin protomers in copolymers with cofilin-actin retained their ability to bind cofilin cooperatively8. These results imply that the inhibition of cofilin or S1 binding by actin-S1 or cofilin-actin fusion protein is specific to a certain class of ABPs, rather than due to general structural disturbance caused by the fusion proteins.
Discussion
Mechanism of mutually exclusive cooperative binding of myosin II and cofilin to actin filaments
We have shown that cooperative bindings of cofilin and the motor domain of myosin II to actin filaments are mutually exclusive (Figs 1 and 4). The mutually inhibitory binding of the motor domain of myosin II and cofilin to actin filaments has been reported in a number of studies, and was attributed to direct competition for binding sites on actin28,29,30,31,32. However, our TIRFM observation clearly indicated that, at least when cofilin cluster formation was inhibited by S1 in the presence of ATP, direct competition of binding sites does not play a major role. This is because in the presence of 0.1 mM ATP, binding of S1 to actin filaments is transient, and the majority of actin protomers in filaments are free of bound S1 at any given moment (ref. 33 and Supplementary Fig. S2 and Supplementary Video S2). Our cosedimentation experiments also showed that the inhibition of cofilin binding to copolymers of actin and actin-S1 fusion protein, and the inhibition of S1 binding to copolymers of actin and cofilin-actin fusion protein do not require direct competition for binding sites on actin protomers. This is because the same concentration of actin protomers was available for binding in both the copolymer and the control actin homopolymer filaments (Fig. 4). Modeling using published coordinates of cofilin-decorated actin filaments and an actin-S1 rigor complex indicated that bound cofilin and bound S1 molecules do not physically interfere with each other’s binding at an adjacent binding site (Supplementary Fig. S6). Rather, this inhibition most likely depends primarily on allosteric and cooperative conformational changes of the actin protomers, which were induced by free cofilin, HMM or S1 in the microscopic binding experiments (Figs 1–3) or by the neighboring fusion protein in the cosedimentation experiments (Fig. 4). Notably, in the case of S1-dependent inhibition of cofilin cluster formation in the presence of ATP, binding of S1 to actin filaments was transient and the majority of actin protomers in filaments were free of bound S1 molecules (Supplementary Fig. S2 and Supplementary Video S2). This suggests that the inhibitory effect of transient binding of S1 propagates over a long distance. Alternatively, each bound S1 molecule may affect fewer neighboring actin protomers, but the inhibitory effect persisted after the S1 molecule dissociated.
Implied in this hypothesis is that the conformational changes of the actin protomers induced by myosin II and cofilin are different. Cofilin binding significantly supertwists the helix of actin filaments4,5,6,7. In contrast, the actin helix slightly untwists when myosin motor domain binds in the absence of ATP34,35. This is consistent with the above prediction, but further analysis is needed to reveal the conformational changes of actin filaments induced by the myosin motor domains in the presence of ATP.
Physiological implications of mutually exclusive cooperative binding of myosin II and cofilin to actin filaments
In the amoeboid cells of the cellular slime mold Dictyostelium discoideum, cofilin is enriched in the lamellipodia36 to recycle actin for continuous polymerization at the leading edge, while myosin II is enriched in the posterior cortex37,38 to drive retraction39. Below is a brief summary of the plausible biochemical signaling involved in the localization of cofilin and myosin II in motile cells.
The activity of mammalian cofilin has been shown to be regulated by three independent biochemical mechanisms. First, activity is suppressed by the phosphorylation of Ser340. However, overexpression of constitutively active S3A cofilin did not inhibit cell motility41,42. Second, cofilin is sequestered to the plasma membrane in an inactive form by phosphatidylinositol 4,5-bisphophate (PIP2) and this inhibition is reversed by phospholipase C, which degrades PIP243. However, phospholipase C is not required for chemotaxis in Dictyostelium44. Finally, the severing activity of cofilin is enhanced at a higher pH, which, at the leading edge of motile cells, may be required for local activation of cofilin40. Consistent with this view, inhibition of the Na+/H+ exchanger (NHE1) in neutrophils and Dictyostelium amoebae impaired the elevation of intracellular pH and chemotaxis45,46. However, the motility defect in NHE1-null Dictyostelium amoeba was suppressed by the overexpression of Aip1, which binds and increases the activity of cofilin47. Thus, there is no compelling evidence to show that Ser3 phosphorylation, inactivation by PIP2 binding, or activation by increased pH is singly essential for localized activation of cofilin in Dictyostelium.
