Abstract
Background
Breast cancer is the main type of cancer in women, and triple-negative breast cancer (TNBC) is a unique subtype of breast cancer. The expression of miR-940 has been shown to play an important role in various cancers; however, the role of miR-940 in TNBC remains unknown.
Material/Methods
The expression of miR-940 in TNBC tissues or cells were tested by qRT-PCR; the expression of miR-940 in cells were overexpressed by miR-940 mimics, and suppressed by anti-miR-940. Bioinformatics algorithms from TargetScanHuman were used to predict the target genes of miR-940. The interaction between miR-940 and ZNF24 was confirmed by dual luciferase assays. The protein level was assayed by Western blot.
Results
TNBC tissues and cells showed lower miR-940 levels.
Conclusions
MiR-940 inhibited cellular proliferation and migration in TNBC.
MeSH Keywords: Cell Migration Assays, Cell Proliferation, MicroRNAs, Triple Negative Breast Neoplasms
Background
In China, the predominant type of cancer detected in males is lung cancer; and the predominant type of cancer detected in females is breast cancer [1]. Triple-negative breast cancer (TNBC) is a unique subtype of breast cancer that is histologically defined by the absence of estrogen receptor (ER) and progesterone receptor (PR), and lacks human epidermal growth factor receptor 2 (HER2) overexpression [2–4]. Recent data show that TNBC has specific molecular features that could be possible targets for new therapies; TNBC is still associated with poor prognosis and a high risk of distant recurrence and death [5].
MicroRNAs are small noncoding RNAs, of 21–24 nucleotides, which are involved in the regulation of the expression of protein-coding genes at the posttranscriptional level [6,7]. Many studies have revealed that several miRNAs take part in the hormone receptor expression in breast cancer. For example, miR-342, miR-299, and miR-218 correspond with ER; miR-520g, miR-377; miR-520f-520c corresponds with PR; miR-520d, miR-181c, and miR-302c; and miR-30e corresponds with HER2 [8].
The function of miR-940 has been investigated in different types of cancers including gastric cancer [9], hepatocellular carcinoma [10], pancreatic ductal adenocarcinoma [11], and prostate cancer [12]. These studies have shown that miR-940 may be a potential therapeutic target in cancer therapy. However, the function of miR-940 in TNBC remains unclear.
In this study, we first assayed the level of miR-940 in TNBC, and then evaluated the effect of miR-940 in vitro. We hope our data elucidates the role of miR-940 in TNBC.
Material and Methods
Patients
Twelve TNBC specimens were collected from The Affiliated Hospital of North Sichuan Medical College via Shengong Company (Shanghai, China). Before this study, the senior pathologists of The Affiliated Hospital of North Sichuan Medical College evaluated and confirmed the histological traits of the specimens. Tissue samples were immediately frozen in liquid nitrogen after isolation. Informed consent was obtained from each patient. The Ethics Commitment of The North Sichuan Medical College and the Ethic Commitment of The North Sichuan Medical College approved this study.
Cell culture
ME16C cell line, TNBC cell lines MDA-MB-231 and BT-549, and HEK293 cells were purchased from the Chinese Academy of Science (Beijing, China). ME16C cells, MDA-MB-231 cells, BT-549 cells, and HEK293 cells were maintained in DMEM with 10% fetal bovine serum (FBS) (Invitrogen Corp, Grand Island, NY, USA).
Detection of miR-940 in TNBC tissues or cells
Total RNAs of TNBC tissues and cells were extracted by TRIzol reagent (Invitrogen Corp, Grand Island, NY, USA) according to the manufacturer’s instructions. Total RNAs were reverse-transcribed to cDNA using All-in-One™ miRNA First-Strand cDNA Synthesis Kit (Invitrogen Corp, Grand Island, NY, USA). The primers were constructed by GenScript Company (GenScript, Nanjing, China). U6 snRNA was used to normalize relative miR-940 levels. Real time PCR assay was performed as described previously [11,13–15].
MiR-940 mimics and anti-miR-940 transfection
MiR-940 antisense oligonucleotides (anti-miR-940), miR-940 mimics, and negative control miRNA were purchased from RiboBio (Guangdong, China). Anti-miR-940 or miR-940 mimics were transfected into cells using Lipofectamine (Invitrogen, Shanghai, China), according to the manufacturer’ s instructions [11].
