Abstract
Identifying interactions between ligands and transmembrane receptors is crucial for understanding the endocrine system. However, the present approaches for this purpose are still not capable of high-throughput screening. In this report, a membrane-anchored ligand and receptor yeast two-hybrid (MALAR-Y2H) system was established. In the method, an extracellular ligand is linked with an intracellular split-ubiquitin reporter system via a chimeric transmembrane structure. Meanwhile, the prey proteins of transmembrane receptors are fused to the other half of the split-ubiquitin reporter system. The extracellular interaction of ligands and receptors can lead to the functional recovery of the ubiquitin reporter system in yeast, and eventually lead to the expression of report genes. Consequently, the system can be used to detect the interactions between extracellular ligands and their transmembrane receptors. To test the efficiency and universality of the method, interactions between several pairs of ligands and receptors of mouse were analyzed. The detecting results were shown to be thoroughly consistent with the present knowledge, indicating MALAR-Y2H can be utilized for such purpose with high precision, high efficiency and strong universality. The characteristics of the simple procedure and high-throughput potential make MALAR-Y2H a powerful platform to study protein-protein interaction networks between secreted proteins and transmembrane proteins.
Identifying interactions between cytokines and transmembrane receptors is important for understanding the endocrine regulation system and drug development. Since receptors are accessible to drugs, many important cytokine pathways, such as PD1 and VEGF/EGFR, are drug targets. However, there are still many unknown interactions that need to be mined. Defining these interactions will absolutely aid us in understanding the interaction networks involving cytokines and transmembrane receptors better and will accelerate drug development.
If the nervous system is a “tele-communications network”, the cytokine-receptor network can be seen as a “broadcasting network” between organs or cells. However, studying such a complex network is a huge challenge. Since the cellular membrane is a prerequisite for spatial construction of receptors, especially multi-transmembrane receptors, most in vitro methods, including surface plasmon resonance1, are not suitable to study these membrane-bound protein systems. Moreover, in vivo methods, such as the Ca2+ flux assay, chemotaxis assay and competitive binding assay2, have major drawbacks, including low-throughput and complex processes, which make these assays unsuitable for high-throughput screening.
Yeast two-hybrid (Y2H) method, as an important protein-protein interaction (PPI) assay, has contributed significantly to proteomics in the post-genomic era. Currently, variant Y2H methods have been used for detecting PPIs between two cytoplasmic proteins, between a cytoplasmic protein and a membrane protein, and between two membrane-associated proteins3. However, all these methods are not suitable for detecting PPIs between secreted cytokines and transmembrane receptors for two reasons: (i) almost all transmembrane proteins require the plasma membrane to form native structures; and (ii) PPIs occurring on the outer cytoplasmic membrane need to turn on an intracellular signal cascade that can be readily detected.
Previously, a membrane protein Y2H system to detect PPIs between two membrane proteins was developed4. In the method, the yeast ubiquitin gene is split into N- and C-terminal fragments (Nub and Cub). The two separated fragments are fused, respectively, with the membrane-associated bait and prey proteins of interest. The interaction of bait and prey brings Nub and Cub together to reconstitute a functional ubiquitin. This reconstituted ubiquitin is recognized by an endogenic ubiquitin specific protease (UBP) that leads to the release of a fused transcription factor from the C-terminus of Cub and the transcription of reporter genes.
Inspired by this strategy, we hypothesized that, if an extracellular ligand of interest (bait) could be linked with an intracellular reporter system, this approach could be used to detect interactions between the bait and candidate transmembrane receptors (prey). The hypothesis was finally verified through a series of experiments. Herein, in the present article, we are going to describe the successful development of the membrane-anchored ligand and receptor yeast two-hybrid (MALAR-Y2H) method to detect interactions between extracellular cytokines and transmembrane receptors.
Results
Experiment Design
CXC-motif chemokine ligand 12 (CXCL12), a classic chemokine, is a member of the CXC motif chemokine gene family. CXCL12 binds two known receptors, CXCR4 and CXCR7. Both receptors belong to the CXCR family, a type of seven-transmembrane domain G-protein-coupled receptors (GPCR)5. To verify the effectiveness of the MALAR-Y2H system, we attempted to examine the interactions between CXCL12 and its known receptors and other candidate receptors.
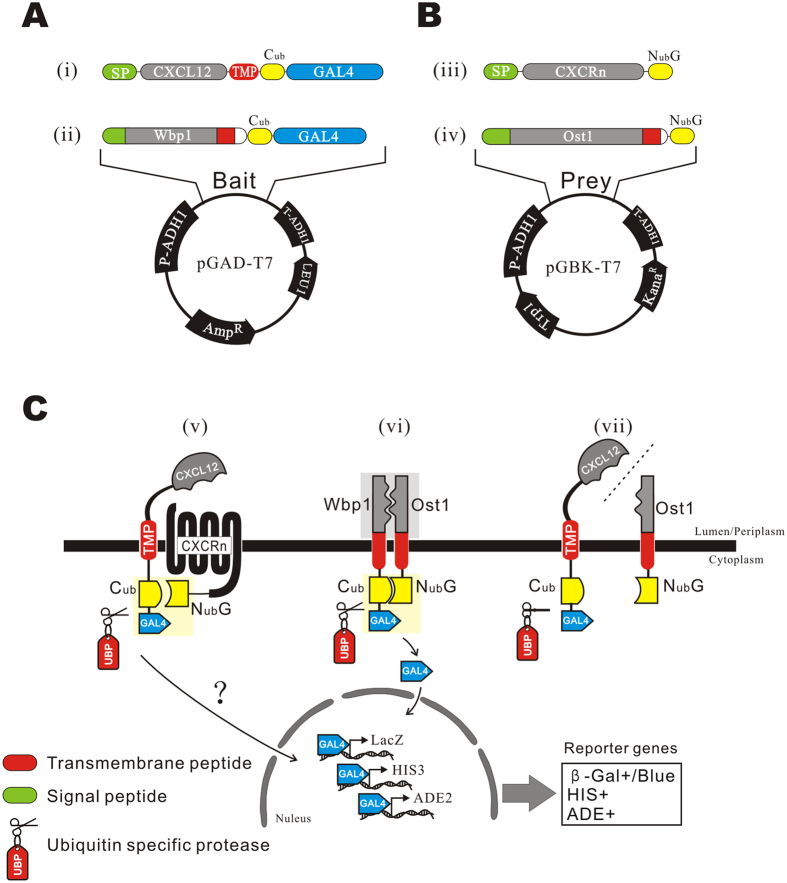
The bait protein was generated by constructing an artificial transmembrane structure (Fig. 1A,i), in which five elements were fused in tandem, including: (1) a signal peptide (SP), from SP of CXCL12, Wbp1 (a yeast transmembrane protein), or MFAL1 (a yeast secretory protein), (2) the bait protein CXCL12, (3) a transmembrane peptide (TMP) from yeast protein Wbp1, (4) a C-terminal fragment of yeast ubiquitin (Cub) and (5) a transcription factor, Regulatory Protein GAL4 (GAL4). The artificial bait protein possesses the same topological structure of Type III membrane proteins6.
Figure 1. Design of bait and prey plasmids and the principles of detecting the interaction.
