Abstract
MicroRNAs (miRNAs) and piwi-interacting RNAs (piRNAs) are two classes of small noncoding RNAs, both of which play roles in regulating tissue development. It is unknown whether these distinct classes of noncoding RNAs can regulate one another. Here we show that ectopic expression of miR-17 inhibited mouse fertility and early embryonic development. Specifically, we found that the piRNA amplification loop was repressed by miR-17-5p, leading to increased levels of transposition mutagenesis. This occurred by suppressing the amplification loop of piRNAs with an identical 5′ sequence and by targeting Mili/Miwi2, an essential component of the piRNA amplification loop, and the DNA methyltransferase, Dnmt3a. We also found that increased levels of piRNAs could compete with miRNAs for target binding, resulting in increased expression of Dnmt3a and Mili. Increased Dnmt3a levels could in turn block miR-17-5p expression, while increased Mili expression could accelerate piRNA amplification and inhibit transposon generation, favoring embryonic development. We report for the first time the reciprocal regulation between miRNAs and piRNAs in mouse embryonic development.
A microRNA (miRNA) is a small non-coding RNA molecule of 18–24 nucleotides in length that are post-transcriptional regulators of gene expression. A miRNA has the capacity to affect the stability of hundreds of unique mRNAs1 and may repress synthesis of hundreds of proteins by base-pairing with the targeting mRNAs.2 The discovery of miRNAs in gene regulation has improved our understanding of post-transcriptional control of tissue development and aging.3, 4, 5 A polycistronic miRNA cluster, miR-17~92, consisting of six mature miRNAs (miR-17, miR-18a, mIR-19a, miR-20a, miR-19-b1, and miR-92a-1), has been reported to play a fundamental role in development and remodeling.6, 7, 8, 9 Furthermore, members of miR-17-92 cluster are found to be commonly down-regulated in aging human cells.10 We have found that ectopic expression of miR-17 retards tissue growth11 and inhibits cell senescence.12
Piwi-interacting RNA (piRNA) is the largest class of small non-coding RNAs expressed by animal cells.13 piRNAs form complexes with piwi proteins, which function post-transcriptionally in silencing retrotransposons and other genetic elements in germ-line cells.14 piRNA activity in gene regulation may lead to silencing of transposon expression,15 as most piRNA sequences are antisense to transposon sequences.16 The activity of piRNAs in transposon silencing appears to be the most important event during embryonic development.17 To facilitate silencing transposons in the testes of mammals or in the germ cells of invertebrates, the piwi proteins play essential roles in interacting with piRNAs. These piwi proteins are members of Argonautes, proteins that are needed for gene silencing in miRNA action, including MIWI, MIWI2 and MILI. The sequences of piRNAs guide the piwi proteins to their transposon targets.17 Decreased expression of these PIWI proteins is correlated with increased expression of transposons, leading to potential infertility.17, 18, 19 We have previously observed that increased expression of miR-17 retards tissue growth.11 Further examination showed that significantly higher incidences of infertility occurred in miR-17 transgenic mice. This study was designed to uncover the role of miR-17 in embryonic development.
Results
miR-17 inhibits mouse fertility and early embryo development
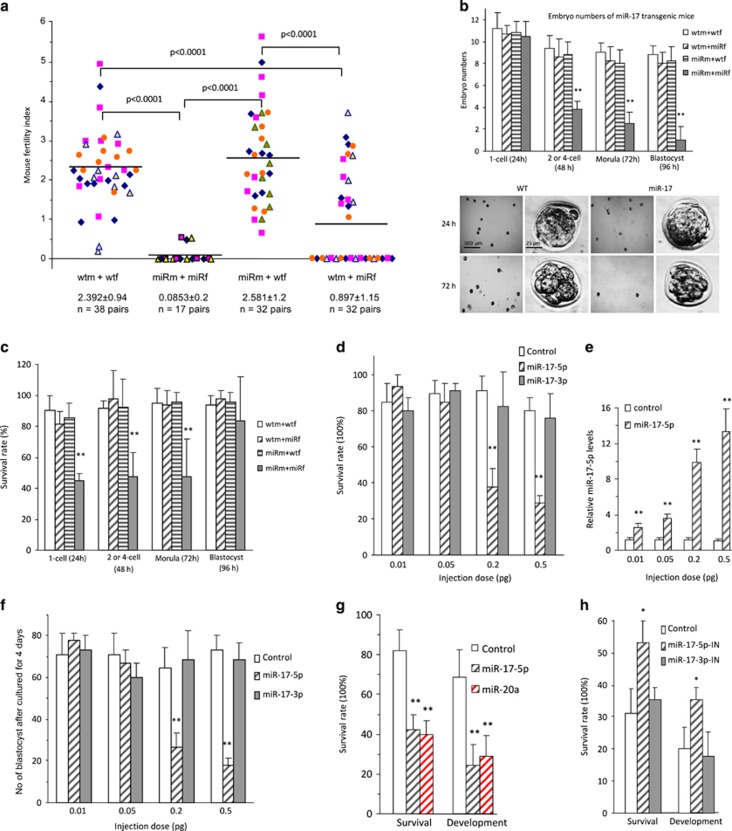
During the course of maintaining miR-17 transgenic mice that were established previously,11 we found that the mice were infertile if both male and female were miR-17 transgenic. We performed a systematic analysis of the fertility of the transgenic mice. Fertility Index was calculated based on cycles of 3 weeks for pregnancy. We found that female miR-17 transgenic mice were infertile if they were matched with male miR-17 transgenic mice, but exhibited normal fertility if matched with wild-type male mice (Figure 1a). The fertility of wild-type female mice was not affected by matching with miR-17 transgenic mice.
Figure 1.
Expression of miR-17 inhibited early embryo development. (a) Fertility rates of miR-17 transgenic (transgenic mating with transgenic, miRm+miRf), wild-type (wtm+wtf) and miR-17 transgenic mice backcrossing with wild-type (miRm+wtf and wtm+miRf) mice, showing infertility of the miR-17 transgenic mice (miRm+miRf). (b) Upper, Embryo numbers in various stages collected from the oviduct or uterus 24, 48, 72, and 96 h after hCG treatment, showing decreased number of embryos in two-cell, morula, and blastocyst stages in miR-17 transgenic mice (miRm+miRf). **P<0.01. Error bars, S.D. (n=6). Lower, Typical pictures of embryos from transgenic (miRm+miRf) and wild-type mice. (c) Survival rates of embryo in various stages cultured for 24 h. Decreased survival rates in zygote, two-cell, and morula stages, but not in blastocyst stage of miR-17 transgenic mice (miRm+miRf). **P<0.01. Error bars, S.D. (n=6). (d) Wild-type zygotes were microinjected with 2 pl miR-17-5p, miR-17-3p (at different doses as marked), or control oligos, and incubated for 96 h. Zygotes injected with miR-17-5p showed decreased survival rates. **P<0.01. Error bars, S.D. (n=4). (e) Zygotes injected with miR-17-5p showed increased levels of miR-17-5p. **P<0.01. Error bars, S.D. (n=4). (f) Zygotes injected with miR-17-5p showed decreased development rates. **P<0.01. Error bars, S.D. (n=4). (g) Zygotes injected with miR-20a showed decreased developmental rates. **P<0.01. Error bars, S.D. (n=4). (h) Transgenic zygotes (miRm+miRf) were microinjected with 2 pl miR-17-5p inhibitor, miR-17-3p inhibitor, or control oligos, and incubated for 96 h. Zygotes injected with miR-17-5p inhibitor showed increased rates of survival and development. **P<0.01. Error bars, S.D. (n=4)
To examine the cause of infertility, we isolated mouse oocytes at various time points from oviduct or uterus after hCG treatment and mating. Each group contained at least six mating pairs. In the four sets of breeding (male miR-17 transgenic: miRm; female miR-17 transgenic: miRf, male wild type: wtm; female wild type: wtf), we detected a significant decrease in embryonic number of 2 or 4-cell, morula and blastocyst in the miRm+miRf set as compared with the other three sets (Figure 1b, Supplementary Figure S1a). The isolated embryos were cultured in M2 medium for 24 h followed by TUNEL straining. We detected significantly decreased survival rates in zygote, two-cell, and morula stages, but not in the blastocyst stage, of miR-17 transgenic mice miRm+miRf compared with the other three sets (Figure 1c, Supplementary Figure S1b). We confirmed expression of miR-17 precursor, mature miR-17-5p, and mature miR-17-3p (Supplementary Figure S1c) in GV, MII, sperm, and embryos in one-cell, two-cell, morula, and blastocyst stages.
