Summary
Haem-regulated eIF2α kinase (HRI) is essential for the regulation of globin gene translation and the survival of erythroid precursors in iron/haem deficiency. This study found that that in iron deficiency, fetal definitive erythropoiesis is inhibited at the basophilic erythroblast stage with increased proliferation and elevated apoptosis. This hallmark of ineffective erythropoiesis is more severe in HRI deficiency. Microarray gene profiling analysis showed that HRI was required for adaptive gene expression in erythroid precursors during chronic iron deficiency. The number of genes with expression affected more than twofold increased, from 213 in iron deficiency and 73 in HRI deficiency, to 3135 in combined iron and HRI deficiencies. Many of these genes are regulated by Gata1 and Fog1. We demonstrate for the first time that Gata1 expression in developing erythroid precursors is decreased in iron deficiency, and is decreased further in combined iron and HRI deficiencies. Additionally, Fog1 expression is decreased in combined deficiencies, but not in iron or HRI deficiency alone. Our results indicate that HRI confers adaptive gene expression in developing erythroblasts during iron deficiency through maintaining Gata1/Fog1 expression.
Keywords: iron and haem, erythroid differentiation, eIF2α kinase, Gata1 and Fog1, gene expression
Haem-regulated eIF2α kinase (HRI, also known as eIF2α kinase 1, gene symbol Eif2ak1) is highly expressed in the erythroid lineage. The kinase activity of HRI is regulated by haem through its two haem-binding domains located in the N-terminus and the kinase insertion (Chen, 2007). In haem deficiency, haem dissociates from the binding site in the kinase insertion and HRI is activated by autophosphorylation. Subsequently, the α-subunit of eukaryotic translation initiation factor 2 (eIF2) is phosphorylated and the recycling of eIF2 for another round of protein synthesis is inhibited. Thus, HRI normally insures that no globin is synthesized in excess of what can be assembled into haemoglobin for the amount of haem available (Chen, 2007).
Iron-deficiency anaemia is one of the most prevalent human diseases (Stoltzfus, 2003), which has long been known to impair physical and mental function. We have demonstrated that HRI is an important physiological regulator of protein synthesis and cell survival of the erythroid lineage during iron deficiency. In the absence of HRI and under iron deficiency, free globins precipitate within the red blood cells (RBCs) and its precursors. Further, in iron deficiency Eif2ak1−/− erythroid precursors have increased apoptosis and Eif2ak1−/− mice exhibit ineffective erythropoieis with splenomegaly (Han et al, 2001). We have shown recently that HRI also reduces the phenotypic severity of β-thalassaemia in mice (Han et al, 2005). While anaemia per se is well tolerated in mice and humans, ineffective erythropoiesis is the cause of major complications in β-thalassaemia (Rund & Rachmilewitz, 2005) because of increased iron absorption (Andrews & Schmidt, 2007) and iron overload in multiple organs.
Gata1 is a key transcriptional regulator of erythropoiesis (Crispino, 2005; Ferreira et al, 2005). Gata1 works in concert with its cofactor, Friend of GATA1 (Fog1, also known as zinc finger protein, multitype 1, gene symbol Zfpm1) (Tsang et al, 1997). Both Gata1 and Fog1 are expressed mainly in haematopoietic cells, and are essential for the production of RBCs and megakaryocytes (Crispino, 2005; Ferreira et al, 2005). Gata1−/− and Zfpm1−/− embryos die from severe anaemia between E10·5 and E11·5 of gestation (Fujiwara et al, 1996; Tsang et al, 1998). Although there are primitive erythroid cells in the Gata1−/− embryonic blood, these cells are arrested in their maturation at the proerythroblast stage. A similar block in erythroid maturation was also observed in Zfpm1−/− embryos (Tsang et al, 1998). In addition, Gata1−/− embryonic stem cells fail to yield mature definitive erythroid cells (Pevny et al, 1991; Weiss et al, 1994). Recently, the study of the conditional knockout of Gata1 in adult mice revealed that Gata1 is essential for both steady-state and stress erythropoiesis (Gutierrez et al, 2008).
This study investigated the mechanisms involved in the ineffective erythropoiesis of chronic iron deficiency and the role of HRI in this process. We report here for the first time the inhibition of erythroid differentiation at basophilic erythroblast stage in iron deficiency. In addition, HRI was found to be necessary for adaptive gene expression in erythroid precursors in iron deficiency. Gata1 expression in developing fetal liver (FL) erythroblasts was shown to be decreased in iron deficiency and further decreased in combination with HRI deficiency. Moreover, Zfpm1 expression is decreased in combined iron and HRI deficiencies. Decreased Gata1 and Zfpm1 expression is most likely to be responsible for a substantial change of gene expression in Eif2ak1−/− erythroblasts in iron deficiency.
Materials and methods
Mouse breeding and sample collection
Mice production and experimentation were approved by the Committee on Animal Care at MIT. Eif2ak1−/− mice, induction of iron deficiency and staining of blood smears were as described previously (Han et al, 2001). Blood samples and FLs were collected from E14·5 embryos.
Erythroid differentiation, cell cycle and apoptosis analyses
Erythroid differentiation of FL cells was examined by fluorescent-activated cell sorting (FACS) analyses using phycoerythrin (PE)-conjugated anti-Ter119 and fluorescein isothiocyanate (FITC)-conjugated anti-CD71 antibodies (BD Biosciences, San Jose, CA, USA) according to Zhang et al (2003). Sorting of FL cells at different stages of differentiation was performed using a BD FACS ARIA machine. Cell cycle status was examined by FACS analysis using propidium iodide (Chan et al, 2006). Apoptosis was analyzed using FITC-conjugated Annexin V as described previously (Han et al, 2001).