(A) The structures of the bait plasmids. (i) CXCL12 was fused with a signal peptide (SP) at the N-terminus and with a transmembrane peptide (TMP) at C-terminus, followed by Cub and GAL4 in tandem. (ii) WBP1 was fused with Cub and GAL4 as a control. Bait genes were cloned into the plasmid pGAD-T7. (B) Structures of prey plasmids. (iii) The prey fusion was composed of a SP, CXCR and NubG in tandem. (iv) OST1 was fused with NubG as a control prey. Prey genes were cloned into the plasmid pGBK-T7. (C) Principles of detecting the interaction. (v) The bait protein is expressed as a transmembrane protein, in which the CXCL12 domain is directed into the lumen (and the periplasm, if mature forms are located on plasmalemma) and linked with the Cub-GAL4 cassette in the cytoplasm by the TMP. CXCRs-NubG is co-expressed on the plasma membrane. The interaction of CXCL12 and CXCRs unites Cub and NubG and leads to the reconstitution of the split ubiquitin protein, which is recognized and cleaved by UBP to release GAL4. The liberated GAL4 protein is transported into the nucleus by the SV40 nuclear localization signal at its N-terminus, and transcription of reporter genes lacZ, HIS3 and ADE2 results in blue clones on a SD/His−Ade− plate in the presence of α-X-Gal. (vi) Coexpression of WBP1-Cub-GAL4 and OST1-NubG was used as the positive interacting proteins control. The two genes are known interacting partners of membrane proteins of yeast. (vii) Coexpression of SP-CXCL12-TMP-Cub-GAL4 and OST1-NubG, two unrelated genes, was used as a negative control.
The candidate prey proteins, CXC motif receptors, were fused with (a) an SP, from yeast gene Wbp1, Ost1 or MFAL1 respectively, at N-terminus; (b) a mutant N-terminal fragment of ubiquitin (NubG) at the C-terminus (Fig. 1B,iii). NubG has an Ile13Gly mutation when compared with the wild-type sequence, which inhibits spontaneous reconstitution between native Nub and Cub4.
In theory, the SP can guide N-terminus of the bait protein targeting to the endoplasmic reticulum (ER) lumen and to the extracellular space. If the mature forms are located on plasmalemma, and the TMP anchors the bait protein to the ER membrane, they can conjugate with the reporter modules, Cub and GAL4, which exist in the cytoplasm (Fig. 1A,i)6. Therefore, preys will be directed to integrate with the ER membrane with a Nub fragment in the cytoplasm (Fig. 1B,iii).
Interaction of SP-CXCL12-TMP-Cub-GAL4 and SP-CXCR-NubG reconstructs the split Cub and NubG into a whole and activated ubiquitin, which can be recognized by ubiquitin-specific protease (UBP) and leads to the release of GAL4 from the C-terminus of Cub (Fig. 1C,v).
Co-expression of WBP1-Cub-GAL4 and OST1-NubG was used as a positive control (Fig. 1A,ii,B,iv). WBP1 and OST1 are transmembrane proteins of S. cerevisiae, and are interacting subunits of the N-oligosaccharyl transferase (OST) complex. Their co-expression leads to the physical reconstitution and functional recovery of ubiquitin via colocalizing Cub and NubG (Fig. 1C,vi). Co-expression of SP-CXCL12-TMP-Cub-GAL4 and OST1-NubG was used as a negative control (Fig. 1C,vii).
Subcellular Localization of Baits and Preys
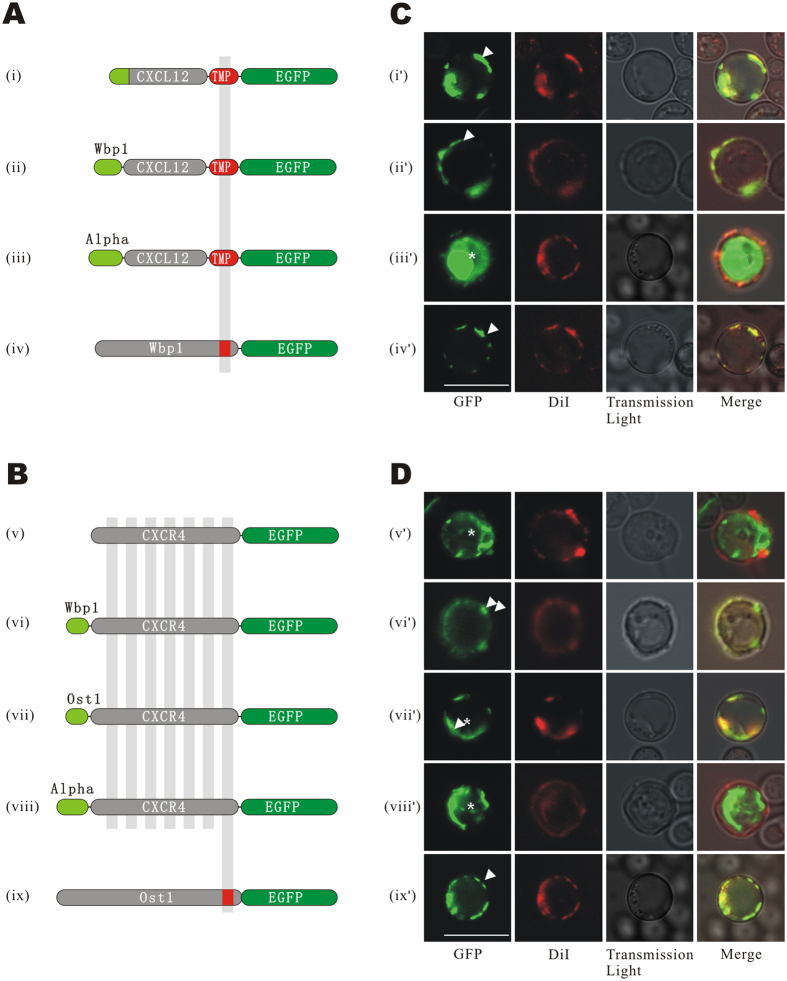
To test the secretory efficacy of different signal peptides, we tried signal peptides derived from three origins, including SPCXCL12 (signal peptide of CXCL12, residues 1–28) (Fig. 2A,i), SPWBP1 (S. cerevisiae WBP1, residues 1–31) (Fig. 2A,ii) and SPMFAL1 (S. cerevisiae MFAL1, residues 1–89) (Fig. 2A,iii), as the signal peptide for the bait protein. The transmembrane peptide TMPWBP1 (WBP1, residues 350–430) was used as the TMP. Enhanced green fluorescence protein (EGFP) was in-frame fused at the C-terminus of the TMP to visualize protein location. WBP1-EGFP was used as positive control (Fig. 2A,iv).
Figure 2. Subcellular localization of baits and preys with different signal peptides.
(A) Structure of bait plasmids. The pGAD-T7 vector was used as the backbone of the bait plasmids, in which bait genes were promoted by the ADH1 promotor. Three different signal peptides, (i) SPCXCL12, (ii) SPWBP1, or (iii) SPMFAL1 were fused at the N-terminal end of CXCL12, and the transmembrane peptide of TMPWBP1 and EGFP were fused to the C-terminus of CXCL12 in tandem. (iv) WBP1-EGFP was used as the positive control. (B) Structures of prey plasmids. The pGBK-T7 vector was used as the backbone of the prey plasmids, in which preys were promoted by the ADH1 promoter. (v) CXCR4 with the native N-terminus or CXCR4 fused with different signal peptides, (vi) SPWBP1, (vii) SPOST1, or (viii) SPMFAL1 were tagged with EGFP at the C-terminus. (ix) OST1-EGFP was used as the control. (C,D) LSCM images of EGFP. Baits (i’-iv’) and preys (v’-ix’) were expressed by GoldY2H and Y187, respectively, and observed with LSCM. Images of EGFP and membrane-specific fluorescent dye DiI and transmission light were captured and merged. The bars indicate 10 micrometers. The arrows direct the membrane localization of baits and preys, and the asterisks indicate the unexpected intracellular localizations.