We dissected the effect of mature miRNA(s) by injecting miR-17-5p, miR-17-3p, or control oligo, into the single-cell zygotes, followed by 96-h incubation and TUNEL staining. We found that injection with miR-17-5p, but not miR-17-3p, at concentrations ≥0.2 pg decreased zygote survival (Figure 1d, Supplementary Figure S1d). The efficiency of injection was confirmed by real-time PCR (Figure 1e). The injected zygotes were also cultured for 4 days, by when zygotes developed to blastocysts, and again, only injection with miR-17-5p decreased embryonic development (Figure 1f). We tested the effect of another miRNA miR-20a that is highly homologous to miR-17-5p and detected a similar function in embryonic development (Figure 1g).
To test the functions of endogenous miR-17-5p, we injected zygotes with inhibitors of miR-17-5p and miR-17-3p, followed by 96-h culture and TUNEL staining. We found that only miR-17-5p inhibitor enhanced zygote survival and development (Figure 1h, Supplementary Figure S1e). In these experiments, 10–12 embryos were injected in each group, and each experiment was repeated four times.
Repression of piRNA expression
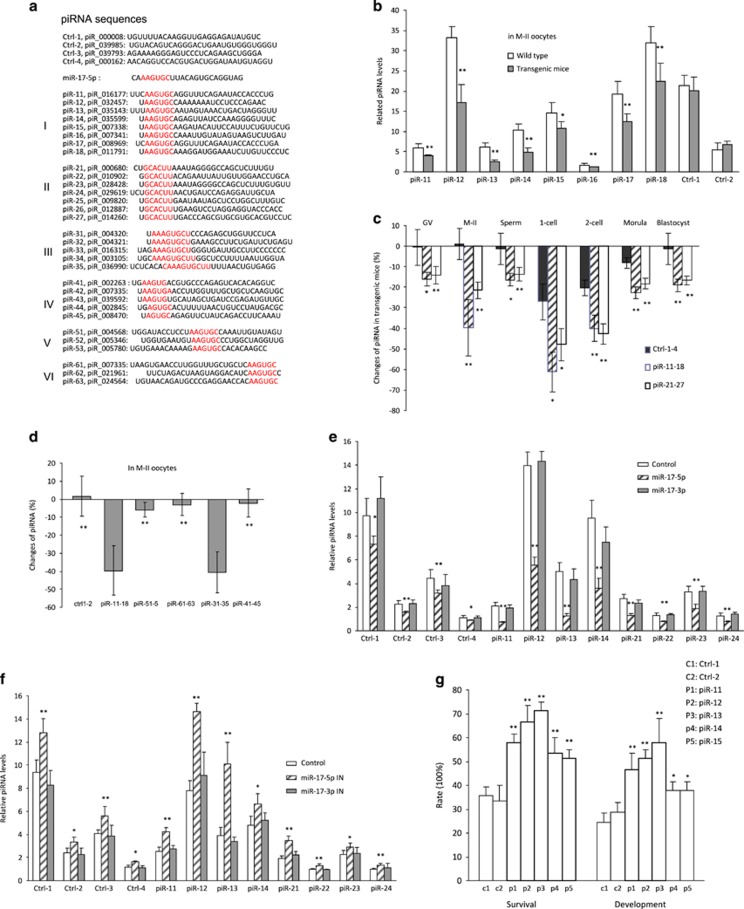
As piRNAs play important roles in the early embryonic development, we analyzed levels of piRNA expression, focusing on the piRNAs with sequences homologous to the 5′ seed region of miR-17-5p (Figure 2a) using the website http://pirnabank.ibab.ac.in. These piRNAs were organized into Group I (having six nucleotides at the 5′ end of the piRNAs that were homologous to miR-17-5p seed region), Group II (having six nucleotides anti-sense to the seed region of miR-17-5p), Group III (having more than six nucleotides homologous to miR-17-5p), Group IV (having five nucleotides homologous to miR-17-5p), Group V (six nucleotides at the middle of the piRNAs), and Group VI (six nucleotides at the 3′ end of the piRNAs). We found that expression of eight piRNAs in Group I was downregulated in the oocytes of miR-17 transgenic mice compared with that of wild type (Figure 2b). Expression of Group II piRNAs was also downregulated in the miR-17 transgenic mice (Supplementary Figure S2a). Analysis of these two groups of piRNAs indicated significant decrease in the number of oocyte, sperm, and embryos in various stages (Figure 2c). Analysis of the average changes of piRNAs indicated that Group III piRNAs had similar fold changes as Group I piRNAs, while Group IV, Group V, and Group VI piRNAs had little change when compared with controls (Figure 2d).
Figure 2.
piRNA expression was repressed in the early embryos of miR-17 transgenic mice. (a) Sequences of piRNAs used in this study. (b) Fifty unfertilized oocytes (M-II) were collected for RNA extraction, followed by real-time PCR. miR-17 transgenic mice expressed decreased piRNA (sense) levels with AAGUGC motif, but not those without AAGUGC motif. *P<0.05. **P<0.01. Error bars, S.D. (n=4). (c) Real-time PCR was performed on RNAs isolated from 50 sperms, unfertilized and fertilized oocytes. miR-17 transgenic mice expressed decreased sense and antisense piRNA levels with AAGUGC motif in sperms, unfertilized and fertilized oocytes in various stages. *P<0.05. **P<0.01. Error bars, S.D. (n=4). (d) Real-time PCR showing that piRNA levels with AAGUGC and AAAGUGCU motif decreased significantly more than those with AGUGC or AAGUG motif, in the unfertilized oocytes (M-II) of miR-17 transgenic mice. **P<0.01. Error bars, S.D. (n=4). (e) Injection with miR-17-5p mimics repressed piRNAs levels in zygotes. *P<0.05. **P<0.01. Error bars, S.D. (n=4). (f) Injection with miR-17-5p-IN increased piRNA levels in zygotes. *P<0.05. **P<0.01. Error bars, S.D. (n=4). (g) Zygotes (miRm+miRf) were microinjected with 2 pl control 1 (PBS), control 2 (piRNA without AAGUGC motif), and piR11-15 (with AAGUGC motif), and incubated for 96 h. Zygotes injected with piRNAs with AAGUGC motif showed increased rates of survival and development. *P<0.05. **P<0.01. Error bars, S.D. (n=4)
To test the effect of miR-17-5p on piRNA expression, we injected oocytes with miR-17-5p and miR-17-3p. We found that injection with miR-17-5p displayed a general decrease of piRNA expression (Figure 2e). Oocytes were also injected with miR-17-5p inhibitor. The injection displayed a general increase in piRNA levels (Figure 2f).
To test the effect of piRNAs on embryonic survival and development, we injected oocytes with five piRNAs in Group I. The injected oocytes were cultured for 4 days, followed by TUNEL staining. We detected a significant increase in oocyte survival and development (Figure 2g, Supplementary Figure S2b).
