Abstract
Background:
Melanoma is a type of cancer that develops from the pigment-containing cells. Until now, its pathological mechanisms remain largely unknown. The aim of this study was to identify metastasis-related microRNA (miRNAs) and gain an understanding of the biological functions in the metastasis of melanoma.
Methods:
We searched the PubMed and Gene Expression Omnibus database to collect miRNA expression profiling datasets about melanoma, with key words of “melanoma”, “miRNA”, “microarray”, and “gene expression profiling”. Only the original experimental works published before June 2016 for analyzing the metastasis of melanoma were retained, other nonhuman studies, reviews, and meta-analyses were removed. We performed a meta-analysis to explore the differentially expressed miRNA between metastatic and nonmetastatic samples. Moreover, we predicted target genes of the miRNAs to study their biological roles for these miRNAs.
Results:
We identified a total of 63 significantly differentially expressed miRNAs by meta-analysis of the melanoma expression profiling data. The regulatory network constructed by using these miRNAs and the predicted targets identified several key genes involved in the metastasis of melanoma. Functional annotation of these genes indicated that they are mainly enriched in some biological pathways such as mitogen-activated protein kinase signaling pathway, cell junction, and focal adhesion.
Conclusions:
By collecting the miRNA expression datasets from different platforms, multiple biological markers were identified for the metastasis of melanoma. This study provided novel insights into the molecular mechanisms underlying this disease, thereby aiding the diagnosis and treatment of the disease.
Keywords: Expression Profiles, Melanoma, Metastasis, MicroRNA
Introduction
Melanoma is the neoplasm of the cells that develops from melanocyte.[1] It could develop in skin or in mouth, eye, and intestine.[2] It has long been considered as resulting from environmental factors such as ultraviolet light exposure.[3] Every year, more than 200,000 people were diagnosed with melanoma and it results in more than 50,000 deaths worldwide.[4] The incidence of melanoma has been increasing at a high speed in recent years. Until now, surgical resection and chemotherapy are still the best curative options for the treatment of melanoma. However, the postoperative 5-year survival rate is only 5–10% due to the high rate of metastasis in patients who have had resection.[5] This study aimed to identify novel biomarkers and to elucidate the molecular mechanisms underlying melanoma metastasis.
In recent years, extensive studies have indicated that microRNAs (miRNAs) are widely involved in the prognosis, metastasis, and overall patient survival of melanoma.[6] miRNA is a kind of small noncoding RNA that regulates gene expression at post-transcriptional level.[7] The major function of miRNA is to regulate the gene expression by binding to the 3’- untranslated region of target mRNAs.[8] Until now, more than 2000 miRNA genes have been identified in the human genome (miRBase release 21).[9] These miRNAs participate in many key cellular processes including proliferation, apoptosis, and metastasis.[10] De-regulation of miRNA expression has been identified in many cancer types, including melanoma. Their functions as etiologic factors in melanoma have been widely revealed. However, these studies are very limited, and the results were inconclusive. The underlying pathogenesis of melanoma, especially for their metastasis, remains far from understood. To derive a more precise estimation of the association between these miRNAs and melanoma, a meta-analysis is needed to help us to better understand the possible risk factor for melanoma metastasis.
In this study, we collected several miRNA expression profiling datasets for melanoma, and then a meta-analysis was performed to identify the differentially expressed miRNAs between metastatic and nonmetastatic samples. We identified the predicted targets of these differentially expressed miRNAs. Functional annotation of the miRNA target genes was further performed to explore the putative links between these miRNAs and melanoma metastasis. We demonstrated that signaling pathways involved in many fundamental biological processes are linked to the metastasis of melanoma, such as focal adhesion pathway and mitogen-activated protein kinase (MAPK) signaling. Our results provide clues for the mechanism of melanoma metastasis and could be used for miRNA-based drug development for preventing this disease.
Methods
Data collection
We first searched the PubMed and Gene Expression Omnibus database to collect miRNA expression profiling datasets about melanoma. Key words including “melanoma”, “miRNA”, “microarray”, and “gene expression profiling” were used for data searching. Only the original experimental works published before June 2016 for analyzing the metastasis of melanoma were retained, other nonhuman studies, reviews, and meta-analysis were removed. In addition, we screened that those expression profiles have more than six arrays. Finally, a total of seven expression profiles which included 82 metastatic and 87 nonmetastatic samples were collected [Table 1].
Table 1.
