Summary
Currently, chlorpyrifos (CP) and carbofuran are often applied together to control major agricultural pests in many developing countries, in most cases, they are simultaneously detected in agricultural soils. Some cost‐effective techniques are required for the remediation of combined pollution caused by multiple pesticides. In this work, we aim at constructing a detectable recombinant microorganism with the capacity to simultaneously degrade CP and carbofuran. To achieve this purpose, CP/carbofuran hydrolase genes and gfp were integrated into the chromosome of a biosafety strain Pseudomonas putida KT2440 using a chromosomal scarless modification strategy with upp as a counter‐selectable marker. The toxicity of the hydrolysis products was significantly lower compared with the parent compounds. The recombinant strain could utilize CP or carbofuran as the sole source of carbon for growth. The inoculation of the recombinant strain to soils treated with carbofuran and CP resulted in a higher degradation rate than in noninoculated soils. Introduced green fluorescent protein can be employed as a biomarker to track the recombinant strain during bioremediation. Therefore, the recombinant strain has potential to be applied for in situ bioremediation of soil co‐contaminated with carbofuran and CP.
Introduction
Chlorpyrifos (CP) is a moderately toxic organophosphate insecticide that exhibits broad‐spectrum activity against major agricultural pests. CP leads to the loss of nerve function by irreversible inhibition of acetylcholinesterase (AChE) (Singh, 2009). Carbofuran is a broad‐spectrum carbamate insecticide that is used widely for disease control in vegetable, fruit and crops. However, the potential health risks to humans posed by excessive use of carbofuran are not ignored because carbofuran is a potent AChE inhibitor (Nguyen et al., 2014). Carbofuran and CP are often applied to agricultural soil together, thus, combined pollution caused by them needs to be resolved by developing new techniques (Rama Krishna and Philip, 2011).
Microbial degradation plays important roles in the removal of various pesticides in soil. Several carbofuran‐ and CP‐degrading bacteria have been isolated from natural environments (Tomasek and Karns, 1989; Feng et al., 1997; Ogramab et al., 2000; Yang et al., 2006; Li et al., 2007; Xu et al., 2008; Anwar et al., 2009). Moreover, carbofuran hydrolase gene (mcd encoding CH) and CP hydrolase gene (mpd encoding MPH) have been cloned from Achromobacter sp. WM111 and Stenotrophomonas sp. YC‐1 respectively (Tomasek and Karns, 1989; Yang et al., 2006). Hydrolysis of carbofuran and CP by the hydrolases reduces their toxicity by several orders of magnitude (Hernándeza et al., 2013), providing a useful enzymatic detoxification method. To date, there are no reports on simultaneous degradation of carbofuran and CP by a single microorganism.
Generally, it takes more effort to simultaneously cultivate multiple pesticide‐degrading bacteria with different culture conditions. When the degraders are applied for in situ bioremediation, the different niches of multiple independent degraders force them to compete limited resources, resulting in the unbalanced growth of the degraders. More importantly, the biosafety of various natural degraders is uncertain, which will give rise to potential risks to environmental safety and human health. An alternative solution is to create a recombinant strain capable of simultaneously degrading carbofuran and CP. Generally, the recombinant strains harbouring degradation genes have higher degradation rates than natural degraders by introducing minimal gene expression regulation elements to promote high‐level expression of degradation genes.
So far, several recombinant strains have been constructed for degradation of different classes of pesticides. Among them, a recombinant strain capable of simultaneously degrading methyl parathion and carbofuran was constructed by random insertion of a methyl parathion hydrolase (mph) gene into the chromosome of a carbofuran‐degrading Sphingomonas sp. CDS‐1 using a mini‐Tn5 transposon system. Unfortunately, antibiotic resistance gene (ARG) left on chromosome has potential risks to environmental safety due to ARG diffusion among bacterial species by horizontal gene transfer (Jiang et al., 2007). In addition, a γ‐hexachlorocyclohexane (γ‐HCH)‐degrading Sphingobium japonicum UT26 was endowed with the capacity to simultaneously degrade γ‐HCH and organophosphates by transforming with a plasmid encoding an organophosphorus hydrolase. However, plasmid instability reduces the efficacy of the recombinant stain for bioremediation of contaminated soil (Yang et al., 2013).
A model microorganism Pseudomonas putida KT2440 was certified by the Recombinant DNA Advisory Committee as a biosafety strain for recombinant DNA manipulation (Nelson et al., 2002). P. putida KT2440 is a robust soil bacterium and is capable of degrading diverse aromatic compounds (Jiménez et al., 2002). The genome of P. putida KT2440 has been sequenced and the chromosomal scarless modification methods have been well established in P. putida KT2440 (Nelson et al., 2002; Graf and Altenbuchner, 2011; Luo et al., 2016), which facilitate the genetic engineering of P. putida KT2440 to acquire novel characteristics for biodegradation and biocatalysis. Recently, P. putida KT2440 has been highlighted as an optimal chassis for metabolic pathway assembly (Nikel et al., 2014).
In this study, P. putida KT2440 was endowed with carbofuran/CP degradation capability and a biomarker by integrating mcd, mpd and gfp into the chromosome using a genomic scarless modification method. This approach allows successive insertion of various degradation genes into the chromosome of P. putida KT2440 to create extended‐spectrum degraders. Moreover, the recombinant strain marked with green fluorescent protein (GFP) may be monitored by fluorescence during bioremediation.