Derepression of transposons
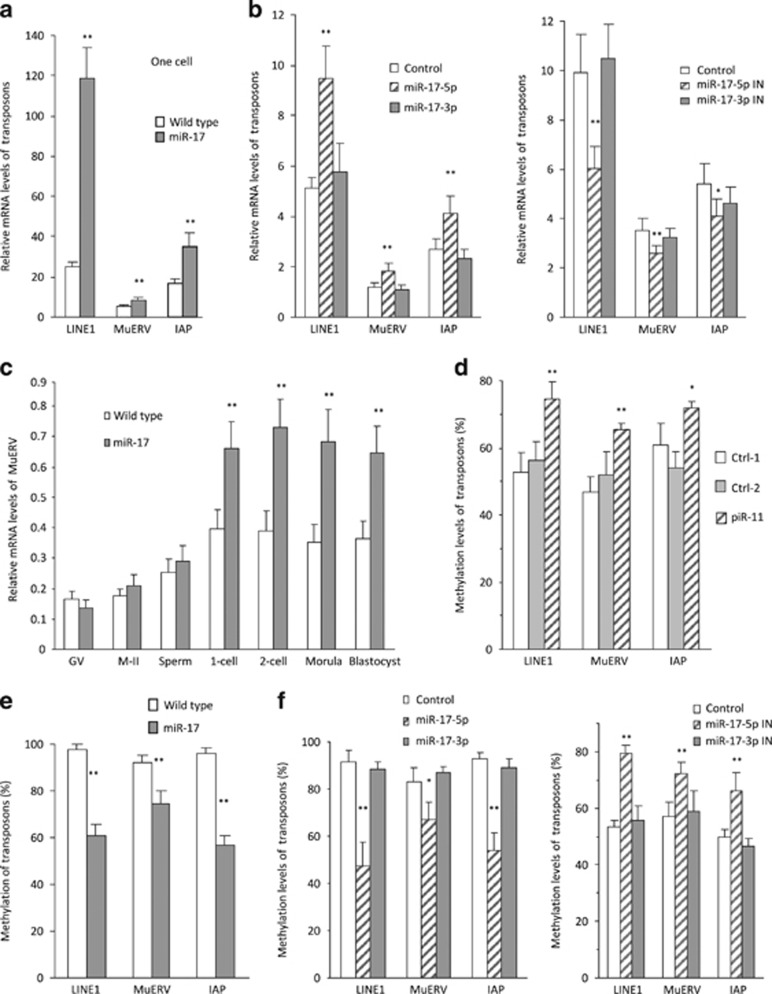
It has been known that piRNAs play roles in repressing transposon activity allowing embryonic development.17 We analyzed transposon levels in the zygotes of miR-17 transgenic and wild-type mice by measuring the levels of transposon markers Line1, MuERV, and IAP.20, 21 Real-time PCR analysis indicated that all of these markers were significantly upregulated in the miR-17 transgenic zygotes compared with wild type (Figure 3a). To dissect the effect of both miR-17-5p and miR-17-3p, we injected wild-type zygotes with miR-17-5p, miR-17-3p, and control oligos, followed by analysis of transposon levels. We found that only miR-17-5p injection increased transposon levels, while injection with miR-17-3p did not produce any effect (Figure 3b).
Figure 3.
Retrotransposons were derepressed in the early embryos of miR-17 transgenic mice. (a) Fifty zygotes were collected for RNA extraction, followed by real-time PCR. miR-17 transgenic zygotes expressed increased mRNA levels of retrotransposon LINE1, MuERV, and IAP. **P<0.01. Error bars, S.D. (n=4). (b) Wild-type zygotes were microinjected with miR-17-5p, miR-17-3p, and control oligos (left), or miR-17-5p inhibitor, miR-17-3p inhibitor, and control oligos (right), followed by incubation for 8 h and real-time PCR. Injection with miR-17-5p enhanced, but injection with miR-17-5p inhibitor inhibited levels of retrotransposon LINE1, MuERV, and IAP. *P<0.05. **P<0.01. Error bars, S.D. (n=4). (c) miR-17 transgenic embryos expressed increased mRNA levels of retrotransposon MuERV in zygote, two-cell, morula, and blastocyst stage. **P<0.01. Error bars, S.D. (n=4). (d) Hundred zygotes were collected for DNA extraction. Methylation-specific PCR (MSP) showed that piR11 injection increased methylation levels of retrotransposon LINE1, MuERV, and IAP. *P<0.05. **P<0.01. Error bars, S.D. (n=4). (e) MSP analysis indicated that methylation levels of retrotransposon LINE1, MuERV, and IAP decreased in the zygotes (100) isolated from miR-17 transgenic mice. **P<0.01. Error bars, S.D. (n=4). (f) MSP analysis indicated that methylation levels of retrotransposon LINE1, MuERV, and IAP decreased in zygotes injected with miR-17-5p (left), but increased in zygotes injected with miR-17-5p inhibitor (right). *P<0.05. **P<0.01. Error bars, S.D. (n=4)
To examine the general effects of miR-17 on derepression of transposon, we isolated RNAs from GV, M-II, sperm, one-cell embryos, two-cell embryos, morulas, and blastocysts. We confirmed that the levels of MuERV (Figure 3c), Line1, IAP, and MT (Supplementary Figure S2c) were significantly upregulated mainly in one-cell to morula stages.
It has been known that piRNAs play roles in repressing transposons.17 We injected zygotes with piRNA (piR-11) and confirmed that injection with piR-11 repressed transposon levels (Supplementary Figure S2d). Further analysis indicated that injection with piR-11 enhanced methylation levels of retrotransposon LINE1, MuERV, and IAP (Figure 3d). To test whether piRNAs might be mediating miR-17's effect on methylation of transposons, we analyzed the transposon levels of zygotes isolated from miR-17 transgenic and wild-type mice. Methylation levels of Line1, MuERV, and IAP were significantly lower in the miR-17 transgenic zygotes than the wild-type zygotes (Figure 3e). To validate the effect of miR-17 on reducing transposon level, isolated zygotes were injected with miR-17-5p, miR-17-3p, and control oligos. The experiment confirmed that injection with miR-17-5p decreased transposon methylation, while injection with miR-17-5p inhibitor increased it (Figure 3f).
miR-17-5p targeted Mili and Dnmt3a
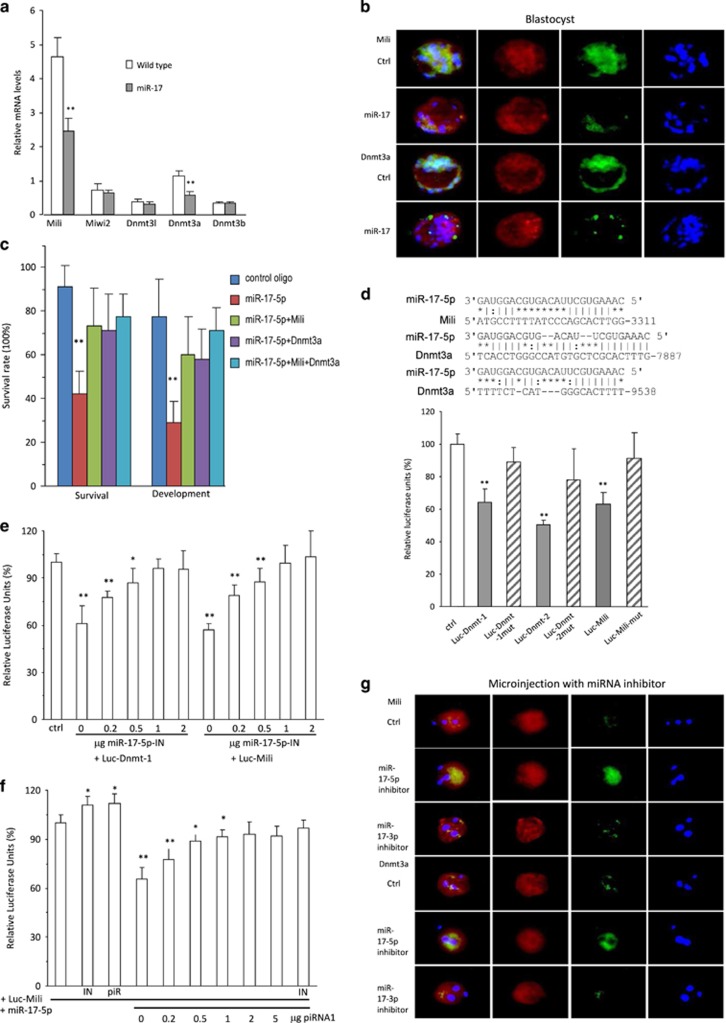
We analyzed expression of a number of enzymes responsible for DNA methylation, including Mili, Mili2, Dnmt3a, Dnmt3b, and Dnmt3I, by real-time PCR. We found that Mili and Dnmt3a were significantly repressed in the miR-17 transgenic zygotes compared with the wild-type zygotes (Figure 4a). We examined expression of these two proteins in blastocysts and found that they were largely repressed in the miR-17 transgenic blastocysts compared with the wild-type blastocysts (Figure 4b) and other stages, including zygotes, two-cell, and morula (Supplementary Figure S3a). Nevertheless, significant repression of Mili and Dnmt3a mRNA was only detected in the one-cell and two-cell stages (Supplementary Figure S3b). Consistent with these results, we did not detect repression of Mili and Dnmt3a proteins in the ovaries and testis of miR-17 transgenic mice (Supplementary Figure S3c). To corroborate these results, we co-injected zygotes with miR-17-5p and purified Dnmt3a or Mili. While injection with miR-17-5p inhibited zygote development, co-injection with Dnmt3a and/or Mili significantly abolished the miR-17-5p effect (Figure 4c).
