Abstract
Cellular senescence is considered as an important tumor-suppressive mechanism. Here, we demonstrated that heparan sulfate (HS) prevents cellular senescence by fine-tuning of the fibroblast growth factor receptor (FGFR) signaling pathway. We found that depletion of 3′-phosphoadenosine 5′-phosphosulfate synthetase 2 (PAPSS2), a synthetic enzyme of the sulfur donor PAPS, led to premature cell senescence in various cancer cells and in a xenograft tumor mouse model. Sodium chlorate, a metabolic inhibitor of HS sulfation also induced a cellular senescence phenotype. p53 and p21 accumulation was essential for PAPSS2-mediated cellular senescence. Such senescence phenotypes were closely correlated with cell surface HS levels in both cancer cells and human diploid fibroblasts. The determination of the activation of receptors such as FGFR1, Met, and insulin growth factor 1 receptor β indicated that the augmented FGFR1/AKT signaling was specifically involved in premature senescence in a HS-dependent manner. Thus, blockade of either FGFR1 or AKT prohibited p53 and p21 accumulation and cell fate switched from cellular senescence to apoptosis. In particular, desulfation at the 2-O position in the HS chain contributed to the premature senescence via the augmented FGFR1 signaling. Taken together, we reveal, for the first time, that the proper status of HS is essential for the prevention of cellular senescence. These observations allowed us to hypothesize that the FGF/FGFR signaling system could initiate novel tumor defenses through regulating premature senescence.
Cellular senescence, which refers to irreversible growth arrest, has recently been considered as an important tumor-suppressive mechanism by which the proliferative potential of cancer cells is prevented.1, 2 Support for a role of cellular senescence as a barrier to tumorigenesis comes from tumor studies in mice and humans.3 Cellular senescence can be prematurely triggered by diverse forms of cellular damage, such as oncogenic mutations, strong mitotic signals, genomic instability, and oxidative stress.4, 5, 6, 7 Senescent cells are characterized by a variety of phenotypes, including enlarged and flattened morphology, senescence-associated β-galactosidase (SA-β-Gal) activity, formation of senescence-associated heterochromatin foci, and altered gene expression and protein processing.8, 9, 10 Senescent cells remain metabolically active and secrete a myriad of factors for cross-talk between senescent cells and their microenvironment, which is called senescence-associated secretory phenotype (SASP).11, 12 Some of the secreted factors are anti-tumorigenic due to the induction and maintenance of senescence, and others are pro-tumorigenic through stimulation of cell growth, migration, and invasion.11, 13, 14 In addition, SASP facilitates tissue repair and remodeling, and elicits immune surveillance to eliminate senescent cells.11, 13, 15 Thus, cellular senescence displays multiple effects on the tumor microenvironment of their neighboring tumor and normal cells. Recent findings suggest that cellular senescence is also involved in embryonic development.16, 17 Although great progress has been made in cellular senescence during the last decade, further elucidation of senescence-associated regulatory genes and mechanisms will improve our understanding regarding the physiological significance and potential therapeutic applications of cellular senescence.
The sulfation of proteoglycans (PGs) is an important post-translational modification that has a regulatory role in cell growth, motility, and metabolism.18, 19, 20 PGs, consisting of a core protein and heavily sulfated glycosaminoglycan chains, are ubiquitous on all animal cell surfaces.18, 21 Major families of membrane-bound PGs bear heparan sulfate (HS), a highly sulfated chain of disaccharide repeating units, as a constant feature18, 22 and are thus called HS proteoglycans (HSPGs). The universal sulfate donor 3′-phosphoadenosine 5′-phosphosulfate (PAPS) is synthesized by PAPS synthetase (3′-phosphoadenosine 5′-phosphosulfate synthetase (PAPSS)) in the cytoplasm.23 In humans, two isoforms of PAPSS show a 77% overlap in amino acid sequence identity,24 but the isoforms display distinct subcellular localization. PAPSS1 is located predominantly in the nucleus, whereas PAPSS2 is mainly located in the cytoplasm.25, 26 The process of sulfation is strictly governed by various enzymes, such as glycosyltransferases, sulfotransferases, and an epimerase.27, 28
Owing to its high negative charge, HS interacts with hundreds of proteins and regulates multiple signaling pathways, including fibroblast growth factor (FGF), hepatocyte growth factor (HGF), vascular endothelial growth factor, Wnt, and BMP signaling pathways.18, 20, 21, 29 The most well-studied functions of HS include its effect on protein conformation, enhancement of protein–protein interactions, role as a co-receptor for growth factors, protection of proteins from degradation, sequestration of protein ligands, and mediation of protein internalization.22 Many of the key paradigms in HS actions have been derived from the protein–protein interactions required for FGF signaling.30, 31, 32 However, the exact role of HS in FGF signaling is still a matter of considerable debate.33 In addition, how FGF signaling is tumor promoting in some contexts, but tumor-suppressive in others remains unclear.34
In this study, we investigate the role of HS sulfation status and FGF signaling in cellular senescence. We show that the HS undersulfation induces augmentation of fibroblast growth factor receptor 1 (FGFR1) activation, ultimately resulting in premature senescence through the p53-p21 signaling pathway.
Results
PAPSS2 depletion induces premature senescence
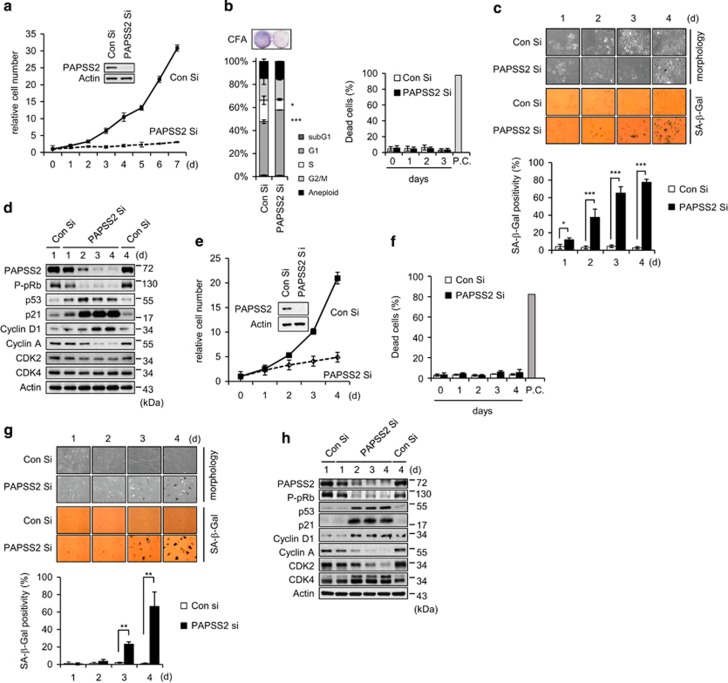
As PAPSS2, a cytoplasmic synthetase of the universal sulfate donor PAPS, was found to be increased in gene expression profile of prematurely senescent cancer cells (Supplementary Table 1), we first investigated the influence of siRNA against PAPSS2 (PAPSS2 Si) on cell proliferation and senescence. MCF7 human breast cancer cells treated with PAPSS2 Si were fewer in number and showed poor colony-forming ability compared with control siRNA (Con Si)-transfected cells (Figure 1a). PAPSS2-depleted cells accumulated in the G1 phase with no increase in the proportion of cells either in the subG1 phase or that were positive for propidium iodide staining (Figure 1b). PAPSS2-depleted cells displayed hallmarks of senescent cells such as enlarged and flattened morphology and SA-β-Gal staining (Figure 1c). As PAPSS2 was depleted, the cells showed a loss of phosphorylated pRb with changes of cell cycle regulatory protein levels and accumulation of p53 and p21 (Figure 1d). To know whether such PAPSS2-mediated premature senescence phenotype could be induced in normal cells, we treated primary human diploid fibroblasts (HDFs) with PAPSS2 Si. PAPSS2 depletion also resulted in fewer cell numbers with no increase of apoptotic cell death (Figures 1e and f), and senescence phenotypes in HDFs (Figure 1g). PAPSS2-depleted HDFs also showed pRb hypophosphorylation, alteration of cell cycle regulatory protein levels, and accumulation of p53 and p21 (Figure 1h). We observed no off-target effect of PAPSS Si on p53 and p21 accumulation or the induction of premature senescence (Supplementary Figure S1). In addition, PAPSS2 depletion contributes to premature senescence in other cell types such as A549 human lung carcinoma cells and MCF10A human mammary epithelial cells (Supplementary Figure S2). However, PAPSS1 depletion did not affect the cellular senescence phenotype (Supplementary Figure S3), most likely due to its localization in the nucleus. Moreover, PAPSS2 overexpression did not affect the rate of cellular proliferation, the amount of cell death, and p53 and p21 levels (Supplementary Figure S4).