Results and discussion
Construction of recombinant P. putida KT2440 for simultaneous degradation of carbofuran and CP
Currently, combined pollution caused by multiple pesticides has become an urgent issue to be resolved. To date, two approaches to remediate multiple pesticides‐contaminated soils have been proposed, including the introduction of different degradation genes into a host strain and the use of multiple natural degraders with different substrate ranges (Singh and Walker, 2006). In this work, a biosafety strain P. putida KT2440 was selected as a host for heterologous expression of carbofuran and CP hydrolase genes. For this purpose, a genome markerless modification method based on the upp counter‐selection system (Gong et al., 2016) was used to construct the recombinant strain with the capacity to simultaneously degrade carbofuran and CP. The synthetic gene cassette (mpd, gfp or mcd) was composed of a P. putida constitutive promoter J23119, a ribosomal binding site and an exogenous gene flanked by the upstream and downstream homologous arms (Fig. S1). Successive insertion of the mpd, gfp and mcd genes into three different target sites on the chromosome of P. putida KT2440 was achieved with the suicide plasmid in combination with the upp counter‐selection system. The successful constructions were verified by PCR detection of the inserted gene cassettes (Fig. S2). The resulting recombination strain was designated as P. putida KTU‐PGC.
Transcription of exogenous genes in P. putida KTU‐PGC
A P. putida constitutive promoter J23119 has been demonstrated to be efficient for the constitutive expression of exogenous genes in P. putida (Shetty et al., 2008). In this study, expression of mpd, gfp and mcd genes was under the control of the constitutive promoter J23119. The transcription of the inserted exogenous genes (mpd, gfp and mcd) in P. putida KTU‐PGC was further analysed by RT‐PCR assays with the extracted mRNA as the template. As a result, the desired target fragments were obtained by PCR with the cDNA or genomic DNA as the template. In contrast, no positive results were observed with the mRNA or ddH2O as the template (Fig. S3). These results indicated that the introduced exogenous genes were successfully transcribed to mRNA in P. putida KTU‐PGC.
Simultaneous degradation of carbofuran and CP by P. putida KTU‐PGC
To demonstrate the capacity of the recombinant strain to simultaneously degrade carbofuran and CP, the degradation experiments were performed by inoculating the recombinant strain into M9 minimal medium supplemented with 100 mg l−1 carbofuran and CP. High‐performance liquid chromatography (HPLC) analysis indicated that 100 mg l−1 carbofuran and CP were completely degraded within 36 and 24 h respectively (Fig. 1). Moreover, the concentration of carbonfuran phenol and 3,5,6‐trichloro‐2‐pyridinol (TCP) in M9 minimal medium increased gradually with the decrease in carbofuran and CP concentration. Both carbofuran and CP were degraded quickly at the first 12 h, which accounted for 60% and 75% of the amount of the initially added pesticides respectively. The reduction in the degradation rate after 12 h may be due to the accumulation of carbonfuran phenol and TCP, which have antimicrobial activity and are toxic to the bacterial growth and metabolism (Singh et al., 2004; Nguyen et al., 2014). In contrast, no decrease in the concentration of carbofuran and CP was detected in M9 minimal medium inoculated with P. putida KTU (data not shown).
Figure 1.
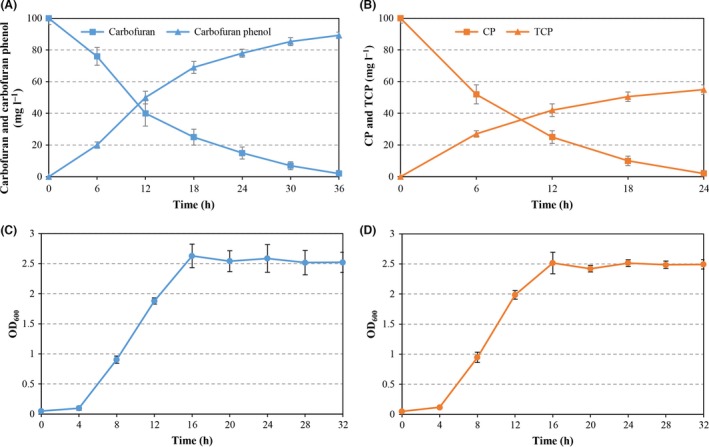
(A and B) Simultaneous degradation of carbofuran and CP by Pseudomonas putida KTU‐PGC. Degradation experiments with P. putida KTU‐PGC were performed at an initial inoculum density of OD 600 = 0.05 in M9 minimal medium supplemented with 100 mg l−1 carbofuran and CP at 30°C. (C and D) Cell growth of P. putida KTU‐PGC at 30°C in M9 minimal medium supplemented with 100 mg l−1 carbofuran or CP as the sole source of carbon.
When the recombinant strain was inoculated at OD600 = 0.05 into M9 minimal medium supplemented with 100 mg l−1 carbofuran or CP, the strain could utilize carbofuran or CP as the sole source of carbon for growth (Fig. 1).
In HPLC analysis of the cultures grown on 100 mg l−1 carbofuran, two peaks with a retension time (RT) of 8.75 and 10.21 min appeared in HPLC separation (Fig. S4), which were further identified as carbofuran and carbonfuran phenol by comparing with their authentic standards. HPLC analysis showed that carbofuran could be degraded by P. putida KTU‐PGC to carbonfuran phenol and methylamine (Fig. S5).