Cell proliferation and migration assay
After 24 hours of transfection of miR-940 mimics and anti-miR-940, cells from MDA-MB-231 and BT-549 cell lines (5×103/well) were seeded into 96-well plates. Then MTT experiments were performed as described in previous articles [16,17]. Absorbance in each well was measured by using a microplate reader set at 570 nm. To measure cell migration, 8-mm pore size-culture inserts (Transwell; Millipore, MA, USA) were placed into wells of 24-well culture plates, separating the upper and lower chambers. In the lower chamber, 600 μL DMEM containing 10% FBS was added. Then 1×105/well cells were added to the upper chamber. After 24 hour incubation, migrated cells were counted by a counting chamber (Shengong, Shanghai, China) [18].
Target prediction
Bioinformatics methods were applied for the prediction of the targeted genes. The online tools for targeted gene prediction from TargetScanHuman were used [19–24].
ZNF24 alteration in various types of cancer
ZNF24 amplification data and graph were obtained from cBioPortal for Cancer Genomics [25,26].
Dual luciferase assays
To confirm direct binding of ZNF24 to miR-940, a dual luciferase assay was performed [27,28]. The 3′UTR of ZNF24 was amplified using PCR from genomic DNA. The product was inserted downstream of ZNF24 3′UTR reporter plasmids (pRL-ZNF24) (Biotech, Chengdu, China), Mutants of ZNF24 3′UTR were generated by Site-Directed Mutagenesis kit (Shanghai, China). Then the whole plasmid was confirmed by sequencing. Mutations in the miR-940 binding site of ZNF24 3′UTR were constructed by Shengong Company (Shengong, Chengdu, China). Luciferase reporter-containing mutants were constructed. For luciferase assays, HEK293 cells were transfected with luciferase reporter plasmid along with miR-940 mimics or negative control using Lipofectamine 2000 (Invitrogen, Shanghai, China). Then 24 hours after transfection, these cells were analyzed using a luciferase assay kit (Promega, Madison, WI, USA) [29].
Western blot
At 48 hours after treatment, cells were washed with cold PBS and subjected to a lysis buffer. Protein lysates were separated using 8% SDS-polyacrylamide gel electrophoresis, then electro-transferred onto nitrocellulose filter membranes. The membranes were blocked with a buffer containing 5% non-fat milk in PBS with 0.05% Tween-20 for 2 hours and incubated with primary antibody (anti-ZNF24) overnight. Then membranes were incubated with peroxidase-conjugated secondary antibodies (Millipore, Darmstadt, Germany) and developed with an enhanced chemiluminescence detection kit (Pierce, Rockford, IL). β-actin was used as blank control.
Statistical analysis
The data are presented as the mean ±SD from three independent experiments. When only two groups were compared, the difference between them was analyzed using a two-tailed Student’s t-test. However, when three or more groups were compared, the difference between them was analyzed using analysis of variance (ANOVA). Statistically significant differences in the expression of miR-940 between matched pairs were detected by the Wilcoxon matched-pairs signed rank test. SPSS software (version 17.0) was used to perform statistical analyses, in which p < 0.05 was considered significant.
Results
The lower expression of miR-940 in TNBC
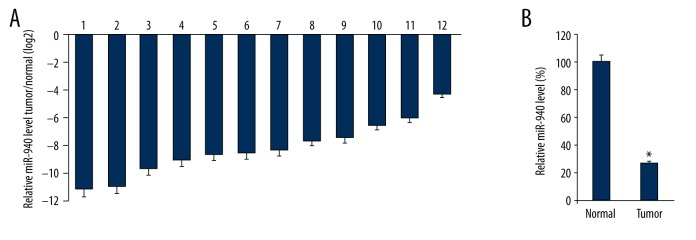
Initially, we collected 12 TNBC clinical species from our hospital. The miR-940 level in the tumor tissues and matched adjacent normal tissues were assayed by qRT-PCR. For every patient selected in the study, we found that the miR-940 levels in tumor tissues were lower than in normal tissues (Figure 1A, Table 1). Furthermore, the mean level of miR-940 in the 12 TNBC tumor tissues was lower than the mean expression of miR-940 in normal tissues (B).
Figure 1.