Western blot and quantitative reverse transcription polymerase chain reaction analyses
Protein extracts were prepared as described previously (Han et al, 2005) except with the supplement of 0·1% sodium dodecyl sulphate (SDS) and sonication. Western blot analyses of protein extracts (20 μg) were performed using antibodies against eIF2α, phosphorylated eIF2α (BioSource, Camarillo, CA, USA), Gata1 and Fog1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Total RNA isolation and quantitative reverse transcription polymerase chain reaction (RT-PCR) were performed as described (Liu et al, 2007). Primers for quantitative RT-PCR (qRT-PCR) were: Gata1 forward primer 5′-GGCAAGACG-GCACTCTACC-3′, reverse primer 5′-CAAGAACGTGTTGTTGCTCTTC-3′; Zfpm1 forward primer 5′-CTGAAGAAG-CCGCCAACTCA-3′, and reverse primer 5′-AAGGCGCACATATAGCAGTCC-3′, eIF2a primers were as described (Liu et al, 2007).
Gene profiling and microarray analysis
Affymetrix mouse expression array 430 A and B chips were used for gene profiling. Total RNAs from E14·5 FLs of iron-sufficient and deficient embryos of wild type (Wt) and Eif2ak1−/− (Ko) (Wt+Fe, Wt-Fe, Ko+Fe and Ko-Fe) were used to prepare cRNA probes according to the Affymetrix protocol. Three replicates of gene chip microarray were performed for each of the four conditions. Raw data were normalized by quantile-based robust multichip average analysis (Jain et al, 2003). A local pooled error test (Jain et al, 2003) was used to identify genes that were expressed with statistical significance among the four groups. P-values were adjusted to reflect the false discovery rate (FDR) using the Benjamini-Hochberg method (Benjamini et al, 2001). Differentially expressed genes were defined by changes of gene expression at either twofold greater or less (with a FDR <0·1) in Wt-Fe, Ko+Fe or Ko-Fe when compared to Wt+Fe. Functional classification of differentially expressed genes and prediction of protein interactions were performed using the Ingenuity Knowledge Base Pathway Analysis Solution software (Ingenuity Systems, Inc. Redwood City, CA, USA). Gene expression data is available at NCBI GEO database, accession no. GSE6808.
Statistical analysis
Two-tailed Student's t-test was used to analyze the statistical significance of FACS, cell cycle and qRT-PCR data; a P-value of <0·05 determined statistical significance.
Results
Impairment of erythroid differentiation in iron and HRI deficiencies
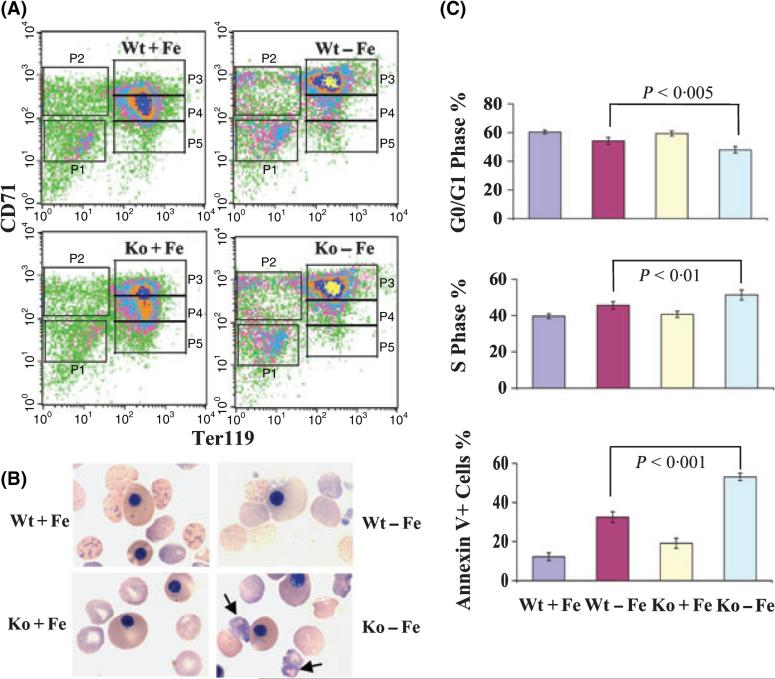
During embryonic development, FL is the main erythropoietic organ for definitive erythropoieis. Erythroid precursors are the predominant cells in the E14·5 FL, which is widely used as a model for studying definitive erythropoiesis. To evaluate the effects of deficiencies of iron and HRI on erythroid differentiation, FACS analyses of E14·5 FL cells from iron sufficient and deficient Eif2ak1+/+ and Eif2ak1−/− embryos (Wt+Fe, Wt-Fe, Ko+Fe and Ko-Fe) were carried out. As shown in Fig 1A, FL cells were divided into five populations based on their expression of CD71 and Ter119. P1, the CD71low TER119low population, contained mainly primitive progenitors. P2, the CD71highTER119low population, was mostly proerythroblasts. P3, the CD71highTER119high population, was mainly composed of basophilic erythroblasts. P4, the CD71medTER119high population, was predominantly chromatophilic and orthochromatophilic erythroblasts and P5, the CD71lowTER119high, comprised late orthochromatophilic erythroblasts and reticulocytes. Most of Wt+Fe FL cells were present at the P4 stage (48·32%, Fig 1 and Table I). Under iron deficiency, however, most FL cells from both Wt-Fe (57·37%) and Ko-Fe (55·65%) were present at the less differentiated P3 stage. These results showed that erythroid differentiation of Wt-Fe and Ko-Fe FL cells was inhibited at the basophilic erythroblast stage. In addition, Ko+Fe FL cells also had a significant increase in percentage of cells in P3 when compared to Wt+Fe (31·72% vs. 20·52%, P = 0·019), albeit to a lesser degree than in iron deficiency.
Fig 1.
Erythroid differentiation of FL in iron deficiency and HRI deficiency. (A) FACS analysis of the erythroid differentiation of E14·5 FL cells. (B) Blood smears of E14·5 embryos. Arrows indicate globin inclusions in Ko-Fe reticulocytes. (C) Cell cycle and apoptosis in E14·5 FL cells. Results are presented as mean ± SD (n = 6).