The interaction between actin filaments and cofilin is influenced by the nucleotide bound to actin protomers. Actin protomers with bound ATP or ADP and phosphate have lower affinity for cofilin than those carrying ADP48,49, so that the newly polymerized actin filaments are less prone to severing by cofilin in vitro. This has been suggested to play a role to protect newly polymerized actin filaments close to the leading edge of lamellipodia50. However, this mechanism cannot explain why cortical actin filaments along the sides and in the posterior region do not bind cofilin.
The assembly of Dictyostelium myosin II into filaments is suppressed by phosphorylation of the heavy chain51,52. However, filaments of mutant myosin IIs, from which this phosphorylation regulation was removed, were still localized in the posterior region52, eliminating the possibility of essential roles of this phosphorylation regulation in the localization of Dictyostelium myosin II.
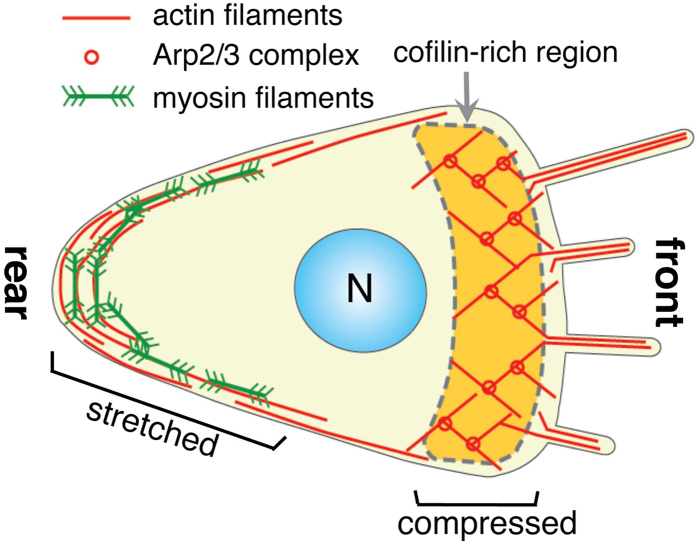
Thus, despite extensive work, the essence of the molecular mechanism of anterior localization of cofilin and posterior localization of myosin II in migrating Dictyostelium cells is still obscure. We propose that different physical conditions of actin filaments combined with the mutually exclusive cooperative binding of cofilin and myosin II to actin filaments contribute to regulating the localization of those ABPs in migrating Dictyostelium cells (Fig. 5), as detailed next.
Figure 5. Schematic drawing of actin structures and forces acting on actin filaments in a migrating Dictyostelium cell.
Note that, unlike in crawling higher animal cells (e.g., ref. 63), myosin II is absent in the anterior region in a migrating Dictyostelium cell38. In higher animal cells, tropomyosin has been implicated in specifying the localization of ABPs64. However, a discernable tropomyosin gene has not been discovered in the fully-sequenced genome of Dictyostelium, suggesting that tropomyosin is dispensable to properly localize ABPs differentially in polarized amoeboid cells of simpler organisms.
Actin filaments in the posterior cortex of migrating cells interact with myosin II filaments to drive contraction of the rear end, so that those actin filaments are mechanically stretched. An X-ray diffraction study of contracting muscle17 and a molecular dynamics simulation16 demonstrated that stretched actin filaments have an untwisted and longer helical pitch than control filaments. Independent of force generation, binding of the myosin motor alone changes the structure of actin filaments and untwists the helical pitch34,35, implying that myosin motors have a higher affinity for untwisted filaments. In the posterior region, therefore, increased tension, an untwisted helical structure, increased binding of myosin II filaments, and the resulting increased tension would form a positive feedback loop, stabilizing the established front-rear cell polarity. Cofilin would be excluded from such established local positive feedback loops, either because cofilin has a lower affinity for stretched actin filaments20 and/or because cofilin cannot bind to actin filaments interacting with myosin motor domains (Figs 1C, 2C, 3C,F and 4B). In the present study, inhibition of actin binding of cofilin by S1 in the presence of saturating concentration of ATP was observed only when the molar ratio of S1 to actin was very large (TIRFM and AFM) or when actin-S1 fusion protein was used (cosedimentation). In vivo, clustering of motor domains in filaments of myosin II would contribute to increase the local concentration of the myosin motor domain near interacting actin filaments.