Figure 4.
miR-17 targeted Mili and Dnmt3a. (a) Real-time PCR analysis showed that miR-17 transgenic zygotes expressed decreased mRNA levels of Mili and Dnmt3a. **P<0.01. Error bars, S.D. (n=4). (b) Embryos were stained with phalloidins (red), DAPI (blue), and green fluorescence, showing expression of Mili or Dnmt3a. miR-17 transgenic embryos expressed decreased levels of Mili and Dnmt3a protein in zygote. (c) Wild-type zygotes were microinjected with control oligos, miR-17-5p, purified Mili, or purified Dnmt3a, and incubated for 96 h. Zygotes injected with miR-17-5p showed decreased survival and development, which could be rescued by Mili and Dnmt3a. **P<0.01. Error bars, S.D. (n=4). (d) Upper, Potential targets of miR-17-5p in the 3′UTRs of Mili and Dnmt3a. Lower, Decreased luciferase activities were observed in cells co-transfected with miR-17-5p and luc-Dnmt-1, luc-Dnmt-2, or luc-Mili, which was reversed when the miR-17-5p binding site was mutated (luc-Dnmt-1mut, luc-Dnmt-2mut, or luc-Mili-mut). **P<0.01. Error bars, S.D. (n=4). (e) Decreased luciferase activities were observed in cells co-transfected with miR-17-5p and Luc-Dnmt-1 or Luc-Mili, which was reversed when co-transfected with miR-17-5p inhibitor. **P<0.01. Error bars, S.D. (n=4). (f) Decreased luciferase activities were observed in cells co-transfected with miR-17-5p and Luc-Mili, which was reversed when co-transfected with piRNA1. IN, miR-17-5p inhibitor; piR, piRNA1. **P<0.01. Error bars, S.D. (n=4). (g) Transgenic zygotes were microinjected with 2 pl control oligos, miR-17-5p inhibitor (IN), or miR-17-3p IN, incubated for 8 h, and processed to immunofluorescence staining. Injection with miR-17-5p IN enhanced Mili and Dnmt3a expression in the zygotes
Computational analysis (http://bio.sz.tsinghua.edu.cn/) indicated that both Mili and Dnmt3a were the potential targets of miR-17-5p (Figure 4d, upper). We generated luciferase constructs harboring the potential target sites and mutant constructs in which the miR-17-5p binding sites were mutated. Luciferase assays showed reduction of luciferase activities when the constructs containing miR-17-5p binding sites were co-transfected with miR-17-5p mimic (Figure 4d, lower). Mutations of the binding sites abolished the repression effect of miR-17-5p.
We confirmed the effect of miR-17-5p on regulating Dnmt3a and Mili by silencing miR-17-5p. Transfection with miR-17-5p inhibitor decreased the effect of endogenous miR-17-5p on the activities of Luc-Dnmt3a-1 and Luc-Mili in a dose-dependent manner (Figure 4e). Co-transfection with piRNA also decreased the effect of miR-17-5p on regulating Luc-Mili activity in a dose-dependent manner (Figure 4f).
To further validate that effect of miR-17-5p, we injected zygotes with miR-17-5p, miR-17-3p, and control oligos, followed by immunostaining with antibodies against Mili and Dnmt3a. We confirmed repression of Mili and Dnmt3a when the zygotes were injected with miR-17-5p (Supplementary Figure S4a). The levels of Mili and Dnmt3a mRNAs were also repressed by miR-17-5p (Supplementary Figure S4b).
We then tested the effect of endogenous miR-17-5p by injecting zygotes with the inhibitors of miR-17-5p, miR-17-3p, and control oligos. Upregulation of Mili and Dnmt3a was detected in zygotes injected with miR-17-5p inhibitor at the protein (Figure 4g) and mRNA (Supplementary Figure S4c) levels. Consistently, zygotes injected with Mili and/or Dnmt3a increased rates of survival and development, while injection with anti-Mili and/or anti-Dnmt3a antibodies decreased these processes (Supplementary Figure S4d). Injection of Mili and Dnmt3a decreased zygote apoptosis (Supplementary Figure S4e, left), but injection of the antibodies increased apoptosis (Supplementary Figure S4e, right).
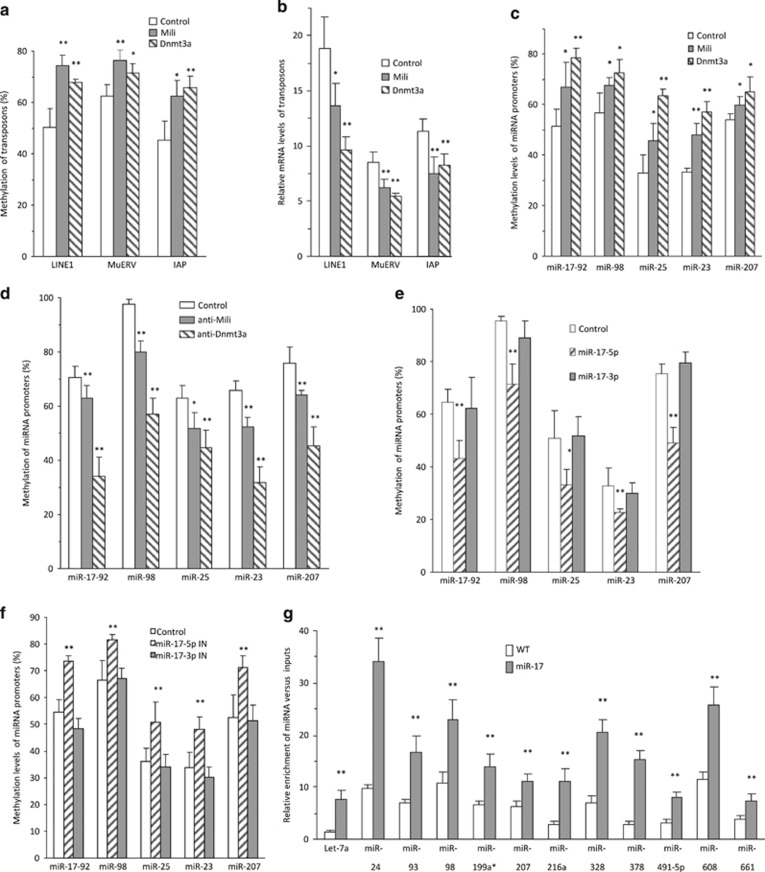
To validate the methylation activity of Mili and Dnmt3a, we injected zygotes with purified Mili and Dnmt3a or antibodies against these proteins, followed by methylation analysis. We confirmed that injection with Mili and Dnmt3a increased methylation of Line1, MuERV, and IAP (Figure 5a), while injection with the antibodies inhibited these processes (Supplementary Figure S5a). As a consequence, injection with Mili and Dnmt3a decreased transposon levels (Figure 5b), while injection with the antibodies increased transposon levels (Supplementary Figure S5b). Injection of Mili and Dnmt3a methylated all miRNA promoters tested (Figure 5c) and decreased expression of miRNAs driven by these promoters (Supplementary Figure S5c). On the other hand, injection with anti-Mili or anti-Dnmt3a antibodies decreased promoter methylation (Figure 5d) and increased miRNA expression (Supplementary Figure S5d).
