Abstract
Protein synthesis in the chloroplast is mediated by the chloroplast ribosome (chloro-ribosome). Overall architecture of the chloro-ribosome is considerably similar to the Escherichia coli (E. coli) ribosome but certain differences are evident. The chloro-ribosome proteins are generally larger because of the presence of chloroplast-specific extensions in their N- and C-termini. The chloro-ribosome harbours six plastid-specific ribosomal proteins (PSRPs); four in the small subunit and two in the large subunit. Deletions and insertions occur throughout the rRNA sequence of the chloro-ribosome (except for the conserved peptidyl transferase center region) but the overall length of the rRNAs do not change significantly, compared to the E. coli. Although, recent advancements in cryo-electron microscopy (cryo-EM) have provided detailed high-resolution structures of ribosomes from many different sources, a high-resolution structure of the chloro-ribosome is still lacking. Here, we present a cryo-EM structure of the large subunit of the chloro-ribosome from spinach (Spinacia oleracea) at an average resolution of 3.5 Å. High-resolution map enabled us to localize and model chloro-ribosome proteins, chloroplast-specific protein extensions, two PSRPs (PSRP5 and 6) and three rRNA molecules present in the chloro-ribosome. Although comparable to E. coli, the polypeptide tunnel and the tunnel exit site show chloroplast-specific features.
Protein translation is a universal process of amino acid polymerization mediated by the ribonucleoprotein complex called the ribosome1,2. The ribosomes are composed of two different subunits, called the large subunit (LSU) and the small subunit (SSU). The SSU is mainly responsible for decoding of mRNA by selecting the correct charged tRNAs (cognate tRNAs)1,3. The LSU catalyzes amino acid polymerization (peptide bond formation)4,5 and provides a tunnel through which nascent polypeptides leave the ribosome. Exit site of the tunnel provide a platform for binding of various factors required for processing, targeting and membrane insertion of the nascent polypeptides5,6,7. In Escherichia coli (E. coli) 70S, the SSU is composed of 16S rRNA and 21 ribosomal proteins while the LSU consists of 5S and 23S rRNA along with 33 ribosomal proteins. The eukaryotic cytoplasmic ribosome (80S) is larger in size than the bacterial 70S ribosome, as it contains more proteins and rRNAs8,9.
The mitochondria and chloroplasts carry their own genome. Genes encoded by these organellar genomes are transcribed by their own transcriptional machineries and translated by the unique ribosomes present in these organelles10,11. The mitochondrial ribosome (mitoribosome) has evolved significantly in terms of composition11, structure and function12 in comparison to their bacterial ancestors. The mitoribosomes contain a number of mitochondria-specific proteins and protein extensions13,14,15,16 and the rRNA content is significantly different both in the composition and size17,18,19,20. These structural changes have enabled the mitoribosome to adapt in mitochondrial environment and specialize in synthesizing mainly membrane proteins of respiratory chain complexes21. Recent high-resolution cryo-electron microscopy (cryo-EM) structures of the mitoribosomes22,23,24,25,26,27 have provided unparalleled details about the structure and function of these unique ribosomes.
According to the endosymbiotic theory28, the chloroplast originated from a free-living photosynthetic prokaryote. During evolution, most of the chloroplast genes were transferred to the nucleus29. The chloro-ribosome synthesizes only a number (~6–7%) of the proteins functioning inside the chloroplast and majority of them are post-translationally imported from the cytoplasm30,31,32,33. The components of some of the protein complexes are encoded by both the chloroplast and nuclear genome34,35 in many instances and therefore, it is no surprise that the gene expression between these two organelles is tightly regulated30,36,37. This is to maintain the stoichiometry of the protein subunits present in these complexes. The light, which changes the redox states of the components involved in photosynthetic electron transport chain, is a major signal controlling the regulation of protein expression in the chloroplast33,38. A total of 146 genes have been identified in the circular genome of the spinach (Spinacia oleracea) chloroplast out of which 98 encodes for proteins31. Majority of these proteins form subunits of the multimeric membrane protein complexes such as ATP synthase, photosystem-1 and −2, cytochrome b6/f complex and RuBisCO (the large subunit), all of which are involved either in photosynthesis or respiration. Other examples of chloroplast-encoded proteins are RNA polymerases, NADH dehydrogenases, ribosomal proteins and translation factors31,33. In general, the chloro-ribosome translates more proteins than the mitoribosome. Unlike the mitoribosome, which mainly synthesizes hydrophobic membrane proteins21,39, the chloro-ribosome synthesizes both soluble and membrane proteins10,40. This can be one of the reasons that chloro-ribosomes have not changed as much as mitoribosomes in comparison to their bacterial ancestor.
The overall composition of the chloro-ribosome is similar to the E. coli 70S but differences occur30,31. Most of the chloro-ribosome proteins (also known as plastid ribosomal proteins or PRPs) are longer due to the presence of chloroplast-specific extensions at their N- and C-termini31. Compared to the E. coli 70S, two of the LSU proteins (bL25 and uL30) are absent and six new plastid-specific ribosomal proteins (PSRPs) are added: four in the SSU and two in the LSU31,40. Deletions and insertions are observed throughout the rRNA sequence except for the conserved peptidyl transferase centre (PTC) region31. Two earlier cryo-EM structures of chloro-ribosomes at 15.530 and 9.4 Å31 resolution provided the first glimpse of their structures. However, molecular insights into the structure of the chloro-ribosome is still lacking due to unavailability of a high-resolution structure.
Here, we report a near-atomic resolution structure of the chloro-ribosome LSU from spinach at 3.5 Å resolution determined using cryo-EM and single particle analysis. High-resolution map allowed de novo modelling of two PSRPs, many of the protein extensions and rRNA insertions. The overall structure resembles the E. coli ribosome LSU but chloroplast-specific features are visible. PTC is conserved but noticeable differences in the structure of proteins lining the polypeptide tunnel and tunnel exit site are present. Our study aims to fill a gap in the field of high-resolution structures of the ribosome available and reveals unique features of the chloro-ribosome at near-atomic resolution.