Along the leading edge of migrating cells, Arp2/3-dependent polymerization of actin filaments pushes the cell membrane forward53, so that those actin filaments experience a compressive force. This compressed state of the filaments would favor cofilin binding20. The abundance of cofilin binding and the supertwisted conformation of the filaments would contribute to exclusion of myosin II from cooperatively binding to actin filaments in the anterior region. Consistent with this suggestion, knockdown of cofilin expression in HeLa cells evoked various abnormalities associated with hyper activation of myosin II32, although these authors attributed this to direct competition for binding sites on actin protomers between cofilin and myosin II.
In summary, this study demonstrated a novel allosteric and cooperative mechanism for a mutually inhibitory relationship between myosin II and cofilin in vitro. In particular, allosteric negative regulation of cofilin activity by myosin II in vivo should be far more potent than the previously assumed direct competition mechanism, in which one molecule of bound motor domain can protect only one actin protomer from cofilin binding. Furthermore, ABPs that bind to the ends of actin filaments also affect the structure of the filament over a long distance54,55,56,57. Thus, ABP-induced allosteric and cooperative modification of the structure of actin filaments, and the resulting functional differentiation of the actin filaments, appear to be widespread.
Methods
Preparation of proteins
His-tagged human cofilin-mCherry was expressed in E. coli and purified using a Ni-affinity column, as detailed in the Supplementary Methods. Preparation of engineered chicken cofilin and labeling with fluorescein26 is also detailed in the Supplementary Methods. The following proteins were purified as described elsewhere: Dd actin58, Dd HMM-GFP15, Dd S159, actin-S1 fusion protein27, cofilin-actin fusion protein8, and His-tagged Dd cofilin60. Rabbit sk actin and S1 were prepared by the method of Spudich and Watt61 and Margossion and Lowey62, respectively. Dd-actin was labeled with Cy3-succinimide and Alexa488-succinimide and sk actin with Alexa555-succinimide as described in the Supplementary Methods.
Fluorescence microscopy
Flow cells coated with a positively charged lipid bilayer were prepared as described in the Supplementary Methods, and blocked with F-buffer 1 (25 mM imidazole, pH 6.5, 4 mM MgCl2, 25 mM KCl, 1 mM DTT, 10 mg/mL BSA). Each fluorescent or unlabeled Dd actin was polymerized for 6 h at 4 °C in F-buffer 1, diluted to 80 nM in F-buffer 1 containing 0.5 μM ATP, and introduced into the flow cell. After incubation for 5 min, excess actin filaments were washed away with F-buffer 1. F-buffer 1 containing 0.2 μM ATP and either or both 80 nM Hs cofilin-mCherry and 7 nM Dd HMM-GFP was then introduced, and incubated for 4 min. Finally, F-buffer 1 containing glucose, glucose oxidase and catalase was introduced and samples were observed under a fluorescence microscope (IX-70, Olympus, Tokyo, Japan) equipped with a Plan-Fluor 100×, 1.30 N.A. oil-immersion objective lens and a sCMOS camera (ORCA-Flash 2.8, Hamamatsu Photonics, Hamamatsu, Japan).
Procedures for TIRF observations are detailed in the Supplementary Methods. Briefly, filaments of Alexa555-labeled sk actin were first loosely immobilized on a positively-charged inner surface of flow cells. After blocking with 2 mg/mL BSA, 0.3 μM fluorescein-chicken cofilin in the observation buffer (40 mM KCl, 20 mM Pipes pH 6.8, 1 mM MgCl2, 0.5 mM EGTA, 10 mM DTT, 0.1 mM ATP, 2 mg/mL BSA, 10 mg/mL glucose, 100 μg/mL glucose oxidase and 20 μg/mL catalase) was gently introduced, and fluorescence images of Alexa555 and fluorescein were imaged simultaneously at a frame rate of 0.1 fps for 10 min. To examine the effects of S1 on binding of cofilin to actin filaments, observation buffer containing 0.3 μM or 1 μM S1 was first introduced into a flow cell with attached actin filaments. After incubation for 5 min, a mixture of 0.3 μM fluorescein-cofilin and either 0.3 μM or 1 μM S1 in the observation buffer was introduced, and time lapse images were taken as indicated above. For semi-quantitative comparison of the binding of fluorescein-cofilin, it was desirable to take snap-shot images of fluorescein without prior illumination with blue excitation light, in order to minimize complications caused by bleaching. Thus, snap-shot TIRF images of fluorescein and Alexa555 were taken at new fields after focusing using Alexa555 fluorescence. Image analyses, including background subtraction, were performed using ImageJ software.