Figure 1.
PAPSS2 depletion suppresses cell growth due to premature senescence in MCF7 cells and HDFs. (a–d) MCF7 cells were transfected with Con Si or PAPSS2 Si. (a) The numbers of viable cells are shown as relative values. Total protein was extracted from cells 3 days after siRNA transfection and was subjected to WB analysis. (b) Cell cycle distributions (left graph) and dead cell populations (right graph) were analyzed by FACS 3 days after siRNA transfection and at the indicated times after siRNA transfection, respectively. Doxorubicin-treated (10 μg/ml) MCF7 cells were used as a positive control (P.C.). A colony-forming assay (CFA) was also performed (left panel) 7 days after siRNA transfection. (c) Cellular morphology and SA-β-Gal positivity were assessed at the indicated times after siRNA transfection (upper panel), and the percentage of senescent cells was quantified (lower graph). (d) Cells were harvested at the indicated times after siRNA transfection and were subjected to WB analysis. (e–h) HDFs were transfected with either Con Si or PAPSS2 Si. (e) Cell viability, (f) the percentage of dead cells, (g) morphological changes and SA-β-Gal positivity, and (h) WB analysis were performed as described in a–d. Error bars indicate the S.D. of three independent experiments. ***P<0.001, **P<0.01, and *P<0.05
HS sulfation status is tightly linked to premature and replicative senescence through the p53-p21 signaling pathway
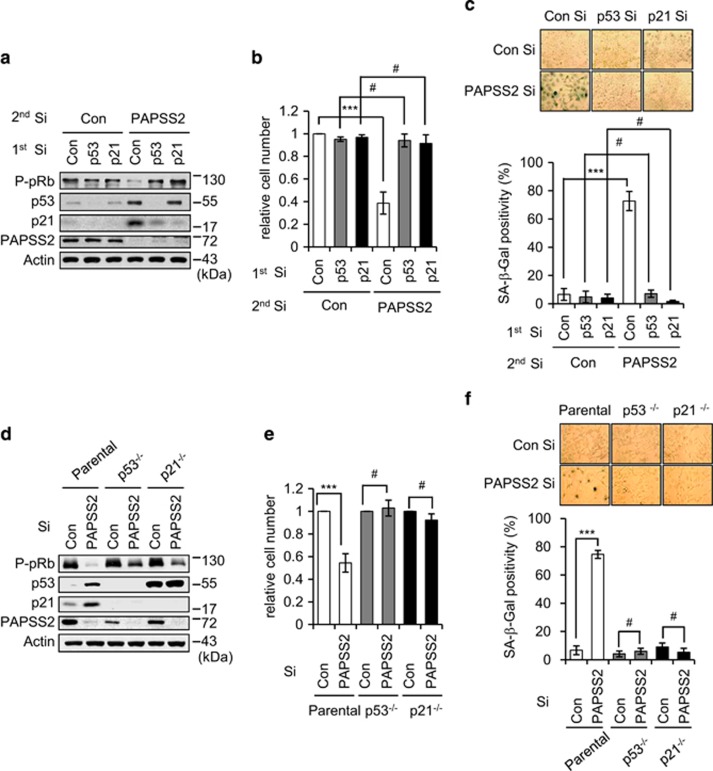
As we observed p53 and p21 accumulation in both cancer and normal cells (Figure 1 and Supplementary Figure S2), we investigated the involvement of p53 and p21 in premature senescence induced by PAPSS2 depletion. In the absence of p53, PAPSS2 depletion failed to induce p21 accumulation or pRb hypophosphorylation (Figure 2a) and had no effect on cell number (Figure 2b) or SA-β-Gal positivity (Figure 2c). The absence of p21 abrogated PAPSS2 depletion-induced premature senescence regardless of the p53 phenotype (Figures 2a and c). Furthermore, PAPSS2-mediated premature senescence, including a loss of phosphorylated pRb, a decrease in cell number, and an increase in SA-β-Gal positivity, was blocked in both p53−/− and p21−/− HCT116 human colon cancer cells (Figures 2d and f). These results demonstrate that the p53-p21 signaling pathway is critical for the induction of premature senescence by PAPSS2 depletion.
Figure 2.
PAPSS2 depletion-mediated premature senescence occurs through the p53-p21 pathway. (a–c) MCF7 cells were transfected with Con Si, p53 Si, or p21 Si (first transfection) 6 h before transfection with Con Si or PAPSS2 Si (second transfection). (a) After 2 days, cells were harvested and subjected to WB analysis using the indicated antibodies. (b) Quantification of relative cell number and (c) SA-β-gal positivity were performed 3 days after transfection. (d–f) HCT116 parental, p53−/−, and p21−/− cells were transfected with Con Si or PAPSS2 Si, and experiments were performed as described in a–c. Cell numbers are relative to that of Con Si-transfected cells. Error bars indicate the S.D. of three independent experiments. ***P<0.001 and #P>0.05
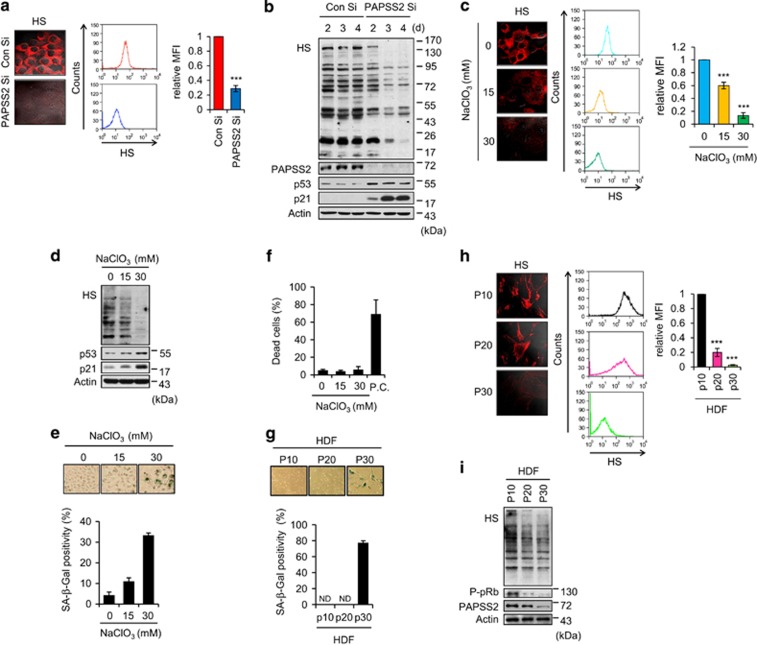
Because PAPSS2 is a major enzyme involved in the sulfation of HS,23 we examined the role of HS sulfation status in PAPSS2-mediated premature senescence. Using an anti-HS antibody, we detected a gradual decline in sulfated HS on the cell surface (Figure 3a) and in cell lysates following PAPSS2 depletion (Figure 3b). These changes were associated with gradual increases in p53 and p21 levels (Figure 3b). To confirm the effect of undersulfation on premature senescence, we treated MCF7 cells with sodium chlorate, a metabolic inhibitor of HS sulfation.35 We found that sodium chlorate treatment induced premature senescence (Figure 3e) with gradual decline in sulfated HS on the cell surface and in cell lysates (Figures 3c and d) as well as p53 and p21 accumulation (Figure 3d). We observed no significant increase in cell death (Figure 3f). We next examined the HS sulfation status in replicative senescence. In association with an increase in SA-β-Gal positivity (Figure 3g), we also detected a decrease in sulfated HS on the cell surface and in cell lysates during the replicative senescence of HDFs (Figures 3h and i). It was closely correlated with reduced PAPSS2 protein level during the replicative senescence (Figure 3i). We found a decrease in PAPSS2 protein levels due to proteosomal degradation during the development of replicative senescence (Supplementary Figure S5). Our results reveal that HS sulfation level is decreased in replicative senescence and decreased level of HS sulfation due to PAPSS depletion induces premature senescence.
Figure 3.