Characteristics of datasets included in this analysis
| GEO ID | Number of controls | Number of cases | GEO platform ID | Year | Reference |
|---|---|---|---|---|---|
| GSE18509 | 8 | 8 | GPL9081 Agilent-016436 Human miRNA Microarray 1.0 G4472A | 2010 | Chen et al.[11] |
| GSE19387 | 21 | 16 | GPL9081 Agilent-016436 Human miRNA Microarray 1.0 G4472A | 2010 | Caramuta et al.[12] |
| GSE24996 | 15 | 8 | GPL6955 Agilent-016436 Human miRNA Microarray 1.0 | 2011 | Chen et al.[13] |
| GSE34460 | 9 | 4 | GPL15019 Agilent-031181 Unrestricted_Human_miRNA_V16.0_Microarray 030840 | 2013 | Sand et al.[14] |
| GSE35579 | 20 | 21 | GPL15183 CRUK/Melton lab-Human melanoma-71-v2-microRNA expression profiling | 2012 | Xu et al.[15] |
| GSE36236 | 6 | 6 | GPL9052 Illumina Genome Analyzer (Homo sapiens) | 2014 | Babapoor et al.[16] |
| GSE54492 | 8 | 19 | GPL10262 Renji Hospital ABI Human TaqMan® MicroRNA Assays V2.0 | 2015 | Bhattacharya et al.[17] |
GEO: Gene Expression Omnibus.
Data preprocessing
For each dataset, the missing values in the original expression values were imputed by using the R package of impute,[18] and then the expression values were logarithmically transformed (base 2) and quantile normalized for each dataset. The average expression value was used if there were multiple probes for a given miRNA in each sample. To make the expression profiles from different comparable studies, the Z-score transformation approach was used to calculate the expression intensities for each gene expression profile to remove the inconsistency and heterogeneity.[19] Z-score was defined as the difference of raw intensity data for each gene and the average gene intensity within a single experiment divided by the standard deviation of all of the measured intensities. Then, all the samples were combined for the subsequent analysis.
MicroRNA target prediction and functional annotation
The predicted targets of miRNAs were obtained from TargetScan,[20] miRanda,[21] and PITA databases.[22] The predicted targets obtained from at least two programs were regarded as the reliable target genes. Based on the interactions of miRNAs and target genes, the miRNA function network was built. The gene ontology (GO) enrichment analysis was performed to investigate the main function of target genes by using the web-based software Gorilla. Hypergeometric test was used to classify the GO category. Adjusted P values were computed for the GO terms by multiple test adjustment of Benjamini-Hochberg (BH). GO terms with an adjusted P < 0.05 were selected. In addition, for identifying the biological pathways associated with the metastasis of melanoma, we also performed the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of target genes.
Statistical analysis
To get the overview of the global shifts of gene expression in metastasis samples, the limma algorithm was used to identify the differentially expressed miRNAs between metastasis and control samples.[23] The BH method was used for multiple test correction. False discovery rate (FDR) <0.01 was selected as the criteria for calculating significant differences. Then, the unsupervised hierarchical clustering analysis of differentially expressed genes was performed.
Results
MicroRNA expression profiles
In recent years, many studies have indicated that miRNAs are involved in the development of melanoma.[24] High throughput methods such as microarray have been widely used to get miRNA expression profiling for many cancer types including the melanoma.[25] However, due to the different platforms used for these researches, only few of the identified miRNAs can be mutually identified. To overcome this problem, we collected seven expression profiles for metastatic samples of melanoma to perform the meta-analysis [Figure 1]. The number of the samples for these studies ranged from 12 to 37, with the total number of 169 samples for all the studies. We collected these expression profiles and performed the quantile normalization for each datum. Then, the Z-score transformation method was used to make the global normalization.
Figure 1.

A schematic workflow of data analysis.
