Abstract
Here we report the genome of a novel rotavirus A (RVA) strain detected in a stool sample collected during routine surveillance by the Centers for Disease Control and Prevention's New Vaccine Surveillance Network. The strain, RVA/human-wt/USA/2012741499/2012/G24P[14], has a genomic constellation of G24-P[14]-I2-R2-C2-M2-A3-N2-T9-E2-H3. The VP2, VP3, VP7 and NSP3 genes cluster phylogenetically with bovine strains. The other genes occupy mixed clades containing animal and human strains. Strain RVA/human-wt/USA/2012741499/2012/G24P[14] most likely is the product of interspecies transmission and reassortment events. This is the second report of the G24 genotype and the first report of the G24P[14] genotype combination in humans.
Keywords: Rotavirus, G24P[14], Bovine, Reassortment
1. Introduction
Rotavirus A (RVA) is a leading cause of diarrheal death and morbidity among children <5 years of age worldwide, particularly in populations not having robust rotavirus vaccination programs (Estes and Greenberg, 2013). RVA are members of the family Reoviridae and possess a genome composed of 11 segments of double-stranded RNA (dsRNA) encoding six viral structural proteins (VP1–VP4, VP6 and VP7) and 5 or 6 non-structural proteins (NSP1–NSP5/6) (Fields et al., 2013). The classification nomenclature for the structural and non-structural proteins of RVA is Gx-P[x]-Ix-Rx-Cx-Mx-Ax-Nx-Tx-Ex-Hx, with x indicating the numbers of the corresponding genotypes (Matthijnssens et al., 2011). Currently, there are 32 distinct G genotypes and 46 distinct P genotypes recognized among RVAs in humans and animals (http://rega.kuleuven.be/cev/viralmetagenomics/virus-classification). The majority of human RVA strains possess either the Wa-like genogroup 1 constellation (Gx-P[x]-I1-R1-C1-M1-A1-N1-T1-E1-H1; porcine origin) or the DS-1-like genogroup 2 constellation (Gx-P[x]-I2-R2-C2-M2-A2-N2-T2-E2-H2; bovine origin) (Matthijnssens et al., 2008a). Occasionally human RVA strains have the AU-1-like genogroup 3 constellation (Gx-P[x]-I3-R3-C3-M3-A3-N3-T3-E3-H3; feline origin) (Matthijnssens et al., 2008b).
RVA strain surveillance and characterization studies from around the world have documented the diversity of RVA strains infecting humans. Whole-genomic analyses are essential for understanding the genetic diversity of RVA strains (Ghosh and Kobayashi, 2011). Uncommon G and P genotypes along with novel genotype combinations are emerging that cause disease in humans; whole-genome analyses of these strains provides evidence of interspecies transmission and reassortment between human and animal RVA strains (Chitambar et al., 2011; Gautam et al., 2015; Martella et al., 2010; Matthijnssens et al., 2009; Matthijnssens and Van Ranst, 2012; Mijatovic-Rustempasic et al., 2015; Tacharoenmuang et al., 2015; Tam et al., 2014). During the 2011–2012 RVA season of the CDC New Vaccine Surveillance Network, one case of pediatric acute gastroenteritis associated with a bovine-like RVA strain was identified in a stool sample from Houston, Texas. Here we report the full genome characterization of the first G24P[14] RVA strain detected in humans.
2. Materials and methods
2.1. Patient history
The patient was a previously healthy two-year-old Hispanic male who presented with a three-day history of diarrhea, a two-day history of non-bilious vomiting, intermittent abdominal pain, subjective fever and one day of decreased urine output. The patient was admitted to a hospital in Houston, Texas and was given fluid resuscitation and antipyretics for fever control. A stool specimen was obtained three days after the onset of illness. The patient had received no doses of RotaTeq® or Rotarix® vaccine. The patient's four-year-old sister was ill with gastrointestinal symptoms one day prior to the patient's symptom onset. Furthermore, the case had household contact with an 82-year old grandfather who had returned from Mexico two months earlier. Of note, at the time of enrollment in this study, the patient's parent reported no history of exposure to animals, including farm animals, or attendance at petting zoos. Institutional review board approvals were obtained from the CDC and from the Houston study site.