Decrease in cellular sulfation is associated with premature and replicative senescence. (a–b) MCF7 cells were transfected with either Con Si or PAPSS2 Si. (a) Transfected cells were stained with an anti-HS (10E4) antibody and visualized (left panel) or subjected to FACS to quantify the HS level (middle graph). Quantified data are shown as the mean fluorescence intensity (MFI) relative to Con Si-transfected cells (right graph). (b) Cells were harvested at the indicated times after siRNA transfection and subjected to WB analysis. (c–f) Undersulfation resulting from NaClO3 treatment induces premature senescence. (c) MCF7 cells were cultured in the absence or presence of NaClO3 for 3 d. The HS level were measured as described in a. (d) Analyses of WB, (e) morphological changes and SA-β-Gal positivity, and (f) the percentage of dead cells were performed. (g–i) Cellular sulfation level is associated with replicative senescence in an HDF system. (g) SA-β-Gal positivity in HDFs at passage 10 (p10), p20, and p30. ND, not determined. (h) The HS level was measured as described in a. (i) HDFs were harvested at the indicated passages and subjected to WB analysis. Error bars indicate the S.D. of three independent experiments. ***P<0.001
PAPSS2-mediated premature senescence is linked to augmented FGFR signaling
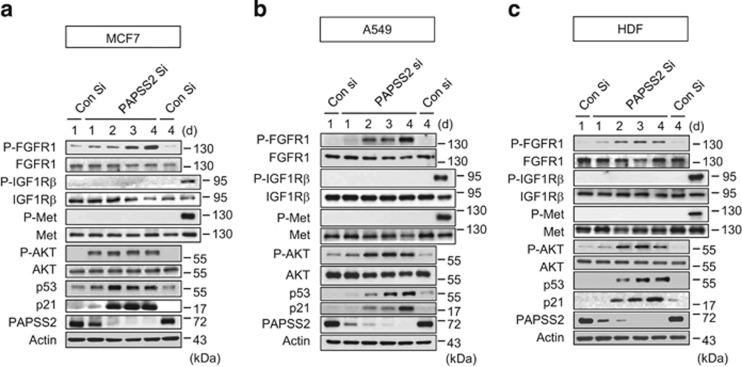
HS has been reported to regulate receptor signaling through the modulation of binding between growth factors (FGF, HGF, and IGF) and their receptors (FGFR and Met) or growth factor binding proteins.30, 31, 32, 36, 37, 38, 39 Thus, we examined the effect of PAPSS2 depletion on receptor signaling pathways such as FGFR1, Met, and beta subunit of IGF-1 receptor (IGF1Rβ). In PAPSS2-depleted MCF7 cells, we observed augmented FGFR phosphorylation. Moreover, we detected augmented AKT phosphorylation in close correlation with sustained p53 and p21 accumulation (Figure 4a). In contrast to FGFR1 phosphorylation, we detected no phosphorylation of either Met or IGF1Rβ in PAPSS2-depleted MCF7 cells (Figure 4a). Similarly, neither IGF1Rβ nor Met phosphorylation was observed after PAPSS2-depletion in A549 or HDF cells (Figures 4b and c). However, we confirmed that PAPSS2 depletion induced augmented FGFR/AKT activation and p53/p21 accumulation in both A549 and HDF cells (Figures 4b and c). These results indicate that FGFR1 signaling pathway is specifically involved in PAPSS2-mediated premature senescence through the regulation of HS sulfation.
Figure 4.
PAPSS2 depletion induces augmented and sustained FGFR/AKT activation. (a) MCF7 cells, (b) A549 cells, and (c) HDF cells were harvested at the indicated times after siRNA transfection and subjected to WB analysis
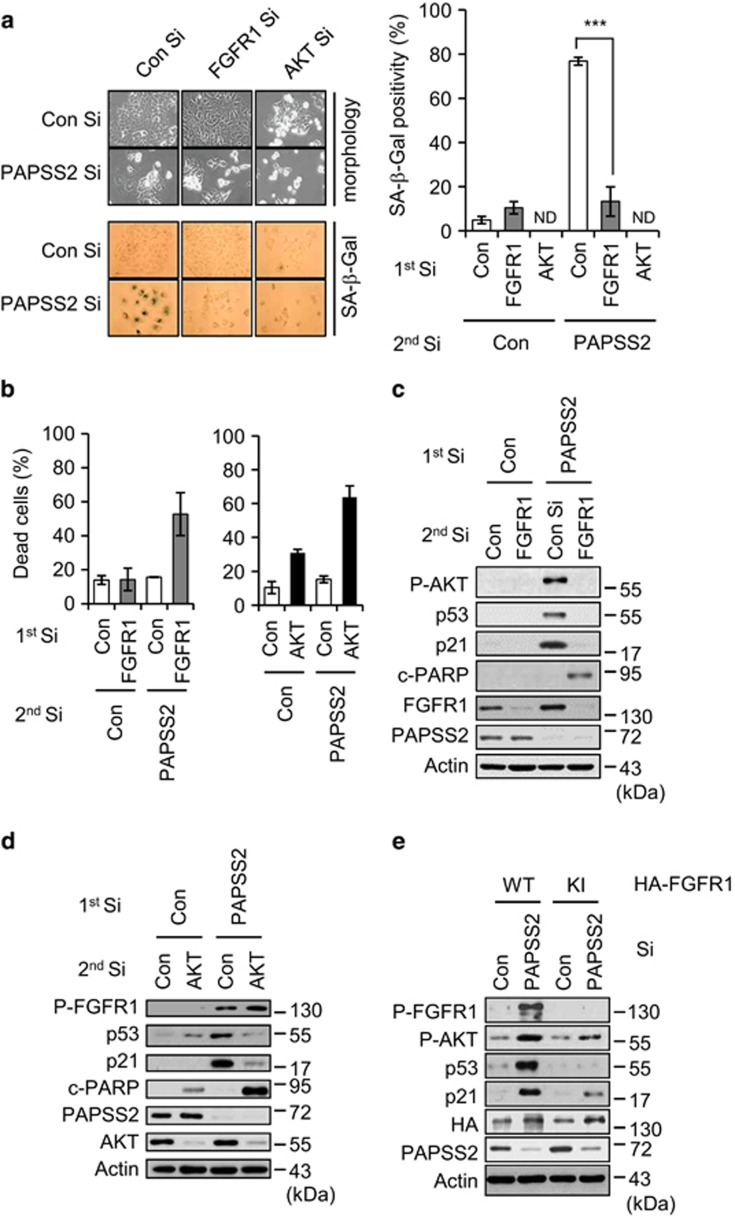
As PAPSS2 depletion evidently affects FGFR1 and AKT phosphorylation status (Figures 4a and c), we further examined the roles of FGFR1 and its downstream signaling molecule AKT. Double-knockdown cells transfected with PAPSS2 Si and either FGFR1 Si or AKT Si exhibited an apoptotic phenotype rather than premature senescence (Figures 5a and d). Specifically, SA-β-Gal positivity was barely detectable and the number of dead cells increased after depletion of either FGFR1 or AKT in PAPSS2-depleted cells (Figures 5a and b). Moreover, in both types of double-knockdown cells, p53/p21 accumulation was disrupted, and poly ADP ribose polymerase (PARP) cleavage was induced (Figures 5c and d). Whereas AKT phosphorylation was decreased by PAPSS2 and FGFR1 depletion, FGFR1 phosphorylation was not affected by PAPSS2 and AKT depletion (Figures 5c and d). Furthermore, the overexpression of a kinase-inactive (KI) mutant of FGFR1 (FGFR1-KI) attenuated AKT phosphorylation and p53/p21 accumulation in PAPSS2-depleted cells (Figure 5e). Altogether, these results demonstrate that PAPSS2 depletion induces premature senescence by increasing the FGFR1-AKT-p53-p21 signal transduction cascade.
Figure 5.