Identification of differentially expressed microRNAs
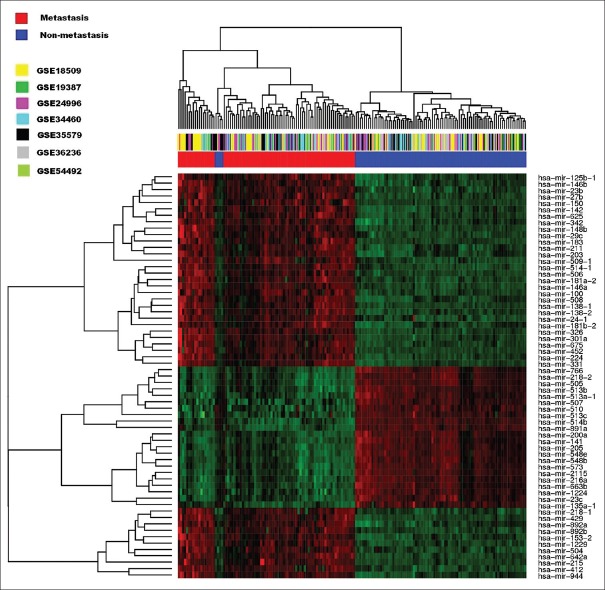
We used the limma package to identify the differentially expressed miRNAs between metastatic and control samples. With the threshold of FDR <0.05, a total of 63 miRNAs were identified as differentially expressed. Among these genes, 35 were upregulated and 28 were downregulated. The top ten upregulated and downregulated miRNAs are listed in Table 2, which include the hsa-mir-205, hsa-mir-203, hsa-mir-508, hsa-mir-514-1, hsa-mir-509-1, and hsa-mir-506. The full miRNA list is provided as Supplementary Table 1. miRNA with the most high upregulation was hsa-mir-508, with a 15-fold higher expression in metastatic samples. We also performed an unsupervised hierarchical clustering analysis of these differentially expressed miRNAs and revealed distinct expression patterns in metastatic and control samples [Figure 2], suggesting the potential of expression profiles of these top differentially expressed genes to distinguish sample status for melanoma.
Table 2.
The top ten upregulated and downregulated microRNAs
| MicroRNAs | logFC | AveExpr | t | P | Adjusted P |
|---|---|---|---|---|---|
| Upregulated microRNA | |||||
| hsa-mir-205 | 3.84074 | 3.24142 | 11.11461 | 7.59E-22 | 7.94E-19 |
| hsa-mir-203 | 3.27052 | 5.96689 | 9.06155 | 3.29E-16 | 1.72E-13 |
| hsa-mir-508 | 1.55319 | 8.35084 | 4.53489 | 1.09E-05 | 6.73E-04 |
| hsa-mir-514-1 | 1.52272 | 5.78308 | 4.66379 | 6.32E-06 | 4.72E-04 |
| hsa-mir-509-1 | 1.44397 | 6.10834 | 4.43348 | 1.67E-05 | 7.61E-04 |
| hsa-mir-506 | 1.35088 | 4.45629 | 4.49817 | 1.28E-05 | 7.03E-04 |
| hsa-mir-211 | 1.33458 | 6.55326 | 3.79426 | 2.07E-04 | 6.00E-03 |
| hsa-mir-200a | 1.30001 | 2.29690 | 7.75183 | 8.35E-13 | 2.91E-10 |
| hsa-mir-141 | 1.26950 | 2.03624 | 6.38541 | 1.62E-09 | 2.82E-07 |
| hsa-mir-513c | 1.127236 | 3.19562 | 4.50259 | 1.25E-05 | 7.03E-04 |
| Downregulated microRNA | |||||
| hsa-mir-326 | −0.47963 | 2.38721 | −3.51696 | 5.62E-04 | 1.22E-02 |
| hsa-mir-766 | −0.54186 | 3.06395 | −4.45824 | 1.51E-05 | 7.17E-04 |
| hsa-mir-153-2 | −0.57756 | 1.01633 | −5.171814 | 6.56E-07 | 6.86E-05 |
| hsa-mir-342 | −0.58209 | 5.68141 | −4.30546 | 2.83E-05 | 1.10E-03 |
| hsa-mir-29c | −0.62039 | 6.12865 | −4.12543 | 5.82E-05 | 2.10E-03 |
| hsa-mir-625 | −0.62962 | 5.73921 | −6.04559 | 9.40E-09 | 1.40E-06 |
| hsa-mir-100 | −0.66570 | 8.64832 | −3.76338 | 2.32E-04 | 6.38E-03 |
| hsa-mir-150 | −0.69533 | 6.69923 | −2.98097 | 3.30E-03 | 4.65E-02 |
| hsa-mir-142 | −0.73181 | 6.46511 | −3.69067 | 3.02E-04 | 7.75E-03 |
| hsa-mir-675 | −1.01107 | 2.71285 | −5.62435 | 7.66E-08 | 1.00E-05 |
E−n = 10−n (n>0).
Supplementary Table 1.