Bait plasmids were transformed into GoldY2H host cells and the co-localization of bait protein and plasma membrane was observed. The fluorescence of EGFP-tagged bait protein and DiI-stained plasma membrane were captured using laser scanning confocal microscopy (LSCM). The result showed that green fluorescence in host cells of SPCXCL12-CXCL12-TMPWBP1-EGFP and SPWBP1-CXCL12-TMPWBP1-EGFP focused accurately to the plasma membrane (Fig. 2C,i’–ii’). In contrast, the fluorescent signal of SPMFAL1-CXCL12-TMPWBP1-EGFP expressing cells was distributed intracellularly (Fig. 2C,iii’). Thus, it was indicated that SPCXCL12 and SPWBP1 guided bait secretion and anchoring to the cell plasma membrane correctly rather than SPMFAL1. The positive control protein WBP1-EGFP was found to locate on the plasma membrane, as expected (Fig. 2C,iv’).
CXCR4, a well-known CXCL12 receptor, was used as a representative to optimize the localization of prey. Signal peptides SPWBP1, SPOST1 (yeast OST1, residues 1–30), or SPMFAL1 were fused at the N-terminus of CXCR4 (Fig. 2B,v–viii). EGFP was fused at the C-terminus as a reporter gene for subcellular localization imaging (Fig. 2B,ix).
Observations revealed that CXCR4 was directed onto the plasma membrane by both SPWBP1 and SPOST1 (Fig. 2D,vi’,vii’), but not by SPMFAL1 or its native N-terminus (Fig. 2D,v’,viii’). The fluorescent signals of the latter two preys were distributed mainly in the cytoplasm or vacuole. The positive control OST1-EGFP was well co-localized with DiI- stained plasma membrane (Fig. 2D,ix’).
Based on the above results, SPCXCL12 and SPOST1 were selected as the optimal signal peptides to direct localization of the baits and preys respectively in the following experiments. The localization assays of other CXCRs are shown in Figure S1.
Construction of Y2H Plasmids
We then introduced the split ubiquitin system to bait and prey proteins to generate the reporter module. The split ubiquitin system was composed of two parts. Part I, containing the Cub fragment and transcript factor GAL4 in tandem, was fused at the C-terminal to the TMP of the bait. Part II, the NubG fragment was fused to the 3′ end of prey genes.
Self-Activation Detection
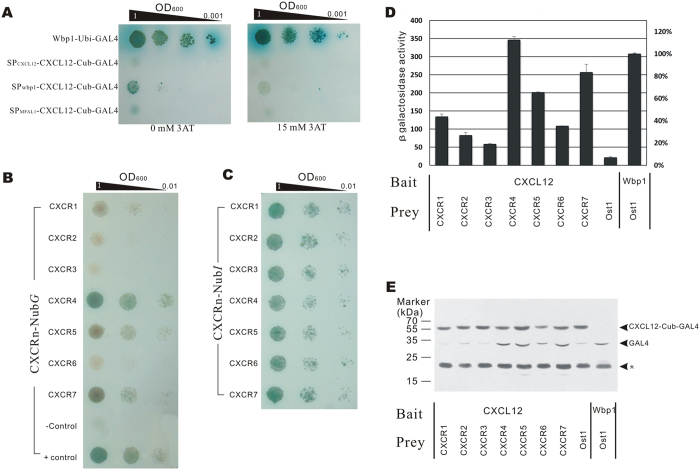
Before detecting the PPIs between baits and preys, we first examined whether there was any self-activation of baits. CXCL12 bait, guided by three different signal peptides, SPCXCL12, SPwbp1 and SP MFAL1 respectively, were expressed in GoldY2H yeast and inoculated on selective plates with α-X-gal. The results indicated that baits with different signal peptides have variant degrees of self-activation, reflecting the existence of UBP-independent release of GAL4. The bait with SPCXCL12 showed lower self-activation (Fig. 3A,i). By adding 15mM 3-AT, a competitive inhibitor of the His3 reporter gene, into the selective medium, the self-activation was mostly inhibited (Fig. 3A,ii).
Figure 3. PPIs of the CXCL12 and CXCR family.
(A) Self-activation detection. GoldY2H cells expressing bait protein with SPCXCL12, SPWBP1 and SPMFAL1 signal peptides were detected to self-activate. Five microliter diluted suspensions were dropped onto SD/Trp−Ade−His− plates with α-X-gal and 3-AT, and grown for 72 h. (B) Cells growth assay. GoldY2H cells expressing the SPCXCL12-CXCL12-TMP-Cub-GAL4 plasmid was hybridized with Y187 cells expressing each SPOST1-CXCR-NubG fusion protein, and diploid cells were dropped onto SD/Leu−Trp−Ade−His− plates containing α-X-gal and 15 mM 3-AT. The growth of cells was observed after 72 h cultivation. SPCXCL12-CXCL12-TMP-Cub-GAL4 × OST1-NubG were used as a negative control, and WBP1-Cub-GAL4 × OST1-NubG were used as a positive control. (C) Growth assay of cells expressing SPCXCL12-CXCL12-TMP-Cub-GAL4 and CXCR-NubI. (D) Quantitative β-Gal assay of diploid cells expressing SP-CXCL12-TMP-Cub-GAL4 and each of the SPOST1-CXCR-NubG proteins. The experiment was repeated three times independently and every sample was tested twice in each experiment (n = 2). (E) Western blot analysis of cleaved GAL4. Diploid cells expressing SPCXCL12-CXCL12-TMP-Cub-GAL4 and each of the CXCR-NubG proteins were lysed and the GAL4 level detected by western blot analysis. The molecular weight (MW) of SPCXCL12-CXCL12-TMP-Cub-GAL4 is 59 kDa, and the MW of cleaved GAL4 is 31 kDa. The experiment was performed three times independently. The asterisk denotes a non-specific band.
Consequently, SPCXCL12 was used as bait signal peptide and 15 mM 3-AT was supplemented in yeast medium in all the following tests.
PPI Detection of CXCL12 and the CXCR Family
Growth Assay
GoldY2H cells harboring bait plasmids and Y187 cells harboring prey plasmids were hybridized and selected on SD/Leu−Trp− plates. Hybrids were spread on SD/Leu−Trp−His−Ade− medium supplemented with α-X-gal. The colors of colonies showed that CXCL12 can interact with CXCRs with different interaction intensities, of which CXCR4 is the strongest interaction partner. CXCR7-CXCL12 and CXCR5-CXCL12 were also found able to interact, but with a lower intensity than CXCR4-CXCL12. CXCR3-CXCL12 and CXCR6-CXCL12 were found to be the weakest interaction partners (Fig. 3B).
System Verification
To exclude bias caused by difference in the expression levels of prey proteins, we substituted the NubG in the prey protein of a wild-type ubiquitin N-terminus, NubI, which could associate with Cub spontaneously and lead to the release of GAL4 independent of the interaction of bait and prey. Therefore, the strength of the reporter genes was dependent on the expression level of the prey protein rather than the interaction between the bait and prey. The result revealed that cells expressing different CXCRs grew at a similar rate, suggesting similar expression levels between preys (Fig. 3C).
