Abstract
A major function of ubiquitylation is to deliver target proteins to the proteasome for degradation. In the apoptotic pathway in Drosophila, the inhibitor of apoptosis protein 1 (Diap1) regulates the activity of the initiator caspase Dronc (death regulator Nedd2-like caspase; caspase-9 ortholog) by ubiquitylation, supposedly targeting Dronc for degradation by the proteasome. Using a genetic approach, we show that Dronc protein fails to accumulate in epithelial cells with impaired proteasome function suggesting that it is not degraded by the proteasome, contrary to the expectation. Similarly, decreased autophagy, an alternative catabolic pathway, does not result in increased Dronc protein levels. However, combined impairment of the proteasome and autophagy triggers accumulation of Dronc protein levels suggesting that autophagy compensates for the loss of the proteasome with respect to Dronc turnover. Consistently, we show that loss of the proteasome enhances endogenous autophagy in epithelial cells. We propose that enhanced autophagy degrades Dronc if proteasome function is impaired.
There are two major catabolic pathways in eukaryotic cells that degrade the bulk of cellular proteins, the ubiquitin–proteasome system (UPS) and macro-autophagy, hereafter referred to as autophagy.1, 2, 3, 4 In the UPS, poly-ubiquitylated proteins are delivered to the 26S proteasome for degradation. The 26S proteasome consists of a 20S catalytic core, flanked by two 19S regulatory complexes.5, 6 The 20S catalytic core is composed of a total of 28 α- and β-type subunits, which are organized in a barrel with four stacked rings. The outer two rings are formed by seven α-type subunits each, the two inner rings by seven β-type subunits each.5 All α- and β-type subunits are needed for structural integrity of the proteasome.7 Three β-type subunits, β1, β2 and β5, have proteolytic activity.6 The 19S regulatory particle is composed of at least 19 subunits involved in recognition of ubiquitin-conjugated substrates, ATP hydrolysis, de-ubiquitination, protein unfolding and feeding of the substrates into the 20S catalytic core for degradation.5 Genetic studies in yeast and Drosophila have revealed that mutations in many subunits of the 20S core and the 19S regulatory domains impair proteasome function.8, 9 Genetic analysis of proteasome function is also of clinical importance as proteasome inhibition may be used as potential antitumor strategy, especially for treatment of multiple myeloma.10, 11, 12, 13
Autophagy is characterized by the formation of double-membrane vesicles termed autophagosomes.14, 15 During autophagosome maturation, cytosolic proteins and entire organelles are trapped and delivered to the lysosome for degradation. Two ubiquitin-like conjugation pathways (autophagy-related-8 (Atg8)/light chain 3 (LC3) and Atg12) are active during maturation of autophagosomes.14, 15 Atg7 is an enzyme 1 (E1)-activating enzyme involved in both conjugation pathways14 and essential for autophagy.16 The incorporation of Atg8 fusion proteins (for example, with green fluorescent protein (GFP) and/or mCherry) into autophagosomes is often used as a marker for autophagosomes17 and autophagic flux.18
Although it was initially assumed that the UPS and autophagy are independent of each other, recent evidence has suggested that there is crosstalk and feedback between the two1, 19, 20, 21, 22, 23, 24, 25, 26, 27 (reviewed by Park and Cuervo,3Wojcik28and Lamark and Johansen29). This is mostly due to the observation that autophagy can also degrade ubiquitylated proteins.30, 31, 32, 33, 34 Mechanistically, adaptor proteins with ubiquitin-binding domains and LC3-interacting regions (LIRs) link poly-ubiquitylated proteins to LC3/Atg8 at the autophagosome.30, 35 Loss of autophagy can lead to the formation of protein aggregates composed of poly-ubiquitylated proteins and ubiquitin-binding proteins. These protein aggregates are frequently associated with neurodegenerative diseases in humans.36, 37, 38
Apoptosis is the major form of cell death and evolutionarily conserved from flies to humans.39, 40 Caspases are highly specific Cys-proteases and are the main effectors of apoptosis. They are produced as inactive zymogens that are activated either through incorporation into large protein complexes, such as the apoptosome (initiator caspases), or by proteolytic processing (effector caspases).41, 42 After activation in the apoptosome, initiator caspases such as caspase-9 and its Drosophila ortholog Dronc (death regulator Nedd2-like caspase), cleave and activate effector caspases, such as caspase-3 and its Drosophila ortholog DrICE (death-related ICE-like protease).40, 43
The activity of caspases is controlled at multiple levels. In addition to zymogen production, apoptosome-mediated activation of caspase-9/Dronc and proteolytic processing of caspase-3/DrICE, caspases are also controlled by ubiquitylation, mediated by inhibitor of apoptosis proteins (IAPs), most notably X-linked IAP (XIAP) in mammals and Drosophila IAP1 (Diap1).43 IAPs carry a RING (really interesting new gene) domain, which has E3 ubiquitin ligase activity.44 In Drosophila, because the RING domain of Diap1 ubiquitylates Dronc,45, 46, 47, 48 it is commonly assumed that this ubiquitylation targets the caspase for proteasome-mediated degradation.45, 46 However, in vivo this has not been observed. On the contrary, we have shown that loss or gain of Diap1 activity does not affect the protein levels of Dronc in surviving cells.49 A similar observation has been reported for DrICE.48 Furthermore, a mouse mutant deleting the RING domain of XIAP does not significantly affect caspase protein levels.50 Therefore, it is currently unclear how the protein levels of Dronc are controlled in living cells to avoid deleterious accumulation and auto-processing of these potentially dangerous proteins.
Here, we report that in epithelial cells of Drosophila eye imaginal discs, the protein levels of Dronc are unaffected by proteasome impairment. Similarly, loss of autophagy by itself does not affect Dronc protein levels. However, simultaneous impairment of the proteasome and autophagy causes accumulation of Dronc suggesting that autophagy can compensate for the loss of the proteasome with respect to Dronc turnover. Consistently, autophagy is enhanced in proteasome mutants in epithelial cells. In summary, these data identify Dronc as a common substrate for both the proteasome and autophagy.