Full lists of the differentially expressed miRNAs
| MicroRNA | logFC | AveExpr | t | P | Adjusted P |
|---|---|---|---|---|---|
| hsa-mir-205 | 3.84074 | 3.24142 | 11.11461 | 7.59E-22 | 7.94E-19 |
| hsa-mir-203 | 3.27052 | 5.96689 | 9.06155 | 3.29E-16 | 1.72E-13 |
| hsa-mir-508 | 1.55319 | 8.35084 | 4.53489 | 1.09E-05 | 6.73E-04 |
| hsa-mir-514-1 | 1.52272 | 5.78308 | 4.66379 | 6.32E-06 | 4.72E-04 |
| hsa-mir-509-1 | 1.44397 | 6.10834 | 4.43348 | 1.67E-05 | 7.61E-04 |
| hsa-mir-506 | 1.35088 | 4.45629 | 4.49817 | 1.28E-05 | 7.03E-04 |
| hsa-mir-211 | 1.33458 | 6.55326 | 3.79426 | 2.07E-04 | 6.00E-03 |
| hsa-mir-200a | 1.30001 | 2.29690 | 7.75183 | 8.35E-13 | 2.91E-10 |
| hsa-mir-141 | 1.26950 | 2.03624 | 6.38541 | 1.62E-09 | 2.82E-07 |
| hsa-mir-513c | 1.12724 | 3.19562 | 4.50259 | 1.25E-05 | 7.03E-04 |
| hsa-mir-891a | 1.04187 | 2.75798 | 3.76366 | 2.31E-04 | 6.38E-03 |
| hsa-mir-514b | 0.93581 | 2.59849 | 3.98210 | 1.02E-04 | 3.54E-03 |
| hsa-mir-513b | 0.85382 | 1.94141 | 4.56446 | 9.66E-06 | 6.31E-04 |
| hsa-mir-513a-1 | 0.83010 | 1.90886 | 4.47301 | 1.42E-05 | 7.17E-04 |
| hsa-mir-944 | 0.77757 | 1.04188 | 5.06242 | 1.08E-06 | 1.03E-04 |
| hsa-mir-510 | 0.63425 | 1.55961 | 3.79819 | 2.04E-04 | 6.00E-03 |
| hsa-mir-507 | 0.621863 | 1.70567 | 3.55528 | 4.91E-04 | 1.14E-02 |
| hsa-mir-183 | 0.54496 | 5.95690 | 3.00999 | 3.02E-03 | 4.41E-02 |
| hsa-mir-224 | 0.51577 | 2.71914 | 3.73860 | 2.54E-04 | 6.80E-03 |
| hsa-mir-429 | 0.50968 | 0.56736 | 5.56833 | 1.01E-07 | 1.17E-05 |
| hsa-mir-146a | 0.50880 | 7.55563 | 3.19641 | 1.66E-03 | 2.75E-02 |
| hsa-mir-27b | 0.50810 | 6.70707 | 4.33356 | 2.52E-05 | 1.02E-03 |
| hsa-mir-138-1 | 0.46358 | 2.74963 | 3.47963 | 6.40E-04 | 1.36E-02 |
| hsa-mir-412 | 0.45386 | 1.42936 | 3.01865 | 2.94E-03 | 4.39E-02 |
| hsa-mir-452 | 0.42736 | 3.19012 | 3.33090 | 1.07E-03 | 1.89E-02 |
| hsa-mir-138-2 | 0.42166 | 2.22355 | 3.47189 | 6.58E-04 | 1.36E-02 |
| hsa-mir-23b | 0.41741 | 6.85683 | 4.46780 | 1.45E-05 | 7.17E-04 |
| hsa-mir-181a-2 | 0.37022 | 8.27776 | 3.00773 | 3.04E-03 | 4.41E-02 |
| hsa-mir-181b-2 | 0.36869 | 2.94543 | 2.97765 | 3.34E-03 | 4.65E-02 |
| hsa-mir-24-1 | 0.36819 | 2.55684 | 3.63169 | 3.74E-04 | 8.89E-03 |
| hsa-mir-892a | 0.34883 | 0.45104 | 2.96809 | 3.44E-03 | 4.67E-02 |
| hsa-mir-892b | 0.24103 | 0.18846 | 3.22879 | 1.50E-03 | 2.57E-02 |