Augmented FGFR1-AKT-p53-p21 signaling is critical for PAPSS2-mediated premature senescence. (a–c) MCF7 cells were transfected with FGFR1 Si or AKT Si (first transfection) 6 h before transfection with Con Si or PAPSS2 Si (second transfection). (a) Analyses of morphological changes (left panel) and SA-β-Gal positivity (right graph), (b) the percentage of dead cells, and (c–d) WB analysis were performed. (e) Cells were transfected with HA-tagged WT or KI FGFR1 6 h before transfection with either Con Si or PAPSS2 Si. After 2 days, the transfected cells were harvested and subjected to WB analysis. Error bars indicate the S.D. of three independent experiments. ***P<0.001. ND, not determined
FGFs are required for augmented FGFR-AKT-p53-p21 signaling in PAPSS2-depleted cells
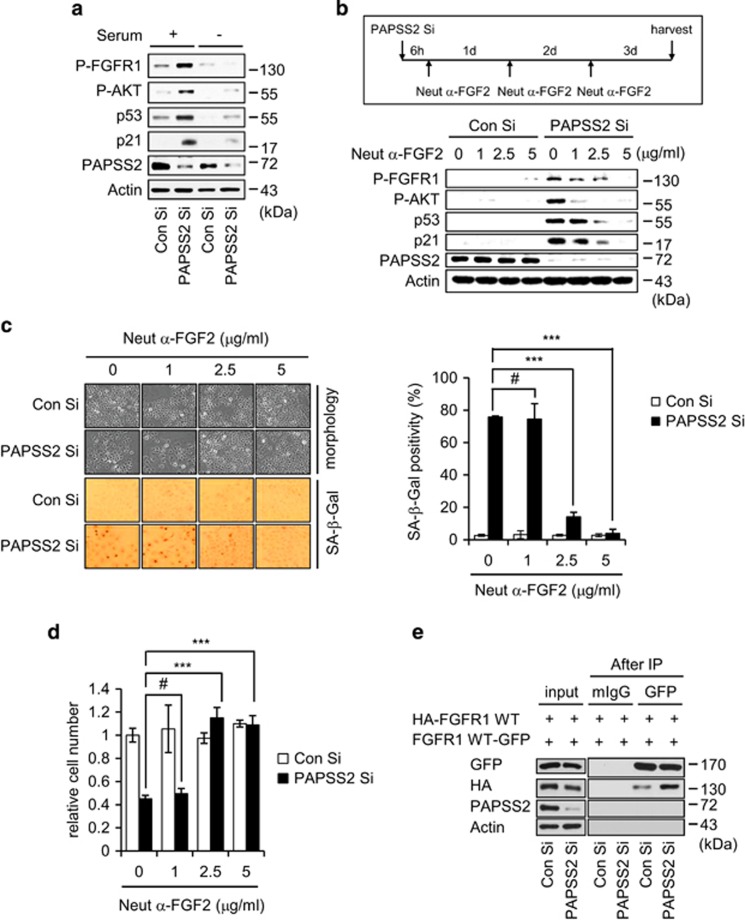
In contrast to previous reports,30, 31, 32 we observed that lack of heparan sulfation augments FGFR signaling, indicating that HS may not be required for FGFR activation. Thus, we wondered whether FGFs are directly involved in the augmentation of FGFR1-AKT-p53-p21 signaling in PAPSS2-depleted cells. We firstly examined the changes in signaling induced by PAPSS2 depletion in the presence or absence of FGF2. When we incubated PAPSS2 Si-transfected cells in serum-free medium, FGFR1-AKT-p53-p21 signaling was blocked (Figure 6a). When we incubated PAPSS2 Si-transfected cells in FGF2-depleted medium using a neutralizing antibody against FGF2 (Neut anti-FGF2), FGFR1-AKT-p53-p21 signaling was diminished in a dose-dependent manner (Figure 6b). The presence of Neut anti-FGF2 also diminished SA-β-Gal positivity (Figure 6c) and restored cell numbers (Figure 6d). In addition, we tested the effect of Neut anti-FGF1, -FGF3, and -FGF4 on PAPSS2-mediated premature senescence and found that FGF1 was also involved in FGFR1-AKT-p53-p21 signaling and senescence phenotypes (Supplementary Figure S6). We observed that PAPSS2 depletion-induced FGFR activation is associated with increased receptor dimerization from a co-immunoprecipitation (IP) assay with two different epitope-tagged FGFR1 constructs (HA-tagged and green fluorescent protein (GFP)-tagged FGFR1) (Figure 6e). These results demonstrate that the absence of HS results in excess binding of the FGF1 subfamily members FGF1/2 to FGFR and consequently excess FGFR dimerization, ultimately leading to premature senescence in PAPSS2-depleted cells.
Figure 6.
FGF2 has an essential role in augmented FGFR1-AKT-p53-p21 signaling in PAPSS2-depleted cells. (a) MCF7 cells transfected with Con Si or PAPSS2 Si for 6 h were cultured in the absence or presence of serum for an additional 36 h; cells were then harvested and subjected to WB analysis. (b) MCF7 cells were transfected with Con Si or PAPSS2 Si 6 h before incubation with or without increasing concentrations of Neut α-FGF2 for 3 days (upper timeline). Analyses of (b) WB (lower panel), (c) morphological changes (left panel) and SA-β-Gal positivity (right graph), and (d) relative cell number were performed. Error bars indicate the S.D. of three independent experiments. Statistical significance was determined using one-factor analysis of variance and Holm–Sidak post hoc tests. ***P<0.001 and #P>0.05. (e) Co-IP assay of epitope-tagged FGFR1. Cells were co-transfected with HA- and GFP-tagged WT FGFR1 24 h before transfection with Con Si or PAPSS2 Si. Transfected cells were immunoprecipitated using an anti-GFP antibody and then blotted with an anti-HA antibody
Sulfation at the 2-O position in HS is critical to prevent cellular senescence via modulation of FGFR1 signaling
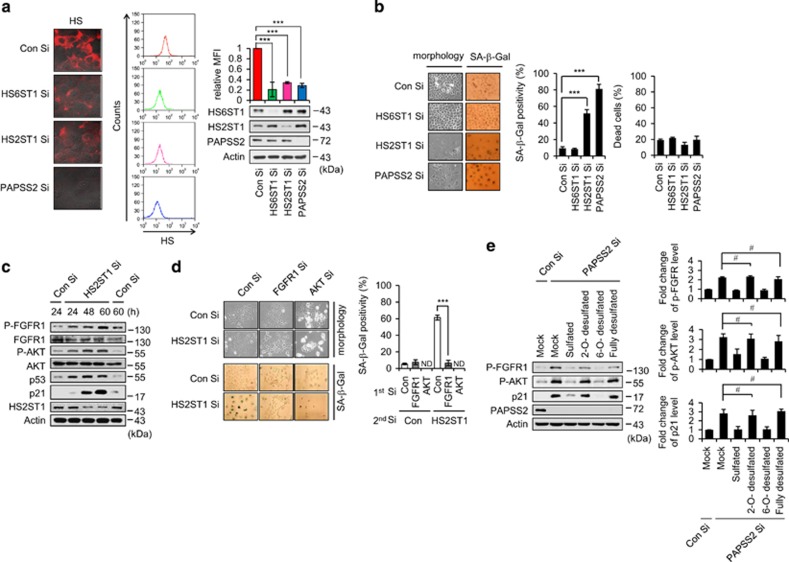
To determine which the sulfation position in HS is directly involved in the induction of cellular senescence through the augmentation of FGFR1 signaling, we transfected MCF7 cells with siRNAs against HS 2-O-sulfotransferase 1 (HS2ST1) and HS6ST1 as both positions are known to be required for FGFR1 activation. Although HS sulfation levels were reduced in both HS2ST1 Si- and HS6ST1 Si-transfected cells (Figure 7a), only HS2ST1-depleted cells exhibited SA-β-Gal positivity with no increase in cell death (Figure 7b), indicating that desulfation at the 2-O position in HS specifically contributes to the induction of cellular senescence. Next, we investigated whether sulfation at the 2-O position is critical for the proper regulation of FGFR signaling. We found that HS2ST1 depletion gradually induced FGFR1 and AKT phosphorylation and increased p53/p21 levels (Figure 7c). In HS2ST1-depleted cells, the blockade of either FGFR1 or AKT prevented a premature senescence phenotype (Figure 7d). We found that the difference in SA-β-gal positivity between PAPSS2- and HS2ST1-depleted cells (Figure 7b) could be partially attributed to other sulfotransferases such as chondroitin 6-O-sulfotransferase 1 (C6ST1) and dermatan 4-O-sulfotransferase 1 (D4ST1) (Supplementary Figure S7). However, there was no direct links among HS3ST1, N-deacetylase/N-sulfotrasferase 1 (NDST1), C6ST1, C4ST1, D4ST1, and FGFR-AKT-p53-p21 signaling. As HS oligosaccharides have the capacity to inhibit FGF signaling via competition with endogenous HS,40 we further examined the competitive effect of octasaccharides on FGFR and AKT phosphorylation in PAPSS2-mediated premature senescence. In contrast to sulfated and 6-O-desulfated octasaccharides, fully desulfated and 2-O-desulfated octasaccharides failed to prevent the activation of the FGFR signaling pathway (Figure 7e). In addition, when we overexpressed SULF1 and SULF2 endosulfatases having restricted substrate specificity toward 6-O sulfate of HS,41, 42 we did not observe senescence phenotypes (Supplementary Figure S8). These results indicate that sulfation at the 2-O position in HS is essential for the prevention of augmented FGFR signaling, which finally prevents premature senescence.
Figure 7.