β-Galactosidase (β-Gal) Activity Assay
We further tested the β-Gal activity of each hybrid cell. The results were consistent with the cell growth test. β-Galactosidase activity in the yeast cells expressing CXCR4-NubG, CXCR7-NubG, or CXCR5-NubG were determined to be 113%, 83% and 65% β-Gal activity compared to the positive control (Fig. 3D).
Western Blot Assay
GAL4 cleavage caused by bait and prey interactions were detected by a western blot assay. The results revealed that GAL4 released from the bait fusion protein is induced mainly by coexpression of CXCR4-, CXCR5- and CXCR7-NubG. This observation indicates that these three receptors can strongly interact with CXCL12 and the result is coincident with the growth assay and β-Gal activity assay (Fig. 3E).
PPIs Detection between CXCL12 and CXCR5 Motifs
Based on the above experiments, we have confirmed the PPIs between CXCL12 and its functional receptor CXCR4, CXCR7 and these two receptors are known to be the of CXCL127,8. CXCR5 was also shown to be capable of strongly interacting with CXCL12, which has not been reported so far.
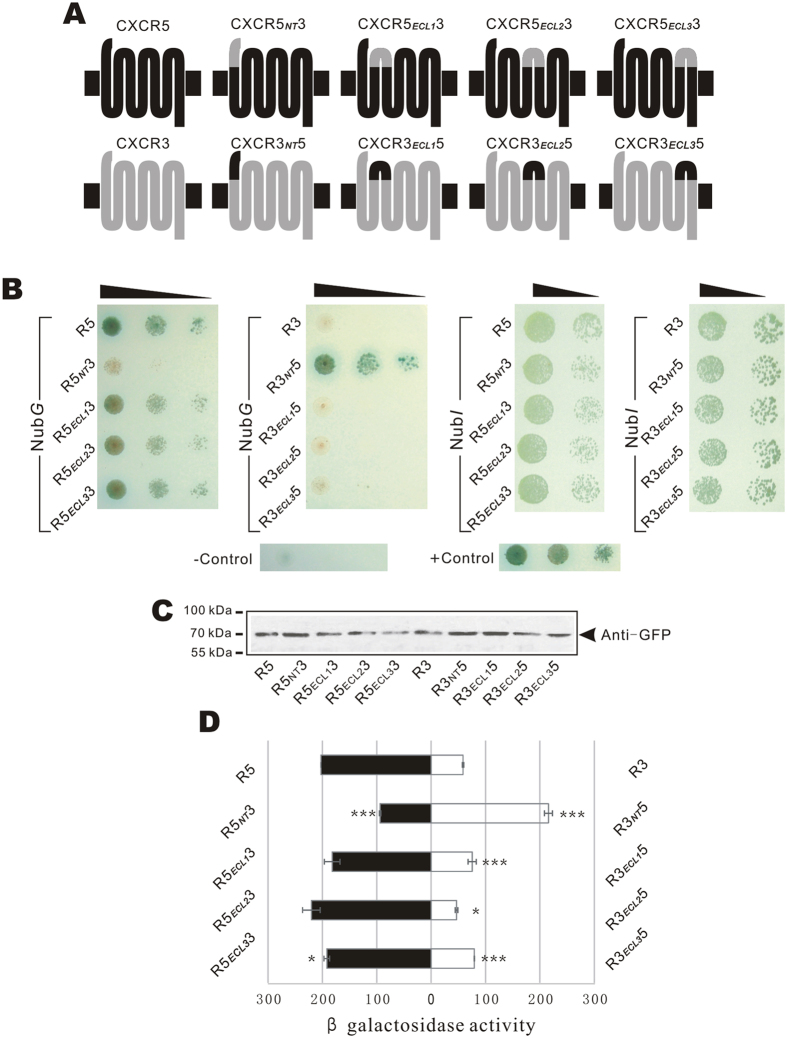
To further understand the interacting features of CXCL12 and CXCR5, we investigated the exact extracellular interaction sites by domain substitution9. According to the UniProt database, four extracellular domains, including an N-terminal fragment (NT) and three extracellular loops (ECLs), were exchanged with homologous fragments of CXCR3, the receptor showing the weakest interaction with CXCL12 (Fig. 4A). The partitions of CXCR5 and CXCR3 sequences are given in Fig. S2.
Figure 4. Detection of PPIs between CXCL12 and chimeric CXCR5.
(A) Structures of chimeric receptors of CXCR5 and CXCR3. Four extracellular fragments of CXCR5 were exchanged with counterparts of CXCR3 to form eight chimeric receptors. (B) Cell growth detection. Diploid cells expressing SPCXCL12-CXCL12-TMP-Cub-GAL4 and CXCR5-NubG, CXCR3-NubG, or NubG tagged chimeric receptors were detected by the growth ability on selection plate (SD-Leu−Trp−Ade−His−) with 15 mM 3AT + α-X-gal (left column). Diploid cells harboring the bait and NubI tagged preys were detected (right column). Positive and negative control cells were the same as Fig. 3B. (C) Expression detection by western blot assay. The chimeric preys labeled with EGFP were detected by western blot. (D) Quantitative β-Gal assay of diploid cells expressing SPCXCL12-CXCL12-TMP-Cub-GAL4 and NubG tagged chimeric receptors. The experiment was repeated three times independently and every sample was tested twice in each experiment (n = 2). * and *** indicate significance p < 0.05 and p < 0.001 of a t-test between chimeric receptors and wild-type counterparts.
Eight chimeric receptors fusing the SPOST1 signal peptide and NubG tags were transformed into Y187 cells, and hybridized with GoldY2H cells expressing SPCXCL12-CXCL12-TMP-Cub-GAL4. Growth of hybrid cells was detected on nutrition selective plates with X-gal and 15 mM 3-AT. The results revealed that cells expressing bait and chimeric receptor CXCR5NT3-NubG (CXCR5-NubG with substituted N-terminus) lost growth ability. In contrast, cells with CXCR3NT5-NubG gained growth ability (Fig. 4B,I,ii). Cells expressing bait and CXCR5ECL13-NubG, CXCR5ECL23-NubG, or CXCR5ECL33-NubG grew slower than cells with CXCR5-NubG. Correspondingly, cells with CXCR3ECL15-NubG, CXCR3ECL25-NubG, or CXCR3ECL35-NubG gained stronger growth ability than their wild-type counterpart, CXCR3-NubG. Using the β-Gal assay, we quantitatively evaluated the interactions between chimeric receptors and CXCL12 (Fig. 4D). The β-Gal activity of cells expressing CXCR5NT3-NubG was 49% relative to the activity of wild-type CXCR5 (p < 0.0001, n = 2). The activity of CXCR3NT5-NubG expression cells was equal to the activity of CXCR5 (106%, p = 0.065). The β-Gal activities of cells with CXCR5ECL13-NubG or CXCR5ECL33-NubG was 90% (p = 0.088) or 95% (p = 0.05) of CXCR5-NubG. In contrast, CXCR3ECL15 or CXCR3ECL35 increased β-Gal activity in their host cells by 28% and 35% (p = 0.046 and 0.0014), respectively. Exchange of ECL2 did not significantly change the β-Gal activities for either CXCR5 (p = 0.14) or CXCR3 (p = 0.013).
Taken together, the result suggests that the N-terminus of CXCR5 is the main interaction motif in its PPI with CXCL12, at the same time ECL1 and ECL3 also contribute to the PPI, but to a lesser extent.