Results
Accumulation of poly-ubiquitylated proteins is a convenient marker for proteasome dysfunction
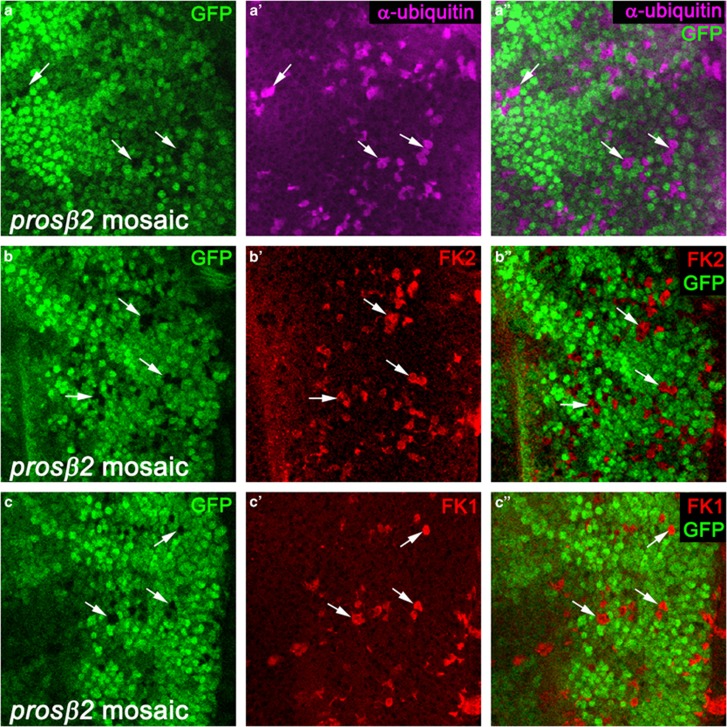
Ubiquitylation of Dronc by Diap1 has previously been observed in vitro.45, 47 To directly test if ubiquitylated Dronc is degraded by the proteasome, we analyzed Dronc protein levels in two mutants affecting the proteasome. The first mutant affects the prosβ2 gene, also known as DTS7 in Drosophila,51 which encodes the β2 subunit of the 20S catalytic core of the proteasome. The proteasome subunit beta 2 (Prosβ2) subunit provides both structural integrity to the proteasome and proteolytic activity.7 The second mutant affects the Mov34 gene (also known as p39B), which encodes a subunit in the 19S regulatory complex, corresponding to regulatory particle non-ATPase-8 (Rpn8) in yeast and S12 in the human regulatory complex.52 Both mutants, prosβ2EP3067 and Mov34k08003, are caused by P element insertions in the first exon, which likely disrupt the transcripts. Both proteasome mutants behave identically in our assays (see below). As these proteasome mutants are homozygous lethal, we induced mutant clones of cells using the ey-Flp/FRT system.53, 54 Not unexpectedly, mutant clones affecting the proteasome are very small and are difficult to identify using negative selection with GFP (Figure 1a–c). However, we were able to positively mark and identify mutant clones using antibodies that recognize ubiquitin or ubiquitin-conjugated proteins. Antibodies raised against ubiquitin display increased immunoreactivity in prosβ2 mutant clones (Figure 1a'). There is a perfect match in the areas lacking GFP, which mark the prosβ2 mutant cells, and increased abundance of ubiquitin labeling (Figure 1a'').
Figure 1.
prosβ2 mutant cells accumulate ubiquitin-conjugated proteins. Shown are high magnification images (x100) of the posterior compartment of prosβ2 mosaic eye imaginal discs labeled for ubiquitin (a), FK2 (b) and FK1 (c). In this and all other figures, posterior is to the right. The FK2 and FK1 antibodies detect ubiquitin-conjugated proteins, but not free unconjugated ubiquitin. The left panels indicate the positions of the proteasome mutant cell clones by absence of GFP, the middle panels show the experiment (in magenta), and the right panels are the merged images of left and middle panels. White arrows mark a few cell clones as examples. Similar data were obtained for Mov34 mosaic discs (Supplementary Figure S1). Wild-type mosaic control discs do not show accumulation of ubiquitin (Supplementary Figure S2). Genotype: ey-FLP; prosβ2EP3067 FRT80/ubi-GFP FRT80
As poly-ubiquitin-conjugated proteins are often subject to proteasome-mediated degradation, we tested whether ubiquitin-conjugated proteins account for the accumulation of ubiquitin in prosβ2 mutants. The FK1 and FK2 antibodies specifically recognize ubiquitin-conjugated proteins, but not unconjugated ubiquitin (FK1 labels poly-ubiquitylated conjugates, FK2 labels mono- and poly-ubiquitylated proteins).55, 56 As shown in Figures 1b' and c', the immunoreactivity of FK1 and FK2 antibodies increases in prosβ2 mutant cells. We also find increased FK1 and FK2 labeling in Mov34 mutant cells (Supplementary Figure S1). In wild-type (wt) control mosaics (wt clones in wt background), an accumulation of ubiquitin and conjugated ubiquitin (FK1 and FK2) is not observed (Supplementary Figure S2) suggesting that ubiquitin-conjugated proteins specifically accumulate in proteasome-deficient cells in vivo, consistent with the expectation.