Figure 5.
Methylation activities of Mili and Dnmt3a. (a) MSP analysis indicated that injection of Mili or Dnmt3a into 100 zygotes enhanced methylation levels of retrotransposon LINE1, MuERV, and IAP. *P<0.05. **P<0.01. Error bars, S.D. (n=4). (b) miR-17 zygotes were microinjected with 2 pl control 1 (PBS), control 2 (mouse Ig), Mili, Dnmt3a, and Mili+Dnmt3a, incubated for 8 h, followed by real-time PCR. Injection with Mili or Dnmt3a repressed mRNA levels of LINE1, MuERV, and IAP. *P<0.05. **P<0.01. Error bars, S.D. (n=4). (c) MSP analysis indicated that injection with Mili or Dnmt3a enhanced methylation levels of the miRNA genes in zygotes. *P<0.05. **P<0.01. Error bars, S.D. (n=4). (d) Injection with anti-Mili or anti-Dnmt3a antibody decreased the methylation levels of the miRNA genes in zygotes. *P<0.05. **P<0.01. Error bars, S.D. (n=4). (e) Injection with miR-17-5p mimic repressed methylation levels of miRNA genes in zygotes. *P<0.05. **P<0.01. Error bars, S.D. (n=4). (f) Injection with miR-17-5p-IN enhanced methylation levels of miRNA genes. *P<0.05. **P<0.01. Error bars, S.D. (n=4). (g) Hundred zygotes were collected, lysed, and precipitated with antibody against AGO2. Precipitated AGO2 pulled down more miRNAs in the zygotes of miR-17 transgenic mice. **P<0.01. Error bars, S.D. (n=4)
To analyze the observed decrease in methylation as a result of repressing Mili and Dnmt3a, we injected zygotes with miR-17-5p mimic and detected decreased methylation of miRNA promoters (Figure 5e) but increased expression of miRNAs associated with these promoters (Supplementary Figure S6a). Injection of miR-17-5p inhibitor increased methylation of these promoters (Figure 5f) but decreased expression of these miRNAs (Supplementary Figure S6b). Consistently, we detected increased expression of a number of miRNAs in the miR-17 transgenic mice as compared with wild-type mice (Supplementary Figure S6c, left), which occurred through decreased methylation of miRNA promoters (Supplementary Figure S6c, right). We examined whether the increased miRNA levels occurred through a normal miRNA biogenesis pathway. The levels of miRNAs in the zygotes of miR-17 mice were upregulated when we performed immunoprecipitation with anti-Ago2 antibody (Figure 5g).
piRNAs compete with miRNAs
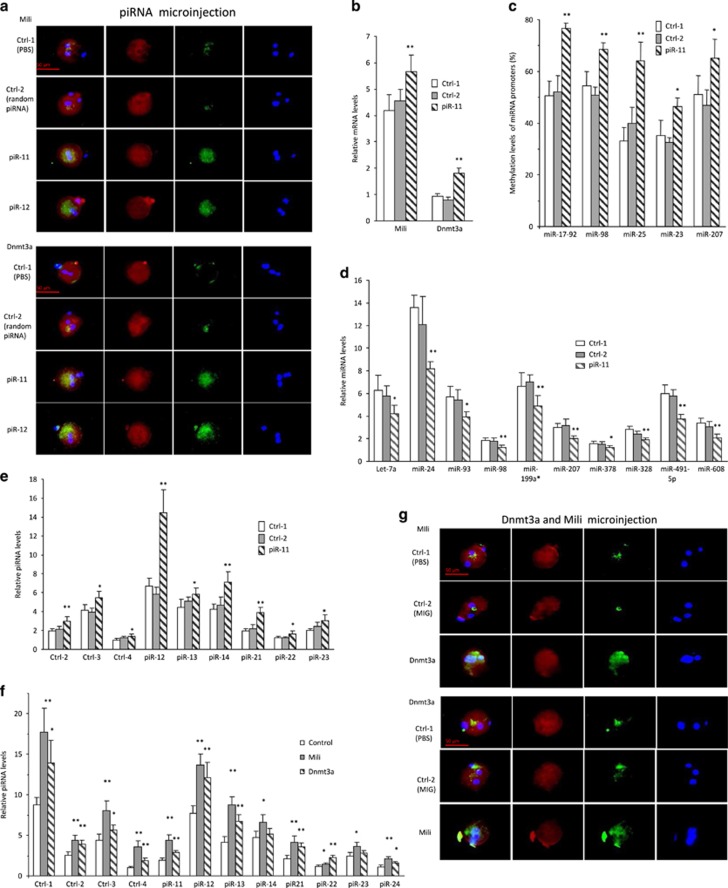
We tested whether piRNAs with sequences homologous to the miR-17-5p seed region could compete with miR-17-5p function in repressing Mili and Dnmt3a expression. Zygotes were injected with piR-11 and piR-12 followed by immunostaining for Mili and Dnmt3a levels. We found that injection with piRNAs increased Mili and Dnmt3a expression (Figure 6a). Their mRNA levels were also found to be higher by injecting zygotes with piR-11 (Figure 6b). Promotion of Dnmt3a expression by piR-11 injection was consequently followed by methylation of miRNA promoters tested (Figure 6c) and downregulation of miRNA expression (Figure 6d). Promotion of Dnmt3a and Mili by piR-11 injection was also found to increase piRNA expression (Figure 6e). To confirm the effects of Mili and Dnmt3a on piRNA expression, we injected zygotes with Mili and Dnmt3a. We confirmed that injection of the zygotes with Mili and Dnmt3a increased not only the piRNAs with sequences homologous to the miR-17-5p seed region, but also other unrelated piRNAs (Figure 6f), suggesting a general effect of these two proteins on piRNA biogenesis. Injection with antibodies against Mili and Dnmt3a decreased piRNA expression (Supplementary Figure S7a). When piRNA expression was upregulated as a consequence of increased Mili and Dnmt3a levels, the increased piRNAs would compete with miR-17-5p function. As a consequence, expression of Mili and Dnmt3a was found to be upregulated at the mRNA (Supplementary Figure S7b) and protein (Figure 6g) levels.
Figure 6.