To avoid the bias caused by different prey expression level, we tested the interactions of the bait and NubI-labeled prey proteins. The result showed that cells harboring NubI-tagged preys had a similar growth ability, indicating an equal expression level of the preys (Fig. 4B,iii,iv). The expression level of the preys were also tested by western blot. To avoid the interruption of intrinsic ubiquitinated proteins, the preys were labeled with EGFP and blotted by anti-EGFP antibody. The result suggested that the expression level of the different chimeric preys are similar (Fig. 4C).
Verification of the CXCL12 and CXCR5NT interaction
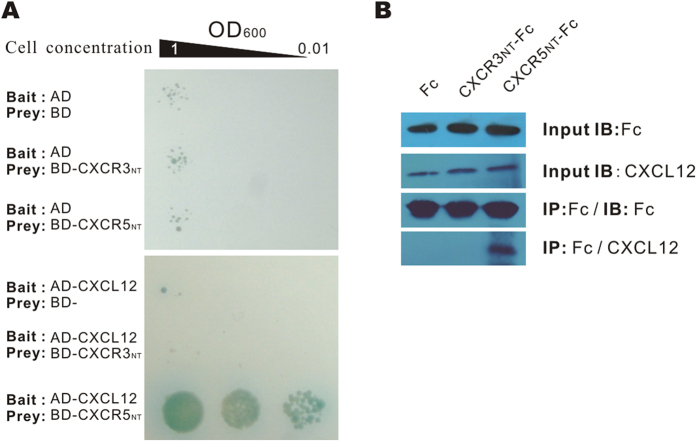
To further verify the interaction between CXCL12 and CXCR5NT, we tested the PPIs between CXCL12 and the N-terminus of CXCR5 by a traditional Y2H assay and a co-immunoprecipitation (CoIP) assay (Fig. 5A,B). Both assays indicated an interaction occurs between CXCL12 and CXCR5NT.
Figure 5. Detection of the interactions between CXCL12 and CXCR5NT.
(A) The interaction between CXCL12 and the N-terminus of CXCR5 or CXCR3 was detected by a traditional Y2H, in which CXCL12 was fused with the activation domain of GAL4 (AD) as the bait, and CXCR5NT or CXCR3NT was fused with the binding domain (BD) of GAL4 as the preys. Only yeast expressing AD-CXCL12 and BD-CXCR5NT could survive the selective medium, indicating the interaction between prey and bait. (B) CoIP of CXCL12 and CXCR5NT. CXCL12 and CXCR5NT-Fc or CXCR3NT-Fc were co-transfected into the 293T cell line. Fc-tagged fusion proteins were precipitated by protein G beads, and the precipitates were separated by SDS-PAGE and blotted with rabbit anti-mouse CXCL12 IgG. The result shows that CXCL12 was precipitated by CXCR5NT-Fc, but not by CXCR3NT-Fc or Fc only, indicating an interaction between CXCR5NT and CXCL12.
General Applicability Test
To test the reliability and robustness of the method, we first examined the PPIs between chemokine ligands CXCL7, CXCL9 and CXCR receptors, which are also members of the CXC chemokine family. In the assays, the ligands were fused with SPwbp1 signal peptide (Figure S3A,B). Interactions between CXCL7 and CXCR2, and CXCL9 and CXCR3 were obviously detected by growth assays (Fig. 6A) and β-Gal activity assay (Figure S3C,D). The result was well coincident with the previous reports10,11,12.
Figure 6. Universality study of the MALAR Y2H method.
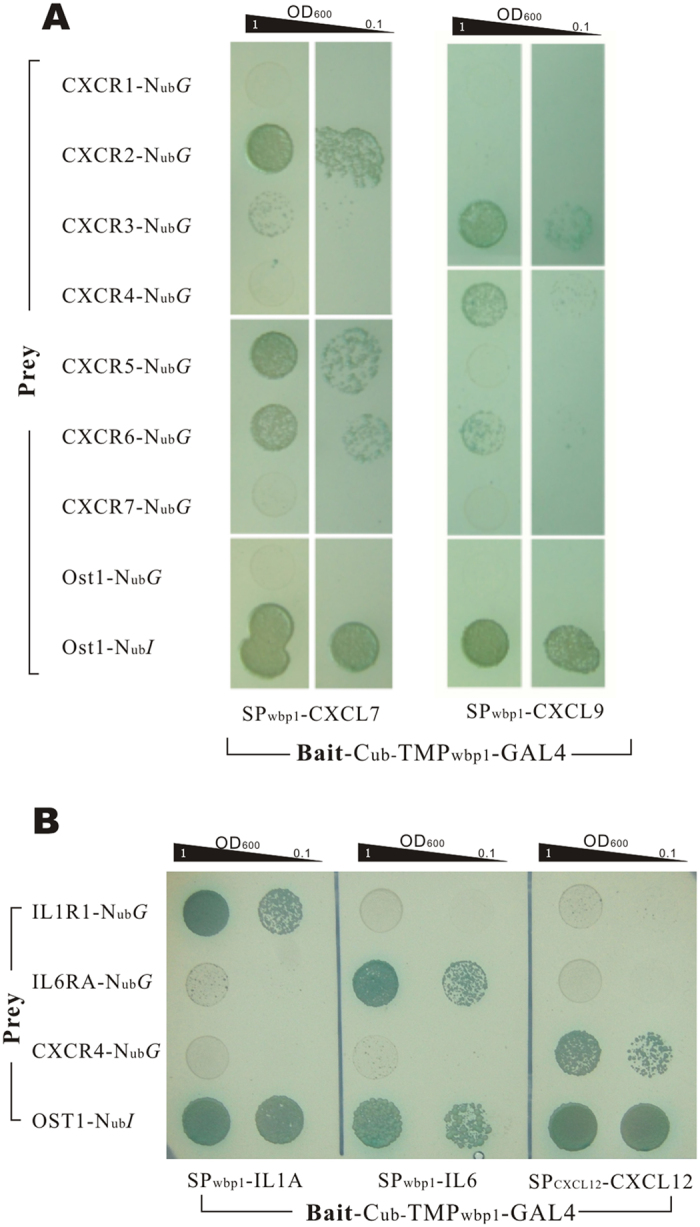
(A) CXCL7 and CXCL9 were detected the PPIs with receptors of CXCR family (CXCRn-NubG) using growth assay, and the result showed different PPIs intensity with each receptors, in which obvious PPIs were detected between CXCL7 and CXCR2 and between CXCL9 and CXCR3. (B) Interleukin ligand/receptor pairs, IL1A/IL1R1 and IL6/IL6RA, were detected the interactions. CXCL12 and CXCR4 was used as negative controls of bait and prey respectively, and Ost1-NubI was used as positive prey control. Through growth assays, the expected interactions of IL1A/IL1R1 and IL6/IL6RA were detected correctly, meanwhile, no crosstalk was detected between unmatched ligand/receptor pairs.
Additionally, the PPIs between IL1A/IL1R1 and IL6/IL6RA were also assayed. IL-1A or IL-6 guided by signal peptide of Wbp1 was detected for their interactions with their receptors respectively (see protein structures in Figure S4A,B). The growth ability assays indicated that the two ligands interacted intensively with their corresponding receptors respectively, while no cross reaction was detected. Meanwhile, CXCL12 and CXCR4, used as negative controls in the assay, did not shown any PPIs with the interleukin ligands or receptors (Fig. 6B) and the same result was also obtained from the β-Gal activity assay (Figure S4C).