In HPLC analysis of the cultures grown on 100 mg l−1 CP, two peaks appeared at a RT of 8.69 and 3.12 min in HPLC separation (Fig. S6), which were further identified as CP and TCP by comparing with their authentic standards. HPLC analysis showed that CP could be degraded by P. putida KTU‐PGC to TCP and diethylthiophosphoric acid (Fig. S5).
Functional expression of GFP in P. putida KTU‐PGC
To easily track the recombinant strain in the field‐scale remediation, a gfp gene was also inserted into the chromosome of P. putida, allowing direct fluorescent detection of the activity of the recombinant strain. Cells of the recombinant strain containing a chromosome‐borne GFP produced bright green fluorescence under a confocal microscope (Fig. 2), indicating the presence of active GFP in this strain. However, no fluorescence was observed with the control wild‐type strain. Therefore, GFP may be used as a biomarker to monitor the survival and movement of the recombinant strain during bioremediation.
Figure 2.
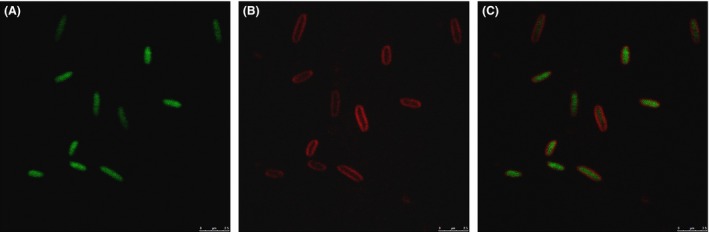
Detection of GFP fluorescence derived from Pseudomonas putida KTU‐PGC using a confocal microscope. A. Green fluorescence within the cell. B. Outline of cell membrane by stain with FM4‐64/L. C. A and B merged together.
Genetic stability of engineered strain
Pseudomonas putida KTU‐PGC was successively subcultured for twenty generations at 30°C in LB medium. These subcultures degraded carbofuran and CP as fast as the parent strain. Results from PCR detection showed that the inserted exogenous genes (mpd, gfp and mcd) stably existed on the chromosome of the twentieth‐generation subcultures of P. putida KTU‐PGC (Fig. S7), which indicated that the engineered strain was genetically stable in the lack of selection pressure.
Nonessential genes of P. putida KT2440 were selected as the insertion sites (Table 1). To check whether the insertion of exogenous genes into the chromosome inhibits cell growth, the cell growth kinetics of P. putida KTU and KTU‐PGC were compared. No growth inhibition was observed for strain KTU‐PGC, which reached the final cell density similar to strain KTU after 48 h of incubation (Fig. S8), indicating that the inactivation of the nonessential genes had no negative influences on cell growth and metabolism.
Table 1.
Information on three exogenous genes and their chromosomal insertion sites
| Gene | Length (bp) | Amino acid residues | Function | Source (GenBank accession no.) | Insertion site |
|---|---|---|---|---|---|
| mpd | 894 | 298 | Chlorpyrifos hydrolase | Stenotrophomonas sp. YC‐1 (DQ677027.1) | PP_5003 (phaA) |
| mcd | 1983 | 661 | Carbofuran hydrolase | Achromobacter sp. WM111 (AF160188.1) | PP_1277/PP_1278 (algA/algF) |
| gfp | 720 | 240 | Green fluorescent protein | Plasmid pEGFP‐N3 (U57609) | PP_3357 (vdh) |
Remediation of soil co‐contaminated with carbofuran and CP by P. putida KTU‐PGC
To examine whether P. putida KTU‐PGC can thrive and effectively degrade carbofuran and CP in soil in the presence of indigenous microbial populations, soils co‐contaminated with carbofuran and CP were inoculated with P. putida KTU‐PGC at the rate of 106 cells g−1 soil. During the 12‐day period, a slight reduction in carbofuran and CP was observed in soils without inoculation, most likely due to the occurrence of abiotic degradation processes. In contrast, carbofuran and CP were completely degraded in soils with inoculation within 12 and 8 days respectively (Fig. 3).
Figure 3.
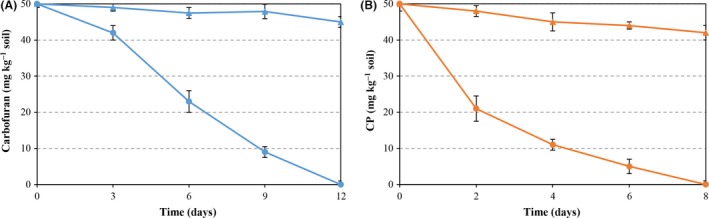
Simultaneous degradation of carbofuran and CP in soils inoculated with Pseudomonas putida KTU‐PGC at the rate of 106 cells/g soil. Symbols: (●) soil, inoculated; (▲) soil, uninoculated.
At the end of soil remediation experiments, soil samples were used for the isolation of inoculated P. putida. Two single colonies that produced clear zones on LB agar plates containing 50 mg l−1 CP were isolated and purified. The two isolates (named SKT‐A and SKT‐B) were identified as P. putida KT2440 by sequencing their 16S rRNA gene. Moreover, mpd, gfp and mcd genes were detected by PCR from the two isolates (Fig. S9). These results indicated that inoculated P. putida truly contributed to the degradation of carbofuran and CP in soil.