HS2ST1 depletion induces activation of FGFR1 signaling, resulting in premature senescence. (a) After transfection of either HS6ST1 Si or HS2ST1 Si, MCF7 cell surface HS levels were visualized (left panel) and quantified (right upper graph), and WB analysis was performed (right lower pannel). (b) Morphological changes (left panel) and SA-β-Gal positivity (middle graph) and percentage of dead cells (right graph) were determined. PAPSS2-depleted MCF7 cells were used as a control. (c) MCF7 cells were harvested at the indicated times after siRNA transfection and subjected to WB analysis. (d) Morphological changes (left panel) and SA-β-Gal positivity (right graph) were determined. (e) MCF7 cells were transfected with Con Si or PAPSS2 Si 6 h before incubation with or without heparan-derived octasaccharides for 3 days. Cells were then lysed and subjected to WB analysis (left panel). Graphs show quantification of WB (right panel). Error bars indicate the S.D. of three independent experiments. ***P<0.001. # indicates statistical indifference, P>0.05. ND, not determined.
PAPSS2 involves in premature senescence in a xenograft tumor mouse model through p53-p21 signaling pathway
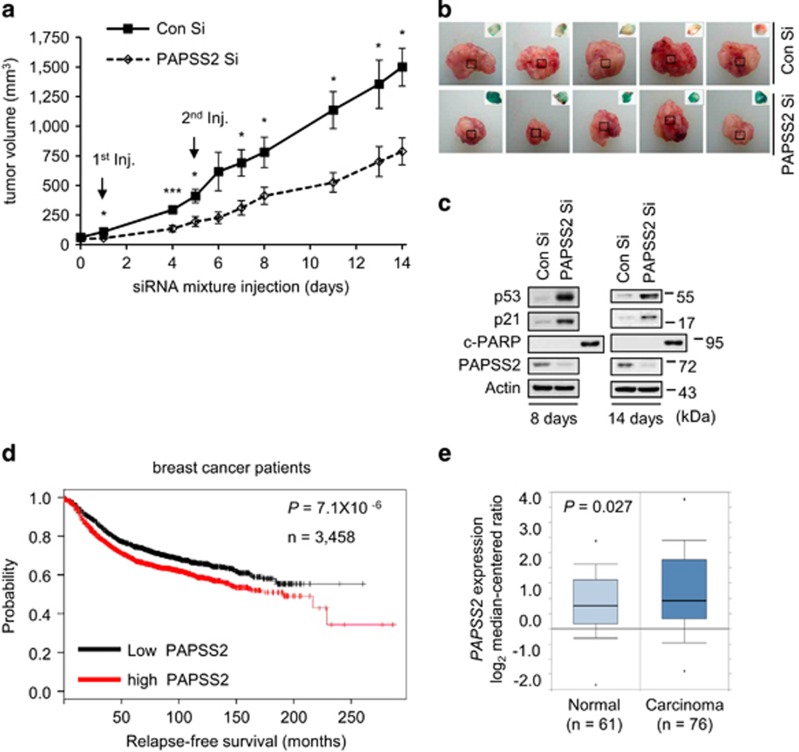
To explore the biological significance of our findings that PAPSS2 is critical to prevent cellular senescence in vivo, we examined the effects of PAPSS2 depletion in a xenograft tumor mouse model. We found that mice administered PAPSS2 Si showed attenuated tumor growth (Figures 8a and b). A western blot (WB) analysis of tumor tissue revealed accumulations of p53 and p21 at 8 and 14 days after PAPSS2 Si administration, with no detectable PARP cleavage (Figure 8c). Next, we evaluated the prognostic value of PAPSS2 expression using microarray data from patients with breast cancer (http://kmplot.com/analysis/). Patients with low PAPSS2 levels showed longer relapse-free survival periods than patients with high PAPSS2 levels (Figure 8d). We also analyzed gene expression data from the Cancer Genome Atlas and found that PAPSS2 expression was upregulated in carcinoma breast tissue compared with normal breast tissue (Figure 8e). Together, these results indicate that PAPSS2 depletion induces premature senescence through p53-p21 signaling pathway in a xenograft tumor mouse model, and that PAPSS2 upregulation may be involved, at least in part, in human breast cancer development via suppression of premature senescence.
Figure 8.
PAPSS2 depletion retards tumor growth due to the induction of cellular senescence in a xenograft tumor mouse model. (a) Tumor volume in xenograft mice (n=5) was measured at the indicated times. Error bars indicate S.E.M. ***P<0.001 and *P<0.05. (b) Pictures of excised and SA-β-Gal-stained tumors 14 days after the first siRNA injection. (c) Tumor tissue lysates 8 or 14 days after the first siRNA injection were subjected to WB analysis. (d) Kaplan–Meier curves of relapse-free survival times of patients with breast cancer. Data were obtained from http://kmplot.com/analysis/. Statistical significance was determined using the log-rank test. (e) Box plots comparing PAPSS2 expression (as log2 median-centered ratios) in normal breast tissue and carcinoma breast tissue. Dots indicate extreme data values. Data were obtained from http://oncomine.org/.
Discussion
Cellular senescence was originally observed in human fibroblasts undergoing replicative exhaustion in vitro.43 As neoplastic transformation involves events that inhibit the senescence program, cancer cells are thought to have lost the capacity to senesce. However, cancer cells can be readily induced to undergo premature senescence by the hyper-activation of oncogenes, the loss of tumor suppressors, and a variety of stresses in vitro and in vivo.44, 45, 46, 47 To our knowledge, no study has elucidated the importance of sulfation status in the regulation of cellular senescence, although tremendous attention has been paid to the role of sulfated PGs in a variety of other biological processes. Here, we demonstrate that the undersulfation of HSPGs, in particular desulfation at the 2-O position of iduronate in HS, prematurely induces cellular senescence through augmented FGFR signaling pathway.
FGF family, which consists of at least 22 distinct protein members and is divided into seven subfamilies in humans, governs many biological and pathological processes.48, 49 Members of each subfamily show similarities in sequence as well as in biochemical and developmental properties.49 FGFs promote angiogenesis, proliferation, apoptosis, differentiation, wound healing, chemotaxis, and motility in several cell types. Because of its angiogenic, mitogenic, and anti-apoptotic properties, FGFs are recognized as potential oncoproteins.34 In particular, some researchers have reported that FGF2 can suppress proliferation via a variety of mechanisms, such as apoptosis50, 51 and cell cycle arrest.52, 53 Although the relationship between FGFs and HSPGs has been the subject of many studies, much remains unknown about the HS-induced potentiation of FGF activity. In this study, in contrast to previous reports,30, 31, 32, 54 we discovered that augmented FGF1/2-FGFR1 signaling triggers cellular senescence due to an undersulfated HSPG status. Moreover, we found that p53-p21 signaling is critical for the induction of HS-dependent premature senescence. Depletion of FGFR1 or AKT inhibits p53/p21 accumulation and finally prohibited premature senescence. Our results demonstrate the importance of the FGFR1-AKT-p53-p21 signal transduction cascade in sulfation status-dependent premature senescence. However, we observed mTOR phosphorylation instead of DNA damage response in PAPSS2-mediated premature senescence (Supplementary Figure S9). This result supports the important role of mTOR in cellular senescence.55, 56 The present study is the first to suggest that the proper modulation of basal FGFR1 activity that is tightly linked to HS sulfation status is critical for preventing cellular senescence both in vitro and in xenograft tumor mouse model. Senescent cells could trigger tumor clearance and limit tissue damage via the innate immune response.57, 58 Thus, we suppose that a PAPSS2-mediated tumor suppressive mechanism in a xenograft tumor mouse model might be attributed to the cooperativity between tumor cell senescence and the innate immune response.