Discussion
Since Y2H technique was first invented, numerous Y2H variants have been designed for different purposes13. These Y2H variants have investigated PPIs between cytosolic proteins13,14, between membrane proteins and cytosolic proteins, and between membrane proteins4,15,16. The powerful high-throughput feature of Y2H has made this technique indispensable in interatomic research of such interactions3,17.
In the present study, we established a MALAR-Y2H system to detect PPIs between secreted proteins and transmembrane proteins. This MALAR-Y2H system enriches the variety of Y2H variants currently available and extends its application range. The system detects PPIs between secreted proteins anchored on the outer suface of the plasma membrane and native transmembrane proteins, thereby facilitating interactions under native physiological conditions. However, this is often not the case for other in vitro assays.
CXCL12/CXCR4 and CXCL12/CXCR7 axes are well-studied chemokine-signaling pathways that are involved in many physical processes7,8. The interactions between CXCL12 and CXCRs are a good sample for verifying the feasibility of the MALAR-Y2H system. In this system, the key to its success was whether S. cerevisiae could recognize the signal peptides and the transmembrane peptides fused with bait and prey proteins. The results proved that yeast, as a lower eukaryotic organism, is compatible with the signal peptide and transmembrane peptides of mammalian genes. Mouse CXCL12, guided by its own signal peptide, can be anchored on the surface of the S. cerevisiae plasma membrane as well as the signal peptide of the yeast gene Wbp1. In addition, S. cerevisiae can recognize the transmembrane peptides of CXCRs and direct their localization to the plasma membrane in the correct orientation. These characteristics, in addition to the simple culture condition and the genetic stability, make yeast a good platform to study PPIs of xenogeneic eukaryotic transmembrane proteins.
Using the presented method, the interactions between chemokines and receptors have been readily assayed. PPIs between CXCL12 and seven CXCRs, and eight chimeric receptor mutants were studied with neither radioactive labels nor protein purification procedures, illustrating the ease and simplicity of the method. The whole experiment was completed within four weeks. Moreover, because the interaction strengths were assayed semi-quantitatively, the system can yield general information that describes the intensity of interactions between CXCL12 and candidate receptors.
Using the Y2H system, high-affinity interactions between CXCL12 and its receptor CXCR4 and CXCR7 were observed, which is in agreement with previous results18. What more important, however, is that a complex crosstalk between CXCL12 and other CXC motif receptors was discovered by the Y2H system during the verification. We observed that CXCR5 and CXCR1 also interacted with CXCL12, suggesting that CXCL12 may function as an agonist or antagonist of CXCR1 and CXCR5. These results are in agreement with current opinions concerning the possible abundant crosstalk between CXC chemokines and receptors19.
During the following experiments, the advantages of the method was further proved by successfully detecting the PPIs between CXCL7, CXCL9 and their CXC receptors, and between IL1A and IL6 and their receptors, IL-1R1 and IL-6RA20,21. The former assays confirmed the reliability of the method in detecting the PPIs between ligands and multiple-pass transmembrane receptors once again, while the latter assays demonstrated that the method is also competent to detecting PPIs involving single-pass transmembrane receptor. The single-pass transmembrane proteins are very different from the multiple-pass ones in topological structures, localization mechanisms, interaction patterns with ligands, and so on6. These successful assays of the PPIs between variant ligands and receptors make us firmly believe the method possesses a good universality.
At present, the MALAR-Y2H system was only used in lower throughput screening, namely one-to-one matching, as described in this article. However, in most cases, we need to figure out the receptors of a ligand, which has no available candidate receptors for use in such screening. Therefore, we need a system which can do a high throughput screening just as performed in other Y2H systems. Herein, we can propose a solution to upgrade the MALAR-Y2H into a high throughput cDNA library screening system.
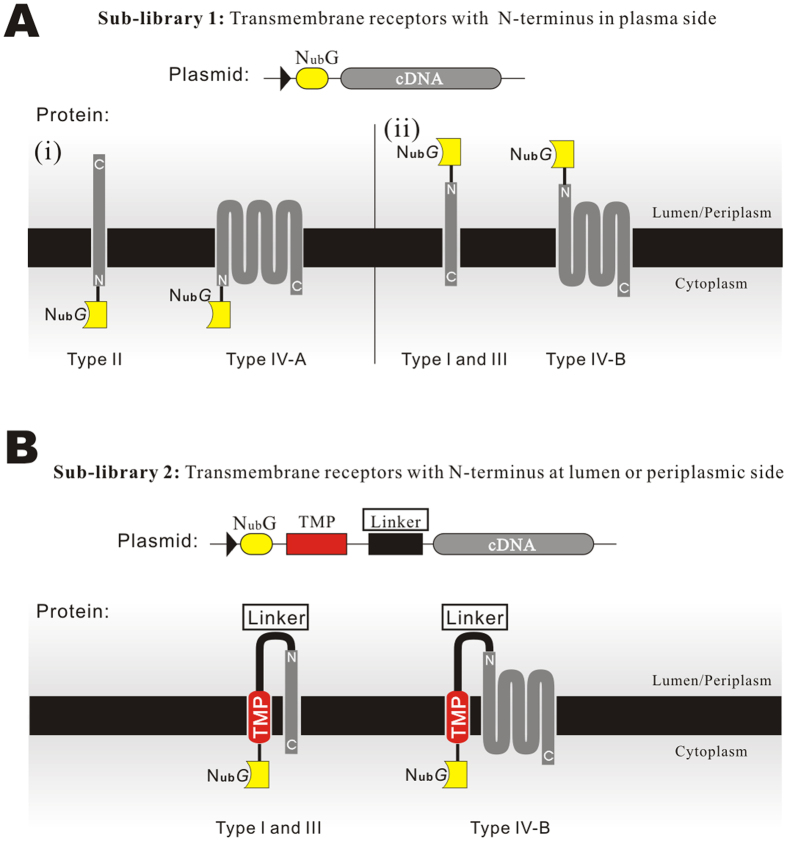
Firstly, just as usual, the bait plasmids can be constructed as described above easily. Then, the prey library can be established with the prey plasmid of MALAR-Y2H, though it is a little bit more complicated due to the different topological structures of transmembrane proteins, which must be taken into consideration. On the one hand, the detection module, NubG, must be fused with cDNAs at their N-terminus rather than C-terminus (because of the termination codon of the cDNA). On the other hand, the fused NubG must be kept into the cytoplasmic side to make sure it able to reconstruct with the Cub on the bait protein once a prey interacts with the bait. Consequently, a single library that simply fuses NubG directly to 5′-end of cDNAs is not enough to satisfy the formation of an efficient reporter system for all types of prey proteins. According to topological classification, type II and IV-A transmembrane proteins have the N-terminus located on the inside of the plasma membrane6 (Fig. 7A). With regard to these proteins, the directly fused NubG motif could be correctly kept on the cytoplasmic side as expected (Fig. 7A-i) and the interaction of baits and preys can unquestionably cause the reconstruction of NubG and Cub. While, for type I, III, and IV-B proteins, with N-terminus natively locating on the outside of the plasma membrane, the NubG will be leaved on the outside surface and the leaved NubG can never contact with the Cub of bait (Fig. 7A-ii) to induce signal cascading.
Figure 7. Strategy of the transmembrane protein library construction.