2.2. Sequencing and genotype assignment
Full-genome next-generation sequencing was performed using an Illumina MiSeq following a previously published protocol (Mijatovic-Rustempasic et al., 2014). The genotype assignment for each gene was done using the RotaC online classification tool (http://rotac.regatools.be/) and BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.3. Phylogenetic and genetic analyses
For phylogenetic analyses, nucleotide sequences of related strains for each gene were retrieved from GenBank and aligned using the MUSCLE program within MEGA version 5 software (http://www.megasoftware.net/). The optimal evolutionary models that best fit the aligned sequence datasets (NSP1-TIM3+I; NSP2-TrN+G; NSP3-TrN+I+G; NSP4-TPM1uf+I+G; NSP5-HKY+I+G; VP1-GTR+I+G; VP2-TIM2+I; VP3-TIM1+I+G; VP4-TIM3+I+G; VP6-TIM2+I; VP7-GTR+G) were determined using the JModelTest 2 program (Posada, 2008). Using the optimal models identified by the corrected Akaike Information Criterion (AICc), maximum likelihood trees were constructed using PhyML 3.0 with aLRT statistics computed for branch support (Guindon et al., 2010). Nucleotide distance matrices were prepared using the p-distance algorithm of MEGA version 5 software.
3. Results and discussion
Here we present the full sequence of strain RVA/human-wt/USA/2012741499/2012/G24P[14] which represents the second report of the G24 genotype and the first G24P[14] RVA strain detected in humans. This strain exhibits close relationships to bovine RVA strains in the majority of genes.
3.1. Sequencing results
A combination of de novo assembly and subsequent mapping to reference strains was used to obtain the full-length genome of strain RVA/human-wt/USA/2012741499/2012/G24P[14]. The sizes of full-length segments 1 to 11 were 3302, 2687, 2591, 2361, 1578, 1356, 1078, 1059, 1067, 751, 667 bp, and the open reading frames (ORFs) lengths for these segments were 3267, 2643, 2508, 2331, 1476, 1194, 942, 954, 987, 528, 597 bp, respectively. The complete ORFs for all 11 genes of G24P[14] were deposited in GenBank under accession numbers KT281120–KT281130 for VP1, VP2, VP3, VP4, NSP1, VP6, NSP3, NSP2, VP7, NSP4, and NSP5, respectively.
3.2. Genetic distances
Nucleotide distance matrices show that strain RVA/human-wt/USA/2012741499/2012/G24P[14] is closely related to bovine strains for the VP7, VP6, VP1, VP2, VP3, NSP1, NSP3, and NSP4 genes (range: 95.3–98.4%) (Table 1). The VP4 gene is closely related to human strain A64 (96.4%), while the NSP2 gene is closely related to simian strain RRV (97.8%) and the NSP5 gene is closely related to guanaco strain ARG Chubut (96.3%) (Table 1).
Table 1.
Comparison of complete ORF sequences between RVA/human-wt/USA/2012741499/2012/G24P[14] and closely related strains.
| Gene | Genotype | Closest strain(s) |
Predominant host of closely related strain(s) |