Table I.
Distribution of Wt and Ko fetal liver cells at different stages of erythroid differentiation under iron-sufficient or iron-deficient conditions.
| P1 (%) | P2 (%) | P3 (%) | P4 (%) | P5 (%) | |
|---|---|---|---|---|---|
| Wt+Fe | 8·95 ± 2·37 | 5·47 ± 1·98 | 20·52 ± 6·74 | 48·32 ± 7·52 | 6·50 ± 2·63 |
| Wt-Fe | 10·47 ± 1·05 | 8·53 ± 2·24* | 57·37 ± 3·20* | 9·96 ± 3·32* | 2·67 ± 0·73* |
| Ko+Fe | 10·27 ± 2·09 | 4·60 ± 1·60 | 31·72 ± 5·19* | 36·80 ± 6·15* | 6·37 ± 1·31 |
| Ko-Fe | 11·46 ± 1·21 | 12·28 ± 2·80*† | 55·65 ± 6·23* | 6·61 ± 1·08*† | 1·79 ± 0·92*† |
Results are presented in mean ± SD (n = 5–6).
P < 0·05, compared with Wt+Fe.
P < 0·05, compared with Wt-Fe.
The blood smears of E14·5 embryos were examined for the effects of HRI and iron deficiencies on embryonic red blood cell (RBC) development. The anaemia was more severe in Ko-Fe embryos with many globin inclusions in definitive embryonic RBCs and reticulocytes. No inclusion was observed in Wt+Fe, Ko+Fe or Wt-Fe embryonic blood cells (Fig 1B). These results underscore the role of HRI in inhibiting globin translation during embryonic red cell development in iron deficiency as seen in the adult mice (Han et al, 2001). Interestingly, the primitive nucleated RBCs were not affected in Ko-Fe embryos; there were no visible globin inclusions or morphological changes (Fig 1B). These results suggested that there was sufficient iron for primitive erythropoiesis under the iron-deficient conditions implemented.
Increased proliferation and accelerated apoptosis in Ko-Fe erythroid cells
To investigate the mechanism of impaired erythropoiesis in HRI and iron deficiencies, cell cycle and apoptosis analyses were performed. Cell cycle status was not affected by HRI deficiency alone (Fig 1C). However, the percentage of cells G0/G1 phase decreased significantly in Wt-Fe (54·17%, P = 0·001) and was further decreased in Ko-Fe (47·91%, P < 0·001) FL cells compared to that in Wt+Fe (60·36%). Concomitantly, there were increase in the percentage of cells in S phase in Wt-Fe (45·56%, P = 0·001) and in Ko-Fe (51·34%, P < 0·001) as compared to Wt+Fe (39·62%) (Fig 1C). These results showed the increased cell cycling in Wt-Fe and Ko-Fe FL cells. There was also an increase in the percentage of Annexin V+ apoptotic cells in the Wt-Fe FL cells (32·51%, P = 0·001), and further increase in Ko-Fe FL cells (53·06%, P < 0·001) compared with that in Wt+Fe cells (12·23%) (Fig 1C). The increase in apoptosis during iron deficiency was accompanied with a significantly decreased expression of antiapoptotic Akt1 (3·25-fold for Wt-Fe and 10·27-fold for Ko-Fe) and Bcl2l1 (2·11-fold for Wt-Fe and 4·99-fold for Ko-Fe) as seen in microarray analyses (data available at NCBI GEO database, no. GSE6808). Together, these results demonstrate that FLs develop ineffective erythropoiesis of increased proliferation and enhanced apoptosis during chronic iron deficiency, and that this ineffective erythropoiesis is more severe in HRI deficiency.
Alternation of gene expression in iron and HRI deficiencies
To gain insights into the protective role of HRI in the ineffective erythropoiesis of iron deficiency, gene profiling was carried out using RNAs from E14·5 FLs of Wt+Fe, Wt-Fe, Ko+Fe and Ko-Fe embryos as probes. The number of differentially expressed genes in Ko+Fe (73) was much smaller than that in Wt-Fe or Ko-Fe, consistent with the rather mild phenotypes in Ko+Fe embryos (Fig 1) and mice (Han et al, 2001). Expression of 213 genes was significantly altered in Wt-Fe. Strikingly, the number of differentially expressed genes increased 15-fold to 3135 in Ko-Fe (data available at NCBI GEO database, no. GSE6808). It is important to note that there was no significant difference between Wt-Fe and Ko-Fe FL cells in cell populations along erythroid differentiation based on CD71 and Ter119 expression (Fig 1 and Table I). Therefore, the difference in gene expression between Wt-Fe and Ko-Fe FL is intrinsic to HRI deficiency in these cells.
Many of differentially expressed genes in Ko-Fe FL were involved in cell growth and survival, protein synthesis and cellular assembly and organization, as would be expected from the increased proliferation and enhanced apoptosis in Ko-Fe erythroid precursors described above (Fig 1). Importantly, 5% of the differentially expressed genes are involved in hematological diseases, such as Akt1, Bcl2l1, Casp8, Epor, Jak2, Myc, Pten, Sod2 etc. Expression of 6 and 34 known erythroid genes were altered significantly in Wt-Fe and Ko-Fe FLs, respectively. Many genes that are upregulated during erythroid differentiation, such as Zfpm1, Abcb10, Epor, Jak2, Eklf, Gypa, Epb 4·1, 4·2 and Stom, Trim10 and Tspan33, were downregulated three- to fourfold in Ko-Fe, and were known to be regulated by Gata1/Fog1 (Table SI). These results strongly support the hypothesis that, in iron deficiency, HRI is necessary to sustain terminal erythroid differentiation by maintaining the expression of many erythroid genes required for differentiation. Furthermore, these results also suggest an impairment of Gata1- and Fog1-induced erythropoiesis in Ko-Fe FL.