Cosedimentation assays
Dd actin (3 μM) or a mixture of 3 μM Dd actin and 1.5 μM actin-S1 fusion protein were polymerized in 70 mM KCl, 20 mM Pipes pH 6.5, 0.4 mM EGTA, 2 mM MgCl2, 2 mM ATP and 1 mM DTT at 22 °C for 1.5 h. Dd cofilin was then added at a final concentration of 3 μM, and incubated for 5 min. In the second experiment, 8 μM Dd actin or mixture of 8 μM actin and 10 μM cofilin-actin fusion protein were polymerized in 100 mM KCl, 2 mM Hepes pH 7.4, 0.5 mM EGTA, 2 mM MgCl2, 0.4 mM ATP, 0.1 mM DTT and 2 mg/mL BSA at 22 °C for 1.5 h. An equal volume of 3 μM Dd S1 in 2 mM ADP and 20 mM Pipes pH 6.5 was then added.
In each experiment, the mixtures were centrifuged at 250,000 × g for 10 min at 22 °C, and the supernatants and pellets were subjected to SDS-PAGE. In some experiments, BSA was included in buffers as a precautionary measure to prevent protein denaturation and absorption to the tube walls.
Additional Information
How to cite this article: Ngo, K. X. et al. Allosteric regulation by cooperative conformational changes of actin filaments drives mutually exclusive binding with cofilin and myosin. Sci. Rep. 6, 35449; doi: 10.1038/srep35449 (2016).
Supplementary Material
Acknowledgments
We thank Dr. Toshio Ando for his support in HS-AFM analyses, Drs Kengo Adachi and Hanako Hayashi for image analysis, and Dr. Makoto Suzuki for his encouragement. This work was supported in part by Grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology to KT (No. 24370069 and 24117008), NK (No. 15H04360), TN (No. 25840057 and 24117008) and TU (No. 24370069 and 24117008), and a PRESTO grant from the Japan Science and Technology Agency to NK.
Footnotes
Author Contributions S.T.K., N.U., K.X.N., N.K., K.T. and T.Q.P.U. designed the experiments, S.T.K., N.U., K.X.N., N.K., H.U., N.F.-U., J.N. T.Q.P.N. and A.N. performed the experiments, S.T.K., N.U. and K.X.N. analyzed the data, and K.T. and T.Q.P.U. wrote the manuscript.
References
- Oosawa F., Fujime S., Ishiwata S. & Mihashi K. Dynamic properties of F-actin and thin filament. Cold Spring Harbor Symp. Quant. Biol. 37, 277–285, doi: 10.1101/SQB.1973.037.01.038 (1973). [DOI] [Google Scholar]
- Miki M., Wahl P. & Auchet J. C. Fluorescence anisotropy of labeled F-actin: influence of divalent cations on the interaction between F-actin and myosin heads. Biochemistry 21, 3661–3665, doi: 10.1021/bi00258a021 (1982). [DOI] [PubMed] [Google Scholar]
- Loscalzo J., Reed G. H. & Weber A. Conformational change and cooperativity in actin filaments free of tropomyosin. Proc Natl Acad Sci USA 72, 3412–3415, doi: 10.1073/pnas.72.9.3412 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin V. E., Orlova A., Lukoyanova N., Wriggers W. & Egelman E. H. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J. Cell Biol. 153, 75–86, doi: 10.1083/jcb.153.1.75 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough A., Pope B., Chiu W. & Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J. Cell Biol. 138, 771–781, doi: 10.1083/jcb.138.4.771 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin V. E. et al. Remodeling of actin filaments by ADF/cofilin proteins. Proc. Natl. Acad. Sci. USA 108, 20568–20572, doi: 10.1073/pnas.1110109108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo K. X., Kodera N., Katayama E., Ando T. & Uyeda T. Q. P. Cofilin-induced unidirectional cooperative conformational changes in actin filaments revealed by high-speed atomic force microscopy. Elife 4:e04806, doi: 10.7554/eLife.04806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeki N., Hirose K. & Uyeda T. Q. P. Cofilin-induced cooperative conformational changes of actin subunits revealed using cofilin-actin fusion protein. Sci Rep 6, 20406, doi: 10.1038/srep20406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobkov A. A. et al. Cooperative effects of cofilin (ADF) on actin structure suggest allosteric mechanism of cofilin function. J. Mol. Biol. 356, 325–334, doi: 10.1016/j.jmb.2005.11.072 (2006). [DOI] [PubMed] [Google Scholar]