FGFs performs pleiotropic functions through their binding to cell surface receptors,34 two of which have been identified: the high-affinity FGFRs and the low-affinity HSPGs.59 The expression and sulfation status of HSPGs differ depending on cell type,60 and HSPGs act as multifunctional regulators of protein activity through a range of different mechanisms that are dependent on specific HS–protein interactions.22 From the many studies subjected to the relationship between HSPGs and FGFs, it has been recognized that the binding of HS to FGFs confer several biological advantages, such as protecting FGFs from proteolysis, serving as a reservoir of FGFs, limiting FGF diffusion, and increasing FGF affinity for its receptors.61 Several previous studies reporting that HS is required for the bindings and activities of FGFs via the formation of the FGF/HS/FGFR ternary complex and that inhibition of the interaction between HS and FGF blocks FGFR signaling led to the acceptance of the notion that HSPGs function as FGF activators, although there was a lack of direct evidence.30, 31, 32, 61, 62, 63 In addition, FGF1/2 binds preferentially to the 2-O position in HS, whereas the formation of the FGF/HS/FGF receptor (FGFR) ternary complex and the mitogenic activity of FGF1/2 require sulfation at the 6-O position in HS.54, 64, 65 However, the exact role of HS in FGF signaling is still a matter of considerable debate.33
In the present study, we found that FGF1 superfamily, FGF1/2 readily binds to FGFR1 and leads the hyper-activation of FGFR1 signaling in the absence of sulfated HSPG. In contrast, HGF/Met signaling, another well-known ligand/receptor signaling regulated by HS, is not affected by HS sulfation status. Our results demonstrated that HSPG has a critical role as a reservoir of FGF to avoid excess activation of receptor signaling, but is not strictly required for FGFR activation. From our results, we suggest that desulfated HS, particularly 2-O-desulfated HS, allows FGF1/2 to bind primarily to FGFR1 instead of HS, and leads to excessive FGFR signaling. Therefore, we propose the hypothesis that HS specifically acts as a fine-tuner of FGFR signaling through modulating FGF and FGFR binding and strictly required for the prevention of cellular senescence. The present study explains how FGF signaling is tumor promoting in some contexts, but tumor-suppressive in others. Our results are consistent with previous studies demonstrating that FGFR1 can bind to FGF2 in the absence of heparin66 and that mutations in the heparin-binding region of FGF2 reduce its affinity for cell-associated HS, but do not affect the affinity for its receptor FGFR1.67
In conclusion, our findings suggest that the potential to regulate FGF activity through HS-dependent interactions can provide an attractive system for regulating cellular responses to FGF in the extracellular environment. Specifically, HSPG undersulfation can render cells incapable of fine-tuning FGF signaling and can thereby induce premature senescence, which can serve as a fail-safe mechanism of defense against tumorigenesis. Fujita et al.68 report that downregulation of telomere repeat binding factor 2 (TRF2) is involved in the onset of cellular senescence. Furthermore, Biroccio et al.69 show that HS3ST4, which is positively regulated by TRF2, inhibits the recruitment of natural killer (NK) cells. Based on these previous reports and our present results, we propose that cell surface HS sulfation could trigger cellular senescence by modulating receptor signaling and could modify innate immune surveillance by affecting sulfated HS recognition by NK cells. Thus, targeting the sulfation pathway of PGs or performing immunotherapy for HS at the cell surface may provide therapeutic benefits to cancer patients.
Materials and Methods
Cell culture
MCF7 and HDF cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM; WelGENE, Daegu, Korea). A549 cells were cultured in RPMI 1640 (WelGENE). HCT116 parental, p53−/−, and p21−/− isogenic cell lines were cultured in McCoy's 5A medium (WelGENE). MCF10A cells were cultured in DMEM/F-12 (WelGENE). All cells were supplemented with 10% fetal bovine serum (Lonza, Basel, Switzerland), 1% penicillin, and streptomycin solution (WelGENE) at 37 °C in a 5% CO2 incubator.
RNA interference and plasmid transfection
All siRNAs (Supplementary Table 2) were obtained from Bioneer Inc. (Daejeon, Korea). Cells were transfected with 100 nM siRNA duplexes using RNAiMAX (Invitrogen, Karlsruhe, Germany). Transfection of plasmids was carried out using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. The plasmid expressing the Flag-tagged PAPSS2 was generated by ligating polymerase chain reaction (PCR) products into p3xFlag CMV7.1 at Hind III and Sal I sites, (Macrogen. Seoul, Korea). The sequences were as follows: 5'-TAAGCTTATGTCGGGGATCAA-3' (forward) and 5′-TGTCGACTTAGTTCTTCTCCA-3′ (reverse). Mammalian expression vectors encoding HA-tagged wild-type (WT) FGFR1 or HA-tagged KI FGFR1, a catalytically inactive mutant of FGFR1 (Y653/654F), were kindly provided by Dr. Daniela Rotin (University of Toronto, Toronto, Canada).70 To create GFP-FGFR1 fusion protein, WT FGFR1 was released by PCR from HA-tagged WT FGFR1 and was inserted to pAcGFP-N1 (Clontech, Mountain View, CA, USA). The primer sequences were as follows: 5′-CCGCTCGAGGCCATGGGCATGTGGAGCTGGAAGTGCCTCCTCTTCTGGGCTGTGCTGGTCACA-3′ (forward) and 5′-CCCAAGCTTGCGGCGTTTGAGTCCGCCATTGGCA AG-3′ (reverse). PCR products were digested and subcloned into the pAcGFP-N1 vector at Xho I and Hind III sites.
WB, IP, and FGFs neutralization
For WB analysis, cell lysates were prepared in RIPA lysis buffer containing protease inhibitors (Roche, Indianapolis, IN, USA) and phosphatase inhibitors (Sigma-Aldrich, St Louis, MO, USA). Lysates were mixed with 2 × Laemmli sample buffer, boiled, and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). After transferring proteins to a nitrocellulose membrane, WB was performed by blocking the membrane with 5% non-fat dried milk and incubating with various primary antibodies (Abs) followed by horseradish peroxidase-conjugated secondary Abs (Supplementary Table 3). WB detection was conducted with ECL reagents (Thermo Scientific, Waltham, MA, USA).
For IP, cell lysates were prepared in 1% NP-40 lysis buffer (25 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1% NP-40) containing protease inhibitors (Roche) and phosphatase inhibitors (Sigma-Aldrich). The lysates were precleared with Protein G- or A-Sepharose beads (Sigma-Aldrich), conjugated with each indicated Ab, and tumbled with Protein G- or A-Sepharose beads. The immune complexes were then subjected to SDS-PAGE, which was followed by WB.
For neutralization of FGFs, 1, 2.5, or 5 μg/ml of a blocking antibody for FGF2 (anti-FGF2 neutralizing antibody; Millipore, Charlottesville, VA, USA) and 5 μg/ml of a blocking antibody for FGF1, FGF3, and FGF4 (anti- FGF1, FGF3, and FGF4 neutralizing antibodies; R&D Systems, Minneapolis, MN, USA) was exogenously added daily after 6 h siRNA transfections and incubated for 3 days. After incubation, WB, SA-β-Gal staining, and quantification of relative cell number were conducted.
Cell growth rate, colony forming assay, and cell cycle analysis
Cell viability was determined using a trypan blue exclusion in which cell suspensions were stained with 0.4% (w/v) trypan blue (GIBCO, grand island, NY, USA) at a 1 : 1 ratio, and viable (unstained) and dead (stained) cells were counted by hemocytometer microscopy. Cell viability was shown as a value relative to the number of Con Si-transfected cells. Clonogenicity was examined using a colony forming assay. Cells were seeded at a density of 1 × 104 cells per 60-mm dish in triplicate and cultured for 7 days. Colonies were stained with Diff-Quick (Sysmex, Kobe, Japan).
For cell cycle analysis, cells were collected by trypsinization, fixed in 95% ethanol, washed in phosphate-buffered saline (PBS), and resuspended in 1 ml PBS containing 1 mg/ml RNase and 50 mg/ml propidium iodide (Sigma-Aldrich). After incubation in the dark for 30 min, cell cycle distributions were analyzed using a fluorescence-activated cell sorting (FACS) Calibur flow cytometer (BD Biosciences, San Diego, CA, USA).
SA-β-Gal staining
Cells and tumors were stained for β-galactosidase activity as described in Dimri et al.71 In brief, cells were washed in PBS and fixed for 5 min in 3.7% formaldehyde. Fixed cells were then incubated at 37 °C (without CO2) with fresh staining solution consisting of 1 mg/ml 5-bromo-4-chloro-3-indolyl β-D-galactoside (X-Gal) in 40 mM citric acid/sodium phosphate (pH 6.0), 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl, and 2 mM MgCl2. Staining was evident after 12–16 h.
Tumor-xenograft mice
Xenograft mice were established as described previously.72 In brief, a single-cell suspension of H460 cells (1 × 106) was injected subcutaneously into the lateral hind leg of 6-week-old BALB/c nude mice (n=5). Average tumor volume was determined as (L × W2)/2; measurements of tumor length (L) and width (W) were taken with a caliper. When the tumor reached an average volume of ~50 mm3, a siRNA mixture with AteloGenes Local Use (KOKEN, Koraku, Tokyo) was injected to wrap up the whole tumor mass. After 5 days, a second injection was performed. Tumor volumes were determined daily after siRNA gel injection. Mice were killed 8 or 14 days after xenograft injection, and tumors were removed for further analysis. All animal studies were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Korea Institute Radiological and Medical Sciences (Approval No. KIRAMS 2012-037).