Results and Discussion
Structure determination of spinach chloro-ribosome LSU
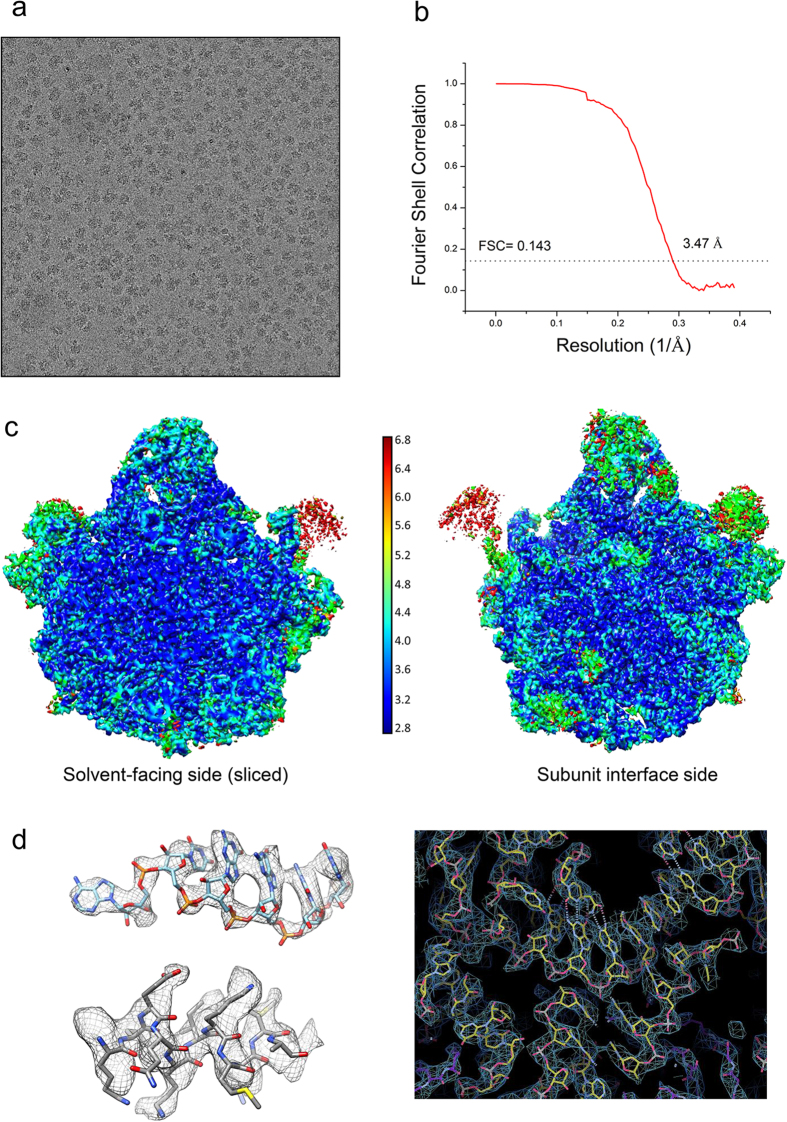
Chloro-ribosomes were purified from spinach leaves using sucrose density gradient centrifugation according to the published procedures31,41. Cryo-EM and single particle analysis were employed to determine the structure of the chloro-ribosome. A representative electron micrograph of the purified chloro-ribosome is shown in Fig. 1a. Attempts to determine a high-resolution structure of the complete chloro-ribosome (70S) were unsuccessful because of the over representation of the 50S particles in our dataset which might have resulted due to subunit dissociation during purification (as revealed by 2D and 3D classification, Supplementary Fig. S1). Therefore, we focused on determining the structure of the chloro-ribosome LSU using masked refinement procedures27. A soft Gaussian mask was created from a 3D reconstruction of the chloro-ribosome LSU generated during 3D classification step (Supplementary Fig. S1). The cryo-EM and single particle reconstruction using this focused mask during 3D refinement step resulted in a cryo-EM density map of the chloroplast LSU at an average resolution of 3.5 Å (Fig. 1b), while best resolved regions of the map contained information up to 2.8 Å resolution as suggested by the local resolution estimation with ResMap42 (Fig. 1c). Most of the protein side chains and rRNA bases were resolved in our map (Fig. 1d), which enabled us to build a near-atomic-resolution model of the spinach chloro-ribosome LSU. Our model comprises of 28 E. coli orthologous proteins (Fig. 2b) with many containing chloroplast-specific N- and C- terminal extensions (Fig. 2a and Supplementary Fig. S2), two PSRPs (Figs 2 and 3) and 4.8S, 5S and 23S rRNAs (Fig. 2). Both the L1 and L7/L12 stalk regions have weak densities in the map due to flexibility and therefore models for the proteins- uL1c, uL10c, uL11c and bL12c could not be built.
Figure 1. Processing of cryo-EM data.
(a) Representative micrograph showing ribosome particles. (b) Fourier Shell Correlation (FSC)61 curve showing the average resolution of the reconstructed density map of the spinach chloro-ribosome LSU. (c) Local resolution estimation of cryo-EM map by ResMap42. Map is coloured according to local resolution of masked map. (d) Representative density showing a 23S rRNA region (nucleotides 2584–2589) in upper left, a helix of PSRP5 protein (amino acid residues 99–109) in lower left and a region around nucleotide G2462 in 23S rRNA on the right, showing quality of the map and fitting of the models.
Figure 2. Overview of the spinach chloro-ribosome LSU.
(a) Cryo-EM reconstructed density of the spinach chloro-ribosome LSU, showing solvent-facing and subunit interface side. All proteins except two PSRPs (red) are coloured yellow. Chloroplast-specific extensions are coloured green. rRNAs are coloured as: 5S (goldenrod), 4.8S (sienna) and 23S (grey). (b) Overview of the proteins and rRNAs modelled in the spinach chloro-ribosome LSU density showing solvent-facing (left) and subunit interface side (right). The protein and rRNA models are differently coloured.
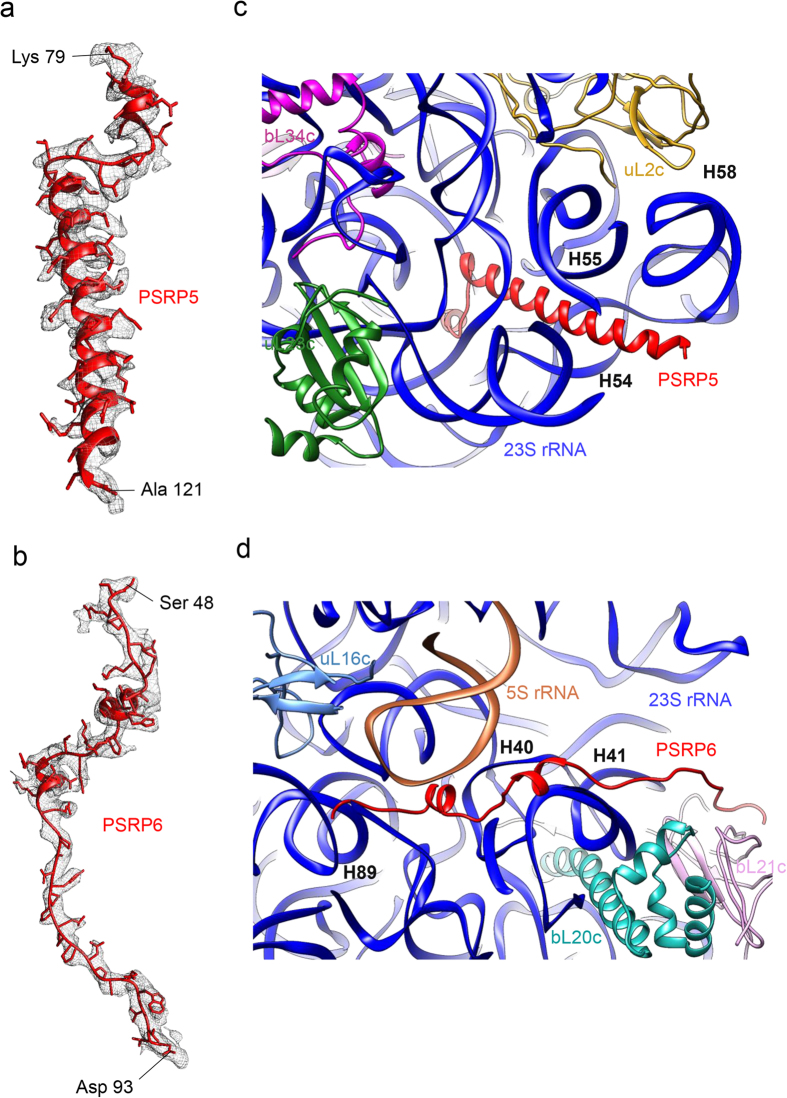
Figure 3. Locations and structures of PSRP5 and PSRP6.
(a,b) Models of the PSRP5 (a) and PSRP6 (b) showing side chains fitted inside density. (c) Location of PSRP5 in an all-rRNA environment. (d) Location of PSRP6 in close association with rRNA and bL21c.