During the soil bioremediation experiments, we used GFP as a biomarker to track the presence and activity of the inoculated recombinant P. putida KT2440. The recombinant P. putida KT2440 was initially screened by PCR detection of the introduced gfp gene. Subsequently, the gfp‐containing strains were further measured for their fluorescence. As expected, the bacterial cells carrying gfp gene emitted green fluorescence under a confocal microscope (Fig. S10). Moreover, the recombinant P. putida KT2440 inoculated in soil could be monitored for its activity by measuring the changes in the fluorescence intensity during bioremediation. These results suggest that GFP may be a powerful real‐time monitoring tool for the visualization of the recombinant P. putida KT2440 during bioremediation.
Potential of P. putida KT2440 for bioremediation of contaminated soil
Previously, a carbofuran‐degrading Sphingomonas sp. CDS‐1 was engineered to simultaneously degrade methyl parathion and carbofuran by randomly inserting a mph gene into the chromosome using a Tn5‐based transposon system (Jiang et al., 2007). However, random insertion on chromosome might disrupt essential genes in host strains, leading to function loss and cell death. In this study, exogenous genes were inserted into desirable regions on the chromosome of P. putida KT2440 by homologous recombination. Obviously, targeted insertion is more advantageous than random insertion. We selected nonessential genes of P. putida KT2440 as the insertion sites; thus, the insertion of exogenous genes neither inhibits cell growth nor affects cell survival. Compared with model strain P. putida KT2440, the biosafety of Sphingomonas sp. CDS‐1 needs to be intensively investigated prior to applying to soil remediation.
P. putida KT2440 possesses diverse catabolic pathways for aromatic compounds (Jiménez et al., 2002). Bacterial aromatic ring‐hydroxylating oxygenase database (RHObase) provides in‐depth information on functionally characterized RHOs from phylogenetically diverse classes of bacteria (Chakraborty et al., 2014), and therefore, RHObase is a useful tool to predict functionality of putative oxygenase from de novo genome or metagenome sequences. Distinctive bacterial communities associated with ectomycorrhizal roots were identified using 454 pyrosequencing (Marupakula et al., 2016). Recently, a rhizoremediation system based on an artificial consortium consisting of plant growth‐promoting rhizobacteria (PGPR) and polycyclic aromatic hydrocarbon‐degrading bacteria was used to remove petroleum wastes (Pizarro‐Tobías et al., 2015). P. putida KT2440 has been identified as PGPR and has an ability to colonize the rhizosphere of crop plants (Espinosa‐Urgel et al., 2002), which may facilitate the development of rhizoremediation systems for soil decontamination.
Synthetic biology has been highlighted as a powerful tool to create novel strains with desired degradative capabilities for bioremediation of contaminated environments (Gong et al., 2016). The recombinant P. putida KT2440 constructed in this study could efficiently degrade carbofuran and CP in soil and utilize carbofuran or CP as the sole source of carbon for growth.
Selection of P. putida KT2440 as host strain has many advantages, including confirmed biosafety, available whole‐genome sequence, and diverse genetic manipulation tools, which should lay a foundation for targeted chromosomal modification and metabolic pathway engineering in P. putida KT2440. P. putida KT2440 has been highlighted as an optimal chassis for implantation of various organic‐degrading genes to create a multifunctional engineered strain for simultaneous degradation of multiple contaminants (Gong et al., 2016). Recently, a two‐step markerless genome modification method was developed for P. putida KT2440, which combined λRed‐mediated homologous recombination with Cre‐recombinase catalysed site‐specific recombination (Luo et al., 2016). Graf and Altenbuchner (2011) developed a chromosomal marker‐free modification method for P. putida KT2440 by combining homologous recombination with upp counter‐selection system. In this work, CP/carbofuran hydrolase genes were inserted scarlessly into target sites on the chromosome of P. putida KT2440 using upp as a counter‐selectable marker. Release of genetically modified organisms (GMO) into the environment is strictly restricted due to serious concerns about the biosafety of GMO. P. putida KT2440 has strong competitiveness in soil microbial community (Espinosa‐Urgel et al., 2002). Most importantly, P. putida KT2440 has been shown to be a biosafety strain for in situ bioremediation (Nelson et al., 2002). The feasibility of applying the recombinant P. putida KT2440 for in situ bioremediation of soil co‐contaminated with carbofuran and CP is currently under investigation.
Experimental procedures
Reagents, strains and culture conditions
Carbofuran, CP, carbonfuran phenol and TCP (99% pure analytical grade) were purchased from Alta Scientific, Tianjin, China. Pesticides were stocked in methanol with a concentration of 10 mg ml−1. All the other chemical reagents were of analytical grade and purchased from Dingguo Biotechnology, Tianjin, China.
Pseudomonas putida strains were grown at 30°C in M9 minimal medium (Graf and Altenbuchner, 2011) as well as LB medium (Sambrook and Russel, 2001) supplemented with kanamycin (50 μg ml−1) or 5‐FU (20 μg ml−1) when required.
DNA manipulation and strain constructions
The strains, plasmids and primers used in this study are listed in Table 2. The gene targeting vector pKU‐P (Gong et al., 2016) used for the integration of mpd gene into the chromosome was transformed into P. putida KTU by electroporation (Cho et al., 1995). Cells were incubated at 30°C for 24 h on LB agar plates containing 50 μg ml−1 kanamycin. Positive recombinants occurring with the integration of the plasmid into the chromosome were identified by colony PCR. To promote the excision of the plasmid from the chromosome, the recombinants were incubated in LB medium at 30°C for 12 h. Then, the culture broths that had been diluted to 10−5 were spread on LB agar plates supplemented with 20 μg ml−1 5‐FU. Positive recombinants occurring with the exogenous gene replacement were identified by colony PCR. The resulting mutant strain was designated as P. putida KTU‐P.