Functions of piRNAs. (a) Transgenic zygotes were microinjected with 2 pl control 1 (PBS), control 2, piR-11, and piR-12, and incubated for 8 h, followed by immunofluorescence staining. Injection of piRNAs enhanced Mili and Dnmt3a expression. (b) Transgenic zygotes were microinjected with 2 pl control 1 (PBS), control 2, and piR-11, and incubated for 8 h. Injection with piR-11 increased mRNA levels of Mili and Dnmt3a. **P<0.01. Error bars, S.D. (n=4). (c) MSP analysis showed that injection with piR-11 enhanced the methylation levels of miR-17-92, miR-98, miR-25, miR-23, and miR-207 in zygotes. *P<0.05. **P<0.01. Error bars, S.D. (n=4). (d) Injection with piR-11 repressed miRNA levels in zygotes. *P<0.05. **P<0.01. Error bars, S.D. (n=4). (e) Injection with piR-11 enhanced piRNA levels in zygotes. *P<0.05. **P<0.01. Error bars, S.D. (n=4). (f) Injection with Mili or Dnmt3a enhanced piRNA expression in zygotes. *P<0.05. Error bars, S.D. (n=4). (g) Injection with Mili increased Dnmt3a expression, while injection with Dnmt3a enhanced Mili expression in the zygotes
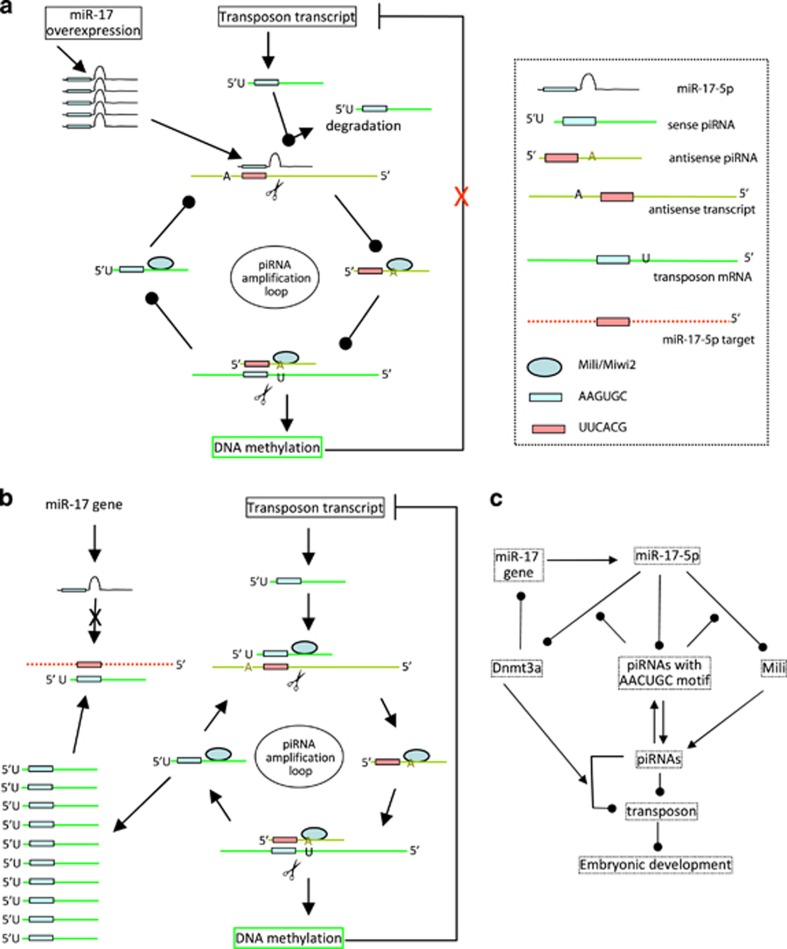
In summary, we demonstrated a relationship between miRNAs and piRNAs in regulating mouse fertility and early embryonic development. Ectopic expression of miR-17-5p suppressed the amplification loop of piRNAs with identical sequences in the 5′ of the piRNAs to the seed region of miR-17-5p. This silenced the effect of piRNAs in the repression of transposons, leading to embryonic infertility (Figure 7a). On the other hand, increased levels of piRNAs blocked the function of miR-17-5p in gene targeting (Figure 7b). For instance, miR-17-5p repressed essential components of the piRNA amplification loop, Mili/Miwi2, and the DNA methytransferase, Dnmt3a, both of which play important roles in regulating piRNA activities and embryonic development (Figure 7c).
Figure 7.
Functions of piRNAs. (a) miR-17-5p interferes with piRNA amplification loop. (b) piRNAs competitively inhibit miRNA expression and function in mouse germ cells or early embryonic development. (c) Pathway by which miR-17-5p inhibits mouse germ cells or early embryonic development
Discussion
As the largest class of small non-coding RNAs, piRNAs can repress the functions of their targets with the assistance of piwi proteins, members of the Argonautes, in a manner similar to miRNAs.22 The piRNAs differ from microRNA in size. While miRNAs have 18–24 nucleotides, piRNAs are 26–31 nucleotides in size. miRNAs are highly conserved across different species, whereas piRNAs lack sequence conservation with increased complexity. The number of unique piRNAs can be as many as 4–5-folds that of miRNAs. The relationship between miRNAs and piRNAs in the early stages of development is summarized in a review article.23 A recent study showed that miRNAs play roles in the biogenesis and stability of Drosophila piRNAs.24 In mammals, it remains unclear how piRNAs are generated, although it is certain that the biogenesis pathway of piRNAs is distinct from that of miRNAs.25 The possible mechanism of the piRNA biogenesis pathway appears to be a ‘Ping-Pong' mechanism. Evidence was obtained from the observation that piRNA sequences have a strand bias: they are the products of one DNA strand, suggesting that they are produced from a single-strand transcript.13 This ‘Ping-Pong' mechanism is also called the piRNA amplification loop (Figure 7b). In the loop, piRNA precursors are transcribed, producing piRNAs with a tendency to target 5′ uridines.26 In addition, it is proposed that the primary piRNAs bind to their complementary targets followed by the recruitment of the Piwi proteins, or the Argonautes. This induces cleavage of the transcript at a point 10 nucleotides away from the 5′ end of the primary piRNA, producing a secondary piRNA.26 This amplification loop can generate a large number of piRNAs with complexity.
In the early stages of embryonic development, it is essential to produce a large number of piRNAs with significant complexity. These piRNAs can induce DNA methylation and inhibit transposon activity, representing an essential step in embryonic development. We have found that ectopic expressed miR-17-5p competed with endogenous piRNAs in the generation of piRNAs in the amplification loop. Specifically, the seed region of miR-17-5p competed with piRNAs whose 5′' segments contain sequences identical to the seed region of miR-17-5p. This does not allow the piRNAs to bind to their target transposons. As a result, the piRNA amplification loop is disrupted by miR-17-5p overexpression, leading to increased activities of transposons and inhibition of embryonic development.
Although our results showed that only the piRNAs with sequences homology to the seed region of miR-17-5p had roles in embryonic development, other piRNAs, besides those used as controls in this study, may also have roles in development by way of targeting developmental element transposons. Furthermore, miR-17-5p may target other molecules that also have roles in embryonic development, as a miRNA can potentially target several hundreds of mRNAs.1, 2 As well, other miRNAs may target Mili and Dnmt3a and modulate embryonic development, as an mRNA can be targeted by many miRNAs.27 Nevertheless, miR-17-3p, a miRNA generated from the same precursor of miR-17, was not able to repress expression of Mili and Dnmt3a and had no effect on embryonic development. This is different from our previous study showing that both miR-17-5p and the passenger strand miR-17-3p could target the same molecule, TIMP3, leading to increased prostate tumor growth and invasion.28 It suggests that the micro-environments are essential in determining the function of a miRNA.
Reciprocally, our results showed that increased levels of piRNAs with sequences identical to the seed region of miR-17-5p could block the activity of miR-17-5p in gene targeting. The results suggest that these piRNAs could bind to the potential targets of miR-17-5p without repressing the expression and functions of the targets. Instead, they interfere with the binding of miR-17-5p to the potential targets. Our results also confirm that the functions of piRNAs are distinct from those of miRNAs, as they use different Argonaute proteins.
In a cell, a miRNA or a piRNA may predominate the function of the others, as the number of Argonaute molecules cannot be unlimitedly expressed. As such, the miRNAs with significantly increased levels may predominantly occupy Argonaute molecules, resulting in the degradation of other miRNAs or piRNAs,29 following the strategy of 'Use it or lose it'. Therefore, a distinct function of infertility could be clearly observed in the miR-17 transgenic mice. In the embryonic injection, we anticipate that the levels of increased miR-17-5p or piRNAs could be much higher than those in physiological conditions, resulting in amplified effects of the microinjection.
In our study, we identified two important targets of miR-17-5p: Miwi2/Mili and Dnmt3a, both of which have essential roles in embryonic development. While MIWI2/Mili is essential for spermatogenesis and repression of transposons in the mouse male germ line,30 Dnmt3a is an enzyme that catalyzes DNA methylation.31 While DNA methylation facilitates the inhibitory effect of piRNAs on transposon expression and has important roles in embryonic development,32 we found that Dnmt3a could methylate the miR-17 promoter, producing a feed-forward loop. This feed-forward loop maintained high levels of miR-17-5p, leading to embryonic infertility. For example, when the level of miR-17-5p is high, it would repress Dnmt3a expression. This would decrease methylation of the miR-17 promoter, allowing increased expression of miR-17-5p. When the level of miR-17-5p is low, its inhibitory effect on Dnmt3a expression is suspended. The increased expression of Dnmt3a could methylate the miR-17 promoter, decreasing miR-17-5p expression. This feed-forward loop may explain the in vivo effect of infertility in the mice expressing miR-17-5p. This feed-forward loop appears to play essential roles in the early stages of development. The clinical significance of this feed-forward loop awaits further investigation.