Overall structure of spinach chloro-ribosome LSU
Since the chloro-ribosome is more similar to the E. coli ribosome than the mitoribosome30,31, we compared our map of the spinach chloro-ribosome LSU to the E. coli 50S ribosome. Atomic model of the E. coli 50S (PDB ID: 4YBB)43 fitted very well in our map (Supplementary Fig. S3) and therefore was used as a starting point to generate models for the spinach chloro-ribosome LSU. Rigid body docking of the E. coli ribosome 50S model into our map lead to the identification and localization of the densities for the E. coli orthologous proteins and rRNAs present in the spinach chloro-ribosome LSU. A combination of secondary structure prediction, homology modelling and tracing of amino acid side chains were used for model building. Details of the protein models are presented in Table 1.
Table 1. Summary of the proteins present in the spinach chloro-ribosome LSU.
| Protein name | Chain ID | Uniprot ID (GenBank ID) | Mature protein (range, aa) | Built residues (range, aa) | Size of homologous E. coli proteins (aa) | Comments |
|---|---|---|---|---|---|---|
| uL1c | Q9LE95 | 73–352 | 234 | Poor density, model could not be built | ||
| uL2c | E | P06509 | 1–272 | 23–270 | 273 | |
| uL3c | F | (KNA04906.1) | 85–305 | 86–297 | 209 | |
| uL4c | G | O49937 | 51–293 | 52–261 | 201 | |
| uL5c | H | (KNA19821.1) | 39–258 | 56–230 | 179 | |
| uL6c | I | (KNA14466.1) | 39–220 | 41–214 | 177 | |
| bL9c | J | (KNA21690.1) | 42–196 | 43–95 | 149 | |
| uL10c | (KNA14105.1) | 53–232 | 165 | Poor density, model could not be built | ||
| uL11c | P31164 | 67–224 | 142 | Poor density, model could not be built | ||
| bL12c | P02398 | 57–189 | 121 | Poor density, model could not be built | ||
| uL13c | L | P12629 | 60–250 | 100–246 | 142 | |
| uL14c | M | P09596 | 1–121 | 1–121 | 123 | |
| uL15c | N | (KNA06817.1) | 80–271 | 93–254 | 144 | |
| uL16c | O | P17353 | 1–135 | 2–135 | 136 | |
| bL17c | P | (KNA19692.1) | 11–126 | 11–126 | 127 | |
| uL18c | Q | (KNA08833.1) | 44–166 | 47–166 | 117 | |
| bL19c | R | P82413 | 78–233 | 118–232 | 115 | |
| bL20c | S | P28803 | 1–128 | 3–117 | 118 | |
| bL21c | T | P24613 | 56–256 | 91–234 | 103 | |
| uL22c | U | P09594 | 1–199 | 26–149 | 110 | |
| uL23c | V | Q9LWB5 | 77–198 | 105–193 | 100 | |
| uL24c | W | P27683 | 47–191 | 50–173 | 104 | |
| bL27c | X | (KNA14420.1) | 58–194 | 68–165 | 85 | |
| bL28c | Y | (KNA16864.1) | 72–148 | 72–145 | 78 | |
| uL29c | Z | (KNA15596.1) | 60–168 | 63–149 | 63 | |
| bL31c | a | (KNA12538.1) | 37–130 | 38–75 | 70 | Poor density |
| bL32c | b | P28804 | 1–57 | 2–47 | 57 | |
| bL33c | c | P28805 | 1–66 | 9–59 | 55 | |
| bL34c | d | P82244 | 92–152 | 92–148 | 46 | |
| bL35c | e | P23326 | 87–159 | 91–157 | 65 | |
| bL36c | f | P12230 | 1–37 | 1–37 | 38 | |
| PSRP5 | g | P27684 | 63–142 | 79–121 | PSRP variant 5α, pI = 10.01 | |
| PSRP6 | h | P82411 | 48–116 | 48–93 | pI = 9.87 |
All protein orthologues of the E. coli ribosome LSU could be identified in the spinach chloroplast LSU map with the exceptions of two proteins, namely bL25 and uL30 (Supplementary Fig. S3). This is consistent with earlier proteomic studies40 where both the bL25 and uL30 proteins could not be identified in the chloro-ribosome. Interestingly, two novel PSRPs (PSRP5 and PSRP6) have been acquired, keeping the total number of proteins in the LSU of spinach chloro-ribosome similar to the E. coli LSU. In comparison to the E. coli ribosome LSU, which contains two rRNA molecules (5S and 23S), the spinach chloro-ribosome LSU contains an additional rRNA molecule named as 4.8S rRNA (Supplementary Fig. S3). Chloroplast-specific N- and C-terminal extensions are located at the solvent exposed side and the two PSRPs are present at novel sites mainly surrounded by rRNA (Figs 2 and 3). Subunit interface side is almost unchanged suggesting a conserved PTC (Fig. 2). Major alterations and remodelling of the rRNAs occur at their flexible end towards the periphery (Fig. 4, Supplementary Figs S4 and S5).
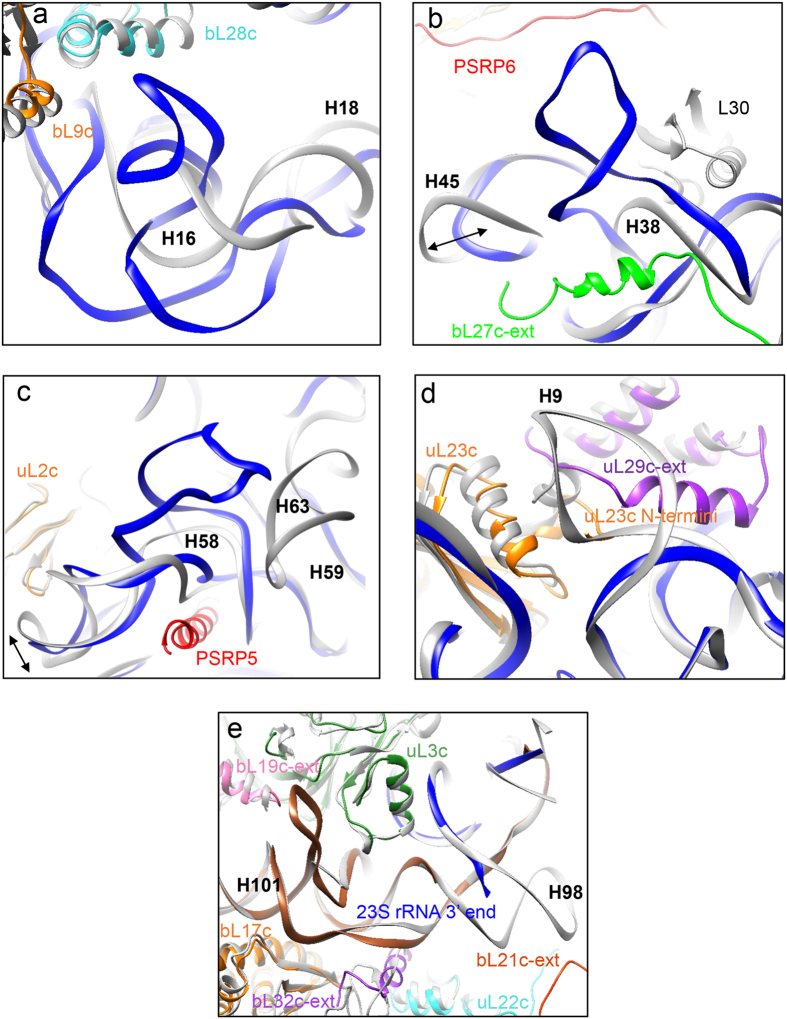
Figure 4. Remodelled rRNAs of the spinach chloro-ribosome LSU.
Regions in the 23S rRNA (a,d) and 4.8S rRNA (e) which have been remodelled because of rRNA insertions or deletions. Models for the spinach chloro-ribosome LSU (built in the current study) and the E. coli (PDB ID: 4YBB) are rigid body fitted into the cryo-EM map of the spinach chloro-ribosome LSU for comparison. The 23S rRNA of the spinach chloro-ribosome LSU is shown in blue in all cases and the chloro-ribosome LSU proteins are coloured and labeled. 4.8S rRNA (e) is shown in sienna. Overlaid E. coli models are indicated in grey.