Diminished Gata1 and Zfpm1 expression in Ko-Fe erythroblasts
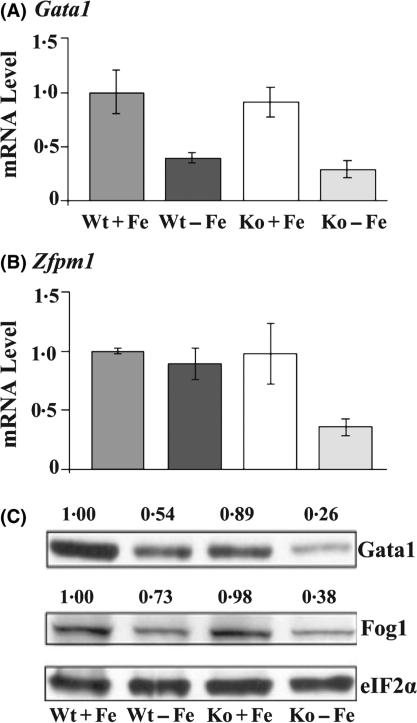
Ingenuity Pathway Analysis Solution software was used to help identifying the potential network of these altered gene expression. Decreased Gata1 and Zfpm1 expression was predicted to be linked to the changes of gene expression observed in Wt-Fe and Ko-Fe FL cells. In addition, our manual analysis of gene profiling results also indicated that Gata1/Fog1 activity might be compromised, particularly in Ko-Fe FLs, as many of the affected genes were regulated by Gata1/Fog1. We therefore examined the expression of Gata1 and Zfpm1. Although the microarray analysis did not reveal significant changes of Gata1 mRNA, qRT-PCR analysis showed a significant decrease of Gata1 mRNA in Wt-Fe and Ko-Fe FL cells (Fig 2A, P < 0·05). The difference in the expression changes of Gata1 between genechip and qRT-PCR data is probably because the genechip data does not fully reflect the gene changes, especially for those genes with only one probe-set, such as Gata1. Consistent with microarray data, Zfpm1 mRNA expression was significantly decreased only in Ko-Fe (Fig 2B, P < 0·05). In addition, both Gata1 and Fog1 proteins were expressed significantly lower in Ko-Fe as compared to Wt-Fe (Fig 2C, P < 0·05 n = 3).
Fig 2.
Expression of Gata1 and Zfpm1 in FL under iron and HRI deficiencies. (A) Expression of Gata1 mRNA. (B) Expression of Zfpm1 mRNA. (C) Expression of Gata1 and Zfpm1 proteins. The results of mRNA expression are presented in mean ± SD (n = 4). The intensities of the autoradiograms in the Western blots were quantified by Alpha EaseFC software. Expression of proteins was normalized with eIF2α. Expression of proteins in Wt+Fe is defined as 1. The normalized values are shown above the autoradiograms.
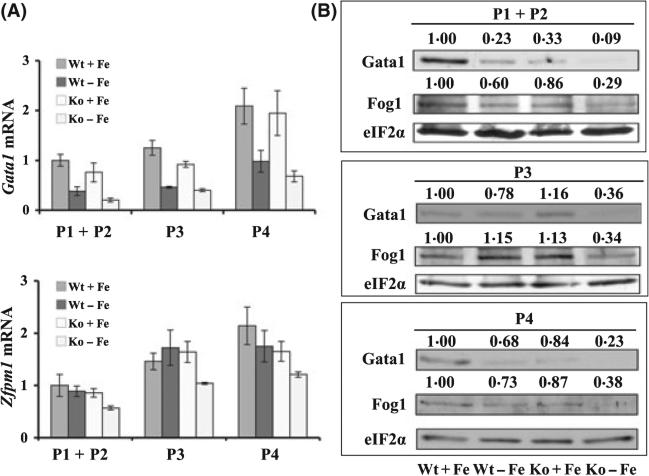
To determine whether the decreased expression of Gata1 and Zfpm1 in iron and HRI deficiencies occurred at specific stages of erythroid differentiation, FL cells were sorted into three populations, Ter119low (P1 + P2), Ter119highCD71high (P3) and Ter119highCD71med (P4). Expression of Gata1 and Fog1 in these three populations was shown in Fig 3. Gata1 mRNA was decreased in all three populations during iron deficiency. In addition, Gata1 mRNA was further decreased upon combined iron and HRI deficiencies in all three populations. Similarly, Gata1 protein was also decreased in iron deficiency in all three populations and further decreased upon HRI deficiency. However, Zfpm1 mRNA and protein were significantly decreased in all three populations only in Ko-Fe (Fig 3). We also found that Gata1 and Zfpm1 expression remained high during normal FL erythroblast maturation (Fig 3A). This result is consistent with reports that Gata1 expression is necessary throughout erythroid maturation (Zheng et al, 2006; Marinkovic et al, 2007). Together, these results demonstrated that the decrease of Gata1 and Zfpm1 expression in combined iron and HRI deficiencies occurred throughout erythroid differentiation.
Fig 3.
Expression of Gata1 and Zfpm1 in sorted cell populations of FL under iron and HRI deficiencies. (A) Expression of Gata1 and Zfpm1 mRNAs in three sorted populations. Results are presented in mean ± SD (n = 4). (B) Expression of GATA1 and Zfpm1 proteins in sorted cells. Quantification of Western blots was as described in the legend of Fig 2.