Mutations in proteasome subunits result in elevated cleaved caspase-3
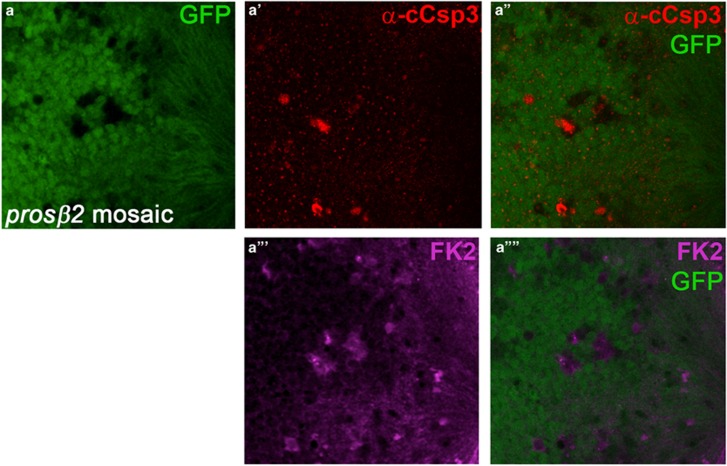
Proteasome inhibitors are used to induce cell death in cancer patients, including those with multiple myeloma.11, 13 Consistently, pharmacological inhibition of the proteasome by Bortezomib in whole flies resulted in DrICE cleavage after 4 days of treatment.27 In contrast, tissue-specific inhibition of the proteasome in Drosophila fat body cells by ribonucleic acid (RNA) interference (RNAi) did not confirm such an apoptotic response.57 As RNAi is known to cause partial loss-of-function phenotypes, we tested the strong prosβ2 mutant allele in mosaic eye imaginal discs for apoptosis induction. We used FK2 labeling to positively mark and identify proteasome mutant cells, and examined the consequence of proteasome dysfunction for the survival of the affected cells. Indeed, we observed increased cleaved Caspase-3 (cCsp-3) antibody labeling in proteasome-deficient cells (Figure 2). However, not all mutant cells contain cCsp-3 staining suggesting that not all of them are apoptotic. Nevertheless, whether apoptotic or not, all proteasome-defective cell clones are very small indicating that the mutant cells do not grow very well.
Figure 2.
Mutations in proteasome subunits result in cell death. Shown are high magnification images (x100) of the posterior compartment of prosβ2 mosaic eye imaginal discs labeled for cCsp-3 (a', a'') and FK2 (a''', a'''') to identify prosβ2 mutant cell clones. cCsp-3 labeling is increased in some, but not all, prosβ2 mutant cells (red in a' and a''). Genotype: ey-FLP; prosβ2EP3067 FRT80/ubi-GFP FRT80
Proteasome subunit mutant cells accumulate Diap1, but do not affect Dronc protein levels
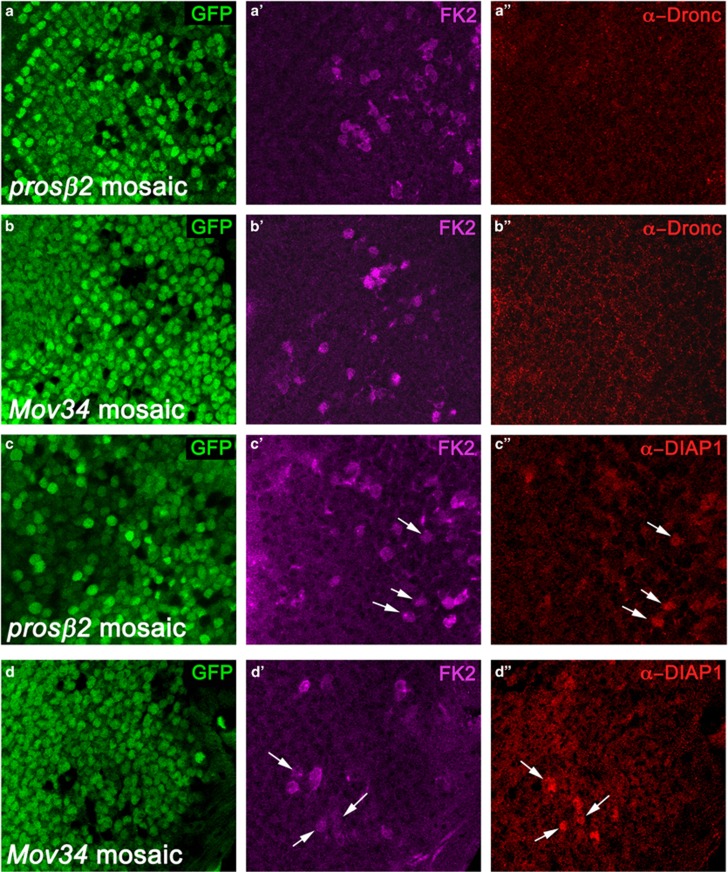
As Diap1 can ubiquitylate Dronc in vitro,45, 47 it was predicted that Dronc is subject to proteasome-mediated degradation in surviving cells.45, 46 Therefore, it would be expected that the protein levels of Dronc would accumulate in proteasome mutants. We tested this expectation by analyzing Dronc protein levels in prosβ2 and Mov34 mutant cells located in the developing posterior eye imaginal disc of third instar larvae. We chose the larval posterior eye imaginal disc in these analyses because under normal conditions, there is no developmental apoptosis in this tissue, thus avoiding complications with physiological apoptosis. Surprisingly, Dronc protein levels are not detectably altered in prosβ2 and Mov34 mutant cells (Figures 3a–b''). We also obtained the same results using a different prosβ2 mutant allele, the commonly used DTS7 allele, which we used in genetic mosaics (Supplementary Figure S3). However, under these conditions, the DTS7 allele appears to be weaker than the allele used in Figure 3a, because it did not cause accumulation of poly-ubiquitylated proteins (FK1 antibody labeling) and the mutant clones grow much larger (Supplementary Figure S3). The specificity of this Dronc antibody was previously established in dronc mosaic eye and wing imaginal discs, and this antibody can also detect accumulation of Dronc protein.49
Figure 3.