Chloroplast-specific protein-extensions
Although the total number of proteins present in the spinach chloro-ribosome LSU is same as in the E. coli 50S ribosome8,40, chloro-ribosome proteins are usually longer in the length. The increment in length is due to the presence of the chloroplast-specific protein extensions at their N- and C-termini. Among the 31 E. coli orthologous proteins present in the spinach chloro-ribosome LSU, 25 contain chloroplast-specific extensions which are variable in their sizes, ranging from as small as 3 residues (in uL18c) to as long as 97 residues (in bL21c). We have localized and partially modelled 16 of these protein extensions, de novo (Supplementary Fig. S2). These extensions are mostly located on the solvent-exposed side of the chloro-ribosome LSU (Fig. 2). Secondary structural elements can be identified in some of the extensions but in most cases extensions maintain an extended conformation which resulted in poorly resolved densities and therefore complete structural modelling of all the extensions could not be possible.
A summary of the modelled chloroplast-specific extensions is presented in Supplementary Table S1. bL21c and uL22c proteins are almost doubled in size compared to their E. coli orthologues because of their N- and C-terminal extensions. The combined extensions at both the N- and C-termini, which are longer than 20 residues in length, are present in L1 (46 residues), L4 (39 residues), L5 (41 residues), L13 (49 residues), L15 (37 residues), L19 (46 residues), L21 (97 residues), L22 (89 residues), L23 (35 residues), L24 (40 residues), L27 (53 residues), L29 (46 residues), L31 (23 residues) and L32 (23 residues). The presence of these extensions in the chloro-ribosome proteins are mainly responsible for altered protein: rRNA ratio in the chloro-ribosome (2:3) as compared to the E. coli (1:3)31,40. Notably, some of these extensions are located in positions where alterations in rRNA structures are visible and are discussed in details in the following section. The close association of these extensions with rRNA is suggestive of their possible role in maintaining the structural integrity of the chloro-ribosome.
Structures of PSRP5 and PSRP6
After assigning the densities for all of the E. coli orthologous proteins (except proteins residing in the L1 and L7/L12 stalks), chloroplast-specific protein extensions and rRNAs, extra densities at two places remain unaccounted. High-resolution map allowed us to identify the unaccounted densities as PSRP5 and PSRP6 and build de novo models for these two proteins (Fig. 3).
PSRP5 is a conserved, nuclear-encoded, plastid-specific protein of 142 amino acids in length (mature protein consists of 80 aa) and basic in nature. As expected from the basic nature of PSRP5, its density is surrounded by rRNA suggesting its close association with rRNA (Fig. 3c). Density for PSRP5 residues ranging from 79 to 121 is clearly visible in our map (Fig. 3a) while densities for the first 16 N-terminal and the last 21 C-terminal residues are missing possibly because of their flexibility. The density for the residues ranging from 92–121 is seen to adopt a helical conformation while residues ranging from 80–91 remains partially disordered with the presence of a small alpha-helical twist between residues 80–85 (Fig. 3a). Structure of the rRNA around PSRP5 is remodelled (discussed in details in the next section) in the chloro-ribosome as compared to the E. coli 50S. Therefore, it seems plausible to hypothesize that chloroplast has specifically evolved to incorporate PSRP5 in their ribosomes to stabilize the altered rRNA structure around its position.
Localization of the PSRP5 in our map does not agree with Sharma et al.31, who had earlier localized PSRP5 near to the tRNA exit site (E-site). Analysis of our map suggests the presence of the chloroplast-specific C-terminal extension of the protein uL15c in this region (Fig. 2b). Mutations in the psrp5 gene have been reported to impair growth and reduce photosynthesis in Arabidopsis44. Subunit accumulation studies using Arabidopsis psrp5 mutants have suggested that PSRP5 might be required for the assembly and stability of the chloro-ribosome LSU44. The close association of PSRP5 with rRNA, as observed in our structure, raises such a possibility. However, the RNA-buried appearance of PSRP5 in our map indicates that it might not be involved in light-dependent regulation of translation in chloroplast as speculated earlier30,31.
PSRP6, the second of the PSRPs present in the spinach chloro-ribosome LSU, is also a conserved, nuclear-encoded, plastid-specific protein of 116 amino acids in length (mature protein consists of 69 aa) and basic in nature. However, in contrast to the PSRP5, density for the PSRP6 is mainly extended with the presence of minimal secondary structural features (Fig. 3b). The density for the residues ranging from 48 to 93 is clearly visible in the map whereas the density for the last 23 C-terminal residues is missing probably due to flexibility. The local environment of the PSRP6 in the LSU is shown in Fig. 3d. The N-terminal of PSRP6 is buried between a groove formed by 5S rRNA and H89 of 23S rRNA while C-terminal extends towards protein bL21c present on the surface of the LSU. Our de novo modelling of the PSRP6 reveals that it is mostly disordered except for two very short stretches (residues ranging from 57 to 62 and 69 to 73) of helical turns in the N-terminal region. Interestingly, in the previous cryo-EM study of spinach chloro-ribosome31, authors could not identify PSRP6 due to the low-resolution of their map and suggested that it might be relatively loosely associated with the ribosome. However, our map suggests that PSRP6 is an integral part of the spinach chloro-ribosome LSU. It has been reported that PSRP6 is dispensable for chloroplast ribosomal assembly and function under normal growth conditions44. However, it is possible that PSRP6 is required under unfavorable growth conditions. An interesting feature of the PSRP6 is the presence of a proline-rich motif (residues 80 to 86) which is located at the solvent exposed side of the LSU. Proline-rich motifs have earlier been suggested to act as ligands of modulator proteins in the eukaryotic signal transduction pathways45. Therefore, it is possible that the proline-rich motif present in PSRP6 is used as an accessible site for the non-ribosomal factors to interact with the chloro-ribosome45. The surface exposed presence of this motif in PSRP6 is reminiscent of such a possibility.
The fact that both bL25 and uL30 are absent from the chloro-ribosome LSU while two new proteins, namely PSRP5 and PSRP6 have been acquired, lead to the assumption that these two PSRPs might have replaced the role of bL25 and uL30 in the chloro-ribosome. However, locations and structures (Fig. 3 and Supplementary Fig. S3) of these two PSRPs suggest that they are not a substitution of bL25 and uL30 and might be required for some other specialized functions during ribosome assembly or protein synthesis in the chloroplast.
Remodelling of rRNA structures
Total length of the rRNA in the spinach chloro-ribosome LSU is 3037 nucleotides, which is 13 nucleotides longer than the E. coli 50S rRNA (3024 nt). Unlike E. coli 50S, where only 5S (120 nt) and 23S rRNAs (2904 nt) are present, chloro-ribosomes contain three molecules of rRNAs named as 4.8S (106 nt), 5S (121 nt) and 23S rRNA (2810 nt).
Global sequence alignment using EMBOSS46 needle tool reveals that 23S rRNA is 64.7% and 5S rRNA is 58.7% identical in sequence between the E. coli LSU and the spinach chloro-ribosome LSU. Nucleotide sequence alignment also reveals that insertions and deletions have occurred in the entire span of rRNA sequence in chloro-ribosome. But deletion in a position is balanced by insertion in a nearby position (and vice versa), keeping the overall structure of the chloro-ribosome rRNA comparable to the E. coli. However, analysis of our map reveals that nucleotide insertions and deletions have altered local rRNA structure significantly at five different places (Fig. 4 and Supplementary Figs S4 and S5). Four of these regions are located in 23S rRNA surrounding nucleotide positions 280–315, 939–968, 1502–1526 (chloroplast 23S rRNA numbering) and 129–153 (E. coli 23S rRNA numbering) and the fifth one is located in the 4.8S rRNA surrounding positions 1–28. All of these alterations in the rRNA structure are located far from the conserved core (PTC forming region of domain V and VI in 23S rRNA).