% identity to closest related strain(s) |
|---|---|---|---|---|
| VP7 | G24 | Dai-10 | Bovine | 96.3 |
| VP4 | P[14] | A64 | Human | 96.4 |
| VP6 | I2 | Dai-10, AzuK-1 |
Bovine | 96.4, 95.9 |
| VP1 | R2 | RF-TOPORF11 | Bovine | 95.3 |
| VP2 | C2 | Sun9 | Bovine | 97.1 |
| VP3 | M2 | Tottori-SG | Bovine | 98.4 |
| NSP1 | A3 | NCDV-Sapporo | Bovine | 96.5 |
| NSP2 | N2 | RRV | Simian | 97.8 |
| NSP3 | T9 | Dai-10, AzuK-1 |
Bovine | 95.1, 94.9 |
| NSP4 | E4 | CHLY | Bovine | 97.0 |
| NSP5 | H3 | ARG Chubut | Guanaco | 96.3 |
3.3. Results of phylogenetic analyses
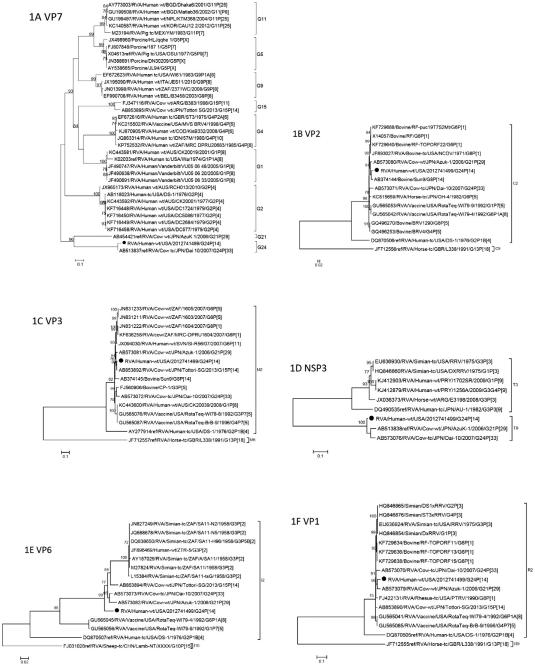
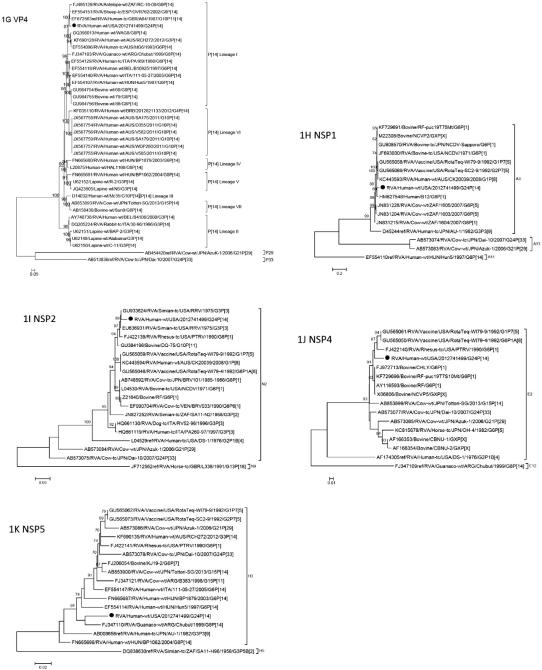
Phylogenetic analyses of the eleven gene segments revealed that genes VP7, VP6, VP1, VP2, VP3, NSP1, NSP3, and NSP4 were related to bovine strains, in particular AzuK-1(G21P[29]-I2-R2-C2-M2-A13-N2-T9-E2-H3) and Dai-10 (G24P[33]-I2-R2-C2-M2-A13-N2-T9-E2-H3) (Fig. 1A-D). The VP6 gene occupies the basal position in a cluster containing bovine, simian, and human strains (Fig. 1E). The VP1 gene occupies a mixed clade with bovine, simian and human strains (Fig. 1F). The NSP1 gene clusters with bovine and human strains (Fig. 1H). The NSP2 gene occupies a mixed clade with simian, bovine, human, and canine strains (Fig. 1I). Also in mixed clades, the NSP4 and NSP5 genes cluster with bovine, simian and equine strains and guanaco (a camelid native to South America), human, bovine, and simian strains, respectively (Fig. 1J-K).
Fig. 1.
A-K. Phylogenetic trees based on the full-length nucleotide sequences of the structural (VP) and non-structural (NSP) genes. Strain RVA/human-wt/USA/2012741499/2012/G24P[14] is identified by the black dots. aLRT values ≥70 are shown adjacent to each node. Each scale bar indicates the number of nucleotide substitutions per site.
The G-genotype of strain RVA/human-wt/USA/2012741499/2012/G24P[14] is G24 and shares 96.3% sequence identity with Dai-10 (Table 1). The genes of Dai-10 possess typical bovine genotypes except for VP7 and VP4, which were assigned new G24 and P[33] genotypes by the RCWG (Abe et al., 2011). Abe and co-workers surveyed asymptomatic calves for RVA infection in Japan from 2006 to 2007 (Abe et al., 2009). It was during the survey that a non-typeable strain (AzuK-1) was detected and sequenced. The AzuK-1 strain demonstrated low nucleotide sequence identity with reference G and P genotypes and was assigned new G21 and P[29] genotypes by the Rotavirus Classification Working Group (RCWG) (Abe et al., 2009). In a subsequent study, the complete ORF sequences of all 11 genes of the AzuK-1 strain and another new strain, Dai-10, were determined (Abe et al., 2011). While AzuK-1 appears to be endemic in cattle in Japan, the Dai-10 strain was detected in only two asymptomatic cows in the survey, a mother and calf, suggesting that this strain is rare in cattle (Abe et al., 2011).