Diap1, but not Dronc, accumulate in proteasome mutant clones. Shown are high magnification images (x100) of the posterior compartment of prosβ2 (a and c) and Mov34 (b and d) mosaic eye imaginal discs labeled for Dronc (a and b) and Diap1 (c and d). FK2 labeling was used to identify mutant clones. The left panels indicate the positions of the proteasome mutant cell clones by absence of GFP. In the middle panels, the proteasome mutant cell clones are positively marked by FK2 labeling (in magenta). The right panels show the Dronc (a'' and b'') and Diap1 labelings (c'' and d'') in red. White arrows mark a few cell clones as examples. See also related Supplementary Figures S3 and S4. Genotype in (a and c): ey-FLP; prosβ2EP3067 FRT80/P[ubi-GFP] FRT80. Genotype in (b and d): ey-FLP; FRT42D Mov34k08003/FRT42D P[ubi-GFP]
To confirm the lack of Dronc accumulation in proteasome mutants, we analyzed the protein levels of Flag-tagged Dronc58 expressed in proteasome mosaic eye imaginal discs. Using the Flag antibody as a tool to monitor Dronc levels, we also did not detect an accumulation of Flag-Dronc in Mov34 mutant cells (Supplementary Figure S4).
As a positive control, we analyzed Diap1 protein levels in proteasome mutant cells as Diap1 degradation has been demonstrated in vivo.59, 60, 61, 62 Consistently, we observe accumulation of Diap1 in prosβ2 and Mov34 mutant cells (Figure 3c–d''). Thus, this assay can detect accumulating proteins in vivo.
Combined, these data indicate that protein levels of Dronc in living cells are either not regulated by the UPS, or there are compensatory mechanisms operating that turn over Dronc upon proteasome impairment.
Loss of the autophagy gene Atg7 does not affect Dronc protein levels
In addition to the UPS, autophagy is a cellular catabolic process that is known to degrade proteins.1, 4, 15 We considered the possibility that autophagy may regulate the protein levels of Dronc. The autophagy gene Atg7 encodes the E1-activating enzyme for the two ubiquitin-like conjugation systems and is an important regulator for autophagy.14, 16 As proteasome impairment does not affect the protein levels of Dronc in epithelial disc cells, we tested the possibility that autophagy may control it. We downregulated Atg7 function by RNAi or inactivated Atg7 in mutant cell clones in eye imaginal discs. However, similar to proteasomal dysfunction, impaired Atg7 function does not affect Dronc protein levels in mosaic eye imaginal discs (Figures 4a–b'').
Figure 4.
Loss of Atg7 alone does not affect Dronc protein levels. Dronc labeling of Atg7 mosaic eye imaginal discs. Atg7 was either downregulated by RNAi (a) or genetically inactivated in mutant cell clones (b). Atg7-deficient clones are marked by absence of GFP (a', b'). White arrows highlight one clone in each panel as example. Dronc protein levels are unaltered in Atg7-deficient clones (a'', b''). Genotypes: (a) yw hsFLP; tub>GFP>Gal4/UAS-Atg7RNAi (>denotes FRT). (b) yw eyFLP; FRT42D Atg7d14/FRT42D P[ubi-GFP]
Simultaneous inactivation of both the proteasome and autophagy triggers accumulation of Dronc protein
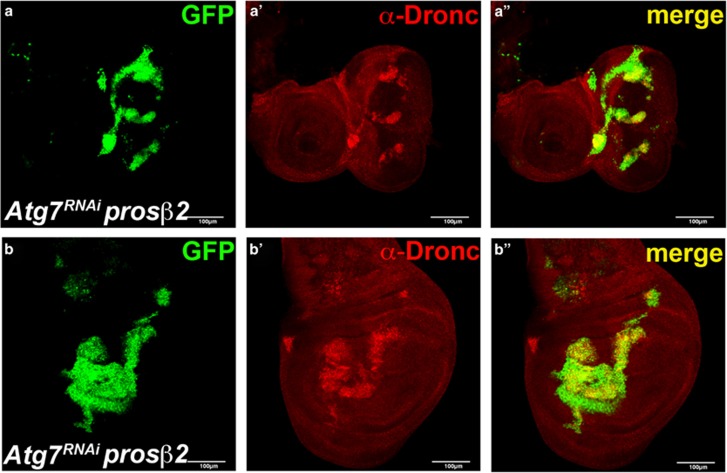
We examined the possibility that the protein levels of Dronc are coordinately regulated by both the proteasome and autophagy. To address this question, we inactivated autophagy by Atg7 RNAi in prosβ2 mutant cell clones using the mosaic analysis with a repressible cell marker (MARCM) method.63 Indeed, simultaneous impairment of both the proteasome and autophagy causes strong accumulation of Dronc protein in epithelial cells of eye imaginal discs (Figures 5a–a''). Similar observations were also made in wing imaginal discs, another epithelial tissue (Figures 5b–b''). The accumulation of Dronc in proteasome/autophagy double-deficient cells is not because of a transcriptional upregulation of dronc transcripts (Supplementary Figure S5). Together, these data suggest that Dronc protein levels are coordinately regulated by both the UPS and autophagy.
Figure 5.