The first region with alterations in rRNA structure results because of a combined 24 nucleotides long insertion in domain I of 23S rRNA between positions 280–315, involving H18 (uppercase H is used to denote rRNA helices and numbering of rRNA helices are based on E. coli 23S rRNA secondary structure diagram obtained from http://rna.ucsc.edu/rnacenter/ribosome_images.html) and H19 (Fig. 4a and Supplementary Figs S4a and S5a). Insertion in this region does not change the structure of H18 and H19 but causes a slight shift in the axis of H18. Surprisingly, topology of neighbouring H16 is perturbed, which forms a variable loop structure and extends further towards the solvent side (Fig. 4a and Supplementary Figs S4a and S5a).
The second region with alterations in rRNA structure results due to a combined 20 nucleotides long insertion in domain II of 23S rRNA between positions 938–969 (H38, Fig. 4b and Supplementary Figs S4b and S5b). This insertion remodels H38 by introducing a protrusion which extends towards the solvent side. Interestingly, this protrusion is located near to a place where uL30 is situated in the E. coli ribosome LSU. Because uL30 is absent in the chloro-ribosome, it is possible that the function of uL30 is being compensated by this variable loop of rRNA. Interestingly, PSRP6 and chloro-specific C-terminal extension of bL27c are present in close vicinity of this protrusion. Variability around this region also includes shortening (indicated by an arrow) of H45 due to a net deletion of 6 nucleotides in the chloro-ribosome 23S rRNA sequence.
The third region with alterations in the rRNA structure results because of a combined 12 nucleotides long insertion in domain III of 23S rRNA between positions 1502–1526 (H58). This insertion creates a protrusion in H58 which extends towards the solvent exposed side (Fig. 4c and Supplementary Figs S4c and S5c left panel). Local changes resulting from this insertion event have remodelled the rRNA causing shortening (indicated by an arrow) of H58 which is located towards uL2c. In E. coli ribosome, H63 extends near to this region but deletion of 26 residues in equivalent position in chloroplast 23S rRNA sequence makes H63 much shorter (Fig. 4c and Supplementary Figs S4c and S5c right panel).
The fourth region with alterations in rRNA structure results because of a combined 14 nucleotides long deletion in domain I (E. coli numbering 129–153) of 23S rRNA and results in the deletion of H9 from the chloroplast 23S rRNA structure (Fig. 4d and Supplementary Figs S4d and S5d). Presence of chloroplast-specific C-terminal extension of uL29c around this region suggest a possible replacement of E. coli H9 with this chloro-specific protein extension.
The last region with significant alterations in rRNA structure results from a deletion and an insertion in the 4.8S rRNA (Fig. 4e and Supplementary Figs S4e and S5e). Deletion of a continuous stretch of 16 nucleotides at the 5′ end of 4.8S rRNA results in the absence of H98 in the spinach chloro-ribosome LSU. Interestingly, this deletion shares the same region where presumably 4.8S rRNA had got cleaved from its precursor 23S rRNA, and evolved as a new rRNA molecule. A 9 nucleotides long insertion at position 28 in chloro 4.8S rRNA (domain VI in the E. coli 23s rRNA) creates a bulge between H100 and H101, which resides in close proximity with chloro-specific N-terminal extension of bL19c.
Sequence analysis indicates that 4.8S rRNA is similar to the 3′ end of the E. coli 23S rRNA. Therefore, it is possible that 4.8S rRNA might have got separated from a 23S rRNA precursor during evolution and thereafter co-existed as a separate rRNA molecule along with the 23S rRNA to facilitate rRNA remodelling by loosing H98 at this cleavage site (Fig. 4e and Supplementary Figs S4e and S5e). The loss of H98 might have been necessary for the structure or function of the chloro-ribosome. Detachment of 4.8S rRNA from 23S rRNA makes chloroplast 23S rRNA shorter in length by 94 nucleotides compared to the E. coli 23S rRNA.
H38 and H58 in the 50S subunit of H. marismortui ribosome have earlier been reported as Kink-turn (K-turn) containing sites by Klein et al.47. K-turns are characterized by the presence of the helix-turn-helix structure, which help in introducing flexibility in rRNA structures48. K-turns act as important rRNA recognition motifs for the ribosomal proteins and form nucleation points during ribosome assembly. Introduction of the protrusions in chloro-ribosome 23S rRNA (H38 and H58) near to the positions where K-turns are present in H. marismortui 23S rRNA (Supplementary Fig. S6) is an interesting feature of these ribosomes that might have structural or functional implications in chloro-ribosomes.
Polypeptide tunnel and tunnel exit site
Nascent polypeptide chains travel through a polypeptide tunnel of about 100 Å in the LSU and interact with a diverse range of protein biogenesis factors involved in protein maturation, folding and targeting at the tunnel exit site. The tunnel exit site acts as a docking platform for these protein biogenesis factors. In E. coli ribosomes, polypeptide exit tunnel is mainly consisted of 23S rRNA along with three ribosomal proteins (uL4, uL22 and uL23) while the tunnel exit site is surrounded by uL22, uL23, uL24 and uL29. Our map of the spinach chloro-ribosome LSU reveals that the overall architecture of the polypeptide tunnel and tunnel exit site is similar to the E. coli 50S. However, certain differences are visible. The density for a conserved beta hairpin loop present in the E. coli uL23, which extends to the polypeptide tunnel is absent in uL23c (Fig. 5 and Supplementary Fig. S7). Additionally, N-termini of uL23c is mainly disordered and extends towards a position where H9 (absent in the chloro-ribosome) is located in the E. coli 50S (Fig. 4d). Moreover, C-termini of uL23c adopts a small helical turn while, it is in extended conformation in E. coli L23 (Supplementary Fig. S7). uL23 has been proposed to act as a universal docking site49 for a number of nascent polypeptide processing factors such as the signal recognition particle (SRP)50, trigger factor (TF)50,51 and SecA52, etc. The Absence of this loop in the chloro-ribosome makes the tunnel wider at this position. In E. coli, this loop extends into the polypeptide tunnel to form a potential interaction site with the nascent chain. Truncation of parts of the beta hairpin loop in E. coli does not impair cell growth under normal growth conditions but has been shown to lower the affinity of SRP binding to the ribosome nascent chain complex (RNC)53. It has also been proposed that the interaction between the nascent chain and this beta hairpin loop in the tunnel induces conformational change in L23 at the tunnel exit site, therefore increasing the affinity of SRP to bacterial RNC53. The SRP system in the chloroplast is unique in both the composition and function54,55. In chloroplast, the RNA component of E. coli SRP (4.8S RNA) is substituted with a protein named as cpSRP43. Unlike the E. coli SRP system which functions only co-translationally, the cpSRP has been suggested to function both co-translationally and post-translationally. The alteration in the structure of uL23c raises a possibility that a different mechanism of SRP recruitment is followed by the chloro-ribosome. However, further investigations are required to confirm this hypothesis.