| hsa-mir-2115 | 0.21607 | 0.29474 | 3.91869 | 1.30E-04 | 4.37E-03 |
| hsa-mir-573 | 0.19097 | 0.30461 | 3.11817 | 2.14E-03 | 3.35E-02 |
| hsa-mir-23c | 0.18185 | 0.33044 | 3.46777 | 6.67E-04 | 1.36E-02 |
| hsa-mir-663b | −0.03914 | 0.01899 | −3.30731 | 1.15E-03 | 2.01E-02 |
| hsa-mir-548e | −0.18424 | 0.42785 | −3.39538 | 8.56E-04 | 1.69E-02 |
| hsa-mir-548b | −0.18499 | 0.38084 | −2.96899 | 3.43E-03 | 4.67E-02 |
| hsa-mir-216a | −0.21139 | 0.23894 | −3.83200 | 0.000180 | 5.69E-03 |
| hsa-mir-1224 | −0.21655 | 0.23499 | −3.52879 | 0.000539 | 1.20E-02 |
| hsa-mir-148b | −0.22071 | 5.67531 | −3.02302 | 2.90E-03 | 4.39E-02 |
| hsa-mir-218-1 | −0.24822 | 0.67965 | −2.95054 | 3.63E-03 | 4.86E-02 |
| hsa-mir-135a-1 | −0.28062 | 0.25828 | −3.89722 | 1.41E-04 | 4.59E-03 |
| hsa-mir-1229 | −0.29592 | 0.90717 | −3.22271 | 1.53E-03 | 2.58E-02 |
| hsa-mir-331 | −0.30701 | 3.22686 | −3.18981 | 1.70E-03 | 2.75E-02 |
| hsa-mir-301a | −0.32842 | 2.17655 | −2.98894 | 3.22E-03 | 4.62E-02 |
| hsa-mir-505 | −0.33969 | 4.04555 | −3.35999 | 9.65E-04 | 1.76E-02 |
| hsa-mir-215 | −0.37497 | 1.44599 | −3.35979 | 9.66E-04 | 1.76E-02 |
| hsa-mir-504 | −0.37641 | 0.94157 | −3.36009 | 9.65E-04 | 1.76E-02 |
| hsa-mir-146b | −0.41183 | 6.82078 | −3.68916 | 3.04E-04 | 7.75E-03 |
| hsa-mir-642a | −0.42094 | 1.53763 | −3.16664 | 1.83E-03 | 2.90E-02 |
| hsa-mir-218-2 | −0.45173 | 3.18601 | −3.80808 | 1.96E-04 | 6.00E-03 |
| hsa-mir-125b-1 | −0.45657 | 6.93287 | −3.46334 | 6.77E-04 | 1.36E-02 |
| hsa-mir-326 | −0.47963 | 2.38721 | −3.51696 | 5.62E-04 | 1.22E-02 |
| hsa-mir-766 | −0.54186 | 3.06395 | −4.45824 | 1.51E-05 | 7.17E-04 |
| hsa-mir-153-2 | −0.57756 | 1.01633 | −5.17181 | 6.56E-07 | 6.86E-05 |
| hsa-mir-342 | −0.58209 | 5.68141 | −4.30546 | 2.83E-05 | 1.10E-03 |
| hsa-mir-29c | −0.62039 | 6.12865 | −4.12543 | 5.82E-05 | 2.10E-03 |
| hsa-mir-625 | −0.62962 | 5.73921 | −6.04559 | 9.40E-09 | 1.40E-06 |
| hsa-mir-100 | −0.66570 | 8.64832 | −3.76338 | 2.32E-04 | 6.38E-03 |
| hsa-mir-150 | −0.69533 | 6.69923 | −2.98097 | 3.30E-03 | 4.65E-02 |
| hsa-mir-142 | −0.73181 | 6.46511 | −3.69067 | 3.02E-04 | 7.75E-03 |
| hsa-mir-675 | −1.01107 | 2.712853 | −5.62435 | 7.66E-08 | 1.00E-05 |
E−n = 10−n (n>0). miRNAs: MicroRNAs.
Figure 2.
Hierarchical clustering of the microRNAs whose expressions were significantly altered in metastatic samples.