(A) Type II and IV-A transmembrane proteins have an N-terminus on the inner side of the cytoplasmic membrane. NubG is directly tagged at the 5′ end of the cDNA as a report module. (B) Type I, III, and IV-B proteins have the N-terminus located on the outside of the cytoplasmic membrane. The NubG was fused to the 5′ end of the cDNA by a transmembrane peptide and a linker. The transmembrane peptide mediates the intracellular localization of NubG, at the same time, the linker provides the N-terminus of preys sufficient flexibility to maintain their native structures.
To resolve the problem, one more cDNA sub-library needs to be generated, in which the total cDNAs are fused at the downstream of the NubG with an additional transmembrane peptide plus a linker between them (Fig. 7B). Through this strategy, the NubG that fused with type I, III and IV-B transmembrane proteins will be translocated to the inner surface of the plasma membrane via the transmembrane peptide. It needs to be noted that the linker should be long enough to give the N-terminus of the prey proteins sufficient freedom to maintain their native structures and, at the same time, ensure the linker does not interact with the bait.
By using the above two kinds of sub-libraries individually or in combination, with all types of transmembrane proteins being tagged with the NubG module locating on the inside of the plasma membrane, the high throughput screening of PPIs of ligand and transmembrane receptors can be eventually realized. In addition, some prey libraries tagged with NubG at the N- or C-terminus are commercially available22.
In summary, the present study elaborated how the MALAR Y2H system had been used to study the PPIs between secreted proteins and transmembrane receptors. The reliability and high efficiency were fully revealed and verified. Through the further research, the method could be potentially developed into a high-throughput library screening method.
Materials and Methods
Cell lines and Plasmids
Eerichia. coli strain DH5α was used as the host for plasmid construction and amplification. Yeast strains GoldY2H (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2:: GAL1UAS-Gal1TATA-His3, GAL2UAS-Gal2TATA-Ade2 URA3::MEL1UAS-Mel1TATA AUR1-C MEL1) and Y187 (MATα, ura3-52, his3-200, ade2-101, trp1-901, leu2-3, 112, gal4Δ, met–, gal80Δ, URA3::GAL1UAS-GAL1TATA-lacZ), plasmids pGAD-T7 and pGBK-T7 were purchased from Clonetech, USA. The 293T cell line was maintained by our lab. The pcDNA3.1(−) plasmid was purchased from Life Technology, USA.
Construction of Bait Plasmids
The bait was expressed by the pGAD-T7 plasmid, in which all elements between the ADH1 promoter and ADH1 terminator were deleted and the target sequences were cloned by a one-step clone kit from Vazyme, Co. China. The bait protein consisted of a signal peptide, a ligand protein, a transmembrane peptide, the C-terminus of yeast ubiquitin (Cub), and reporter genes. A signal peptide from one of three genes, SPCXCL12 (signal peptide of CXCL12, residues 1–28), SPWBP1 (WBP1, residues 1–31) or SPMFAL1 (MFAL1, residues 1–89), was fused at the N-terminus of ligand in different experiments. The WBP1 transmembrane peptide and flanking sequences (residues 350–430) were used as the transmembrane peptide and linkers, and fused between CXCL12 and the reporter genes. EGFP was used as a reporter gene to exam the subcellular localization. The Cub-GAL4 cassette was used as a reporter module for the detection of PPIs (Fig. 1A).
Construction of Prey Plasmids
Prey genes were cloned into the pGBK-T7 vector, in which the original ORF between the ADH1 promoter and terminator was deleted, and replaced with mouse CXC motif receptors and reporter genes. EGFP was used as a reporter gene to examine the subcellular localization or NubG was used as a reporter module for the detection of PPIs (Fig. 1B).
Membrane Staining and Co-localization Assay
A lipophilic dye, DiI (Beyotime, China (C1036)), was solved by DMSO to 1 mM. Cells (0.5 ml, OD600 = 2.0) were paraformaldehyde fixed on ice for 10 minutes, and washed twice and resuspended with 0.5 ml phosphate buffered solution (PBS), and stained for 10 minutes with 10 μM DiI, and washed twice and mounted with PBS. Fluorescent light of EGFP and Dil (λex: 543 nm, λem: 565 nm) and transmission light were captured by Leica confocal system.
Yeast Plasmid Transformation and Mating
Plasmids were transformed into yeast cells using the LiAc method, as described in the commercial kit manual. Bait plasmids were transformed into GoldY2H cells, whereas prey plasmids were transformed into Y187 cells. Transformants were selected with synthetic dropout nutrient medium/plate (SD medium) without leucine or tryptophan (SD/Leu− or SD/Trp−). Mating was performed by suspending 2–3 clones of each cell into 0.5 ml YPD medium, and incubating overnight with shaking. Cells were collected by centrifugation and spread onto SD/Leu−Trp− media.
Predicting Transmembrane Topological Structures
The transmembrane topological structure of all fusion transmembrane proteins was predicted by an online tool: (http://www.cbs.dtu.dk/services/TMHMM/, a hidden Markov model for predicting transmembrane helices in protein sequences).
Self-Activating Assay
Self-activation of bait proteins was detected by testing cell growth and a color shift after culturing cell suspensions on SD/Leu−His−Ade− media with or without 3-AT.
Interaction Strength Detection
Growth assay
Five microliters of 10-fold diluted cells suspension, OD600 = 1, 0.1 and 0.01 series, were dropped on SD/Leu−Trp−His−Ade− medium plates. Cell growth was recorded by photographing the plates after 72 h incubation.
β-Galactosidase (β-Gal) Activity Assay
Cells were grown in liquid SD/Leu−Trp− medium to an OD600 of 0.6–0.8. A one milliliter culture was harvested by centrifugation and suspended in 100 μl Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, pH 7.0). Cells were lysed through three freezing and thawing cycles in liquid nitrogen and a 37 °C water bath. Seven-hundred microliters of Z buffer containing 0.27% (v/v) 2-mercaptoethanol and 160 μl Z buffer containing 0.4% 2-nitrophenyl-β-D-galactopyranoside were added and incubated for 30–60 min at 30 °C. Four hundred microliters of 1 M Na2CO3 was added to stop the reaction. The sample was centrifuged and the supernatant absorbance was measured at 420 nm. β-Gal activity was calculated by the equation (1).
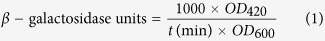 |
Western Blot
Cells were grown in 1 ml liquid SD/Leu−Trp− medium at 30 °C overnight to an OD600 of 0.6–1.0. Proteins were extracted according to a modified post-alkaline extraction method23. Briefly, 2 ml culture of yeast cells (OD600 of 0.6–1.0) were harvested by centrifugation and resuspended in 100 μl distilled water and 100 μl of 0.2 M NaOH was added. This solution was incubated for 5 min at room temperature. Cells were pelleted by centrifugation and resuspended in 50 μl SDS-PAGE sample buffer and incubated at 95 °C in a water bath for 5 min. Samples were separated by SDS-PAGE and transferred to a PVDF membrane. GAL4 was detected by a rabbit anti-GAL4 IgG (abcam, ab1396), and CXCL12 by a rabbit anti-mouse CXCL12 IgG (abcam, 9797). The Fc-tagged CXCR5NT and CXCR3NT were detected by a HRP-labeled donkey anti-human IgG.
Construction of Chimeric Receptors
Extracellular fragments of CXCR5 and CXCR3 were determined according to the UniProt database. The sequences are shown in Figure S2. Insert fragments were cloned seamlessly into recipient genes using the one-step clone kit from Vazyme, Co. China (C112-01). All plasmids were confirmed by sequencing.