The P-genotype of strain RVA/human-wt/USA/2012741499/2012/G24P[14] is P[14], lineage I (Tam et al., 2014), and like the P[14] strains sequenced by Matthijnssens and co-workers (Matthijnssens et al., 2009), phylogenetic analysis shows the close genomic relatedness of this strain to ovine (OVR762) and antelope (RC-18/08) strains and human strain AG4 (Fig. 1G). The P[14] genotype has been identified in humans, rabbits, and members of the order Artiodactyla (sheep, guanaco, antelope or cattle) (Matthijnssens et al., 2009; Matthijnssens and Van Ranst, 2012). Matthijnssens and Van Ranst compared the genotype constellations of human P[14] strains with those of animal strains and reported that the human strains are probably examples of interspecies transmissions from the order Artiodactyla (Matthijnssens and Van Ranst, 2012).
The VP4 structural protein, specifically the VP8* spike protein, binds to host cells and there is evidence that the VP8* spike in P[14] strains, among others, can recognize human blood group antigens (HBGAs) (Hu et al., 2012; Liu et al., 2012). Liu and colleagues performed phylogenetic analyses of 35 P genotypes of humans and animals and grouped them into five P genogroups based on sequences of VP8*: groups P[I], P[IV], and P[V] infect animals; group P[II] infects humans; and, group P[III] infects both humans and animals and includes P[9], P[14], and P[25] (Liu et al., 2012). They observed that VP8* proteins in P[III] rotaviruses, including P[14] strains, recognize not only human blood group antigens, but bovine and porcine mucins as well, suggesting a mechanism for some rotavirus strains to infect both humans and animals (Liu et al., 2012).
RVA/human-wt/USA/2012741499/2012/G24P[14] is the third reported strain in the T9 genotype. The strain exhibited less than 80% nucleotide sequence identity for NSP3 with other RVA strains, but shared 95% identity with AzuK-1 and Dai-10 (Table 1). Both AzuK-1 and Dai-10 were assigned to a new NSP3 genotype, T9. The new T9 genotype clusters close to T2, T3, and T5, genotypes that are found in humans, canines, felines, and simians, but not in bovines (Abe et al., 2011).
4. Conclusion
Here we report the complete genomic sequence of strain RVA/human-wt/USA/2012741499/2012/G24P[14]. This is the second report of genotype G24 and the first detection of the G24P[14] genotype combination in humans. This strain exhibits close relationships to bovine RVA strains in the majority of genes and most likely is the product of interspecies transmission and reassortment events that include bovine, canine/feline and simian strains. Continued surveillance and full-genome sequencing is required to elucidate the origins and prevalence of novel RVA strains in humans and animals.
Acknowledgment
We wish to thank Rashi Gautam for her constructive comments on the manuscript and Jon Gentsch for his scientific guidance.
Funding source
Funding for this study was provided by the Centers for Disease Control and Prevention.
Footnotes
Institution where work was completed: National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention, 1600 Clifton Rd. NE, Atlanta, GA 30329-4027, USA.
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. Names of specific vendors, manufacturers, or products are included for public health and informational purposes; inclusion does not imply endorsement of the vendors, manufacturers, or products by the Centers for Disease Control and Prevention or the US Department of Health and Human Services.