Determination of the sulfation level of HS
For determination of the level of HS by a confocal microscopy, cells were fixed, blocked, and then incubated with mouse anti-human HS (10E4 epitope; US Biological, Salem, MA, USA). After washing, cells were stained with PE-conjugated goat anti-mouse IgG+IgM (Jackson ImmunoResearch, Suffolk, UK), mounted, and then visualized using a LSM-710 confocal microscope and × 63 objective lens (Carl Zeiss, Jena, Germany). For quantification of the level of HS, stained cells were measured using a FACS Calibur flow cytometer (BD Biosciences).
FGFR1 dimerization assay and competition assay of heparin-derived oligosaccharides
MCF7 cells were co-transfected with plasmids encoding HA-tagged or GFP-tagged WT FGFR1 6 h before transfection with Con Si or PAPSS2 Si. Transfected cells were lysed with 1% NP-40 lysis buffer containing protease and phosphatase inhibitors, and cell lysates were immunoprecipitated using mouse anti-GFP antibody. The precipitates were separated by 7.5% SDS-PAGE and then blotted with anti-HA antibody.
For competition assay of heparin-derived oligosaccharides, MCF7 cells were transfected with Con Si or PAPSS2 Si 6 h before incubation for 3 days with or without 5 nmol/ml heparin-derived octasaccharides. Cells were lysed with RIPA buffer containing protease and phosphatase inhibitors and subjected to WB. All heparin-derived octasaccarides were obtained from Neoparin Inc. (Alameda, CA, USA). In brief, 2-O-desulfated heparin is a derivative in which only the O-sulfated groups on C-2 of uronic acid residues have been removed. 6-O-desulfated heparin is a derivative in which most O-sulfate groups on C-6 of glucosamine residues have been removed but most of the 2-O-sulfate/3-O-sulfate groups remain intact. Fully desulfated heparin is a derivative in which all O-sulfate and N-sulfate esters of heparin have been removed without changing the backbone structure. Oversulfated heparin is a derivative in which all primary hydroxyls in glucosamine residues and a large proportion of secondary hydroxyl groups in disaccharide units have been substituted by O-sulfate esters.
Statistical analysis
Unless otherwise stated, comparisons between groups were performed using two-tailed Student's t-tests. Values of P<0.05 were considered statistically significant.
Acknowledgments
This work was supported by the Nuclear Research and Development Program (Grant No. 2012M2B2B1055637) and Medical Research Center (MRC) (Grant No. 2014009392) of the National Research Foundation of Korea (NRF), funded by the Korean government (MSIP).
Glossary
- FACS
Fluorescence activated cell sorter
- FGF
fibroblast growth factor
- FGFR
fibroblast growth factor receptor
- HDF
human diploid fibroblast
- HGF
hepatocyte growth factor
- HS6ST1
heparan sulfate 6-O-sulfotransferase 1
- HSPG
heparan sulfate proteoglycan
- IGF1Rβ
insulin-like growth factor 1 receptor β
- IP
immunoprecipitation
- KI
kinase inactive
- PAPSS2
3′-phosphoadenosine 5′-phosphosulfate synthetase 2
- PARP
poly ADP ribose polymerase
- PG
proteoglycan
- SA-β-Gal
senescence-associated β-galactosidase
- SASP
senescence associated secretory phenotype
- WB
western blot
- WT
wild type
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd).
Edited by J Bartek
Supplementary Material
References
- Narita M, Lowe SW. Senescence comes of age. Nat Med 2005; 11: 920–922. [DOI] [PubMed] [Google Scholar]
- Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol 2013; 75: 685–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer 2010; 10: 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 2007; 8: 729–740. [DOI] [PubMed] [Google Scholar]
- Prieur A, Peeper DS. Cellular senescence in vivo: a barrier to tumorigenesis. Curr Opin Cell Biol 2008; 20: 150–155. [DOI] [PubMed] [Google Scholar]
- Ohtani N, Mann DJ, Hara E. Cellular senescence: its role in tumor suppression and aging. Cancer Sci 2009; 100: 792–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira V, Park Y, Chen CC, Xu PZ, Chen ML, Tonic I et al. Akt determines replicative senescence and oxidative or oncogenic premature senescence and sensitizes cells to oxidative apoptosis. Cancer Cell 2008; 14: 458–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Roninson IB. Hallmarks of senescence in carcinogenesis and cancer therapy. Oncogene 2004; 23: 2919–2933. [DOI] [PubMed] [Google Scholar]
- Schmitt CA. Cellular senescence and cancer treatment. Biochim Biophys Acta 2007; 1775: 5–20. [DOI] [PubMed] [Google Scholar]
- Lee M, Lee JS. Exploiting tumor cell senescence in anticancer therapy. BMB Rep 2014; 47: 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer 2009; 9: 81–94. [DOI] [PubMed] [Google Scholar]
- Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol 2010; 5: 99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F, Campisi J. Four faces of cellular senescence. J Cell Biol 2011; 192: 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Mancera PA, Young AR, Narita M. Inside and out: the activities of senescence in cancer. Nat Rev Cancer 2014; 14: 547–558. [DOI] [PubMed] [Google Scholar]
- Campisi J. Cellular senescence: putting the paradoxes in perspective. Curr Opin Genet Dev 2011; 21: 107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Espin D, Canamero M, Maraver A, Gomez-Lopez G, Contreras J, Murillo-Cuesta S et al. Programmed cell senescence during mammalian embryonic development. Cell 2013; 155: 1104–1118. [DOI] [PubMed] [Google Scholar]
- Storer M, Mas A, Robert-Moreno A, Pecoraro M, Ortells MC, Di Giacomo V et al. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell 2013; 155: 1119–1130. [DOI] [PubMed] [Google Scholar]
- Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J et al. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem 1999; 68: 729–777. [DOI] [PubMed] [Google Scholar]
- Hacker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nat Rev Mol Cell Biol 2005; 6: 530–541. [DOI] [PubMed] [Google Scholar]
- Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 2007; 446: 1030–1037. [DOI] [PubMed] [Google Scholar]
- Couchman JR. Transmembrane signaling proteoglycans. Annu Rev Cell Dev Biol 2010; 26: 89–114. [DOI] [PubMed] [Google Scholar]
- Turnbull J, Powell A, Guimond S. Heparan sulfate: decoding a dynamic multifunctional cell regulator. Trends Cell Biol 2001; 11: 75–82. [DOI] [PubMed] [Google Scholar]
- Li H, Deyrup A, Mensch Jr JR, Domowicz M, Konstantinidis AK, Schwartz NB. The isolation and characterization of cDNA encoding the mouse bifunctional ATP sulfurylase-adenosine 5'-phosphosulfate kinase. J Biol Chem 1995; 270: 29453–29459. [DOI] [PubMed] [Google Scholar]
- Xu ZH, Otterness DM, Freimuth RR, Carlini EJ, Wood TC, Mitchell S et al. Human 3'-phosphoadenosine 5′-phosphosulfate synthetase 1 (PAPSS1) and PAPSS2: gene cloning, characterization and chromosomal localization. Biochem Biophys Res Commun 2000; 268: 437–444. [DOI] [PubMed] [Google Scholar]
- Besset S, Vincourt JB, Amalric F, Girard JP. Nuclear localization of PAPS synthetase 1: a sulfate activation pathway in the nucleus of eukaryotic cells. FASEB J 2000; 14: 345–354. [DOI] [PubMed] [Google Scholar]
- Schroder E, Gebel L, Eremeev AA, Morgner J, Grum D, Knauer SK et al. Human PAPS synthase isoforms are dynamically regulated enzymes with access to nucleus and cytoplasm. PLoS One 2012; 7: e29559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem 2002; 71: 435–471. [DOI] [PubMed] [Google Scholar]
- Kusche-Gullberg M, Kjellen L. Sulfotransferases in glycosaminoglycan biosynthesis. Curr Opin Struct Biol 2003; 13: 605–611. [DOI] [PubMed] [Google Scholar]
- Zhang L. Glycosaminoglycan (GAG) biosynthesis and GAG-binding proteins. Prog Mol Biol Transl Sci 2010; 93: 1–17. [DOI] [PubMed] [Google Scholar]
- Spivak-Kroizman T, Lemmon MA, Dikic I, Ladbury JE, Pinchasi D, Huang J et al. Heparin-induced oligomerization of FGF molecules is responsible for FGF receptor dimerization, activation, and cell proliferation. Cell 1994; 79: 1015–1024. [DOI] [PubMed] [Google Scholar]
- Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell 1991; 64: 841–848. [DOI] [PubMed] [Google Scholar]
- Rapraeger AC, Krufka A, Olwin BB. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science 1991; 252: 1705–1708. [DOI] [PubMed] [Google Scholar]