miRNA regulatory network
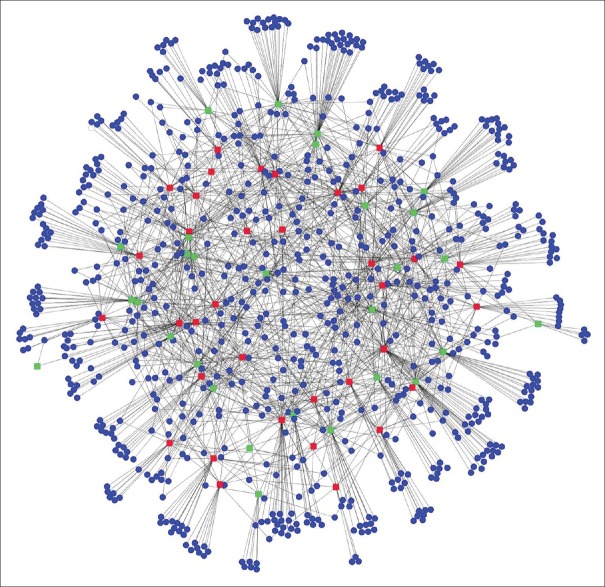
To get a more comprehensive understanding of the biological roles of the differentially expressed miRNAs in melanoma metastasis, the targets of these differentially expressed miRNAs were predicted by using TargetScan, PITA, and miRanda. A total of 816 protein-coding genes were predicted to be regulated by the miRNAs identified above. Then, we constructed the miRNA regulatory by using the differentially expressed miRNAs and the predicted targets. This network consisted of 880 nodes and 1576 edges [Figure 3]. Within this network, 9 miRNAs were associated with only one kind of downstream gene, while 18 miRNAs were associated with more than 30 target genes. The top-ranked miRNA included hsa-mir-548b, hsa-mir-181b-2, hsa-mir-141, hsa-miR-766, and hsa-miR-203. For miRNA targets, KSR2, SUDS3, SH3TC2, MVB12B, and LONRF2 were all critical genes that demonstrated to have the highest connectivity in the miRNA regulatory network, suggesting that these genes could be of functional importance for the metastatic processes.
Figure 3.
MicroRNA regulatory network based on the differentially expressed microRNA and predicted targets. Red nodes represent the upregulated microRNAs and green nodes represent the downregulated microRNAs, whereas the blue nodes represent microRNA target genes.
Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses of predicted microRNA targets
To get a more comprehensive understanding of the biological roles of these differentially expressed miRNAs in melanoma, we also performed the GO categories enrichment analysis for the miRNA target genes by using the Gorilla online software. GO category provides a descriptive framework of functional annotation for gene set analysis. With the FDR <0.05, we found GO terms for biological processes significantly enriched in nuclear division, cell division, mitotic cell cycle, etc. [Figure 4]. Whereas for molecular functions, the enriched GO terms mainly included hydrolase activity, transmembrane transporter activity, and protein kinase binding [Supplementary Figure 1 (1MB, tif) ]. In addition, we also performed the KEGG pathway enrichment analysis to further evaluate the biological significance for the differentially expressed genes. The top-enriched biological pathways mainly included pathways in cancer, MAPK signaling pathway, cell junction, focal adhesion, and cell adhesion [Table 3].
Figure 4.

Enriched gene ontology terms (biological process) of predicted targets of differentially expressed microRNAs in metastatic samples.
Table 3.
Enriched KEGG pathways of differentially expressed genes
| Gene set name | Number of genes in overlap (k) | P | FDR Q-value |
|---|---|---|---|
| Pathways in cancer | 23 | ||
| Endocytosis | 14 | 1.48E-08 | 2.75E-06 |
| MAPK signaling pathway | 6 | 3.52E-06 | 3.27E-04 |
| Tight junction | 11 | 8.10E-06 | 5.02E-04 |
| Gap junction | 9 | 2.00E-05 | 7.75E-04 |
| Melanoma | 8 | 2.36E-05 | 7.75E-04 |
| Glycosaminoglycan biosynthesis - chondroitin sulfate | 5 | 2.81E-05 | 7.75E-04 |
| Nonsmall cell lung cancer | 7 | 2.92E-05 | 7.75E-04 |
| Focal adhesion | 13 | 3.56E-05 | 8.27E-04 |
| Glycosaminoglycan biosynthesis - heparan sulfate | 5 | 4.60E-05 | 9.52E-04 |
| Acute myeloid leukemia | 7 | 6.89E-05 | 1.20E-03 |
| Regulation of actin cytoskeleton | 13 | 7.10E-05 | 1.20E-03 |
| Glioma | 7 | 9.58E-05 | 1.48E-03 |
| Cell adhesion | 7 | 1.19E-04 | 1.70E-03 |
| Endometrial cancer | 6 | 2.48E-04 | 3.00E-03 |
| Calcium signaling pathway | 11 | 2.49E-04 | 3.00E-03 |
| GnRH signaling pathway | 8 | 2.58E-04 | 3.00E-03 |
| Axon guidance | 9 | 3.41E-04 | 3.73E-03 |
| Small cell lung cancer | 7 | 3.82E-04 | 3.94E-03 |
| Insulin signaling pathway | 9 | 5.86E-04 | 5.51E-03 |
E−n = 10−n (n>0). FDR: False discovery rate; KEGG: Kyoto Encyclopedia of Genes and Genomes; MAPK: Mitogen-activated protein kinase.