The levels of miR-940 in TNBC cells. The miR-940 expression level in 12 TNBC tissues and tumor adjacent normal tissues were assayed by qRT-PCR. U6 snRNA was used as the blank control (A). The mean expression of miR-940 in the 12 TNBC tissues and matched adjacent normal cancer tissues were calculated; the mean level of 12 normal tissues are arbitrarily defined as 100% (B). The qRT-PCR experiments were performed three times. Data are presented as mean ± SD, * p<0.05.
Table 1.
The miR-940 expression values in 12 TNBC tissues and tumor adjacent normal tissues.
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tumor | 4.56E-04 | 4.88E-04 | 1.59E-03 | 1.95E-03 | 2.58E-03 | 2.96E-03 | 3.17E-03 | 5.15E-03 | 5.52E-03 | 1.18E-02 | 1.56E-02 | 5.08E-02 |
| Normal | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Tumor/Normal (log2) | −11.1 | −11.0 | −9.3 | −9.0 | −8.6 | −8.4 | −8.3 | −7.6 | −7.5 | −6.4 | −6.0 | −4.3 |
MiR-940 inhibited cell growth and migration
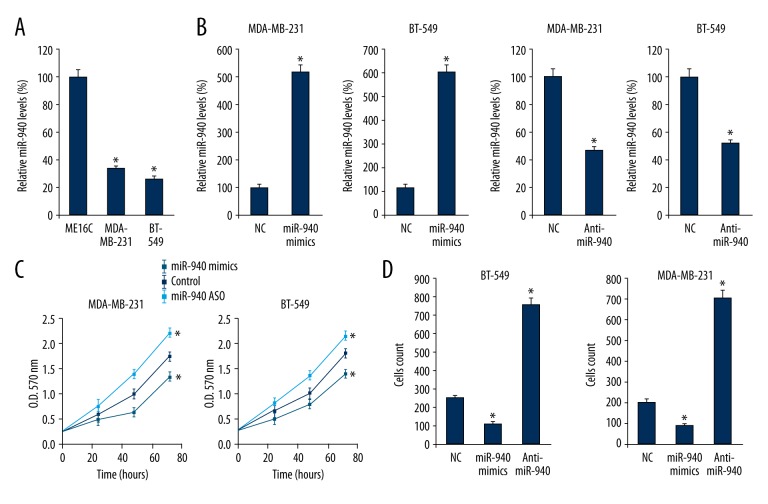
Two TNBC cell lines, MDA-MB-231 and BT-549, were chosen for investigating the role of miR-940 in TNBC. The miR-940 expression levels in MDA-MB-231 and BT-549 cells were analyzed by qRT-PCR. We found that the miR-940 expression levels in MDA-MB-231 and BT-549 cells were higher than the miR-940 expression levels in ME16C cells, which were used as a negative control (Figure 2A). Then we overexpressed the miR-940 levels in MDA-MB-231 and BT-549 cells by transfection of miR-940 mimics, and inhibited the miR-940 levels by anti-miR-940. We found that miR-940 mimics and anti-miR-940 altered the miR-940 expression effectively (Figure 2B). Next the effects of miR-940 on cell growth and migration were tested. MTT analysis revealed that overexpression of miR-940 suppressed cellular proliferation, and on the contrary, inhibition of miR-940 promoted cellular proliferation (Figure 2C). As expected, miR-940 mimics resulted in fewer migrated cells and anti-miR-940 led to more migrated cells (Figure 2D).
Figure 2.
Overexpression of miR-940 inhibited cells growth and migration and vice versa. After total RNA extraction, the miR-940 levels in ME16C, MDA-MB-231, and BT-549 cells were assayed by qRT-PCR. The miR-940 level in ME16C cells was deliberately defined as 100% (A). MDA-MB-231 and BT-549 cells (1×106/well) were transfected with miR-940 mimics or anti-miR-940, and 24 hours later, the miR-940 levels were assayed by qRT-PCR. The miR-940 level in the negative control was deliberately treated as 100% (B). After miR-940 mimics or anti-miR-940 transfection, the cellular proliferation of MDA-MB-231 and BT-549 cells was tested by MTT analysis (C). Then 24 hours after miR-940 mimics or anti-miR-940 transfection, MDA-MB-231 and BT-549 cells were collected for migration tests (D). The experiments were performed three times. Data are presented as mean ±SD, * p<0.05.