References
- Abe M, Ito N, Morikawa S, Takasu M, Murase T, Kawashima T, Kawai Y, Kohara J, Sugiyama M. Molecular epidemiology of rotaviruses among healthy calves in Japan: isolation of a novel bovine rotavirus bearing new P and G genotypes. Virus Res. 2009;144:250–257. doi: 10.1016/j.virusres.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Abe M, Ito N, Masatani T, Nakagawa K, Yamaoka S, Kanamaru Y, Suzuki H, Shibano K, Arashi Y, Sugiyama M. Whole genome characterization of new bovine rotavirus G21P[29] and G24P[33] strains provides evidence for interspecies transmission. J. Gen. Virol. 2011;92:952–960. doi: 10.1099/vir.0.028175-0. [DOI] [PubMed] [Google Scholar]
- Chitambar SD, Arora R, Kolpe AB, Yadav MM, Raut CG. Molecular characterization of unusual bovine group a rotavirus G8P[14] strains identified in western India: emergence of P[14] genotype. Vet. Microbiol. 2011;148:384–388. doi: 10.1016/j.vetmic.2010.08.027. [DOI] [PubMed] [Google Scholar]
- Estes M, Greenberg H. Rotaviruses. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. 6th Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 1347–1401. [Google Scholar]
- Gautam R, Mijatovic-Rustempasic S, Roy S, Esona MD, Lopez B, Mencos Y, Rey-Benito G, Bowen MD. Full genomic characterization and phylogenetic analysis of a zoonotic human G8P[14] rotavirus strain detected in a sample from Guatemala. Infect. Genet. Evol. 2015;33:206–211. doi: 10.1016/j.meegid.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Kobayashi N. Whole-genomic analysis of rotavirus strains: current status and future prospects. Future Microbiol. 2011;6:1049–1065. doi: 10.2217/fmb.11.90. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hu L, Crawford SE, Czako R, Cortes-Penfield NW, Smith DF, Le Pendu J, Estes MK, Prasad BV. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature. 2012;485:256–259. doi: 10.1038/nature10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Huang P, Tan M, Liu Y, Biesiada J, Meller J, Castello AA, Jiang B, Jiang X. Rotavirus VP8*: phylogeny, host range, and interaction with histo-blood group antigens. J. Virol. 2012;86:9899–9910. doi: 10.1128/JVI.00979-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V, Banyai K, Matthijnssens J, Buonavoglia C, Ciarlet M. Zoonotic aspects of rotaviruses. Vet. Microbiol. 2010;140:246–255. doi: 10.1016/j.vetmic.2009.08.028. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Van Ranst M. Genotype constellation and evolution of group a rotaviruses infecting humans. Curr. Opin. Virol. 2012;2:426–433. doi: 10.1016/j.coviro.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, Heiman E, Arijs I, Delbeke T, McDonald SM, Palombo EA, Iturriza-Gomara M, Maes P, Patton JT, Rahman M, Van Ranst M. Full genome-based classification of rotaviruses reveals a common origin between human Wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 2008a;82:3204–3219. doi: 10.1128/JVI.02257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, Rahman M, Attoui H, Bányai K, Estes MK, Gentsch JR, Iturriza-Gómara M, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Patton JT, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Desselberger U, Van Ranst M. Recommendations for the classification of group a rotaviruses using all 11 genomic RNA segments. Arch. Virol. 2008b;153:1621–1629. doi: 10.1007/s00705-008-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Potgieter CA, Ciarlet M, Parreño V, Martella V, Bányai K, Garaicoechea L, Palombo EA, Novo L, Zeller M, Arista S, Gerna G, Rahman M, Van Ranst M. Are human P[14] rotavirus strains the result of interspecies transmissions from sheep or other ungulates that belong to the mammalian order Artiodactyla? J. Virol. 2009;83:2917–2929. doi: 10.1128/JVI.02246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Bányai K, Brister JR, Buesa J, Esona MD, Estes MK, Gentsch JR, Iturriza-Gómara M, Johne R, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Parreño V, Rahman M, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Patton JT, Desselberger U, Van Ranst M. Uniformity of rotavirus strain nomenclature proposed by the rotavirus classification working group (RCWG) Arch. Virol. 2011;156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijatovic-Rustempasic S, Roy S, Sturgeon M, Rungsrisuriyachai K, Esona MD, Degroat D, Qin X, Cortese MM, Bowen MD. Full-genome sequence of a rare human G3P[9] rotavirus strain. Genome Announc. 2014:2. doi: 10.1128/genomeA.00143-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijatovic-Rustempasic S, Roy S, Sturgeon M, Rungsrisuriyachai K, Reisdorf E, Cortese MM, Bowen MD. Full-genome sequence of the first G8P[14] rotavirus strain detected in the United States. Genome Announc. 2015:3. doi: 10.1128/genomeA.00677-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Tacharoenmuang R, Komoto S, Guntapong R, Ide T, Haga K, Katayama K, Kato T, Ouchi Y, Kurahashi H, Tsuji T, Sangkitporn S, Taniguchi K. Whole genomic analysis of an unusual human G6P[14] rotavirus strain isolated from a child with diarrhea in Thailand: evidence for bovine-to-human interspecies transmission and reassortment events. PLoS One. 2015;10:e0139381. doi: 10.1371/journal.pone.0139381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam KI, Roy S, Esona MD, Jones S, Sobers S, Morris-Glasgow V, Rey-Benito G, Gentsch JR, Bowen MD. Full genomic characterization of a novel genotype combination, G4P[14], of a human rotavirus strain from Barbados. Infect. Genet. Evol. 2014;28:524–529. doi: 10.1016/j.meegid.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]