Enriched gene ontology terms (molecular functions) of differentially expressed microRNAs in metastatic samples.
Discussion
Metastasis is the major cause of the poor outcome of melanoma. To date, the cause of melanoma metastasis is still far from understood. A better understanding of the pathogenesis of metastasis could be of paramount importance for drug development and treatment of this disease. In recent years, gene expression profiling analysis has been shown to be a useful tool to investigate the pathophysiology of complex genetic tracts, such as cancer. A large number of miRNAs involved in many critical cellular pathways of cancer progression have also been identified to have an aberrant expression in melanoma.
Until now, studies focusing on the miRNA expression profiling analysis on the metastasis of melanoma are of comparatively limited value. In this study, we collected those miRNA expression profiling datasets of melanoma and performed a systemic meta-analysis to retrieve metastasis-associated miRNAs. A total of 63 differentially expressed miRNAs could successfully classify the different types of samples. Many of these miRNAs identified are known to be involved in a variety of cancer types. Specifically, miR-23 has been identified as upregulated in many cancer types that represent putative oncogene,[26,27] whereas miR-625 could function as a tumor suppressor that cause downregulation in esophageal squamous cell carcinoma[28] and colorectal cancers.[29] In addition, many predicted targets of the differentially expressed miRNA have also been suggested to have an association with cancer development. For instance, KSR2 was essential to tumor cell energy homeostasis and was critical to the integration of mitogenic and metabolic signaling pathways.[30]
The followed functional implication analysis indicated several physiological impact pathways for the metastasis of these diseases. We constructed the miRNA regulatory network based on the differentially expressed miRNA. Moreover, we utilized GO and KEGG pathway enrichment analyses to further interpret their biological functions. This systematic meta-analysis of gene profiling took a step in investigating the mechanisms underlying the metastasis of melanoma. These findings have dramatically expanded our knowledge of pathophysiology of this disease.
In conclusion, by collecting the miRNA expression datasets from different platforms, multiple biological markers were identified for the metastasis of melanoma. This work was important to characterize the specific roles of those genes involved in the pathogenesis of melanoma. Functional analysis of these genes may provide additional insights into the complex process of these diseases. In addition, this analysis may help improve the diagnosis and treatment of this disease.
Supplementary information is linked to the online version of the paper on the Chinese Medical Journal website.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Xin Chen
References
- 1.McComiskey M, Iavazzo C, Datta M, Slade R, Winter-Roach B, Lambe G, et al. Balloon cell urethral melanoma: Differential diagnosis and management. Case Rep Obstet Gynecol. 2015;2015:919584. doi: 10.1155/2015/919584. doi: 10.1155/2015/919584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakkal FK, Basman A, Kizil Y, Ekinci Ö, Gümüsok M, Ekrem Zorlu M, et al. Mucosal melanoma of the head and neck: Recurrence characteristics and survival outcomes. Oral Surg Oral Med Oral Pathol Oral Radiol. 2015;120:575–80. doi: 10.1016/j.oooo.2015.06.038. doi: 10.1016/j.oooo2015.06.038. [DOI] [PubMed] [Google Scholar]
- 3.Kanavy HE, Gerstenblith MR. Ultraviolet radiation and melanoma. Semin Cutan Med Surg. 2011;30:222–8. doi: 10.1016/j.sder.2011.08.003. doi: 10.1016/j.sder2011.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Azoury SC, Lange JR. Epidemiology, risk factors, prevention, and early detection of melanoma. Surg Clin North Am. 2014;94:945–62. doi: 10.1016/j.suc.2014.07.013. vii. doi: 10.1016/j.suc.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 5.Shaikh WR, Dusza SW, Weinstock MA, Oliveria SA, Geller AC, Halpern AC. Melanoma thickness and survival trends in the United States 1989 to 2009. J Natl Cancer Inst. 2015:108. doi: 10.1093/jnci/djv294. pii: Djv294. doi: 10.1093/jnci/djv294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jayawardana K, Schramm SJ, Tembe V, Mueller S, Thompson JF, Scolyer RA, et al. Identification, review, and systematic cross-validation of microRNA prognostic signatures in metastatic melanoma. J Invest Dermatol. 2016;136:245–54. doi: 10.1038/JID.2015.355. doi: 10.1038/JID.2015.355. