Abstract
Intracellular bacteria exploit host cytosolic signals to upregulate virulence genes. In this issue of Cell Host & Microbe, Wong et al. (2015) show that Burkholderia pseudomallei senses host cytosolic glutathione, a low-molecular-weight thiol, through the membrane-bound histidine sensor kinase VirA, highlighting the importance of inter-kingdom signaling in bacterial pathogenesis.
Bacterial pathogens have to tightly and precisely regulate expression of their virulence repertoire to establish a successful interaction with the host. The ability to sense and respond to a signal as an identifier of a specific environment is paramount to bacterial-host associations. The multitude of environments intracellular pathogens encounter poses a challenging scenario for such gene regulation. Burkholderia pseudomallei is a saprophyte and a facultative intracellular pathogen that can cause melioidosis in humans. It invades host cells, and escapes from the phagosome reaching the cytoplasm, where it spreads to neighboring cells, inducing the formation of a multinucleated giant cell (MNGC) (Galyov et al., 2010). In this issue of Cell Host & Microbe, Wong et al. (2015) show that B. pseudomallei senses the abundant cytosolic molecule glutathione (L-γ-glutamyl-L-cysteinyl-glycine; GSH) in mammalian cells to upregulate and time virulence gene expression. GSH is a low-molecular-mass thiol ubiquitously present in eukaryotic and many prokaryotic organisms. GSH has important functions in multiple physiological processes, including maintenance of intracellular redox status and antioxidant defense, metabolism, gene regulation, cell proliferation, and apoptosis (Sies, 1999; Wu et al., 2004).
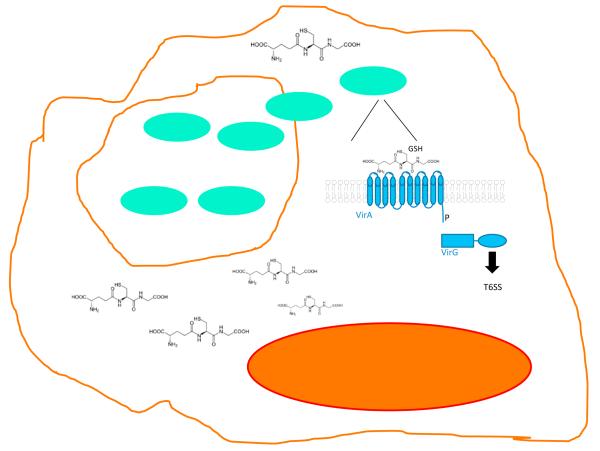
Mammalian-produced GSH is largely found in the cytosol where its concentrations range from 1 to 10 mM (Wu et al., 2004). Wong et al. (2015) report that B. pseudomallei specifically senses host cytosolic GSH through a membrane-bound histidine sensor kinase VirA. Bacteria sense environmental cues largely through two-component signaling systems (TCS) to regulate their gene expression (DiGiuseppe and Silhavy, 2003). A TCS consists of a histidine kinase (HK) sensor protein that autophosphorylates in response to environmental cues and transfers this phosphate to a response regulator (RR) protein, which is generally a transcription factor. TCSs allow bacteria to control cellular functions and respond to various environments (DiGiuseppe and Silhavy, 2003). VirA is a dimeric HK, and reduction of its periplasmic exposed cysteine (C62) by GSH promotes monomerization of this kinase leading to its activation. VirA then initiates a phosphorelay cascade within the bacterial cell, leading to the transcriptional activation of genes encoding for a type six secretion system (T6SS) that is exclusively expressed within mammalian cells and is crucial for B. pseudomallei infection (Burtnick et al., 2011; Chen et al., 2011; Pilatz et al., 2006). Depletion of host GSH abrogates T6SS expression in B. pseudomallei, preventing MNGC formation (Figure 1).
Figure 1. Burkholderia pseudomallei Senses GSH in the Mammalian Cytoplasm.