Upregulation of Eif2ak1 during erythroid differentiation
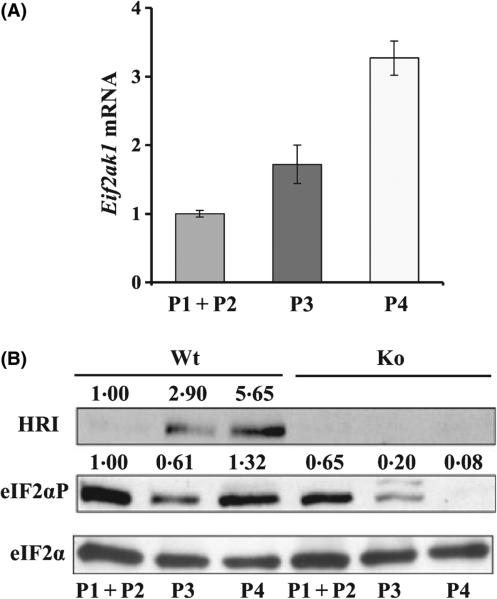
To further understand the relationship of HRI and Gata1/Zfpm1 expression, we examined the expression of Eif2ak1 during erythroid differentiation in the three populations of FL cells described above. Eif2ak1 expression, both mRNA and protein, were increased during erythroid differentiation with higher expression in Ter119high cells (Fig 4). This higher expression of Eif2ak1 in late erythroblasts, in which globin proteins are actively made in large quantities, is consistent with its known function in regulating globin translation. The contribution of HRI to eIF2α phosphorylation among the family of eIF2α kinases was also increased during erythroid maturation, from 35% in Ter119− cells (P1 + P2) to 67·2% Ter119highCD71high cells (P3) and to 93·9% in Ter119highCD71med cells (P4) (Fig 4B, comparing Eif2ak1+/+ to Eif2ak1−/− cells). It is important to note that the temporal expression of Gata1 and Zfpm1 (Fig 3A) is similar to that of Eif2ak1 (Fig 3), consistent with the possibility of the regulation of Gata1 and Zfpm1 expression by HRI activated in iron deficiency.
Fig 4.
Upregulation of Eif2ak1 during erythroid differentiation. (A) Expression of Eif2ak1 mRNA in three sorted populations. (B) Western blot analyses of HRI, eIF2αP, and eIF2α. Expression of proteins in P1 + P2 of Wt is defined as 1. Quantification of Western blots was as described in the legend of Fig 2.
Comparison of gene profiling of Ko-Fe with G1E-Er4 cells
To evaluate the significance of reduced expression of Gata1/Zfpm1 on gene expression in FL cells, the gene profiling data of Ko-Fe erythroid precursors were compared with that of G1E-Er4 cells (Welch et al, 2004). G1E-Er4 cells were derived from Gata1−/− ES cells and expressed Gata1-ER fusion protein, which is activated upon induction of oestradiol. G1E-Er4 cells proliferate as immature erythroblasts, but undergo terminal differentiation upon restoration of Gata1 function (Weiss et al, 1997). At 30 h after Gata1 activation, most of G1E-Er4 cells resembled basophilic erythroblasts expressing haemoglobin (Weiss et al, 1997). Given that Gata1 expression was decreased in Ko-Fe cells, we would expect that a number of differentially expressed genes were in common but regulated in opposite directions between Ko-Fe and Gata1-activated G1E-Er4 cells. The number of these genes increased from 15 at 3 h to 120 at 30 h after the induction of G1E-Er4 cells by oestradiol, with a total of 133 genes throughout the 30 h of Gata1 activation. The majority of theses genes (79·7%) were upregulated by Gata1 in G1E-Er4 cells and downregulated in Ko-Fe. Some genes of interest relating to erythroid maturation are summarized in Table II.
Table II.
Comparison of gene expression between G1ER and Ko-Fe FL cells.
| Gene symbol | Gene name | 3h | 7h | 14 h | 21 h | 30 h | Ko-Fe |
|---|---|---|---|---|---|---|---|
| Erythroid genes | |||||||
| Add1 | Adducin 1 | ↑ | ↑ ↑ | ↑ ↑ ↑ | ↑ ↑ ↑ | ↑ ↑ ↑ ↑ | ↓ ↓ ↓ |
| Eif2ak1 | Haem-regulated eIF2α kinase | – | ↑ | ↑ ↑ | ↑ ↑ ↑ | ↑ ↑ ↑ | N/A |
| Gypa | Glycophorin A | – | ↑ | ↑ ↑ ↑ | ↑ ↑ ↑ | ↑ ↑ ↑ | ↓ ↓ |
| Stom (Epb7·2) | Erythrocyte protein band 7·2 | – | ↑ | ↑ | ↑ | ↑ | ↓ |
| Zfpm1 | Zinc finger protein, multitype 1 | – | ↑ | ↑ | ↑ | ↑ ↑ | ↓ |
| Klf1 | Kruppel-like factor 1 (erythroid) | – | – | ↑ | ↑ | ↑ | ↓ |
| Pklr | Pyruvate kinase liver and red blood cell | – | – | ↑ | ↑ | – | ↓ |
| Bcl2l1 | Bcl2-like (Bcl2l1) 1 | – | – | – | ↑ | ↑ ↑ | ↓ ↓ |
| Aqp1 | Aquaporin 1 | – | – | – | – | ↑ | ↓ ↓ |
| Bpgm | 2,3-bisphosphoglycerate mutase | – | – | – | – | ↑ | ↓ ↓ |
| Iron/haem genes | |||||||
| Trfr2 | Transferrin receptor 2 | – | ↓ | ↓ | ↓ ↓ | ↓ ↓ | ↑ |
| Urod | Uroporphyrinogen decarboxylase | – | – | ↑ | ↑ | ↑ | ↓ |
| Fech | Ferrochelatase | – | – | – | – | ↑ | ↓ ↓ |
| B2m | β-2 microglobulin | – | – | – | – | ↑ | ↓ ↓ ↓ |
| Proliferation, apoptosis and survival-related genes | |||||||
| Pim1 | Proviral integration site 1 | ↓ | – | ↑ | ↑ | ↑ | ↓ ↓ |
| Btg2 | B-cell translocation gene 2, anti-proliferative | – | ↑ | ↑ ↑ | ↑ ↑ | ↑ ↑ ↑ | ↓ ↓ ↓ |
| Cdkn2c | Cyclin-dependent kinase inhibitor 2C | – | – | ↑ | ↑ | ↑ | ↓ |
| Cdkn1a | Cyclin-dependent kinase inhibitor 1A | – | – | – | – | ↑ | ↓ |
| Ptdss2 | Phosphatidylserine synthase 2 | ↑ | ↑ ↑ ↑ | ↑ ↑ ↑ ↑ | ↑ ↑ ↑ ↑ | ↑ ↑ ↑ ↑ | ↓ ↓ |
| Antioxidant genes | |||||||
| Selenbp1 | Selenium binding protein 1 | – | ↑ | ↑ | ↑ | ↑ ↑ | ↓ |
| Ddt | D-dopachrome tautomerase | – | – | ↑ | ↑ | ↑ ↑ ↑ ↑ | ↓ |
| Txnrd2 | Thioredoxin reductase 2 | – | – | ↑ | ↑ ↑ | ↑ | ↓ ↓ |
| Others | |||||||
| Tcf4 | Transcription factor 4 | ↓ ↓ ↓ | ↓ ↓ | – | – | ↓ | ↑ ↑ |
| Slc31a1 | Solute carrier family 31, member 1 | – | – | ↑ | ↑ | ↑ ↑ | ↓ |
Hr 3–30: time course of G1ER cells upon induction of Gata1 in hours.