Dronc accumulates in clones simultaneously mutant for the proteasome and autophagy. Mosaic eye (a) and wing (b) imaginal discs doubly deficient for the proteasome and autophagy labeled for Dronc (red). Atg7 knockdown was induced by RNAi in prosβ2 mutant cell clones using the MARCM method. Mutant cell clones are positively labeled by GFP. Dronc accumulates in double-deficient cell clones. The cross was performed at 25 °C. Genotype: hs-FLP UAS-GFP tub-Gal4; UAS-Atg7RNAi/+prosβ2DTS7 FRT80B/tub-Gal80 FRT80B
Loss of proteasome function enhances autophagy in epithelial cells
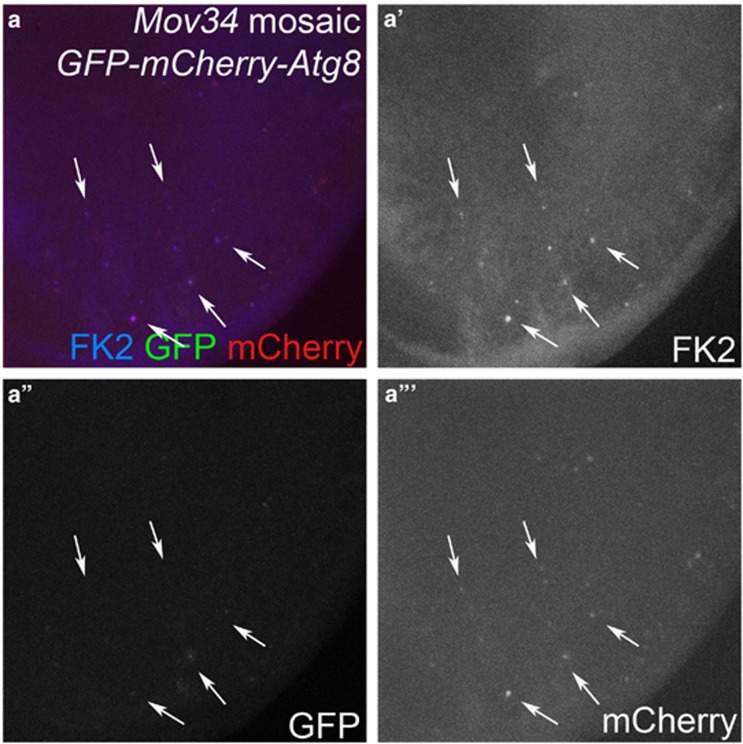
To explain the synergistic control of Dronc protein levels by the UPS and autophagy, we considered that, because the UPS and autophagy are mechanistically linked, impairment of the UPS can enhance autophagy, which is often referred to as compensatory autophagy1, 19, 20, 21, 22, 23, 24, 25, 26, 27 (reviewed by Park and Cuervo,3Wojcik28and Lamark and Johansen29). For example, in Drosophila, compensatory autophagy after proteasome impairment has been reported in neurons, in fat body cells and in adult flies.22, 27, 57 To examine this possibility in epithelial cells of eye imaginal discs, we monitored autophagy using a tandem fusion protein GFP-mCherry-Atg8a as reporter for autophagic flux.18 This reporter is incorporated into autophagosomes, which mature into autolysosomes. In autolysosomes, fluorescence of the GFP moiety of the reporter is quenched, whereas mCherry signals persist. Therefore, this reporter is suited to monitor autophagic flux from autophagosomes into autolysosomes. Indeed, while weak GFP signals are present in a subset of Mov34 mutant cell clones, there are also many clones where mCherry signals exist alone (Figures 6a'' and a''') suggesting that at least in these clones autophagic flux from autophagosomes to autolysosomes is induced in response to proteasome impairment.
Figure 6.
Impaired proteasome function induces autophagic flux. Mov34 mosaic eye imaginal discs expressing GFP-mCherry-Atg8a as marker for autophagic flux. Mov34 mutant cells were identified by FK2 labeling (blue in a; gray in a'). White arrows mark representative Mov34 mutant cells as examples. Although there is little to no GFP labeling in Mov34 mutant cell clones (a''), all clones contain increased mCherry labeling (a''') suggesting that autophagic flux is enhanced in proteasome-deficient cells. Genotype: ey-FLP; FRT42D Mov34k08003 /FRT42D ; patg8a>GFP-mCherry-Atg8a
Discussion
This is the first report in which a clonal analysis of strong proteasome mutants was performed. Usually, dominant temperature sensitive (DTS) alleles of proteasome subunits (DTS5, DTS7, etc.), RNAi or pharmacological inhibition have been used to study proteasome function.8, 22, 27, 51, 57, 64 In other approaches, whole embryos mutant for proteasome subunits were characterized for defects in dendrite pruning in sensory neurons in Drosophila.65, 66, 67 However, a specific analysis characterizing recessive alleles for defects in proteasome activity has not been reported. The reasons for this omission are obvious. Mutant animals are homozygous lethal and mutant clones in otherwise heterozygous animals are very small and difficult to identify. We found that cells mutant for proteasome function accumulate ubiquitin-conjugated proteins (Figure 1), consistent with the expectation. We used markers detecting ubiquitin-conjugated proteins to positively identify mutant clones. That enabled us to identify Diap1 as substrate of the proteasome, whereas control of Dronc protein levels appears to be independent of the proteasome (Figure 3). The proteasome alleles used in this study and our approach to identify mutant clones will be of general use for future analysis of proteasome function.
Dronc also does not accumulate in autophagy-deficient cells in which the E1 encoding gene Atg7 was mutant or downregulated by RNAi (Figure 4). However, simultaneous inactivation of the proteasome and Atg7 resulted in strong accumulation of Dronc (Figure 5). In addition, autophagy is enhanced in proteasome mutants (Figure 6).
There are two possibilities to explain these results. First, both the UPS and autophagy degrade Dronc independently of each other and if one pathway is lost, the other one maintains Dronc at normal protein levels. Or second, under normal conditions, Dronc is degraded only by the UPS, but upon proteasome impairment, enhanced autophagy can compensate for this impairment and degrades Dronc instead. Although we cannot distinguish between these two possibilities, the lack of accumulation of Dronc protein in proteasome and autophagy mutants suggests a fail–safe mechanism that avoids accumulation of this potentially deleterious protein in cells.
Our data indicate that Dronc is a shared substrate for degradation by both the UPS and autophagy. There are not many substrates known which are common to both the UPS and autophagy. α-Synuclein is one substrate and mutant forms of this protein appear to poison both the UPS and autophagy causing Parkinson's disease.68, 69, 70 Another shared substrate is inhibitor kappa B alpha (IĸBα), the inhibitor of the transcription factor nuclear factor kappa B.71 IĸBα appears to be degraded by the UPS and autophagy in different cellular compartments with different rates.71 In addition to these specific shared substrates, misfolded proteins are common substrates for both the UPS and autophagy.29, 72 Whether this relates to Dronc, is currently unknown.