Table 2.
Strains, plasmids and primers used in this study
| Strain, plasmid or primer | Relevant characteristics | Source or reference |
|---|---|---|
| Escherichia coli | ||
| Trans1 T1 | F‐, φ80 (lacZ), ΔM15, ΔlacX74, hsdR (rK ‐, mK +), ΔrecA1398, endA1, tonA | Transgen |
| Pseudomonas putida | ||
| KT2440 | Wild type | ATCC 47054 |
| KTU | upp‐deficient KT2440 | Gong et al. (2016) |
| KTU‐P | KT2440 mutant (Δupp, ΔphaA, mpd +) | This study |
| KTU‐PG | KT2440 mutant (Δupp, ΔphaA, Δvdh, mpd +, gfp +) | This study |
| KTU‐PGC | KT2440 mutant (Δupp, ΔphaA, Δvdh, ΔalgA&algF, mpd +, gfp +, mcd +) | This study |
| Plasmids | ||
| pK18mobsacB | Kanr, suicide plasmid for gene knockout | Gong et al. (2016) |
| pKU | Kanr, pK18mobsacB containing upp gene | This study |
| pKU‐P | Kanr, pK18mobsacB containing upp and mpd | This study |
| pKU‐G | Kanr, pK18mobsacB containing upp and gfp | This study |
| pKU‐C | Kanr, pK18mobsacB containing upp and mcd | This study |
| Primers | ||
| mpd‐1 | 5′‐TGGCCTGGAGCTGAAGAACG‐3′ | This study |
| mpd‐2 | 5′‐CAGTGCAACCACCAGGAGTC‐3′ | This study |
| gfp‐1 | 5′‐TGGCAGGCGCTGATCTGTTG‐3′ | This study |
| gfp‐2 | 5′‐TGGCAGATACCCGACTCCAC‐3′ | This study |
| mcd‐1 | 5′‐AGACTTCCATTGCCAAAGCCCTCAC‐3′ | This study |
| mcd‐2 | 5′‐ACTGCGCGATGGTCTTCACCGAAAC‐3′ | This study |
| mpd‐F | 5′‐GATGCTGCTGGGCGACTTCGAAATC‐3′ | This study |
| mpd‐R | 5′‐AAGGCTTGAACTTGCCGGCCTTCAC‐3′ | This study |
| gfp‐F | 5′‐ATGGTGAGCAAGGGCGAGGAGCTGT‐3′ | This study |
| gfp‐R | 5′‐TTACTTGTACAGCTCGTCCATGCCG‐3′ | This study |
| mcd‐F | 5′‐GGGCTCAAGATCTATGTGCCCGAAG‐3′ | This study |
| mcd‐R | 5′‐CGCCTTGGTCGATTTGGTCCGATAG‐3′ | This study |
The construction processes of P. putida KTU‐PG and KTU‐PGC were the same as P. putida KTU‐P. All the constructed strains were validated by PCR and DNA sequencing. The nucleotide sequences of three synthetic gene cassettes (mpd, gfp and mcd) are shown in Fig. S1. The three synthetic gene cassettes were successively inserted into three different target sites on the chromosome of P. putida KT2440. The detailed information on three exogenous genes and their chromosomal insertion sites is shown in Table 1.
Detection of transcription of exogenous genes by RT‐PCR
Cells of P. putida KTU‐PGC were grown overnight in LB medium and harvested. A RNApure Bacteria kit (Cwbio, Beijing, China) was used to acquire total RNA. DNA contamination was eliminated by processing with a DNase I at 37°C for 50 min. RNA integrity was checked by agarose gel electrophoresis. cDNA was prepared using purified total RNA as template and a PrimeScript RT Master Mix kit (Takara, Dalian, China). PCR was carried out with the PrimeSTAR HS DNA polymerase (Takara) using primers listed in Table 2. mRNA and ddH2O were used as the templates in control reactions to make sure that DNA products were amplified from cDNA rather than DNA contamination. PCR products were separated by electrophoresis at 80 V on 0.8% Tris Borate EDTA agarose gel stained by ethidium bromide.
Degradation of carbofuran and CP by P. putida KTU‐PGC
Cells of P. putida KTU‐PGC were harvested by centrifugation after incubation in 100 ml of LB medium at 30°C for 12 h, washed twice with M9 minimal medium and resuspended (OD600 = 1.0) in the same medium. Subsequently, 5 ml of cell suspensions were inoculated into 95 ml of M9 minimal medium supplemented with 100 mg l−1 carbofuran and/or 100 mg l−1 CP. The samples were incubated at 30°C and 200 rpm in a shaker and withdrawn at regular time intervals. Analysis for the pesticides and their hydrolysis products was performed by HPLC as described below. Degradation experiments were performed with P. putida KTU under the same conditions.