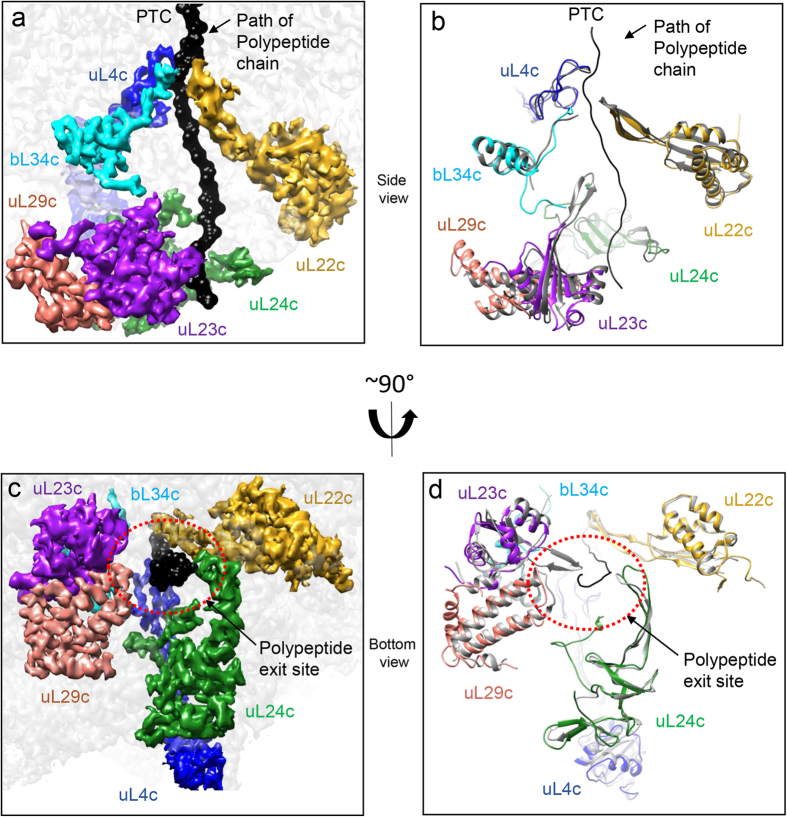
Figure 5. The polypeptide exit tunnel and polypeptide tunnel exit site of the spinach chloro-ribosome LSU.
(a) Polypeptide exit tunnel of the spinach chloro-ribosome indicated by the path of the simulated nascent chain (in black surface). Densities for the proteins lining the tunnel wall are isolated and shown in surface as indicated. (b) Models of the chloro-ribosome proteins lining the tunnel wall are overlaid with the E. coli protein models to show the chloro-specific features of the exit tunnel. (c) View as in (a) is rotated to show the polypeptide tunnel exit site in the spinach chloro-ribosome LSU. (d) Rotated view of (b) showing chloro-specific features of the polypeptide tunnel exit site in comparison to the E. coli tunnel exit site. Protein models and densities are labeled as indicated. For clarity rRNA is omitted.
The density for 15 amino acids of the chloroplast-specific N-terminal extension of uL24c is clearly visible in our map which extends towards the polypeptide tunnel suggesting a possible chloroplast-specific interaction between uL24c and nascent polypeptide (Fig. 5). Density for 24 amino acids of the chloroplast-specific C-terminal extension of uL29c can also be clearly seen in the map. This extension is located parallel to the N-terminal extension of uL23c. In the C-terminal extension of uL29c, the residues spanning 130 to 141 adopt a helical structure, while the residues ranging from 142 to 149 are seen to be extended in conformation. Taken together, it can be postulated that the significant differences present in three (uL23c, uL24c and uL29c) out of the four proteins (uL22c, uL23c, uL24c and uL29c) present at the tunnel exit site might reflect a chloroplast-specific mode of recruitment of protein biogenesis factors on the chloro-ribosome.
Methods
Purification of chloro-ribosomes
Purification of chloro-ribosomes was performed as previously described31,41, with some modifications in the protocols. Briefly, six kilograms of baby spinach (Spinacia oleracea) leaves bought from the local market were deveined, washed (using distilled water) and homogenized using 0.7 M sorbitol in buffer A (10 mM Tris·HCl, pH 7.6, 50 mM KCl, 10 mM MgOAc, and 7 mM 2-mercaptoethanol) using a blender (two shots of 10 sec each). The homogenized leaves were filtered through a double-layer of cheesecloth followed by one layer of Miracloth (Calbiochem). Resultant filtrate was centrifuged at 1200 × g for 15 min. The pelleted chloroplast was resuspended in 0.4 M sorbitol in buffer A and thereafter centrifuged again at 1200 × g for 15 min. The pellet was resuspended in 2% (vol/vol) Triton X-100 in buffer A and incubated on ice for 30 min. The lysed suspension was centrifuged at 26,000 × g for 30 min to achieve clarification and thereafter crude ribosomes were pelleted by centrifugation at 86,000 × g for 17 hr through a sucrose cushion (1 M sucrose in buffer A). The pellet was washed and gently resuspended in buffer B (buffer A with 10% glycerol). The suspension (crude ribosomes) was clarified by centrifugation at 26,000 × g for 15 min and was loaded onto 10–40% sucrose gradient in buffer A (with RNase inhibitor) and centrifuged at 111,000 × g for 4 hr. Fraction corresponding to 70S chloro-ribosomes was collected and further centrifuged through a sucrose cushion (0.75 M sucrose in buffer A, with RNase inhibitor) at 84,000 × g for 2.5 hr. The chloro-ribosome pellet was suspended in a grid buffer containing 20 mM Tris·HCl, pH 7.6, 100 mM KCl, 10 mM MgOAc, 100 mM sucrose, 7 mM 2-mercaptoethanol, 1 unit/ml RNase inhibitor and 0.1% protease inhibitor.
Electron microscopy
4 μl of purified chloro-ribosome at final concentrations of 4 OD260/ml were applied onto glow-discharged 2-nm carbon coated holey grids (Quantifoil R2/2) and incubated for 30 sec. Grids were blotted for 3 sec in 100% humidity at 4 °C and flash frozen in liquid N2-cooled liquid ethane using FEI Vitrobot. Grids were loaded to an FEI Tecnai Arctica cryo-transmission electron microscope, operated at 200 kV and equipped with a back-thinned Falcon III direct electron detection device. Electron micrographs were collected automatically by using FEI’s automatic single particle acquisition software EPU. A total of 1,729 micrographs were recorded in a movie mode as a set of 7 frames (total dose of 26 electrons per Å2) at a calibrated magnification of 109,375 resulting in a pixel size of 1.28 Å on the object scale and at defocus values from 0.2–2.5 μm.
Image processing
After data collection and screening for good micrographs, 1590 micrographs were selected for data processing. Whole-image drift correction was carried out using motioncorr56. e2boxer.py from EMAN 2.157 was used to semi-automatically pick particles from motion-corrected micrographs and CTF parameters were estimated using CTFFIND358. Data was processed with RELION 1.459. In total, 338,305 particles were subjected to reference-free 2D classification to discard bad particles. 261,052 particles selected from the 2D classes were directly used for 3D refinement using a 60 Å filtered E. coli 70S map60 and thereafter 3D classified without image alignment. 3D refinement of the particles from the 3D classes representing 70S particles did not yield a high-resolution map for the complete chloro-ribosome most probably because of the number of the particles being limited and the presence of conformational heterogeneity between the ribosomal subunits. Therefore, particles from both the 70S and 50S 3D classes were combined and further processed using a soft mask on the 50S subunit (Supplementary Fig. S1) to obtain a high-resolution map of the LSU. 202,766 particles from the high-resolution 3D classes obtained with a 50S mask were sorted according to the defocus values of the micrographs. 3D refinement of 174,949 particles with defocus values lower than 1.5 μm yielded a 50S density map at 3.5 Å resolution (Supplementary Fig. S1). ResMap42 was used to calculate local resolutions. Resolutions are calculated according to the gold-standard FSC = 0.143 criterion61. The density map was corrected for the modulation transfer function (MTF) and sharpened using an automatically calculated B-factor (−104.3 Å2), prior to visualization.