It is unclear what distinguishes Dronc from Diap1 with respect to proteasome-mediated degradation and enhanced autophagy. The alternative question would be why Diap1 is not degraded by enhanced autophagy in proteasome-deficient cells. There may be specific ubiquitylation marks or other post-translational modifications that distinguish between these possibilities. Additional work is necessary to answer these questions in the future.
We observed that proteasome dysfunction triggers apoptosis in some, but not all, mutant cells (Figure 2). Given the accumulation of the anti-apoptotic protein Diap1 in proteasome-deficient cells, it is somewhat surprising that some of them undergo apoptosis. However, many other proteins likely also accumulate in proteasome-deficient cells, which combined will tilt the fate of the affected cells to either survival or death, depending on relative ratios. This consideration may explain why some cells are apoptotic and others are not. Another possibility is that the cCsp-3-negative cells in mutant cell clones will die later or are already dead. Consistent with the latter notion, we have previously shown that dead cells lack protein and DNA,73 hence the cCsp-3 antibody would not label these cells. Importantly also, although not all proteasome-deficient cell clones may be apoptotic at the time of investigation, they do not grow very well and remain small. These considerations are important for the potential clinical use of proteasome inhibitors for treatment of cancer.
Materials and Methods
Fly stocks
prosβ2EP3067 is a P element transposon insertion in the first exon at base pair 63.74 Mov34k08003 is a P element transposon insertion in the first exon at base pair 263.65, 67 Both insertions disrupt the transcripts. prosβ2DTS7 encodes a DTS allele of prosβ2; however, in this work, we used it in genetic mosaics using FRT80B DTS7, not applying a temperature-shift. Atg7d14 is a mutant allele as described.16 UAS-Atg7RNAi targets Atg7 by RNAi.17, 75 UAS-Flag-dronc encodes a Dronc protein with an N-terminal Flag-tag.58 It was expressed using daughterless-Gal4 (da-Gal4) in Mov34 mosaic background (Supplementary Figure S4).
Generation of mutant cell clones
Mutant clones of prosβ2, Mov34 and Atg7 were induced in eye imaginal discs using the FLP/FRT-induced mitotic recombination system using ey-FLP.53, 54 For this purpose, mutant alleles of prosβ2 and Mov34 were recombined on FRT80B and FRT42D bearing chromosomes, respectively. FRT42D Atg7d14 was used as described.16 To generate wild-type control mosaics (Supplementary Figure S2), ey-Flp; FRT42D P[ubi-GFP] was used. Clones are marked by loss of GFP. To induce Atg7 RNAi in prosβ2 mutant clones, the MARCM method was used.63 In this case, mutant cell clones are positively marked by GFP (Figure 5).
Immunohistochemistry
Eye imaginal discs from third instar larvae were dissected using standard protocols76 and labeled with antibodies raised against the following antigens: ubiquitin (Sigma Aldrich, St. Louis, MO, USA); FK1 and FK2 (Biomol, Hamburg, Germany); cCsp-3 (Cell Signaling Technology, Danvers, MA, USA); Dronc (kind gift of Pascal Meier), Diap1 (a kind gift of Hermann Steller and Hyung Don Ryoo) and Flag (Sigma Aldrich). Cy3-conjugated and Cy-5 fluorescently conjugated secondary antibodies are obtained from Jackson ImmunoResearch (West Grove, PA, USA) and were used at dilutions of 1 : 400. In each experiment, multiple clones in 10–20 eye imaginal discs were analyzed, unless otherwise noted. Images were captured using Olympus Optical FV500 (Waltham, MA, USA) or Zeiss LSM700 confocal microscopes (Peabody, MA, USA).
Generation of the GFP-mCherry-Atg8a tandem reporter
A transgene encoding the tandem protein GFP-mCherry-Atg8a was generated and used to determine autophagic flux.18 A region 2-kb upstream of Atg8a (CG32672) was inserted upstream of GFP-mCherry-Atg8a in the pCaSpeR4 Drosophila transformation vector, as was previously described for similar GFP-Atg8a and mCherry-Atg8a reporter lines.77, 78 The resulting plasmid pCaSpeR4-promoter-GFP-mCherry-Atg8a was used to generate transgenic Drosophila lines using standard procedures.
qPCR of dronc transcripts
Late 3rd larval stage (L3) larvae of control (w1118) and experimental genotype (w; UAS-Atg7RNAi; da-Gal4/DTS7) were shifted to 29 °C until eclosion of adult flies. mRNA was extracted from 1 to 3 days old flies using RNAeasy (Qiagen, Hilden, Germany) and quantitative polymerase chain reaction (qPCR) was performed with the following dronc-specific primers: Nedd2-like caspase forward primer (NcF) 5'-CTCGCTAAACGAACGGAGAAC-3' and Nedd2-like caspase reverse primer (NcR) 5'-CAACGACACCCACATAAGGG-3', as described.79 Tubulin was used for normalization.
Acknowledgments
We thank Katja Köhler, Sally Kornbluth, Pascal Meier, Hyung Don Ryoo, Hermann Steller and the Bloomington Stock Center for antibodies and fly stocks. This project was supported by grants from the National Institute of General Medical Sciences (NIGMS) to EHB and AB. The content is solely the responsibility of the authors and does not represent the official views of the NIGMS or the NIH.