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. doi: 10.1016/S0092-8674(04) 00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci U S A. 2003;100:9779–84. doi: 10.1073/pnas.1630797100. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozomara A, Griffiths-Jones S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–7. doi: 10.1093/nar/gkq1027. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Feilotter HE, Paré GC, Zhang X, Pemberton JG, Garady C, et al. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J Pathol. 2010;176:2520–9. doi: 10.2353/ajpath.2010.091061. doi: 10.2353/ajpath.2010.091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caramuta S, Egyházi S, Rodolfo M, Witten D, Hansson J, Larsson C, et al. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol. 2010;130:2062–70. doi: 10.1038/jid.2010.63. doi: 10.1038/jid.2010.63. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Zhang X, Lentz C, Abi-Daoud M, Paré GC, Yang X, et al. miR-193b Regulates Mcl-1 in melanoma. Am J Pathol. 2011;179:2162–8. doi: 10.1016/j.ajpath.2011.07.010. doi: 10.1016/j.ajpath.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sand M, Skrygan M, Sand D, Georgas D, Gambichler T, Hahn SA, et al. Comparative microarray analysis of microRNA expression profiles in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases, and benign melanocytic nevi. Cell Tissue Res. 2013;351:85–98. doi: 10.1007/s00441-012-1514-5. doi: 10.1007/s00441-012-1514-5. [DOI] [PubMed] [Google Scholar]
- 15.Xu Y, Brenn T, Brown ER, Doherty V, Melton DW. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer. 2012;106:553–61. doi: 10.1038/bjc.2011.568. doi: 10.1038/bjc.2011.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babapoor S, Fleming E, Wu R, Dadras SS. A novel miR-451a isomiR, associated with amelanotypic phenotype, acts as a tumor suppressor in melanoma by retarding cell migration and invasion. PLoS One. 2014;9:e107502. doi: 10.1371/journal.pone.0107502. doi: 10.1371/journal.pone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhattacharya A, Schmitz U, Raatz Y, Schönherr M, Kottek T, Schauer M, et al. miR-638 promotes melanoma metastasis and protects melanoma cells from apoptosis and autophagy. Oncotarget. 2015;6:2966–80. doi: 10.18632/oncotarget.3070. doi: 10.18632/oncotarget.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–5. doi: 10.1093/bioinformatics/17.6.520. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 19.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–84. doi: 10.1038/ng2135. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 23.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 24.Mirzaei H, Gholamin S, Shahidsales S, Sahebkar A, Jaafari MR, Mirzaei HR, et al. MicroRNAs as potential diagnostic and prognostic biomarkers in melanoma. Eur J Cancer. 2016;53:25–32. doi: 10.1016/j.ejca.2015.10.009. doi: 10.1016/j.ejca.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 25.Leidinger P, Keller A, Borries A, Reichrath J, Rass K, Jager SU, et al. High-throughput miRNA profiling of human melanoma blood samples. BMC Cancer. 2010;10:262. doi: 10.1186/1471-2407-10-262. doi: 10.1186/1471-2407-10-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azizmohammadi S, Azizmohammadi S, Safari A, Kosari N, Kaghazian M, Yahaghi E, et al. The role and expression of miR-100 and miR-203 profile as prognostic markers in epithelial ovarian cancer. Am J Transl Res. 2016;8:2403–10. [PMC free article] [PubMed] [Google Scholar]
- 27.Guzel E, Karatas OF, Semercioz A, Ekici S, Aykan S, Yentur S, et al. Identification of microRNAs differentially expressed in prostatic secretions of patients with prostate cancer. Int J Cancer. 2015;136:875–9. doi: 10.1002/ijc.29054. doi: 10.1002/ijc.29054. [DOI] [PubMed] [Google Scholar]
- 28.Li C, Li DC, Che SS, Ma K, Wang YJ, Xia LH, et al. The decreased expression of miR-625 predicts poor prognosis of esophageal squamous cell carcinoma. Int J Clin Exp Med. 2015;8:9560–4. [PMC free article] [PubMed] [Google Scholar]
- 29.Lou X, Qi X, Zhang Y, Long H, Yang J. Decreased expression of microRNA-625 is associated with tumor metastasis and poor prognosis in patients with colorectal cancer. J Surg Oncol. 2013;108:230–5. doi: 10.1002/jso.23380. doi: 10.1002/jso.23380. [DOI] [PubMed] [Google Scholar]
- 30.Fernandez MR, Henry MD, Lewis RE. Kinase suppressor of Ras 2 (KSR2) regulates tumor cell transformation via AMPK. Mol Cell Biol. 2012;32:3718–31. doi: 10.1128/MCB.06754-11. doi: 10.1128/MCB.06754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Enriched gene ontology terms (molecular functions) of differentially expressed microRNAs in metastatic samples.