ZNF24 was targeted by miR-940
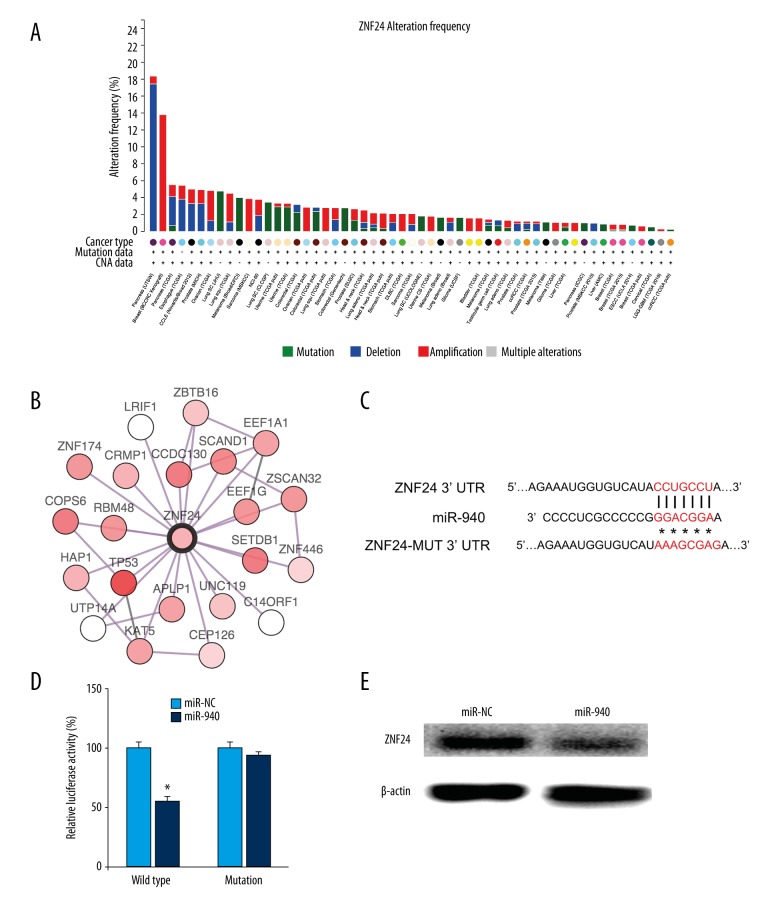
Next we investigate the potential target of miR-940. As a recent study showed that ZNF24 was targeted by miR-940 [30], we chose ZNF24 for further experimentation. To reveal the exact role of ZNF24, we explored The Cancer Genome Atlas (TCGA). TCGA Project is a large-scale collaborative effort to characterize the genomic changes that occur in cancer. This project has profiled and analyzed large numbers of human tumors to discover and catalog molecular aberrations at the DNA, RNA, protein, and epigenetic levels [31]. Interestingly, we found that ZNF24 showed amplification mutation in breast cancer (Figure 3A), and ZNF24 could interact with TP53 (Figure 3B). Next we identified the binding sites between miR-940 and ZNF24, and found that the mutated version of ZNF24 was also displayed (Figure 3C). Next the wild type or mutant of 3′UTR of ZNF24 and miR-940 were co-transfected into HEK293 cells and the effect of miR-940 on the ZNF24 translation was tested by a luciferase reporter assay, and data showed that miR-940 targeted the 3′UTR of ZNF24 (Figure 3D). Next we tested the ZNF24 protein level in HEK293 cells, and found the ZNF24 protein level was suppressed following miR-940 transfection (Figure 3E).
Figure 3.
Alterations of ZNF24 were visualized by cBioPortal for Cancer Genomics. Mutation, deletion, amplification, and other alterations are shown in different colors. The most frequent alteration of ZNF24 in breast cancer is amplification. The CAN data stand for copy number alteration data (A). The network of ZNF24 was visualized by cBioPortal (B). The binding site of miR-940 in ZNF24 was mutated (C). HEK293 cells were co-transfected with miR-940 mimics or control and reporter plasmid or the mutant 3′UTR of ZNF24, together with the controls. Then 48 hours after transfection, the luciferase activity was measured (C). MiR-940 mimics were transfected into HEK293 cells; 48 hours later, the ZNF24 protein level was tested by Western blot (D). The experiments were performed three times. Data are presented as mean ±SD, * p<0.05.
Discussion
In this study, we investigated the role of miR-940 in TNBC and found that miR-940 showed a protection role in TNBC, and the possible targeted gene was ZNF24.