Imaging bacteria
Cells of P. putida KTU‐PGC were grown to the plateau stage at 30°C, harvested, washed with PBS buffer (pH 7.2) twice, and then stained with 10 μM FM4‐64/L for 15 min. Cells were fixed with 2% glycerol on a slice, and the observed by a confocal microscope (Leica, Heidelberg, Germany) fitted with a Leica 100 × 10 numerical aperture objective lens, using parameters appropriate for the fluorescence excitation. Expression levels of the GFP were reliably predicted from the fluorescence intensity.
Soil remediation experiments
Soil samples were collected from the campus of Nankai University, Tianjin, China, which were never exposed to any pesticides before. The soil had a pH of 6.8. Subsamples (100 g) of the soil were treated under aseptic condition with carbofuran (50 mg kg−1 soil) and CP (50 mg kg−1 soil). Soil samples in triplicate were inoculated with P. putida KTU‐PGC (106 cells g−1), and soil samples without inoculation were kept as the controls. The inoculum was thoroughly mixed into the soils under sterile condition. The soil moisture was adjusted by the addition of distilled water to 40% of its water‐holding capacity. Soil samples were incubated at 30°C for 12 days in the dark. Soil samples (5 g) were withdrawn for quantifying carbofuran and CP at different time intervals. Carbofuran and CP were extracted from the soil samples using the previous methods described in Jiang et al. (2007) and Singh et al. (2004). The concentration of carbofuran and CP in the organic extracts was quantified by HPLC as described below.
HPLC analysis
The culture broths were extracted twice with equal volumes of ethyl acetate. The organic layers containing the pesticides and their hydrolytic products were pooled and then dried over anhydrous Na2SO4. Aliquots of 20 μl were injected directly to a reversed‐phase HPLC, which was obtained on an Alltech system controller (Alltech Associates, Lexington, Kentucky, USA) equipped with an Innoval ODS‐2 column (Agel, Tianjin, China) and an Alltech UVIS 200 detector. Carbofuran and carbonfuran phenol were detected at 280 nm with methanol/water (50:50, v/v) containing 0.1% phosphoric acid as the mobile phase at a flow rate of 1 ml min−1. CP and TCP were analysed at 290 nm with methanol/water (90:10, v/v) as the mobile phase at a flow rate of 1 ml min−1. The retention times of the pesticides and their hydrolytic products in HPLC analysis were determined using the authentic standards of each chemical as the reference. The concentration of carbofuran and CP was determined by comparing the peak areas with the calibration curves.
Supporting information
Fig. S1. The nucleotide sequences of the synthetic gene cassettes. (A) mpd gene cassette; (B) gfp gene cassette; (C) mcd gene cassette.
Fig. S2. Detection of the introduced exogenous genes in Pseudomonas putida KT2440 by agarose gel electrophoresis of PCR products. PCR amplifications were performed with chromosomal DNA as template using primers listed in Table 2. Lane M: DNA marker; lane 1: mpd gene cassette; lane 2: gfp gene cassette; lane 3: mcd gene cassette.
Fig. S3. RT‐PCR assays for detecting the transcription of three inserted exogenous genes in Pseudomonas putida KT2440. Panels A‐C are the detection results of mpd, gfp and mcd respectively. Lane M: DNA marker; lane 1: control reaction in which genomic DNA was used as template; lane 2: reaction in which cDNA was used as template; lane 3: control reaction in which mRNA was used as template; lane 4: control reaction in which ddH2O was used as template.
Fig. S4. HPLC analysis of degradation products of carbofuran. Pseudomonas putida KTU‐PGC was incubated at 30°C and 200 rpm in a shaker in M9 minimal medium supplemented with 100 mg l−1 carbofuran as the sole source of carbon. Carbofuran and carbofuran phenol had a retention time (RT) of 8.75 and 10.21 min respectively. Top, carbofuran degradation detected by HPLC at 0 h; middle, carbofuran degradation detected by HPLC at 6 h; bottom, carbofuran degradation detected by HPLC at 36 h.
Fig. S5. Products of degradation of carbofuran and CP by Pseudomonas putida KTU‐PGC.
Fig. S6. HPLC analysis of degradation products of CP. Pseudomonas putida KTU‐PGC was incubated at 30°C and 200 rpm in a shaker in M9 minimal medium supplemented with 100 mg l−1 CP as the sole source of carbon. CP and TCP had a retention time (RT) of 8.69 and 3.12 min respectively. Top, CP degradation detected by HPLC at 0 h; middle, CP degradation detected by HPLC at 10 h; bottom, CP degradation detected by HPLC at 24 h.
Fig. S7. PCR detection of mpd, gfp and mcd genes in the twentieth‐generation subcultures of Pseudomonas putida KTU‐PGC.
Fig. S8. Time‐courses for the growth of Pseudomonas putida KTU and KTU‐PGC. Cells were incubated in LB medium at 30°C for 48 h. The cell concentration was determined by measuring the OD600 of the culture broth.
Fig. S9. PCR detection of mpd, gfp and mcd genes in Pseudomonas putida SKT‐A (top) and SKT‐B (bottom).
Fig. S10. Monitoring of inoculated Pseudomonas putida KTU‐PGC by GFP fluorescence using a confocal microscope during soil bioremediation. (A) Green fluorescence within the cell; (B) outline of cell membrane by stain with FM4‐64/L; (C) panels A and B merged together.
Microbial Biotechnology (2016) 9(6), 792–800
Funding Information
This study was supported by National Key Technology Support Program of China (no. 2015BAD16B04), National Natural Science Foundation of China (nos. 31300032 and 31570035), Open Fund of State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences (no. SKLMR‐20130604), Open Fund of State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology and Ph.D. Candidate Research Innovation Fund of Nankai University.