B. pseudomallei is an intracellular pathogen (visualized in green). It invades and replicates within host cells. Initially it replicates in a vacuole. Subsequently, B. pseudomallei escapes the vacuole into the cytoplasm where it senses the host glutathione (GSH) as a signal through a sensor kinase named VirA. Upon sensing GSH, VirA activates a signaling cascade in the bacterium leading to upregulation of the expression of the type six secretion system, which is key for infection and cell-to-cell spread of this pathogen.
It is fascinating that different intracellular pathogens such as B. pseudomallei (a Gram-negative bacterium) and Listeria monocytogenes (a Gram-positive bacterium) convergently evolved to sense GSH within host cells to initiate signaling cascades involved in intracellular replication and cell-to-cell spread (Reniere et al., 2015). L. monocytogenes is also a saprophyte that is ubiquitous in the environment, and a food-borne pathogen that typically causes self-limiting gastroenteritis; however, in susceptible individuals, L. monocytogenes can cause systemic infections that result in high levels of morbidity and mortality (Freitag et al., 2009). L. monocytogenes invades immune and epithelial cells, replicates in the host cell cytosol, and spreads from cell to cell. Recently, it was demonstrated that the L. monocytogenes cytoplasmic transcription factor PrfA directly binds GSH to activate virulence gene expression (Reniere et al., 2015), demonstrating that this may be a mechanism for L. monocytogenes to specifically recognize the intracellular environment. However, while L. monocytogenes cytoplasmic GSH receptor senses both host- and bacterial-derived GSH, the B. pseudomallei VirA membrane-bound GSH receptor strictly senses host-derived GSH.
In order to survive within a specific environment, bacteria must sense many cues to adapt accordingly. From a bacterium’s perspective, the host is yet another environment to grow and thrive in, although potentially one actively trying to kill it. Hence, recognition of distinct host environments by bacteria is key for adaptation and survival. The most efficient way to acquire such information is by sensing various host-derived molecules. These cell-to-cell signaling interactions are commonly referred to as inter-kingdom signaling. The ability to sense and respond to host signals has been identified in most major pathogens. The prevalence of these signaling systems suggests that inter-kingdom signaling may be extremely common, and many more systems will be uncovered in the future (Hughes and Sperandio, 2008). Given the central role they play in virulence regulation, these signaling pathways make for potential novel targets for new anti-microbial drug development.
REFERENCES
- Burtnick MN, Brett PJ, Harding SV, Ngugi SA, Ribot WJ, Chantratita N, Scorpio A, Milne TS, Dean RE, Fritz DL, et al. Infect. Immun. 2011;79:1512–1525. doi: 10.1128/IAI.01218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wong J, Sun GW, Liu Y, Tan GY, Gan YH. Infect. Immun. 2011;79:3064–3073. doi: 10.1128/IAI.05148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiuseppe PA, Silhavy TJ. J. Bacteriol. 2003;185:2432–2440. doi: 10.1128/JB.185.8.2432-2440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag NE, Port GC, Miner MD. Nat. Rev. Microbiol. 2009;7:623–628. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galyov EE, Brett PJ, DeShazer D. Annu. Rev. Microbiol. 2010;64:495–517. doi: 10.1146/annurev.micro.112408.134030. [DOI] [PubMed] [Google Scholar]
- Hughes DT, Sperandio V. Nat. Rev. Microbiol. 2008;6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilatz S, Breitbach K, Hein N, Fehlhaber B, Schulze J, Brenneke B, Eberl L, Steinmetz I. Infect. Immun. 2006;74:3576–3586. doi: 10.1128/IAI.01262-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reniere ML, Whiteley AT, Hamilton KL, John SM, Lauer P, Brennan RG, Portnoy DA. Nature. 2015;517:170–173. doi: 10.1038/nature14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H. Free Radic. Biol. Med. 1999;27:916–921. doi: 10.1016/s0891-5849(99)00177-x. [DOI] [PubMed] [Google Scholar]
- Wong J, Chen Y, Gan Y-H. Cell Host Microbe. 2015;18(this issue):38–48. doi: 10.1016/j.chom.2015.06.002. [DOI] [PubMed] [Google Scholar]
- Wu G, Fang YZ, Yang S, Lupton JR, Turner ND. J. Nutr. 2004;134:489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]