Glossary
- Atg-7,-8a
autophagy-related-7, -8a
- β2
beta 2
- cCsp-3
cleaved caspase-3
- da-Gal4
daughterless-Gal4
- Diap1
Drosophila inhibitor of apoptosis protein
- DrICE
death-related ICE-like protease
- Dronc
death regulator Nedd2-like caspase
- DTS7
dominant temperature sensitive 7
- E1
enzyme 1
- ey
eyeless
- Flp
Flippase
- FRT
Flp recombination target
- GFP
green fluorescent protein
- IAP
inhibitor of apoptosis protein
- IκBα
inhibitor kappa B alpha
- L3
3rd larval stage
- LC3
light chain 3
- LIR
LC3-interacting region
- MARCM
mosaic analysis with a repressible cell marker
- NcF
Nedd2-like caspase forward primer
- NcR
Nedd2-like caspase reverse primer
- Prosβ2
proteasome subunit beta 2
- qPCR
quantitative polymerase chain reaction
- RING
really interesting new gene
- RNAi
ribonucleic acid (RNA) interference
- Ubi-GFP
ubiquitous GFP
- UPS
ubiquitin–proteasome system
- XIAP
X-linked IAP
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by H-U Simon
Supplementary Material
References
- Wong E, Cuervo AM. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb Perspect Biol 2010; 2: a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G, Mayor T. Perilous journey: a tour of the ubiquitin-proteasome system. Trends Cell Biol 2014; 24: 352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Cuervo AM. Selective autophagy: talking with the UPS. Cell Biochem Biophys 2013; 67: 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol 2010; 129: 814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol 2004; 5: 177–187. [DOI] [PubMed] [Google Scholar]
- Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol 2009; 10: 104–115. [DOI] [PubMed] [Google Scholar]
- Kurucz E, Ando I, Sumegi M, Holzl H, Kapelari B, Baumeister W et al. Assembly of the Drosophila 26 S proteasome is accompanied by extensive subunit rearrangements. Biochem J 2002; 365(Pt 2): 527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belote JM, Fortier E. Targeted expression of dominant negative proteasome mutants in Drosophila melanogaster. Genesis 2002; 34: 80–82. [DOI] [PubMed] [Google Scholar]
- Ghaboosi N, Deshaies RJ. A conditional yeast E1 mutant blocks the ubiquitin-proteasome pathway and reveals a role for ubiquitin conjugates in targeting Rad23 to the proteasome. Mol Biol Cell 2007; 18: 1953–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoy A, Soane T, Welchman R, Mayer RJ. The ubiquitin-proteasome system and cancer. Essays Biochem 2005; 41: 187–203. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Development of proteasome inhibitors as research tools and cancer drugs. J Cell Biol 2012; 199: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov 2006; 5: 596–613. [DOI] [PubMed] [Google Scholar]
- Suraweera A, Munch C, Hanssum A, Bertolotti A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell 2012; 48: 242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehrecke EH. Autophagy: dual roles in life and death? Nat Rev Mol Cell Biol 2005; 6: 505–510. [DOI] [PubMed] [Google Scholar]
- Zhang H, Baehrecke EH. Eaten alive: novel insights into autophagy from multicellular model systems. Trends Cell Biol 2015; 25: 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev 2007; 21: 3061–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 2004; 7: 167–178. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8: 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaarur N, Meriin AB, Bejarano E, Xu X, Gabai VL, Cuervo AM et al. Proteasome failure promotes positioning of lysosomes around the aggresome via local block of microtubule-dependent transport. Mol Cell Biol 2014; 34: 1336–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 2010; 584: 1393–1398. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, Rubinsztein DC. A novel link between autophagy and the ubiquitin-proteasome system. Autophagy 2009; 5: 862–863. [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 2007; 447: 859–863. [DOI] [PubMed] [Google Scholar]
- Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol 2007; 171: 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Dunner K Jr. McConkey DJ. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene 2010; 29: 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama S, Sou YS, Uemura T, Kametaka S, Saito T, Ishimura R et al. Proteasome dysfunction activates autophagy and the Keap1-Nrf2 pathway. J Biol Chem 2014; 289: 24944–24955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janen SB, Chaachouay H, Richter-Landsberg C. Autophagy is activated by proteasomal inhibition and involved in aggresome clearance in cultured astrocytes. Glia 2010; 58: 1766–1774. [DOI] [PubMed] [Google Scholar]
- Velentzas PD, Velentzas AD, Mpakou VE, Antonelou MH, Margaritis LH, Papassideri IS et al. Detrimental effects of proteasome inhibition activity in Drosophilamelanogaster: implication of ER stress, autophagy, and apoptosis. Cell Biol Toxicol 2013; 29: 13–37. [DOI] [PubMed] [Google Scholar]
- Wojcik S. Crosstalk between autophagy and proteasome protein degradation systems: possible implications for cancer therapy. Folia Histochem Cytobiol/Pol Acad Sci Pol Histochem Cytochem Soc 2013; 51: 249–264. [DOI] [PubMed] [Google Scholar]
- Lamark T, Johansen T. Autophagy: links with the proteasome. Curr Opin Cell Biol 2010; 22: 192–198. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282: 24131–24145. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Johansen T. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy 2006; 2: 138–139. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171: 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezis IP, Stenmark H. p62 at the interface of autophagy, oxidative stress signaling, and cancer. Antioxidants Redox Signal 2012; 17: 786–793. [DOI] [PubMed] [Google Scholar]
- Bartlett BJ, Isakson P, Lewerenz J, Sanchez H, Kotzebue RW, Cumming RC et al. p62, Ref(2)P and ubiquitinated proteins are conserved markers of neuronal aging, aggregate formation and progressive autophagic defects. Autophagy 2011; 7: 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell 2014; 53: 167–178. [DOI] [PubMed] [Google Scholar]
- Frake RA, Ricketts T, Menzies FM, Rubinsztein DC. Autophagy and neurodegeneration. J Clin Invest 2015; 125: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif M, Hetz C. Autophagy impairment: a crossroad between neurodegeneration and tauopathies. BMC Biol 2012; 10: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf DJ, Garcia-Arencibia M, Hochfeld WE, Rubinsztein DC. Autophagy and misfolded proteins in neurodegeneration. Exp Neurol 2012; 238: 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann A, Steller H. Apoptosis, stem cells, and tissue regeneration. Sci Signal 2010; 3: re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y, Steller H. Live to die another way: modes of programmed cell death and the signals emanating from dying cells. Nat Rev Mol Cell Biol 2015; 16: 329–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. Caspase function in programmed cell death. Cell Death Differ 2007; 14: 32–43. [DOI] [PubMed] [Google Scholar]