- Prochniewicz E., Janson N., Thomas D. D. & De La Cruz E. M. Cofilin increases the torsional flexibility and dynamics of actin filaments. J Mol Biol 353, 990–1000, doi: 10.1016/j.jmb.2005.09.021 (2005). [DOI] [PubMed] [Google Scholar]
- Dedova I. V., Nikolaeva O. P., Mikhailova V. V., dos Remedios C. G. & Levitsky D. I. Two opposite effects of cofilin on the thermal unfolding of F-actin: a differential scanning calorimetric study. Biophys. Chem. 110, 119–128, doi: S0301462204000341 (2004). [DOI] [PubMed] [Google Scholar]
- Hayakawa K., Sakakibara S., Sokabe M. & Tatsumi H. Single-molecule imaging and kinetic analysis of cooperative cofilin-actin filament interactions. Proc Natl Acad Sci USA 111, 9810–9815, doi: 10.1073/pnas.1321451111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez C. et al. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr Biol 21, 862–868, doi: 10.1016/j.cub.2011.03.064 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova A. & Egelman E. H. Cooperative rigor binding of myosin to actin is a function of F-actin structure. J. Mol. Biol. 265, 469–474, doi: 10.1006/jmbi.1996.0761 (1997). [DOI] [PubMed] [Google Scholar]
- Tokuraku K., Kurogi R., Toya R. & Uyeda T. Q. P. Novel mode of cooperative binding between myosin and Mg2+ -actin filaments in the presence of low concentrations of ATP. J Mol Biol 386, 149–162, doi: 10.1016/j.jmb.2008.12.008 (2009). [DOI] [PubMed] [Google Scholar]
- Matsushita S., Inoue Y., Hojo M., Sokabe M. & Adachi T. Effect of tensile force on the mechanical behavior of actin filaments. J. Biomech. 44, 1776–1781, doi: 10.1016/j.jbiomech.2011.04.012 (2011). [DOI] [PubMed] [Google Scholar]
- Wakabayashi K. et al. X-ray diffraction evidence for the extensibility of actin and myosin filaments during muscle contraction. Biophys. J. 67, 2422–2435, doi: 10.1016/S0006-3495(94)80729-5 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin V. E., Orlova A. & Egelman E. H. Actin filaments as tension sensors. Curr Biol 22, R96–R101, doi: 10.1016/j.cub.2011.12.010 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda T. Q. P., Iwadate Y., Umeki N., Nagasaki A. & Yumura S. Stretching actin filaments within cells enhances their affinity for the myosin II motor domain. PLoS One 6, e26200, doi: 10.1371/journal.pone.0026200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa K., Tatsumi H. & Sokabe M. Actin filaments function as a tension sensor by tension dependent binding of cofilin to the filament. J. Cell Biol. 195, 721–727, doi: 10.1083/jcb.201102039 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelot A. & Drubin D. G. Building distinct actin filament networks in a common cytoplasm. Curr Biol 21, R560–R569, doi: 10.1016/j.cub.2011.06.019 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenenberger C. A., Mannherz H. G. & Jockusch B. M. Actin: from structural plasticity to functional diversity. Eur J Cell Biol 90, 797–804, doi: 10.1016/j.ejcb.2011.05.002 (2011). [DOI] [PubMed] [Google Scholar]
- Hayden S. M., Miller P. S., Brauweiler A. & Bamburg J. R. Analysis of the interactions of actin depolymerizing factor with G- and F-actin. Biochemistry 32, 9994–10004, doi: 10.1021/bi00089a015 (1993). [DOI] [PubMed] [Google Scholar]
- Yonezawa N., Nishida E. & Sakai H. pH control of actin polymerization by cofilin. J. Biol. Chem. 260, 14410–14412 (1985). [PubMed] [Google Scholar]
- Hawkins M., Pope B., Maciver S. K. & Weeds A. G. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry 32, 9985–9993, doi: 10.1021/bi00089a014 (1993). [DOI] [PubMed] [Google Scholar]
- Nagaoka R., Kusano K., Abe H. & Obinata T. Effects of cofilin on actin filamentous structures in cultured muscle cells. Intracellular regulation of cofilin action. J Cell Sci 108 (Pt 2), 581–593 (1995). [DOI] [PubMed] [Google Scholar]
- Yokoyama K. et al. Design and functional analysis of actomyosin motor domain chimera proteins. Biochem. Biophys. Res. Commun. 299, 825–831, doi: 10.1016/S0006-291X(02)02758-4 (2002). [DOI] [PubMed] [Google Scholar]