Contributor Information
Cunjiang Song, Email: songcj@nankai.edu.cn.
Yanping Liu, Email: lyp5588@sohu.com.
Chao Yang, Email: yang_chao2008@hotmail.com.
References
- Anwar, S. , Liaquat, F. , Khan, Q.M. , Khalid, Z.M. , and Iqbal, S. (2009) Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6‐tricholoro‐2‐pyridinol by Bacillus pumilus strain C2A1. J Hazard Mater 168: 400–405. [DOI] [PubMed] [Google Scholar]
- Chakraborty, J. , Jana, T. , Saha, S. , and Dutta, T.K. (2014) Ring‐Hydroxylating Oxygenase database: a database of bacterial aromatic ring‐hydroxylating oxygenases in the management of bioremediation and biocatalysis of aromatic compounds. Environ Microbiol Rep 6: 519–523. [DOI] [PubMed] [Google Scholar]
- Cho, J.H. , Kim, E.K. , and So, J.S. (1995) Improved transfomation of Pseudomonas putida KT2440 by electroporation. Biotechnol Tech 9: 41–44. [Google Scholar]
- Espinosa‐Urgel, M. , Kolter, R. , and Ramos, J.L. (2002) Root colonization by Pseudomonas putida: love at first sight. Microbiology 148: 341–343. [DOI] [PubMed] [Google Scholar]
- Feng, X. , Ou, L.T. , and Ogram, A. (1997) Plasmid‐mediated mineralization of carbofuran by Sphingomonas sp. strain CF06. Appl Environ Microbiol 63: 1332–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, T. , Liu, R. , Zuo, Z. , Che, Y. , Yu, H. , Song, C. , and Yang, C. (2016) Metabolic engineering of Pseudomonas putida KT2440 for complete mineralization of methyl parathion and γ‐hexachlorocyclohexane. ACS Synth Biol 5: 434–442. [DOI] [PubMed] [Google Scholar]
- Graf, N. , and Altenbuchner, J. (2011) Development of a method for markerless gene deletion in Pseudomonas putida . Appl Environ Microbiol 77: 5549–5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernándeza, A.F. , Parrónb, T. , Tsatsakisd, A.M. , Requenab, M. , Alarcónc, R. , and López‐Guarnidoa, O. (2013) Toxic effects of pesticide mixtures at a molecular level: their relevance to human health. Toxicology 307: 136–145. [DOI] [PubMed] [Google Scholar]
- Jiang, J. , Zhang, R. , Li, R. , Gu, J.D. , and Li, S. (2007) Simultaneous biodegradation of methyl parathion and carbofuran by a genetically engineered microorganism constructed by mini‐Tn5 transposon. Biodegradation 18: 403–412. [DOI] [PubMed] [Google Scholar]
- Jiménez, J.I. , Miñambres, B. , García, J.L. , and Díaz, E. (2002) Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ Microbiol 4: 824–841. [DOI] [PubMed] [Google Scholar]
- Li, X. , He, J. , and Li, S. (2007) Isolation of chlorpyrifos degrading bacterium Sphingomonas sp strain Dsp‐2 and cloning of the mpd gene. Res Microbiol 158: 143–149. [DOI] [PubMed] [Google Scholar]
- Luo, X. , Yang, Y. , Ling, W. , Zhuang, H. , Li, Q. , and Shang, G. (2016) Pseudomonas putida KT2440 markerless gene deletion using a combination of λRed recombineering and Cre/loxP site‐specific recombination. FEMS Microbiol Lett 363: fnw014. [DOI] [PubMed] [Google Scholar]
- Marupakula, S. , Mahmood, S. , and Finlay, R.D. (2016) Analysis of single root tip microbiomes suggests that distinctive bacterial communities are selected by Pinus sylvestris roots colonized by different ectomycorrhizal fungi. Environ Microbiol 18: 1470–1483. [DOI] [PubMed] [Google Scholar]
- Nelson, K.E. , Weinel, C. , Paulsen, I.T. , Dodson, R.J. , Hilbert, H. , Martins dos Santos, V.A. , et al (2002) Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol 4: 799–808. [DOI] [PubMed] [Google Scholar]
- Nguyen, T.P. , Helbling, D.E. , Bers, K. , Fida, T.T. , Wattiez, R. , Kohler, H.P. , et al (2014) Genetic and metabolic analysis of the carbofuran catabolic pathway in Novosphingobium sp. KN65.2. Appl Microbiol Biotechnol 98: 8235–8252. [DOI] [PubMed] [Google Scholar]
- Nikel, P.I. , Martínez‐García, E. , and de Lorenzo, V. (2014) Biotechnological domestication of pseudomonads using synthetic biology. Nat Rev Microbiol 12: 368–379. [DOI] [PubMed] [Google Scholar]
- Ogramab, A.V. , Duana, Y. , Trabuea, S.L. , Fengc, X. , Castroa, H. , and Oua, L. (2000) Carbofuran degradation mediated by three related plasmid systems. FEMS Microbiol Ecol 32: 197–203. [DOI] [PubMed] [Google Scholar]
- Pizarro‐Tobías, P. , Niqui, J.L. , Roca, A. , Solano, J. , Fernández, M. , Bastida, F. , et al (2015) Field trial on removal of petroleum‐hydrocarbon pollutants using a microbial consortium for bioremediation and rhizoremediation. Environ Microbiol Rep 7: 85–94. [DOI] [PubMed] [Google Scholar]