CoIP of CXCL12 and CXCR5NT
The N-terminal fragment of the CXCR5 fragment (residues 1–55) was fused with the human IgG1 Fc fragment (CXCR5NT-Fc) and inserted into vector pcDNA3.1(−). The CXCL12 mature protein CDS was cloned into pcDNA3.1(−). Plasmids were co-transfected into the 293T cell line, and lysed at 48 h for CoIP. Co-transfection of CXCR3NT (1–52)-Fc and CXCL12 was used as the negative control.
Fifty microliter protein A-coupled Sepharose beads were blocked by PBS containing 0.1% BSA and incubated with cell lysis buffer at 4 °C for 4 h, and washed with the lysis buffer three times. The beads were suspended with 50 μl 2×SDS-PAGE loading buffer and boiled for 5 min. Samples were separated by SDS-PAGE and blotted to PVDF membranes.
Classic Intracellular Y2H of CXCL12 and CXCR5NT
Mature CXCL12 was fused to the activation domain of GAL4 in the pGAD-T7 plasmid, and CXCR5NT or CXCR3NT was fused to the binding domain of GAL4 in the pGBK-T7 plasmid, according to the instructions from the manufacturer. Transformation, mating and detection were performed according to the above method [Yeast Plasmid Transformation and Mating].
Additional Information
How to cite this article: Li, J. et al. Development of a membrane-anchored ligand and receptor yeast two-hybrid system for ligand-receptor interaction identification. Sci. Rep. 6, 35631; doi: 10.1038/srep35631 (2016).
Supplementary Material
Acknowledgments
This work was supported by the Shanghai Municipal Natural Science Foundation (grant 14ZR1421200) and SJTU Young Researcher Start-up Funding (grant 14 × 100040015). We thank Tina Dugan, Gonzaga University, for her crucial reviewing of the manuscript.
Footnotes
Author Contributions J.L. designed this work, did LSCM experiments, and prepared all figures, J.G. and Y.Z. performed the yeast two hybrid assays, L.H. and L.Z. performed all gene cloning works, W.G. worked on CoIP experiments, Y.Y. and W.H. wrote the main manuscript text. All authors reviewed the manuscript.
References
- Phillips K. S., Wilkop T., Wu J. J., Al-Kaysi R. O. & Cheng Q. Surface plasmon resonance imaging analysis of protein-receptor binding in supported membrane arrays on gold substrates with calcinated silicate films. Journal of the American Chemical Society 128, 9590–9591, 10.1021/ja0628102 (2006). [DOI] [PubMed] [Google Scholar]
- Lasagni L. et al. An alternatively spliced variant of CXCR3 mediates the inhibition of endothelial cell growth induced by IP-10, Mig, and I-TAC, and acts as functional receptor for platelet factor 4. The Journal of experimental medicine 197, 1537–1549, 10.1084/jem.20021897 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter B., Kittanakom S. & Stagljar I. Two-hybrid technologies in proteomics research. Curr Opin Biotechnol 19, 316–323, 10.1016/j.copbio.2008.06.005 (2008). [DOI] [PubMed] [Google Scholar]
- Stagljar I., Korostensky C., Johnsson N. & te Heesen S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci USA 95, 5187–5192 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzaskowski B. et al. Action of molecular switches in GPCRs–theoretical and experimental studies. Current medicinal chemistry 19, 1090–1109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H., Berk A. & Kaiser C. A. Molecular Cell Biology. 6th edn, (Freeman Custom Publishing, 2010). [Google Scholar]
- Liekens S., Schols D. & Hatse S. CXCL12-CXCR4 axis in angiogenesis, metastasis and stem cell mobilization. Current pharmaceutical design 16, 3903–3920 (2010). [DOI] [PubMed] [Google Scholar]
- Yamada K. et al. CXCL12-CXCR7 axis is important for tumor endothelial cell angiogenic property. International journal of cancer. Journal international du cancer 137, 2825–2836, 10.1002/ijc.29655 (2015). [DOI] [PubMed] [Google Scholar]
- Xanthou G., Williams T. J. & Pease J. E. Molecular characterization of the chemokine receptor CXCR3: evidence for the involvement of distinct extracellular domains in a multi-step model of ligand binding and receptor activation. Eur J Immunol 33, 2927–2936, 10.1002/eji.200324235 (2003). [DOI] [PubMed] [Google Scholar]
- Ghasemzadeh M. et al. The CXCR1/2 ligand NAP-2 promotes directed intravascular leukocyte migration through platelet thrombi. Blood 121, 4555–4566, 10.1182/blood-2012-09-459636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desurmont T. et al. Overexpression of chemokine receptor CXCR2 and ligand CXCL7 in liver metastases from colon cancer is correlated to shorter disease-free and overall survival. Cancer Sci 106, 262–269, 10.1111/cas.12603 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin R. A., Campanella G. S., Sun J. & Luster A. D. Intracellular domains of CXCR3 that mediate CXCL9, CXCL10, and CXCL11 function. J Biol Chem 279, 30219–30227, 10.1074/jbc.M403595200 (2004). [DOI] [PubMed] [Google Scholar]
- Fields S. & Song O. A novel genetic system to detect protein-protein interactions. Nature 340, 245–246, 10.1038/340245a0 (1989). [DOI] [PubMed] [Google Scholar]
- Mockli N. et al. Yeast split-ubiquitin-based cytosolic screening system to detect interactions between transcriptionally active proteins. Biotechniques 42, 725–730 (2007). [DOI] [PubMed] [Google Scholar]
- Thaminy S., Auerbach D., Arnoldo A. & Stagljar I. Identification of novel ErbB3-interacting factors using the split-ubiquitin membrane yeast two-hybrid system. Genome Res 13, 1744–1753, 10.1101/gr.1276503 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumi C. M., Chuk M., Chevelev I., Stagljar I. & Michaelis S. Negative regulation of the yeast ABC transporter Ycf1p by phosphorylation within its N-terminal extension. The Journal of biological chemistry 283, 27079–27088, 10.1074/jbc.M802569200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petschnigg J., Snider J. & Stagljar I. Interactive proteomics research technologies: recent applications and advances. Current opinion in biotechnology 22, 50–58, 10.1016/j.copbio.2010.09.001 (2011). [DOI] [PubMed] [Google Scholar]
- Sun X. et al. CXCL12/CXCR4/CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev 29, 709–722, 10.1007/s10555-010-9256-x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields D. C. Molecular evolution of CXC chemokines and receptors. Trends in immunology 24, 355; author reply 356–357 (2003). [DOI] [PubMed] [Google Scholar]
- Dower S. K. et al. The cell surface receptors for interleukin-1 alpha and interleukin-1 beta are identical. Nature 324, 266–268, 10.1038/324266a0 (1986). [DOI] [PubMed] [Google Scholar]
- Skiniotis G., Boulanger M. J., Garcia K. C. & Walz T. Signaling conformations of the tall cytokine receptor gp130 when in complex with IL-6 and IL-6 receptor. Nat Struct Mol Biol 12, 545–551, 10.1038/nsmb941 (2005). [DOI] [PubMed] [Google Scholar]
- Snider J. et al. Detecting interactions with membrane proteins using a membrane two-hybrid assay in yeast. Nature protocols 5, 1281–1293, 10.1038/nprot.2010.83 (2010). [DOI] [PubMed] [Google Scholar]
- Lentze N. & Auerbach D. The yeast two-hybrid system and its role in drug discovery. Expert Opin Ther Targets 12, 505–515, 10.1517/14728222.12.4.505 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.