Model building and refinement
Coordinates for the 23S, 5S and 4.8S rRNAs were extracted from the reported coordinates of the spinach chloro-ribosome (PDB ID: 4V61)31. To model the missing (in PDB ID: 4V61) nucleotide residues of the 23S rRNA, sequence of the chloroplast 23S rRNA was obtained from the complete chloroplast genome sequence in NCBI (Accession no. AJ400848.1) and a homology model was generated for the complete 23S rRNA using ModeRNA server62. The missing nucleotide residues of the 23S rRNA were extracted from the generated homology model and connected to the 23S rRNA coordinates from PDB ID of 4V6131 and thereafter manually rebuilt in COOT63 guided by the predicted secondary structure from the Comparative RNA Website (CRW)64. To optimize the fitting in the density, base-pairing restraints were generated using ‘PDB to 3D Restraints’ server (http://rna.ucsc.edu/pdbrestraints/index.html) and used throughout the refinement cycles of the rRNA models using phenix.real_space_refine65.
To model chloro-ribosome LSU proteins, sequence information for the mature proteins were obtained from UniProt66 and GenBank67. Protein coordinates isolated from recently solved high resolution structure of the E. coli ribosome (PDB ID: 4YBB)43 were used as templates to guide homology modelling using ITASSER server68. Homology models were rigid body fitted into their respective densities (guided by positioning of the proteins in PDB ID: 4YBB) using ‘Fit in Map’ functionality in UCSF Chimera69 and were manually adjusted in COOT63, to achieve optimal fitting. Most of the chloroplast-specific extensions of the chloro-ribosome proteins didn’t fit into the density and were deleted from the homology models. These extensions were manually built using a combination of ‘C-alpha baton mode’ and ‘add terminal residue’ functionalities in COOT63. Extensions which assumed well defined helical structures, were de novo modelled using the tool ‘place helix here’ in COOT63. In all stages of modelling, ‘Real Space Refine Zone’ and ‘Regularize Zone’ were used to locally fit model into density while maintaining the geometry. PSRP5 and PSRP6 were modelled de novo using the toolsets mentioned above. Optimal fitting of individual proteins in the density was achieved by running phenix.real_space_refine65 on individual protein coordinates using local rotamer fitting, morphing, simulated annealing and secondary structure restraints applied throughout the refinement cycles. At the final stage, the coordinates for the rRNAs and the proteins were merged together in COOT63 and phenix.real_space_refine65 was run again using default parameters with secondary structure and base pair restraints in order to improve the map fitting and avoid clashes. In spite of the high-quality of our cryo-EM map, register shifts at poorly resolved parts and peripheral regions could not be excluded.
Model validation
The entire structure for the spinach chloro-ribosome LSU was validated using Molprobity70. A detailed account of model validation and refinement statistics is presented in Supplementary Table S2.
Figures
UCSF Chimera69, PyMOL71 and COOT63 are used to render all the figures. The simulated path of the nascent chain is built in COOT63 using the cryo-EM reconstruction reported by Bhushan et al. (EMDB ID: 1829)72. Jalview73 is used to generate the sequence alignment for the protein L23.
Additional Information
Accession codes: The 3.5 Å cryo-EM map of the spinach chloro-ribosome LSU has been deposited in the Electron Microscopy Data Bank with accession code EMD-9572, the coordinates of the atomic model have been deposited in the Protein Data Bank under accession code 5H1S.
How to cite this article: Ahmed, T. et al. Cryo-EM structure of the large subunit of the spinach chloroplast ribosome. Sci. Rep. 6, 35793; doi: 10.1038/srep35793 (2016).
Supplementary Material
Acknowledgments
We thank Andrew See Weng Wong for electron microscopy, Rikky Wenang Purbojati and Darshan Chandrakant Redij for help in computing. This work was supported by Nanyang Technological University Start Up Grant and Ministry of Education (MOE) of Singapore, AcRF Tier 1, 2014-T1-001-019 (RG32/14) to S.B.
Footnotes
Author Contributions S.B. conceived the project. T.A. and S.B. purified chloro-ribosomes. S.B. collected the cryo-EM data. T.A., Z.Y. and S.B. carried out the cryo-EM data processing. T.A. modelled the ribosomal proteins and rRNAs. T.A. and S.B. prepared all the figures and wrote the manuscript. All authors reviewed manuscript.
References
- Schmeing T. M. & Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature 461, 1234–1242 (2009). [DOI] [PubMed] [Google Scholar]
- Steitz T. A. A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol 9, 242–253 (2008). [DOI] [PubMed] [Google Scholar]
- Ogle J. M. et al. Recognition of Cognate Transfer RNA by the 30S Ribosomal Subunit. Science 292, 897–902 (2001). [DOI] [PubMed] [Google Scholar]
- Nissen P., Hansen J., Ban N., Moore P. B. & Steitz T. A. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 (2000). [DOI] [PubMed] [Google Scholar]
- Ban N., Nissen P., Hansen J., Moore P. B. & Steitz T. A. The Complete Atomic Structure of the Large Ribosomal Subunit at 2.4 Å Resolution. Science 289, 905–920 (2000). [DOI] [PubMed] [Google Scholar]
- Jenni S. & Ban N. The chemistry of protein synthesis and voyage through the ribosomal tunnel. Curr Opin Struct Biol 13, 212–219 (2003). [DOI] [PubMed] [Google Scholar]
- Kramer G., Boehringer D., Ban N. & Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol 16, 589–597 (2009). [DOI] [PubMed] [Google Scholar]
- Wilson D. N. & Doudna Cate J. H. The structure and function of the eukaryotic ribosome. Cold Spring Harb Perspect Biol 4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter H., Myasnikov A. G., Natchiar S. K. & Klaholz B. P. Structure of the human 80S ribosome. Nature 520, 640–645 (2015). [DOI] [PubMed] [Google Scholar]
- Harris E. H., Boynton J. E. & Gillham N. W. Chloroplast ribosomes and protein synthesis. Microbiological Reviews 58, 700–754 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien T. W. Evolution of a protein-rich mitochondrial ribosome: implications for human genetic disease. Gene 286, 73–79 (2002). [DOI] [PubMed] [Google Scholar]
- Sharma M. R. et al. Structure of the Mammalian Mitochondrial Ribosome Reveals an Expanded Functional Role for Its Component Proteins. Cell 115, 97–108 (2003). [DOI] [PubMed] [Google Scholar]
- Koc E. C. et al. The Large Subunit of the Mammalian Mitochondrial Ribosome: Analysis of the Complement of Ribosomal Proteins Present. Journal of Biological Chemistry 276, 43958–43969 (2001). [DOI] [PubMed] [Google Scholar]
- Cavdar Koc E., Burkhart W., Blackburn K., Moseley A. & Spremulli L. L. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J Biol Chem 276, 19363–19374 (2001). [DOI] [PubMed] [Google Scholar]
- Suzuki T. et al. Structural Compensation for the Deficit of rRNA with Proteins in the Mammalian Mitochondrial Ribosome: Systematic Analysis of Protein Components of the Large Ribosomal Subunit from Mammalian Mitochondria. Journal of Biological Chemistry 276, 21724–21736 (2001). [DOI] [PubMed] [Google Scholar]
- Suzuki T. et al. Proteomic Analysis of the Mammalian Mitochondrial Ribosome: Identification of Protein Components in the 28 S Small Subunit. Journal of Biological Chemistry 276, 33181–33195 (2001). [DOI] [PubMed] [Google Scholar]
- Stegeman W. J., Cooper C. S. & Avers C. J. Physical characterization of ribosomes from purified mitochondria of yeast. Biochemical and Biophysical Research Communications 39, 69–76 (1970). [DOI] [PubMed] [Google Scholar]
- Kuntzel H. & Noll H. Mitochondrial and Cytoplasmic Polysomes from Neurospora crassa. Nature 215, 1340–1345 (1967). [DOI] [PubMed] [Google Scholar]
- van der Sluis E. O. et al. Parallel Structural Evolution of Mitochondrial Ribosomes and OXPHOS Complexes. Genome Biology and Evolution 7, 1235–1251 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S. et al. Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 (1981). [DOI] [PubMed] [Google Scholar]
- Ott M. & Herrmann J. M. Co-translational membrane insertion of mitochondrially encoded proteins. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1803, 767–775 (2010). [DOI] [PubMed] [Google Scholar]