Materials and Methods
Materials
The monoclonal antibodies against Mili and Ago2 were obtained from Cell Signaling (Danvers, MA, USA). The monoclonal antibodies against Dnmt3a and β-actin were obtained from Abcam (Cambridge, MA, USA). RNA and DNA extract kits, miRNA RT and PCR kits, and EpiTect Bisulfite kit were obtained from Qiagen (Hilden, Germany). M2 embryo culture medium, pregnant mare serum gonadotropin (PMSG) and human chronic gonadotropin (hCG) were from Sigma-Aldrich (St. Louis, MO, USA), and CZB medium was obtained from EMD Millipore (Billerica, MA, USA).
Constructs and primers
Plasmid miR-17 and control plasmid were generated previously. Both plasmids contain a Bluescript backbone, a CMV promoter driving green fluorescent protein (GFP) expression, and a human H1 promoter driving pre-miR-17 or a non-related sequence serving as a control. The pMir-Report (Ambion, Frederick, MD, USA), luciferase reporter vector, was used to generate the luciferase constructs. A fragment of the 3'-untranslated region (3'UTR) of mouse Mili and Dnmt3a was cloned by RT-PCR. Two primers, musMili-SacI and musMili-MluI, were synthesized to clone the 3'UTR fragment of Mili. All primer sequences used are listed in Supplementary Figure S7c. Two 3'UTR fragments of Dnmt3a were cloned with the primers musDnmt3a-1-SacI and musDnmt3a-1-MluI, and primers musDnmt3a-2-SacI and musDnmt3a-2-MluI. The PCR products were digested with SacI and MluI and the fragments were inserted into the luciferase vector to obtain Luc-Mili, Luc-Dnmt3a-1, and Luc-Dnmt3a-2. The mutant constructs Luc-Mili-mut mutation of the miR-17-5p binding site were generated by using the PCR approach with primers musMili-SacI and musMili-MluI-mut. Primers musDnmt3a-1-ScaI and musDnmt3a-1-MluI-mut, and primers musDnmt3a-2-ScaI and musDnmt3a-2-MluI-mut were used to generate the Luc-Dnmt3a-1-mut and Luc-Dnmt3a-2-mut constructs. A non-related sequence, amplified from the coding sequence of the chicken versican G3 domain, was used as a negative control, as previously.
Transgenic mice
The miR-17 transgenic mice were generated previously.11 Transgenic lines were maintained by backcrossing with F1 (C57BL/6XCBA) wild-type mice. Male mice used in this study were between 8 to 12 weeks of age, and the female between 3 to 4 weeks. They were maintained under 12 h light/ 12 h dark photoperiod at a room temperature of 22–23 °C and relative humidity 50%. All animal experiments were approved by the Animal Care Committee of Sunnybrook Research Institute, Ontario, Canada. Genotyping was performed by PCR, using primers EGFP-347F pairing with EGFP-668R for CMV promoter and EGFP981F pairing with EGFP-CApaI for GFP coding sequence, with tail snip or ear punch DNA as template. Mouse GAPDH served as a control using primers muGapdh1F and muGapdh250R.
Collection of epididymal sperms and oocytes
The cauda epididymidis was removed from male mice and the epididymal fluid was squeezed out and placed on the bottom of a 1.5-ml tube containing 0.5 ml CZB medium for use. Fully grown GV oocytes were collected from the ovaries of female mice and cultured under mineral oil (Sigma-Aldrich) in M2 or CZB medium at 37 °C, 5% CO2. For collection of unfertilized or fertilized oocytes, females were induced to superovulate with intraperitoneal injection of 5 IU PMSG and 5 IU hCG given 48 h apart. To obtain unfertilized oocytes, the PMSG and hCG were injected, and the oviducts were removed 14 h after the injection of hCG. The cumulus−oocyte complexes were released from the oviducts into 0.1% bovine testicular hyaluronidase in M2 medium to disperse cumulus cells. The cumulus-free oocytes were washed with M2 medium and cultured in M2 or CZB medium. To collect fertilized oocytes (embryos), PMSG and hCG were injected. Females were mated with males after hCG injection, and the oviducts and uteruses were removed 24, 48, 72 or 96 h after hCG injection. All the fertilized oocytes (embryos) were collected from oviducts or uteruses under the dissecting microscope, washed with M2 medium, and in M2 or CZB medium.
Immunofluorescent staining
The early embryos were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) for 20 min. After washing three times in PBS, embryos were treated with 0.5% Triton X-100 (Sigma-Aldrich) in PBS for 30 min, blocked with 10% goat serum for 1 h at room temperature, and then incubated with primary antibody in PBS containing 10% goat serum albumin overnight. The embryos were washed and stained with goat anti-rabbit Alexa 488 (Life Technologies, Carlsbad, CA, USA) at room temperature for 1 h. Fluorescent phalloidins, Acti-stain 555 (Sigma-Aldrich), was used to stain F-actin to show cell structure. DNA staining was performed with DAPI. Confocal laser scanning microscopy was performed using an LSM 510 Meta microscope (Carl Zeiss, Oberkochen, Germany).
Immunohistochemistry
We followed the techniques as described.33 Briefly, tissues were freshly excised and fixed in 10% formalin over 24 h, immersed in 70% ethanol, embedded in wax, and sectioned. The sections were de-paraffinized with xylene and ethanol and then boiled in a pressure cooker. After blocked with 10% goat serum for half an hour, all samples were incubated with primary antibody at 4 °C overnight. The sections were incubated with biotinylated secondary antibody for 1–2h, followed by avidin conjugated horseradish peroxidase provided by the Vectastain ABC kit (Vector, PK-4000, Burlingame, CA, USA). The slides were then stained with DAB followed by Mayer's Hematoxylin counterstaining.
Protein extract and purification
To cross-link antibodies to protein A-Sepharose, a slurry of Protein A-Sepharose was incubated with antibodies to destined proteins at 4 °C for 2 h, and incubated in borate/NaCl buffer (0.04 M boric acid, 40 mM sodium tetraborate decahydrate, 3M NaCl, pH 9.0) with 20 mM DMP at room temperature for 30 min. Unreacted DMP was quenched with ethanolamine buffer (200 mM ethanolamine, pH 8.0) for 2 h, and the cross-linked beads were kept in PBS with 0.01% merthiolate (Sigma-Aldrich) at 4 °C. The ovary tissues were lysed and incubated with antibodies (against Mili or Dnmt3a) cross-linked with protein A-Sepharose at 4 °C for 2 h, mixed with 3 volumes of 0.2 M glycine (pH 2.6) and centrifuged. The eluted supernatant was neutralized by equal volume of 1 M Tris buffer (pH 8.0). The precipitated proteins were purified and concentrated with Amicon Ultra Centrifugal Filters (Merck Millipore, Darmstadt, Germany) and quantified before microinjection.
Microinjection
Microinjection was performed with Eppendorf microinjector 5246. Zygotes collected in M2 medium at 24 h after hCG injection were microinjected with 2 pl of 20–100 ng/μl miRNA mimics, miRNA antisenses, or piRNA mimics. Approximately 1–2 pl of 200 ng/μl proteins or antibodies were microinjected into the cytoplasm of the above early zygotes. After injection, the embryos were cultured in CZB medium under mineral oil at 37 oC in an atmosphere of 5% CO2 for 8 or 96 h.