- Qu X, Carbe C, Tao C, Powers A, Lawrence R, van Kuppevelt TH et al. Lacrimal gland development and Fgf10-Fgfr2b signaling are controlled by 2-O- and 6-O-sulfated heparan sulfate. J Biol Chem 2011; 286: 14435–14444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 2010; 10: 116–129. [DOI] [PubMed] [Google Scholar]
- Safaiyan F, Kolset SO, Prydz K, Gottfridsson E, Lindahl U, Salmivirta M. Selective effects of sodium chlorate treatment on the sulfation of heparan sulfate. J Biol Chem 1999; 274: 36267–36273. [DOI] [PubMed] [Google Scholar]
- Qiu H, Jiang JL, Liu M, Huang X, Ding SJ, Wang L. Quantitative phosphoproteomics analysis reveals broad regulatory role of heparan sulfate on endothelial signaling. Mol Cell Proteomics 2013; 12: 2160–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund J, Sondergaard MT, Conover CA, Overgaard MT. Heparin-binding mechanism of the IGF2/IGF-binding protein 2 complex. J Mol Endocrinol 2014; 52: 345–355. [DOI] [PubMed] [Google Scholar]
- Arai T, Busby Jr W, Clemmons DR. Binding of insulin-like growth factor (IGF) I or II to IGF-binding protein-2 enables it to bind to heparin and extracellular matrix. Endocrinology 1996; 137: 4571–4575. [DOI] [PubMed] [Google Scholar]
- Harmer NJ, Chirgadze D, Hyun Kim K, Pellegrini L, Blundell TL. The structural biology of growth factor receptor activation. Biophys Chem 2003; 100: 545–553. [DOI] [PubMed] [Google Scholar]
- Ashikari-Hada S, Habuchi H, Sugaya N, Kobayashi T, Kimata K. Specific inhibition of FGF-2 signaling with 2-O-sulfated octasaccharides of heparan sulfate. Glycobiology 2009; 19: 644–654. [DOI] [PubMed] [Google Scholar]
- Frese MA, Milz F, Dick M, Lamanna WC, Dierks T. Characterization of the human sulfatase Sulf1 and its high affinity heparin/heparan sulfate interaction domain. J Biol Chem 2009; 284: 28033–28044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine S, Tamba M, Ishimine H, Araki K, Shiomi K, Okada T et al. Organ-specific sulfation patterns of heparan sulfate generated by extracellular sulfatases Sulf1 and Sulf2 in mice. J Biol Chem 2012; 287: 9579–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res 1961; 25: 585–621. [DOI] [PubMed] [Google Scholar]
- Byun HO, Han NK, Lee HJ, Kim KB, Ko YG, Yoon G et al. Cathepsin D and eukaryotic translation elongation factor 1 as promising markers of cellular senescence. Cancer Res 2009; 69: 4638–4647. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Kim BC, Park MJ, Lee YS, Kim YN, Lee BL et al. PTEN status switches cell fate between premature senescence and apoptosis in glioma exposed to ionizing radiation. Cell Death Differ 2011; 18: 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Weinberg RA. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol 2005; 37: 961–976. [DOI] [PubMed] [Google Scholar]
- Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell 2008; 132: 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Ornitz DM. Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J Biochem 2011; 149: 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends Genet 2004; 20: 563–569. [DOI] [PubMed] [Google Scholar]
- Westwood G, Dibling BC, Cuthbert-Heavens D, Burchill SA. Basic fibroblast growth factor (bFGF)-induced cell death is mediated through a caspase-dependent and p53-independent cell death receptor pathway. Oncogene 2002; 21: 809–824. [DOI] [PubMed] [Google Scholar]
- Williamson AJ, Dibling BC, Boyne JR, Selby P, Burchill SA. Basic fibroblast growth factor-induced cell death is effected through sustained activation of p38MAPK and up-regulation of the death receptor p75NTR. J Biol Chem 2004; 279: 47912–47928. [DOI] [PubMed] [Google Scholar]
- Wang H, Rubin M, Fenig E, DeBlasio A, Mendelsohn J, Yahalom J et al. Basic fibroblast growth factor causes growth arrest in MCF-7 human breast cancer cells while inducing both mitogenic and inhibitory G1 events. Cancer Res 1997; 57: 1750–1757. [PubMed] [Google Scholar]
- Krejci P, Bryja V, Pachernik J, Hampl A, Pogue R, Mekikian P et al. FGF2 inhibits proliferation and alters the cartilage-like phenotype of RCS cells. Exp Cell Res 2004; 297: 152–164. [DOI] [PubMed] [Google Scholar]
- Pye DA, Vives RR, Hyde P, Gallagher JT. Regulation of FGF-1 mitogenic activity by heparan sulfate oligosaccharides is dependent on specific structural features: differential requirements for the modulation of FGF-1 and FGF-2. Glycobiology 2000; 10: 1183–1192. [DOI] [PubMed] [Google Scholar]
- Leontieva OV, Blagosklonny MV. DNA damaging agents and p53 do not cause senescence in quiescent cells, while consecutive re-activation of mTOR is associated with conversion to senescence. Aging 2010; 2: 924–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnichenko M, Hong L, Liao R, Vogt PK, Sun P. Attenuation of TORC1 signaling delays replicative and oncogenic RAS-induced senescence. Cell Cycle 2012; 11: 2391–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007; 445: 656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C et al. Senescence of activated stellate cells limits liver fibrosis. Cell 2008; 134: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov AN, Schlessinger J, Hubbard SR, Mohammadi M. Structural basis for FGF receptor dimerization and activation. Cell 1999; 98: 641–650. [DOI] [PubMed] [Google Scholar]
- Carey DJ. Syndecans: multifunctional cell-surface co-receptors. Biochem J 1997; 327: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo I, Kimura-Yoshida C. Extracellular modulation of Fibroblast Growth Factor signaling through heparan sulfate proteoglycans in mammalian development. Curr Opin Genet Dev 2013; 23: 399–407. [DOI] [PubMed] [Google Scholar]
- Richard C, Liuzzo JP, Moscatelli D. Fibroblast growth factor-2 can mediate cell attachment by linking receptors and heparan sulfate proteoglycans on neighboring cells. J Biol Chem 1995; 270: 24188–24196. [DOI] [PubMed] [Google Scholar]
- Roghani M, Mansukhani A, Dell'Era P, Bellosta P, Basilico C, Rifkin DB et al. Heparin increases the affinity of basic fibroblast growth factor for its receptor but is not required for binding. J Biol Chem 1994; 269: 3976–3984. [PubMed] [Google Scholar]
- Ai X, Do AT, Lozynska O, Kusche-Gullberg M, Lindahl U, Emerson CP Jr. QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J Cell Biol 2003; 162: 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemth P, Kreuger J, Kusche-Gullberg M, Sturiale L, Gimenez-Gallego G, Lindahl U. Biosynthetic oligosaccharide libraries for identification of protein-binding heparan sulfate motifs. Exploring the structural diversity by screening for fibroblast growth factor (FGF)1 and FGF2 binding. J Biol Chem 2002; 277: 30567–30573. [DOI] [PubMed] [Google Scholar]
- Kiefer MC, Baird A, Nguyen T, George-Nascimento C, Mason OB, Boley LJ et al. Molecular cloning of a human basic fibroblast growth factor receptor cDNA and expression of a biologically active extracellular domain in a baculovirus system. Growth Factors 1991; 5: 115–127. [DOI] [PubMed] [Google Scholar]
- Presta M, Statuto M, Isacchi A, Caccia P, Pozzi A, Gualandris et al. Structure-function relationship of basic fibroblast growth factor: site-directed mutagenesis of a putative heparin-binding and receptor-binding region. Biochem Biophys Res Commun 1992; 185: 1098–1107. [DOI] [PubMed] [Google Scholar]
- Fujita K, Horikawa I, Mondal AM, Jenkins LM, Appella E, Vojtesek B et al. Positive feedback between p53 and TRF2 during telomere-damage signalling and cellular senescence. Nat Cell Biol 2010; 12: 1205–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biroccio A, Cherfils-Vicini J, Augereau A, Pinte S, Bauwens S, Ye J et al. TRF2 inhibits a cell-extrinsic pathway through which natural killer cells eliminate cancer cells. Nat Cell Biol 2013; 15: 818–828. [DOI] [PubMed] [Google Scholar]
- Persaud A, Alberts P, Hayes M, Guettler S, Clarke I, Sicheri F et al. Nedd4-1 binds and ubiquitylates activated FGFR1 to control its endocytosis and function. EMBO J 2011; 30: 3259–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 1995; 92: 9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Lee HC, Lee JJ, Choi CM, Kim DK, Lee JC et al. Wig1 prevents cellular senescence by regulating p21 mRNA decay through control of RISC recruitment. EMBO J 2012; 31: 4289–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.