Fold changes: ↓↓↓↓, −32 to −16; ↓↓↓, −16 to −8; ↓↓, −8 to −4; ↓, −4 to −2; −, −2 to2; ↑, 2 to 4; ↑↑, 4–8; ↑↑↑, 8–16; ↑↑↑↑, 16–32; N/A, not applicable.
Consistent with our results (Fig 4), Eif2ak1 expression was increased during differentiation of G1E-Er4 cells upon activation of Gata1 starting at 7 h [Table II and (Welch et al, 2004)]. This result provides further support that Gata1 activity was diminished in Ko-Fe FL cells and suggests that HRI is required to maintain Gata1 expression and function in these cells during chronic iron deficiency.
Discussion
We have previously shown that HRI is important in protecting erythroid precursors during iron/haem deficiency and in β-thalassaemia (Han et al, 2001, 2005). HRI exerts this protective function in part by inhibiting protein synthesis to reduce globin aggregates. However, it is also established that activation of eIF2α kinases upon stress also increases translation of specific mRNAs to alter gene expression for the adaptation to stress (Holcik & Sonenberg, 2005; Chen, 2007). We report here the alternation of gene expression in developing FL erythroblasts upon iron and HRI deficiencies. We demonstrate that in chronic iron deficiency HRI is also required for adaptive gene expression in the cellular processes of proliferation, apoptosis, erythroid differentiation and iron/haem homeostasis.
This study demonstrated for the first time that definitive erythropoiesis is inhibited at the basophilic erythroblast stage during iron deficiency in both Eif2ak1+/+ and Eif2ak1−/− FLs. However, Eif2ak1−/− embryos exhibit more severe ineffective erythropoiesis and anaemia with reticulocytosis and pronounced globin inclusions in the reticulocytes of embryonic blood. These phenotypic changes in fetal definitive erythropoiesis induced by iron deficiency correlated very well with the gene profiling data of E14·5 FL cells under these conditions. The extent of differentially expressed genes in iron deficiency was increased 15-fold upon HRI deficiency. These results underline the significant role of HRI in erythroid development during iron deficiency. Although Ko+Fe FL cells displayed a mild defect in erythroid differentiation in vivo, these cells showed a significant block of maturation at the basophilic erythroblast stage when cultured in vitro where conditions may not be as optimal as in vivo (R.N.V.S. Suragani and J.-J. Chen unpublished observations). We have also reported earlier that the survival of Eif2ak1−/− mice is severely compromised upon phenylhydrazine induced acute hemolytic anaemia as compared to Wt mice (Han et al, 2001). Together, these results underscore a functional role of HRI in stress erythropoiesis and are consistent with the fact that HRI and other eIF2α kinases are stress-activated kinases (Holcik & Sonenberg, 2005; Chen, 2007).
Our microarray data also demonstrated changes of expression of many of Gata1/Fog1 target genes in Ko-Fe FL. We show for the first time that Gata1 expression is decreased in developing erythroblasts during iron deficiency. Interestingly, Zfpm1 expression is reduced only under combined iron and HRI deficiencies. Consistent with the compromised Gata1/Fog1 functional activity in Ko-Fe erythroblasts, we demonstrated that more than 100 genes were downregulated more than twofold in Ko-Fe FL cells, but were upregulated in G1E-ER4 cells by Gata1 activation.
Both Gata1 and Fog1 are essential for the proliferation, survival and differentiation of the erythroid lineage (Cantor & Orkin, 2005). Previous studies of knockdown mutations of Gata1, Gata1low and Gata1·05/X, have revealed that erythroid maturation is dependent on the level of Gata1 (McDevitt et al, 1997; Takahashi et al, 1998). Moreover, expression of Gata1 at late-stage maturation is essential for the terminal differentiation of primitive and definitive erythroblasts (Zheng et al, 2006). It should be noted that the level of Gata1 in Ko-Fe (26%) FL is similar to that of Gata1low. Therefore, the attenuation of phenotypic severity and Gata1 expression by HRI in iron deficiency reported here provides further support for such a dose-dependent effect of Gata1 on erythroid maturation. In addition, a reduced level of Gata1 is observed in Lyn−/− J2E-NR cells, which fail to differentiate upon exposure to erythropoietin (Tilbrook et al, 1997). Like Gata1low and Ko-Fe mice, Lyn−/− mice (Tilbrook et al, 1997) also display an impairment of erythropoiesis with severe splenomegaly. Thus, reduced Gata1 activity in erythroid precursors may be a general mechanism responsible for ineffective erythropoiesis. Physical interaction of Gata1 and Fog1 is necessary for erythroid maturation (Cantor & Orkin, 2005; Crispino, 2005). Mutations of Gata1 that disrupt its binding to Fog1 have been documented in several families with X-linked congenital anaemia and thrombocytopenia (Cantor & Orkin, 2005; Crispino, 2005). Thus, decreased Zfpm1 expression in HRI deficiency also contributes to ineffective erythropoiesis in iron deficiency.