- Elam W. A., Kang H. & De La Cruz E. M. Competitive displacement of cofilin can promote actin filament severing. Biochem Biophys Res Commun 438, 728–731, doi: 10.1016/j.bbrc.2013.07.109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida E., Maekawa S. & Sakai H. Cofilin, a protein in porcine brain that binds to actin filaments and inhibits their interactions with myosin and tropomyosin. Biochemistry 23, 5307–5313, doi: 10.1021/bi00317a032 (1984). [DOI] [PubMed] [Google Scholar]
- Mabuchi I. Effects of muscle proteins on the interaction between actin and an actin-depolymerizing protein from starfish oocytes. J Biochem 92, 1439–1447 (1982). [DOI] [PubMed] [Google Scholar]
- Abe H. & Obinata T. An actin-depolymerizing protein in embryonic chicken skeletal muscle: purification and characterization. J Biochem 106, 172–180 (1989). [DOI] [PubMed] [Google Scholar]
- Wiggan O., Shaw A. E., DeLuca J. G. & Bamburg J. R. ADF/cofilin regulates actomyosin assembly through competitive inhibition of myosin II binding to F-actin. Dev Cell 22, 530–543, doi: 10.1016/j.devcel.2011.12.026 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelstein R. S. & Eisenberg E. Regulation and kinetics of the actin-myosin-ATP interaction. Annu Rev Biochem 49, 921–956, doi: 10.1146/annurev.bi.49.070180.004421 (1980). [DOI] [PubMed] [Google Scholar]
- Tsaturyan A. K. et al. Strong binding of myosin heads stretches and twists the actin helix. Biophys. J. 88, 1902–1910, doi: 10.1529/biophysj.104.050047 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K. C., Angert I., Kull F. J., Jahn W. & Schroder R. R. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature 425, 423–427, doi: 10.1038/nature02005 (2003). [DOI] [PubMed] [Google Scholar]
- Aizawa H., Fukui Y. & Yahara I. Live dynamics of Dictyostelium cofilin suggests a role in remodeling actin latticework into bundles. J Cell Sci 110, 2333–2344 (1997). [DOI] [PubMed] [Google Scholar]
- Yumura S., Mori H. & Fukui Y. Localization of actin and myosin for the study of ameboid movement in Dictyostelium using improved immunofluorescence. J Cell Biol 99, 894–899, doi: 10.1083/jcb.99.3.894 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores S. L., Sabry J. H. & Spudich J. A. Myosin dynamics in live Dictyostelium cells. Proc Natl Acad Sci USA 93, 443–446 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujioka M. et al. Talin couples the actomyosin cortex to the plasma membrane during rear retraction and cytokinesis. Proc Natl Acad Sci USA 109, 12992–12997, doi: 10.1073/pnas.1208296109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. W. & Bamburg J. R. ADF/cofilin: a functional node in cell biology. Trends Cell Biol 20, 187–195, doi: 10.1016/j.tcb.2010.01.001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M. et al. Control of growth cone motility and morphology by LIM kinase and Slingshot via phosphorylation and dephosphorylation of cofilin. J Neurosci 23, 2527–2537 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popow-Wozniak A., Mazur A. J., Mannherz H. G., Malicka-Blaszkiewicz M. & Nowak D. Cofilin overexpression affects actin cytoskeleton organization and migration of human colon adenocarcinoma cells. Histochem Cell Biol 138, 725–736, doi: 10.1007/s00418-012-0988-2 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rheenen J. et al. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J Cell Biol 179, 1247–1259, doi: 10.1083/jcb.200706206 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drayer A. L., Van der Kaay J., Mayr G. W. & Van Haastert P. J. Role of phospholipase C in Dictyostelium: formation of inositol 1,4,5-trisphosphate and normal development in cells lacking phospholipase C activity. EMBO J 13, 1601–1609 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simchowitz L. & Cragoe E. J. Jr. Regulation of human neutrophil chemotaxis by intracellular pH. J Biol Chem 261, 6492–6500 (1986). [PubMed] [Google Scholar]