- Rama Krishna, K. , and Philip, L. (2011) Bioremediation of single and mixture of pesticide‐contaminated soils by mixed pesticide‐enriched cultures. Appl Biochem Biotechnol 164: 1257–1277. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , and Russel, D.W. (2001) Molecular Cloning: A Laboratory Manual, 3rd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Shetty, R.P. , Endy, D. , and Knight, T.F. Jr (2008) Engineering BioBrick vectors from BioBrick parts. J Biol Eng 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, B.K. (2009) Organophosphorus‐degrading bacteria: ecology and industrial applications. Nat Rev Microbiol 7: 156–164. [DOI] [PubMed] [Google Scholar]
- Singh, B.K. , and Walker, A. (2006) Microbial degradation of organophosphorus compounds. FEMS Microbiol Rev 30: 428–471. [DOI] [PubMed] [Google Scholar]
- Singh, B.K. , Walker, A. , Morgan, J.A.W. , and Wright, D.J. (2004) Biodegradation of chlorpyrifos by enterobacter strain B‐14 and its use in bioremediation of contaminated soils. Appl Environ Microbiol 70: 4855–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasek, P.H. , and Karns, J.S. (1989) Cloning of a carbofuran hydrolase gene from Achromobacter sp. strain WM111 and its expression in gram‐negative bacteria. J Bacteriol 171: 4038–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G. , Zheng, W. , Li, Y. , Wang, S. , Zhang, J. , and Yan, Y. (2008) Biodegradation of chlorpyrifos and 3,5,6‐tricholoro‐2‐pyridinol by a newly isolated Paracoccus sp. strain TRP. Int Biodeterior Biodegrad 62: 51–56. [Google Scholar]
- Yang, C. , Liu, N. , Guo, X. , and Qiao, C. (2006) Cloning of mpd gene from a chlorpyrifos‐degrading bacterium and use of this strain in bioremediation of contaminated soil. FEMS Microbiol Lett 265: 118–125. [DOI] [PubMed] [Google Scholar]
- Yang, C. , Liu, R. , Yuan, Y. , Liu, J. , Cao, X. , Qiao, C. , and Song, C. (2013) Construction of a green fluorescent protein (GFP)‐marked multifunctional pesticide‐degrading bacterium for simultaneous degradation of organophosphates and γ‐hexachlorocyclohexane. J Agr Food Chem 61: 1328–1334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The nucleotide sequences of the synthetic gene cassettes. (A) mpd gene cassette; (B) gfp gene cassette; (C) mcd gene cassette.
Fig. S2. Detection of the introduced exogenous genes in Pseudomonas putida KT2440 by agarose gel electrophoresis of PCR products. PCR amplifications were performed with chromosomal DNA as template using primers listed in Table 2. Lane M: DNA marker; lane 1: mpd gene cassette; lane 2: gfp gene cassette; lane 3: mcd gene cassette.
Fig. S3. RT‐PCR assays for detecting the transcription of three inserted exogenous genes in Pseudomonas putida KT2440. Panels A‐C are the detection results of mpd, gfp and mcd respectively. Lane M: DNA marker; lane 1: control reaction in which genomic DNA was used as template; lane 2: reaction in which cDNA was used as template; lane 3: control reaction in which mRNA was used as template; lane 4: control reaction in which ddH2O was used as template.
Fig. S4. HPLC analysis of degradation products of carbofuran. Pseudomonas putida KTU‐PGC was incubated at 30°C and 200 rpm in a shaker in M9 minimal medium supplemented with 100 mg l−1 carbofuran as the sole source of carbon. Carbofuran and carbofuran phenol had a retention time (RT) of 8.75 and 10.21 min respectively. Top, carbofuran degradation detected by HPLC at 0 h; middle, carbofuran degradation detected by HPLC at 6 h; bottom, carbofuran degradation detected by HPLC at 36 h.
Fig. S5. Products of degradation of carbofuran and CP by Pseudomonas putida KTU‐PGC.
Fig. S6. HPLC analysis of degradation products of CP. Pseudomonas putida KTU‐PGC was incubated at 30°C and 200 rpm in a shaker in M9 minimal medium supplemented with 100 mg l−1 CP as the sole source of carbon. CP and TCP had a retention time (RT) of 8.69 and 3.12 min respectively. Top, CP degradation detected by HPLC at 0 h; middle, CP degradation detected by HPLC at 10 h; bottom, CP degradation detected by HPLC at 24 h.
Fig. S7. PCR detection of mpd, gfp and mcd genes in the twentieth‐generation subcultures of Pseudomonas putida KTU‐PGC.
Fig. S8. Time‐courses for the growth of Pseudomonas putida KTU and KTU‐PGC. Cells were incubated in LB medium at 30°C for 48 h. The cell concentration was determined by measuring the OD600 of the culture broth.
Fig. S9. PCR detection of mpd, gfp and mcd genes in Pseudomonas putida SKT‐A (top) and SKT‐B (bottom).
Fig. S10. Monitoring of inoculated Pseudomonas putida KTU‐PGC by GFP fluorescence using a confocal microscope during soil bioremediation. (A) Green fluorescence within the cell; (B) outline of cell membrane by stain with FM4‐64/L; (C) panels A and B merged together.