Cleavage of Gata1 by caspase 3 has been implicated as a negative regulatory mechanism of erythropoiesis (De Maria et al, 1999). Recently, hsp70 has been shown to regulate erythropoiesis by preventing Gata1 cleavage by caspase 3 (Ribeil et al, 2007). The present study found that, in iron deficiency, decreased Gata1 protein expression correlated well with decreased mRNA expression. These results suggest that regulation of Gata1 in FL erythroblasts during chronic iron deficiency probably occurs at the RNA level rather than at the protein stability. While the mechanism of decreased Gata1 and Zfpm1 expression in HRI deficiency remains to be further investigated, it is possible that activation of HRI in iron deficiency may inhibit the translation of some unknown short-lived proteins that are necessary to maintain Gata1 mRNA level. Such a translational repression has recently been documented for Mcl-1 upon stress activation of eIF2α kinases (Fritsch et al, 2007). Little is known about the regulation of Gata1 mRNA besides auto-regulation (Vyas et al, 1999; Nishimura et al, 2000; Yu et al, 2002a). It was reported that the BMP/Smad pathway could direct the onset of Gata1 expression during erythroid differentiation of the embryonic body (Adelman et al, 2002). We recently found that the serum level of BMP2 was decreased threefold in Ko-Fe, but not in Wt-Fe mice (S.J. Liu and J.-J. Chen, unpublished observations). It is possible that the effect of HRI on Gata1 expression in iron deficiency may be indirect through BMPs. These possibilities remains to be investigated further.
The rapid induction of Eif2ak1 expression upon activation of Gata1 in G1E-ER cells (Table II) and the temporal of Eif2ak1 and Gata1 expression during normal erythroid differentiation (Fig 3) also raised the possibility of the potential regulation of Eif2ak1 expression by Gata1, which would confer the high level expression of Eif2ak1 in the erythroid lineage (Chen, 2007). Mutation of Gata1 with either reduced interaction with Fog1 (V205G) (Crispino et al, 1999) or decreased DNA binding (R216N) (Yu et al, 2002b) has been shown to result in reduced expression of Eif2ak1 in G1E-ER cells, consistent with the potential regulation of Eif2ak1 expression by Gata1.
In summary, our study demonstrates the impairment of erythroid differentiation and Gata1 expression in iron deficiency. Furthermore, HRI is required to maintain the Gata1/Fog1 function during stress erythropoiesis in the setting of iron deficiency.
Supplementary Material
Acknowledgements
We would like to thank Dr Manlin Luo, the BioMicro Center MIT, for the expert help in genechip experiments. This work was supported in part by grants from NIH DK16272 (JJC) and grants from Cooley's Anemia Foundation (SJL, APH and RNVS).
Footnotes
Author contribution
SJL and JJC designed the research, analyzed the data and wrote the paper; SJL, APH, RNS and WTZ performed experiments; SB and SJL performed the genechip data analysis; SB and RCF participated in writing the paper.
Conflict-of-interest disclosure
The authors declare no competing financial interests.
Data deposition
Microarray data are available at NCBI GEO data base accession no. GSE6808.
Supplementary material
The following supplementary material is available for this article online:
Table SI. Differentially expressed erythroid and iron/haem homeostasis genes in FL cells during iron and HRI deficiencies.
The material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2141.2008.07293.x
(This link will take you to the article abstract).
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Adelman CA, Chattopadhyay S, Bieker JJ. The BMP/BMPR/Smad pathway directs expression of the erythroid-specific EKLF and GATA1 transcription factors during embryoid body differentiation in serum-free media. Development. 2002;129:539–549. doi: 10.1242/dev.129.2.539. [DOI] [PubMed] [Google Scholar]
- Andrews NC, Schmidt PJ. Iron homeostasis. Annual Review of Physiology. 2007;69:69–85. doi: 10.1146/annurev.physiol.69.031905.164337. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behavioural Brain Research. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH. Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins. Seminars in Cell & Developmental Biology. 2005;16:117–128. doi: 10.1016/j.semcdb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Chan JY, Siu KP, Fung KP. Effect of arsenic trioxide on multidrug resistant hepatocellular carcinoma cells. Cancer Letter. 2006;236:250–258. doi: 10.1016/j.canlet.2005.05.017. [DOI] [PubMed] [Google Scholar]
- Chen JJ. Regulation of protein synthesis by the haem-regulated eIF2alpha kinase: relevance to anemias. Blood. 2007;109:2693–2699. doi: 10.1182/blood-2006-08-041830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino JD. GATA1 in normal and malignant hematopoiesis. Seminars in Cell & Developmental Biology. 2005;16:137–147. doi: 10.1016/j.semcdb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Crispino JD, Lodish MB, MacKay JP, Orkin SH. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Molecular Cell. 1999;3:219–228. doi: 10.1016/s1097-2765(00)80312-3. [DOI] [PubMed] [Google Scholar]
- De Maria R, Zeuner A, Eramo A, Domenichelli C, Bonci D, Grignani F, Srinivasula SM, Alnemri ES, Testa U, Peschle C. Negative regulation of erythropoiesis by caspase-mediated cleavage of GATA-1. Nature. 1999;401:489–493. doi: 10.1038/46809. [DOI] [PubMed] [Google Scholar]
- Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 function, a paradigm for transcription factors in hematopoiesis. Molecular and Cellular Biology. 2005;25:1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch RM, Schneider G, Saur D, Scheibel M, Schmid RM. Translational repression of MCL-1 couples stress-induced eIF2 alpha phosphorylation to mitochondrial apoptosis initiation. Journal of Biological Chemistry. 2007;282:22551–22562. doi: 10.1074/jbc.M702673200. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Tsukamoto S, Suzuki M, Yamamoto-Mukai H, Yamamoto M, Philipsen S, Ohneda K. Ablation of Gata1 in adult mice results in aplastic crisis, revealing its essential role in steady-state and stress erythropoiesis. Blood. 2008;111:4375–4385. doi: 10.1182/blood-2007-09-115121. [DOI] [PubMed] [Google Scholar]
- Han AP, Yu C, Lu L, Fujiwara Y, Browne C, Chin G, Fleming M, Leboulch P, Orkin SH, Chen JJ. Haem-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO Journal. 2001;20:6909–6918. doi: 10.1093/emboj/20.23.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han AP, Fleming MD, Chen JJ. Haem-regulated eIF2alpha kinase modifies the phenotypic severity of murine models of erythropoietic protoporphyria and beta-thalassemia. Journal of Clinical Investigation. 2005;115:1562–1570. doi: 10.1172/JCI24141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nature Reviews. Molecular Cell Biology. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Jain N, Thatte J, Braciale T, Ley K, O'Connell M, Lee JK. Local-pooled-error test for identifying differentially expressed genes with a small number of replicated microarrays. Bioinformatics. 2003;19:1945–1951. doi: 10.1093/bioinformatics/btg264. [DOI] [PubMed] [Google Scholar]
- Liu S, Suragani RN, Wang F, Han A, Zhao W, Andrews NC, Chen JJ. The function of haem-regulated eIF2alpha kinase in murine iron homeostasis and macrophage maturation. Journal of Clinical Investigation. 2007;117:3296–3305. doi: 10.1172/JCI32084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinkovic D, Zhang X, Yalcin S, Luciano JP, Brugnara C, Huber T, Ghaffari S. Foxo3 is required for the regulation of oxidative stress in erythropoiesis. Journal of Clinical Investigation. 2007;117:2133–2144. doi: 10.1172/JCI31807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt MA, Shivdasani RA, Fujiwara Y, Yang H, Orkin SH. A ’knockdown’ mutation created by cis-element gene targeting reveals the dependence of erythroid cell maturation on the level of transcription factor GATA-1. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:6781–6785. doi: 10.1073/pnas.94.13.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S, Takahashi S, Kuroha T, Suwabe N, Nagasawa T, Trainor C, Yamamoto M. A GATA box in the GATA-1 gene hematopoietic enhancer is a critical element in the network of GATA factors and sites that regulate this gene. Molecular and Cellular Biology. 2000;20:713–723. doi: 10.1128/mcb.20.2.713-723.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L, Simon MC, Robertson E, Klein WH, Tsai S-F, D'Agati V, Orkin SH, Costantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- Ribeil JA, Zermati Y, Vandekerckhove J, Cathelin S, Kersual J, Dussiot M, Coulon S, Moura IC, Zeuner A, Kirkegaard-Sorensen T, Varet B, Solary E, Garrido C, Hermine O. Hsp70 regulates erythropoiesis by preventing caspase-3-mediated cleavage of GATA-1. Nature. 2007;445:102–105. doi: 10.1038/nature05378. [DOI] [PubMed] [Google Scholar]
- Rund D, Rachmilewitz E. Beta-thalassemia. New England Journal of Medicine. 2005;353:1135–1146. doi: 10.1056/NEJMra050436. [DOI] [PubMed] [Google Scholar]
- Stoltzfus RJ. Iron deficiency: global prevalence and consequences. Food and Nutrition Bulletin. 2003;24:S99–S103. doi: 10.1177/15648265030244S206. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Komeno T, Suwabe N, Yoh K, Nakajima O, Nishimura S, Kuroha T, Nagasawa T, Yamamoto M. Role of GATA-1 in proliferation and differentiation of definitive erythroid and megakaryocytic cells in vivo. Blood. 1998;92:434–442. [PubMed] [Google Scholar]
- Tilbrook PA, Ingley E, Williams JH, Hibbs ML, Klinken SP. Lyn tyrosine kinase is essential for erythropoietin-induced differentiation of J2E erythroid cells. EMBO Journal. 1997;16:1610–1619. doi: 10.1093/emboj/16.7.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- Tsang AP, Fujiwara Y, Hom DB, Orkin SH. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes & Development. 1998;12:1176–1188. doi: 10.1101/gad.12.8.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas P, McDevitt MA, Cantor AB, Katz SG, Fujiwara Y, Orkin SH. Different sequence requirements for expression in erythroid and megakaryocytic cells within a regulatory element upstream of the GATA-1 gene. Development. 1999;126:2799–2811. doi: 10.1242/dev.126.12.2799. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Keller G, Orkin SH. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1− embryonic stem cells. Genes & Development. 1994;8:1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Molecular and Cellular Biology. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, Orkin SH. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. The Journal of Experimental Medicine. 2002a;195:1387–1395. doi: 10.1084/jem.20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Niakan KK, Matsushita M, Stamatoyannopoulos G, Orkin SH, Raskind WH. X-linked thrombocytopenia with thalassemia from a mutation in the amino finger of GATA-1 affecting DNA binding rather than FOG-1 interaction. Blood. 2002b;100:2040–2045. doi: 10.1182/blood-2002-02-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Socolovsky M, Gross AW, Lodish HF. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood. 2003;102:3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]
- Zheng J, Kitajima K, Sakai E, Kimura T, Minegishi N, Yamamoto M, Nakano T. Differential effects of GATA-1 on proliferation and differentiation of erythroid lineage cells. Blood. 2006;107:520–527. doi: 10.1182/blood-2005-04-1385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.