- Greber B. J. et al. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science 348, 303–308 (2015). [DOI] [PubMed] [Google Scholar]
- Amunts A., Brown A., Toots J., Scheres S. H. W. & Ramakrishnan V. The structure of the human mitochondrial ribosome. Science 348, 95–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber B. J. et al. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature 505, 515–519 (2014). [DOI] [PubMed] [Google Scholar]
- Greber B. J. et al. The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature 515, 283–286 (2014). [DOI] [PubMed] [Google Scholar]
- Brown A. et al. Structure of the large ribosomal subunit from human mitochondria. Science 346, 718–722 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A. et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L. Origin of eukaryotic cells: evidence and research implications for a theory of the origin and evolution of microbial, plant, and animal cells on the Precambrian earth. (Yale University Press New Haven, 1970). [Google Scholar]
- Martin W. & Herrmann R. G. Gene Transfer from Organelles to the Nucleus: How Much, What Happens, and Why? Plant Physiology 118, 9–17 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuell A. L., Quispe J. & Mayfield S. P. Structure of the chloroplast ribosome: novel domains for translation regulation. PLoS biology 5, e209 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M. R. et al. Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc Natl Acad Sci USA 104, 19315–19320 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuell A., Beligni M. V., Yamaguchi K. & Mayfield S. P. Regulation of chloroplast translation: interactions of RNA elements, RNA-binding proteins and the plastid ribosome. Biochem Soc T 32, 601–605 (2004). [DOI] [PubMed] [Google Scholar]
- Navarro M. J., Manuell A. L., Quispe J., Wu J. & Mayfield S. P. Chloroplast translation regulation. Photosynthesis Research 94, 359–374 (2007). [DOI] [PubMed] [Google Scholar]
- Cullis C. A., Vorster B. J., Van Der Vyver C. & Kunert K. J. Transfer of genetic material between the chloroplast and nucleus: how is it related to stress in plants? Annals of Botany 103, 625–633 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y. & Wollman F.-A. Translational regulations as specific traits of chloroplast gene expression. FEBS Letters 529, 39–42 (2002). [DOI] [PubMed] [Google Scholar]
- Barkan A. & Goldschmidt-Clermont M. Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572 (2000). [DOI] [PubMed] [Google Scholar]
- Eberhard S., Drapier D. & Wollman F.-A. Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. The Plant Journal 31, 149–160 (2002). [DOI] [PubMed] [Google Scholar]
- Surpin M., Larkin R. M. & Chory J. Signal Transduction between the Chloroplast and the Nucleus. The Plant Cell 14, s327–s338 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber B. J. & Ban N. Structure and Function of the Mitochondrial Ribosome. Annu Rev Biochem (2016). [DOI] [PubMed] [Google Scholar]
- Yamaguchi K. & Subramanian A. R. The Plastid Ribosomal Proteins: Identification of all the Proteins in the 50 S Subunit of an Organelle Ribosome (Chloroplast). Journal of Biological Chemistry 275, 28466–28482 (2000). [DOI] [PubMed] [Google Scholar]
- Bartsch M., Kimura M. & Subramanian A.-R. Purification, primary structure, and homology relationships of a chloroplast ribosomal protein. Proceedings of the National Academy of Sciences 79, 6871–6875 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A., Sigworth F. J. & Tagare H. D. Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeske J. et al. High-resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol 22, 336–341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N. et al. The plastid-specific ribosomal proteins of Arabidopsis thaliana can be divided into non-essential proteins and genuine ribosomal proteins. The Plant Journal 69, 302–316 (2012). [DOI] [PubMed] [Google Scholar]
- Yamaguchi K. & Subramanian A. R. Proteomic identification of all plastid-specific ribosomal proteins in higher plant chloroplast 30S ribosomal subunit. European Journal of Biochemistry 270, 190–205 (2003). [DOI] [PubMed] [Google Scholar]
- Rice P., Longden I. & Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
- Klein D. J., Schmeing T. M., Moore P. B. & Steitz T. A. The kink-turn: a new RNA secondary structure motif. EMBO J 20, 4214–4221 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. & Lilley D. M. The Kink Turn, a Key Architectural Element in RNA Structure. J Mol Biol 428, 790–801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G., Boehringer D., Ban N. & Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol 16, 589–597 (2009). [DOI] [PubMed] [Google Scholar]
- Bornemann T., Holtkamp W. & Wintermeyer W. Interplay between trigger factor and other protein biogenesis factors on the ribosome. Nat Commun 5 (2014). [DOI] [PubMed] [Google Scholar]
- Wegrzyn R. D. et al. A conserved motif is prerequisite for the interaction of NAC with ribosomal protein L23 and nascent chains. J Biol Chem 281, 2847–2857 (2006). [DOI] [PubMed] [Google Scholar]
- Singh R. et al. Cryo-electron microscopic structure of SecA protein bound to the 70S ribosome. J Biol Chem 289, 7190–7199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornemann T., Jockel J., Rodnina M. V. & Wintermeyer W. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat Struct Mol Biol 15, 494–499 (2008). [DOI] [PubMed] [Google Scholar]
- Stengel K. F., Holdermann I., Wild K. & Sinning I. The structure of the chloroplast signal recognition particle (SRP) receptor reveals mechanistic details of SRP GTPase activation and a conserved membrane targeting site. FEBS Letters 581, 5671–5676 (2007). [DOI] [PubMed] [Google Scholar]
- Nussaume L. Chloroplast SRP takes another road. Nat Chem Biol 4, 529–531 (2008). [DOI] [PubMed] [Google Scholar]
- Li X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Meth 10, 584–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J. M., Chen M., Baldwin P. R. & Ludtke S. J. High resolution single particle refinement in EMAN2.1. Methods 100, 25–34 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell J. A. & Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of Structural Biology 142, 334–347 (2003). [DOI] [PubMed] [Google Scholar]
- Scheres S. H. W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. Journal of Structural Biology 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N. et al. Structure of the E. coli ribosome-EF-Tu complex at <3 A resolution by Cs-corrected cryo-EM. Nature 520, 567–570 (2015). [DOI] [PubMed] [Google Scholar]
- Rosenthal P. B. & Henderson R. Optimal Determination of Particle Orientation, Absolute Hand, and Contrast Loss in Single-particle Electron Cryomicroscopy. Journal of Molecular Biology 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- Rother M. et al. ModeRNA server: an online tool for modeling RNA 3D structures. Bioinformatics 27, 2441–2442 (2011). [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W. G. & Cowtan K. Features and development of Coot. Acta Crystallographica Section D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone J. J. et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3, 2 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P. V., Headd J. J., Terwilliger T. C. & Adams P. D. New tool: phenix.real_space_refine. Computational Crystallography Newsletter 4, 43–44 (2013). [Google Scholar]
- Apweiler R. et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Research 32, D115–D119 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D. A. et al. GenBank. Nucleic Acids Research 41, D36–D42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 1–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F. et al. UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- Chen V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D: Biological Crystallography 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger L. L. C. Py-MOL, The PyMOL Molecular Graphics System, V1.8 (2010).
- Bhushan S. et al. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS biology 9, e1000581 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M. & Barton G. J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.