- Van Duijn B. & Inouye K. Regulation of movement speed by intracellular pH during Dictyostelium discoideum chemotaxis. Proc Natl Acad Sci USA 88, 4951–4955 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C. H., Patel H. & Barber D. L. Expression of actin-interacting protein 1 suppresses impaired chemotaxis of Dictyostelium cells lacking the Na+-H+ exchanger NHE1. Mol Biol Cell 21, 3162–3170, doi: 10.1091/mbc.E09-12-1058 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M. F. et al. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136, 1307–1322, doi: 10.1083/jcb.136.6.1307 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L. & Pollard T. D. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J Biol Chem 274, 15538–15546 (1999). [DOI] [PubMed] [Google Scholar]
- Svitkina T. M. & Borisy G. G. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 31, 1009–1026 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck V. D., Schleicher M., Grabatin B., Wippler J. & Gerisch G. Replacement of threonine residues by serine and alanine in a phosphorylatable heavy chain fragment of Dictyostelium myosin II. Febs Lett 269, 239–243, doi: 10.1016/0014-5793(90)81163-I (1990). [DOI] [PubMed] [Google Scholar]
- Egelhoff T. T., Lee R. J. & Spudich J. A. Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell 75, 363–371, doi: 10.1016/0092-8674(93)80077-R (1993). [DOI] [PubMed] [Google Scholar]
- Mullins R. D. & Pollard T. D. Structure and function of the Arp2/3 complex. Curr Opin Struct Biol 9, 244–249, doi: 10.1016/S0959-440X(99)80034-7 (1999). [DOI] [PubMed] [Google Scholar]
- Prochniewicz E., Zhang Q., Janmey P. A. & Thomas D. D. Cooperativity in F-actin: binding of gelsolin at the barbed end affects structure and dynamics of the whole filament. J Mol Biol 260, 756–766, doi: 10.1006/jmbi.1996.0435 (1996). [DOI] [PubMed] [Google Scholar]
- Orlova A., Prochniewicz E. & Egelman E. H. Structural dynamics of F-actin: II. Cooperativity in structural transitions. J. Mol. Biol. 245, 598–607, doi: 10.1006/jmbi.1994.0049 (1995). [DOI] [PubMed] [Google Scholar]
- Bugyi B. et al. Formins regulate actin filament flexibility through long range allosteric interactions. J Biol Chem 281, 10727–10736, doi: 10.1074/jbc.M510252200 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp G. et al. Conformational changes in actin filaments induced by formin binding to the barbed end. Biophys J 91, 2564–2572, doi: 10.1529/biophysj.106.087775 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T. Q. P., Kanzaki N., Ueno H., Hirose K. & Uyeda T. Q. P. A novel system for expressing toxic actin mutants in Dictyostelium and purification and characterization of a dominant lethal yeast actin mutant. J Biol Chem 282, 27721–27727, doi: 10.1074/jbc.M703165200 (2007). [DOI] [PubMed] [Google Scholar]
- Uyeda T. Q. P., Tokuraku K., Kaseda K., Webb M. R. & Patterson B. Evidence for a novel, strongly bound acto-S1 complex carrying ADP and phosphate stabilized in the G680V mutant of Dictyostelium myosin II. Biochemistry 41, 9525–9534, doi: 10.1021/bi026177i (2002). [DOI] [PubMed] [Google Scholar]
- Umeki N. et al. Rapid nucleotide exchange renders Asp-11 mutant actins resistant to depolymerizing activity of cofilin, leading to dominant toxicity in vivo. J Biol Chem 288, 1739–1749, doi: 10.1074/jbc.M112.404657 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. A. & Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 (1971). [PubMed] [Google Scholar]
- Margossian S. S. & Lowey S. Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol 85, 55–71, doi: 10.1016/0076-6879(82)85009-X (1982). [DOI] [PubMed] [Google Scholar]
- Verkhovsky A. B., Svitkina T. M. & Borisy G. G. Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J Cell Biol 131, 989–1002, doi: 10.1083/jcb.131.4.989 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vindin H. & Gunning P. Cytoskeletal tropomyosins: choreographers of actin filament functional diversity. J Muscle Res Cell Motil 34, 261–274, doi: 10.1007/s10974-013-9355-8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.