- Shalini S, Dorstyn L, Dawar S, Kumar S. Old new and emerging functions of caspases. Cell Death Differ 2015; 22: 526–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell 2011; 147: 742–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux DL, Silke J. IAPs RINGs and ubiquitylation. Nat Rev Mol Cell Biol 2005; 6: 287–297. [DOI] [PubMed] [Google Scholar]
- Chai J, Yan N, Huh JR, Wu JW, Li W, Hay BA et al. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat Struct Biol 2003; 10: 892–898. [DOI] [PubMed] [Google Scholar]
- Muro I, Hay BA, Clem RJ. The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J Biol Chem 2002; 277: 49644–49650. [DOI] [PubMed] [Google Scholar]
- Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J et al. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol 2002; 4: 445–450. [DOI] [PubMed] [Google Scholar]
- Ditzel M, Broemer M, Tenev T, Bolduc C, Lee TV, Rigbolt KTG et al. Inactivation of effector caspases through non-degradative polyubiquitylation. Mol Cell 2008; 32: 540–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TV, Fan Y, Wang S, Srivastava M, Broemer M, Meier P et al. Drosophila IAP1-mediated ubiquitylation controls activation of the initiator caspase DRONC independent of protein degradation. PLoS Genet 2011; 7: e1002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schile AJ, Garcia-Fernandez M, Steller H. Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev 2008; 22: 2256–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth KA, Belote JM. The dominant temperature-sensitive lethal DTS7 of Drosophila melanogaster encodes an altered 20S proteasome beta-type subunit. Genetics 1999; 151: 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzl H, Kapelari B, Kellermann J, Seemuller E, Sumegi M, Udvardy A et al. The regulatory complex of Drosophilamelanogaster 26S proteasomes. Subunit composition and localization of a deubiquitylating enzyme. J Cell Biol 2000; 150: 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 2000; 127: 851–860. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 1993; 117: 1223–1237. [DOI] [PubMed] [Google Scholar]
- Fujimuro M, Sawada H, Yokosawa H. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett 1994; 349: 173–180. [DOI] [PubMed] [Google Scholar]
- Fujimuro M, Sawada H, Yokosawa H. Dynamics of ubiquitin conjugation during heat-shock response revealed by using a monoclonal antibody specific to multi-ubiquitin chains. Eur J Biochem 1997; 249: 427–433. [DOI] [PubMed] [Google Scholar]
- Low P, Varga A, Pircs K, Nagy P, Szatmari Z, Sass M et al. Impaired proteasomal degradation enhances autophagy via hypoxia signaling in Drosophila. BMC Cell Biol 2013; 14: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CS, Thomenius MJ, Gan EC, Tang W, Freel CD, Merritt TJ et al. Metabolic regulation of Drosophila apoptosis through inhibitory phosphorylation of Dronc. EMBO J 2010; 29: 3196–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays R, Wickline L, Cagan R. Morgue mediates apoptosis in the Drosophila melanogaster retina by promoting degradation of DIAP1. Nat Cell Biol 2002; 4: 425–431. [DOI] [PubMed] [Google Scholar]
- Holley CL, Olson MR, Colon-Ramos DA, Kornbluth S. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat Cell Biol 2002; 4: 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol 2002; 4: 432–438. [DOI] [PubMed] [Google Scholar]
- Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL et al. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol 2002; 4: 416–424. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 2001; 24: 251–254. [DOI] [PubMed] [Google Scholar]
- Schweisguth F. Dominant-negative mutation in the beta2 and beta6 proteasome subunit genes affect alternative cell fate decisions in the Drosophila sense organ lineage. Proc Natl Acad Sci USA 1999; 96: 11382–11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Jan LY, Jan YN. Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc Natl Acad Sci USA 2005; 102: 15230–15235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Zhu S, Younger S, Jan LY, Jan YN. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron 2006; 51: 283–290. [DOI] [PubMed] [Google Scholar]
- Watts RJ, Hoopfer ED, Luo L. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron 2003; 38: 871–885. [DOI] [PubMed] [Google Scholar]
- Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-synuclein is degraded by both autophagy and the proteasome. J Biol Chem 2003; 278: 25009–25013. [DOI] [PubMed] [Google Scholar]
- Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA et al. Alpha-synuclein impairs macroautophagy: implications for Parkinson's disease. J Cell Biol 2010; 190: 1023–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 2004; 305: 1292–1295. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Hu W, Lim B, Dice JF. IkappaB is a substrate for a selective pathway of lysosomal proteolysis. Mol Biol Cell 1998; 9: 1995–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy 2008; 4: 141–150. [DOI] [PubMed] [Google Scholar]
- Werz C, Lee TV, Lee PL, Lackey M, Bolduc C, Stein DS et al. Mis-specified cells die by an active gene-directed process, and inhibition of this death results in cell fate transformation in Drosophila. Development 2005; 132: 5343–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 2004; 167: 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 2007; 131: 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty CE, Bergmann A. Detecting caspase activity in Drosophila larval imaginal discs. Methods Mol Biol 2014; 1133: 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol 2009; 19: 1741–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton D, Chang TK, Nicolson S, Shravage B, Simin R, Baehrecke EH et al. Relationship between growth arrest and autophagy in midgut programmed cell death in Drosophila. Cell Death Differ 2012; 19: 1299–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihry RJ, Sapiro AL, Bashirullah A. Translational control by the DEAD box RNA helicase belle regulates ecdysone-triggered transcriptional cascades. PLoS Genet 2012; 8: e1003085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.