The role of miR-940 has been studied in many cancers: miR-940 was found to play an anti-oncogenic role in pancreatic ductal adenocarcinoma and prostate cancer [32,33]; and miR-940 promoted tumor cell invasion and metastasis by downregulating ZNF24 in gastric cancer [30]. Our data showed that miR-940 inhibited cellular proliferation and migration, and the potential targeted gene was ZNF24. This discrepancy indicates that miR-940 plays different roles in various cancers.
ZNF24 has mainly been identified as a repressor of vascular endothelial growth factor (VEGF) [34]. An inverse correlation between the expression of VEGF and ZNF24 was observed in a series of independent studies. In gastric cancer, ZNF24 served as a tumor suppressor [30], the related mechanism suggested was that ZNF24 negatively regulated VEGF expression and acted as a transcriptional repressor of VEGF gene. However, in another study, data showed that knockdown of ZNF24 in microvascular endothelial cells led to decreased cell migration and attenuated VEGFR2 signaling [35]. Our data also indicated the ZNF24 served as a promoter of TNBC.
Interestingly, we found that ZNF24 showed amplification of mutations in breast cancer cells (Figure 3A), which was consistent with the oncogenic role of ZNF24 in TNBC. In addition, the network analysis indicated that ZNF24 was connected with TP53, which we will investigate in future studies.
Conclusions
In conclusion, our study found that miR-940 inhibited cellular proliferation and migration in TNBC cells, and the potential targeted gene was ZNF24.
MiR-940 showed a tumor suppressor role via ZNF24 in TNBC.
Footnotes
Source of support: This work was supported by grants from the National Natural Science Foundation of China (Grants No. 81172496), grants from The North Sichuan Medical College (Grants No. CBY12-A-QN07), grants from Education Department of Sichuan Province (Grants No.13ZA0228) and grants from Science and Technology Department of Sichuan Province (Grants No.2016060)
Conflict of interest
The authors declare no conflict interest
Reference
- 1.Thummuri D, Kumar S, Surapaneni SK, Tikoo K. Epigenetic regulation of protein tyrosine phosphatase PTPN12 in triple-negative breast cancer. Life Sci. 2015;130:73–80. doi: 10.1016/j.lfs.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 2.Humphries B, Wang Z, Oom AL, et al. MicroRNA-200b targets protein kinase Calpha and suppresses triple-negative breast cancer metastasis. Carcinogenesis. 2014;35(10):2254–63. doi: 10.1093/carcin/bgu133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Criscitiello C, Azim HA, Schouten P, et al. Understanding the biology of triple-negative breast cancer. Ann Oncol. 2012;23(Suppl 6):vi13–vi18. doi: 10.1093/annonc/mds188. [DOI] [PubMed] [Google Scholar]
- 4.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. New Engl J Med. 2010;363(20):1938–48. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 5.Tomao F, Papa A, Zaccarelli E, et al. Triple-negative breast cancer: New perspectives for targeted therapies. OncoTargets Ther. 2015;8:177. doi: 10.2147/OTT.S67673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho WC. OncomiRs: The discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu J, Xu J, Wu Y, et al. Identification of microRNA-93 as a functional dysregulated miRNA in triple-negative breast cancer. Tumour Biol. 2015;36(1):251–58. doi: 10.1007/s13277-014-2611-8. [DOI] [PubMed] [Google Scholar]
- 8.Lowery AJ, Miller N, Devaney A, et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009;11(3):R27. doi: 10.1186/bcr2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Kwong A, Sihoe A, Chu KM. Plasma miR-940 may serve as a novel biomarker for gastric cancer. Tumour Biol. 2016;37(3):3589–97. doi: 10.1007/s13277-015-4019-5. [DOI] [PubMed] [Google Scholar]
- 10.Yuan B, Liang Y, Wang D, Luo F. MiR-940 inhibits hepatocellular carcinoma growth and correlates with prognosis of hepatocellular carcinoma patients. Cancer Sci. 2015;106(7):819–24. doi: 10.1111/cas.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song B, Zhang C, Li G, et al. MiR-940 inhibited pancreatic ductal adenocarcinoma growth by targeting MyD88. Cell Physiol Biochem. 2015;35(3):1167–77. doi: 10.1159/000373941. [DOI] [PubMed] [Google Scholar]