Real-time PCR
For mature miRNA, piRNA and endogenous siRNA analysis, total RNAs were extracted from 30−50 embryos with mirVana miRNA Isolation Kit (Ambion) according to the manufacturer's instructions. cDNA synthesis was performed with miScript Reverse Transcription Kit (Qiagen), using 1 μg RNA as template as described.34 PCRs were performed with miScriptSYBR GreenPCR Kit (Qiagen) using 1 μl cDNA as templates. Mouse U6 was used as a real-time PCR control.
DNA methylation assay
DNA methylation levels were assessed by Methylation-Specific PCR (MSP). Genomic DNA was extracted from the harvested embryos (80–100) using the QIAmp DNA mini kit (Qiagen). Bisulfite treatment of genomic DNA was performed using the EpiTect Bisulfite Kit (Qiagen) according to the manufacturer's protocol. The MSP primers were designed using the MethPrimer software (http://www.urogene.org/methprimer). PCRs were performed with miScriptSYBR GreenPCR Kit (Qiagen) using 1 μl DNA as templates, with mouse GAPDH as a real-time PCR control.
RNA-binding protein immunoprecipitation assay (RIP)
A total of 80–100 embryos were washed in ice-cold PBS, lysed in 200 μl lysis buffer, and incubated with 5 μg primary antibody at 4 °C for 2 h. Forty microliters of 50% slurry of protein A-Sepharose was added to each sample, and the mixtures were incubated at 4 °C for 4 h. The pellets were washed three times with PBS and resuspended in 0.5 ml Tri Reagent (Sigma-Aldrich). Eluted, coprecipitated RNA in the aqueous was subjected to qRT-PCR analysis to demonstrate the presence of the binding using respective primers.
Luciferase activity assay
A dual-luciferase reporter system (Promega) was used to perform luciferase activity assay as described.35 Briefly, MEF cells were cultured on 12-well tissue culture plates at a density of 2 × 105 cells per well. Cells were co-transfected with the luciferase reporter constructs, corresponding miRNA mimics, and Renilla luciferase construct for 5–6 h. After culturing for 24 h, the transfected cells were lysed by 150 μl of passive lysis buffer. Thirty microliters of lysates was mixed with 50 μl of LAR II, and then firefly luciferase activity was measured by a luminometer. Fifty microliters of Stop & Glo reagent was added to the sample as the internal control.
Statistical analysis
All experiments were performed in triplicate and numerical data were subjected to independent-samples t test. The levels of significance were set at *P<0.05 and **P<0.01.
Acknowledgments
The authors thank Drs. Chengyan He and Ling Fang (China-Japan Union Hospital of Jilin University) and Dr. Xiangling Yang (The Sixth Affiliated Hospital, Sun Yat-sen University) for technical assistance. This work was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC; 227937-2012) to BBY and a grant from the Canadian Institutes of Health Research (CHRP 462341-2014) and Natural Sciences and Engineering Research Council of Canada (CPG 134747) to SNK and BBY. BBY is the recipient of a Career Investigator Award (CI 7418) from the Heart and Stroke Foundation of Ontario. WWD is supported by a fellowship from the Canadian Breast Cancer Foundation of Ontario.
Glossary
- DMEM
Dulbecco's modified Eagle's medium
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- siRNA
small interfering RNA
- miRNA
microRNA
- 3′-UTR
3′-untranslated region
- Dnmt3a
DNA (cytosine-5)-methyltransferase 3A
- Piwi
P-element induced wimpy testis
- MIWI2
mouse piwi 2
- MILI
miwi-like
- piRNA
piwi-interacting RNA
- Line-1
long interspersed elements
- MuERV
murine endogenous retrovirus
- IAP
intracisternal A-particle
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Calin
Supplementary Material
References
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005; 433: 769–773. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008; 455: 58–63. [DOI] [PubMed] [Google Scholar]
- Noren Hooten N, Fitzpatrick M, Wood WH 3rd, De S, Ejiogu N, Zhang Y et al. Age-related changes in microRNA levels in serum. Aging 2013; 5: 725–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang R, Bates DJ, Wang E. Epigenetic control of microRNA expression and aging. Curr Genomics 2009; 10: 184–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Yang X, Fang L, Rutnam ZJ, Yang BB. Misprocessing and functional arrest of microRNAs by miR-Pirate: roles of miR-378 and miR-17. Biochem J 2013; 450: 375–386. [DOI] [PubMed] [Google Scholar]
- Bonauer A, Dimmeler S. The microRNA-17-92 cluster: still a miRacle? Cell Cycle 2009; 8: 3866–3873. [DOI] [PubMed] [Google Scholar]
- Dakhlallah D, Batte K, Wang Y, Cantemir-Stone CZ, Yan P, Nuovo G et al. Epigenetic regulation of miR-17~92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med 2013; 187: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian S, Hong J, Li Q, Schebelle L, Pollock A, Knauss JL et al. MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep 2013; 3: 1398–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh T, Aprea J, Nardelli J, Engel H, Selinger C, Mombereau C et al. MicroRNAs establish robustness and adaptability of a critical gene network to regulate progenitor fate decisions during cortical neurogenesis. Cell Rep 2014; 7: 1779–1788. [DOI] [PubMed] [Google Scholar]
- Hackl M, Brunner S, Fortschegger K, Schreiner C, Micutkova L, Muck C et al. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell 2010; 9: 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW et al. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol 2009; 11: 1031–1038. [DOI] [PubMed] [Google Scholar]
- Rutnam ZJ, Du WW, Yang W, Yang X, Yang BB. The pseudogene TUSC2P promotes TUSC2 function by binding multiple microRNAs. Nat Commun 2014; 5: 2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Kingston RE, Lau NC. The coming of age for Piwi proteins. Mol Cell 2007; 26: 603–609. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 2011; 12: 246–258. [DOI] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S et al. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 2007; 13: 1911–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell 2009; 136: 656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 2008; 31: 785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development 2008; 135: 3–9. [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell 2007; 129: 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrusan G, Zhang Y, Szilagyi A. Structure prediction and analysis of DNA transposon and LINE retrotransposon proteins. J Biol Chem 2013; 288: 16127–16138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet 2006; 2: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda M, Cifuentes D, Izumi N, Sakaguchi Y, Suzuki T, Giraldez AJ et al. Poly(A)-specific ribonuclease mediates 3'-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep 2013; 5: 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Blelloch R. Small RNAs in early mammalian development: from gametes to gastrulation. Development 2011; 138: 1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugat B, Akkouche A, Serrano V, Armenise C, Li B, Brun C et al. MicroRNA-dependent transcriptional silencing of transposable elements in Drosophila follicle cells. PLoS Genet 2015; 11: e1005194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Marinov GK, Aravin AA. A transgenerational process defines piRNA biogenesis in Drosophila virilis. Cell Rep 2014; 8: 1617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007; 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ et al. Combinatorial microRNA target predictions. Nat Genet 2005; 37: 495–500. [DOI] [PubMed] [Google Scholar]
- Yang X, Du WW, Li H, Liu F, Khorshidi A, Rutnam ZJ et al. Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res 2013; 41: 9688–9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai ZS, Pasquinelli AE. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol 2010; 17: 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 2007; 12: 503–514. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 1998; 19: 219–220. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999; 99: 247–257. [DOI] [PubMed] [Google Scholar]
- Shan SW, Fang L, Shatseva T, Rutnam ZJ, Yang X, Du W et al. Mature miR-17-5p and passenger miR-17-3p induce hepatocellular carcinoma by targeting PTEN, GalNT7 and vimentin in different signal pathways. J Cell Sci 2013; 126(Pt 6): 1517–1530. [DOI] [PubMed] [Google Scholar]
- Du WW, Fang L, Li M, Yang X, Liang Y, Peng C et al. MicroRNA miR-24 enhances tumor invasion and metastasis by targeting PTPN9 and PTPRF to promote EGF signaling. J Cell Sci 2013; 126(Pt 6): 1440–1453. [DOI] [PubMed] [Google Scholar]
- Li H, Yang BB. Stress response of glioblastoma cells mediated by miR-17-5p targeting PTEN and the passenger strand miR-17-3p targeting MDM2. Oncotarget 2012; 3: 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.