- 12.Rajendiran S, Parwani AV, Hare RJ, et al. MicroRNA-940 suppresses prostate cancer migration and invasion by regulating MIEN1. Mol Cancer. 2014;13:250. doi: 10.1186/1476-4598-13-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li D, Liu X, Lin L, et al. MicroRNA-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma. J Biol Chem. 2011;286(42):36677–85. doi: 10.1074/jbc.M111.270561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou X, Deng S, Liu H, et al. Knockdown of ubiquitin protein ligase E3A affects proliferation and invasion, and induces apoptosis of breast cancer cells through regulation of annexin A2. Mol Med Rep. 2015;12(1):1107–13. doi: 10.3892/mmr.2015.3549. [DOI] [PubMed] [Google Scholar]
- 15.Zhou X, Deng S, Liu H, et al. Knockdown of ubiquitin protein ligase E3A affects proliferation and invasion, and induces apoptosis of breast cancer cells through regulation of annexin A2. Mol Med Rep. 2015;12(1):1107–13. doi: 10.3892/mmr.2015.3549. [DOI] [PubMed] [Google Scholar]
- 16.Liu C, Gao F, Li B, et al. TLR4 knockout protects mice from radiation-induced thymic lymphoma by downregulation of IL6 and miR-21. Leukemia. 2011;25(9):1516–19. doi: 10.1038/leu.2011.113. [DOI] [PubMed] [Google Scholar]
- 17.Liu C, Zhou C, Gao F, et al. MiR-34a in age and tissue related radio-sensitivity and serum miR-34a as a novel indicator of radiation injury. Int J Biol Sci. 2011;7(2):221–33. doi: 10.7150/ijbs.7.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Dong Y, Zhu N, et al. microRNA-139-5p exerts tumor suppressor function by targeting NOTCH1 in colorectal cancer. Mol Cancer. 2014;13:124. doi: 10.1186/1476-4598-13-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 20.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimson A, Farh KK, Johnston WK, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia DM, Baek D, Shin C, et al. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18(10):1139–46. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Human T. Prediction of microRNA targets. 2009 [Google Scholar]
- 24.Lee S, Paulson KG, Murchison EP, et al. Identification and validation of a novel mature microRNA encoded by the Merkel cell polyomavirus in human Merkel cell carcinomas. J Clin Virol. 2011;52(3):272–75. doi: 10.1016/j.jcv.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin L, Liang H, Wang Y, et al. microRNA-141 inhibits cell proliferation and invasion and promotes apoptosis by targeting hepatocyte nuclear factor-3beta in hepatocellular carcinoma cells. BMC Cancer. 2014;14:879. doi: 10.1186/1471-2407-14-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu N, Zhang C, Bai C, et al. MiR-4782-3p inhibited non-small cell lung cancer growth via USP14. Cell Physiol Biochem. 2014;33(2):457–67. doi: 10.1159/000358626. [DOI] [PubMed] [Google Scholar]
- 29.Grentzmann G, Ingram JA, Kelly PJ, et al. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4(4):479–86. [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Ge X, Zhang Z, et al. MicroRNA-940 promotes tumor cell invasion and metastasis by downregulating ZNF24 in gastric cancer. Oncotarget. 2015;6(28):25418–28. doi: 10.18632/oncotarget.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research, N. Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajendiran S, Parwani AV, Hare RJ, et al. MicroRNA-940 suppresses prostate cancer migration and invasion by regulating MIEN1. Mol Cancer. 2014;13(1):250. doi: 10.1186/1476-4598-13-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song B, Zhang C, Li G, et al. MiR-940 Inhibited pancreatic ductal adenocarcinoma growth by targeting MyD88. Cell Physiol Biochem. 2015;35(3):1167–77. doi: 10.1159/000373941. [DOI] [PubMed] [Google Scholar]
- 34.Harper J, Yan L, Loureiro RM, et al. Repression of vascular endothelial growth factor expression by the zinc finger transcription factor ZNF24. Cancer Res. 2007;67(18):8736–41. doi: 10.1158/0008-5472.CAN-07-1617. [DOI] [PubMed] [Google Scholar]
- 35.Jia D, Huang L, Bischoff J, Moses MA. The endogenous zinc finger transcription factor, ZNF24, modulates the angiogenic potential of human microvascular endothelial cells. FASEB J. 2015;29(4):1371–82. doi: 10.1096/fj.14-258947. [DOI] [PMC free article] [PubMed] [Google Scholar]