Abstract
Endoplasmic reticulum (ER) stress, defective autophagy and genomic instability in the central nervous system are often associated with severe developmental defects and neurodegeneration. Here, we reveal the role played by Rint1 in these different biological pathways to ensure normal development of the central nervous system and to prevent neurodegeneration. We found that inactivation of Rint1 in neuroprogenitors led to death at birth. Depletion of Rint1 caused genomic instability due to chromosome fusion in dividing cells. Furthermore, Rint1 deletion in developing brain promotes the disruption of ER and Cis/Trans Golgi homeostasis in neurons, followed by ER-stress increase. Interestingly, Rint1 deficiency was also associated with the inhibition of the autophagosome clearance. Altogether, our findings highlight the crucial roles of Rint1 in vivo in genomic stability maintenance, as well as in prevention of ER stress and autophagy.
Neurons homeostasis is dependent on a proper regulation of protein synthesis, transportation and degradation. Biological mechanisms controlling protein secretory pathways such as endoplasmic reticulum (ER)-Golgi trafficking are essential for cell homeostasis. Defective ER-Golgi trafficking leads to ER stress1 and is involved in neurodegenerative disorders including Alzheimer's disease.1, 2 Also protein and organelle turnover by autophagy is crucial in post-mitotic neurons to prevent protein accumulation-associated toxicity. The autophagic protein degradation pathway is tightly regulated and its disruption leads to neurodegeneration.2, 3, 4 ER-Golgi trafficking and autophagy are related biological mechanisms. In the both processes, the fusion of membranes ensures the transport of proteins to their destination. Autophagy requires the clearance of autophagosomes and their cargo upon fusion with lysosomes.5, 6 It is suggested that this fusion is regulated by several Rab proteins and soluble N-ethylmaleimidine-sensitive factor attachment protein receptors (SNAREs).4, 6, 7 Similarly in case of bi-directional ER-Golgi transport the tethering of the COPII-coated vesicles at Golgi membrane takes place in anterograde transport, when in retrograde transport COPI-coated vesicles are tethered at ER membrane.8, 9 Membrane fusion is again mediated by Rab GTPases and SNAREs and SNARE-associated tethering complexes. One of them is the ZW10 complex composed of RINT1, ZW10 and NAG that has a pivotal role in the retrograde transport in association with ER SNAREs Syntaxin 18, p31 and Sec22b.10, 11, 12 The complex integrity is regulated by an autophagy factor UVRAG, which interacts with RINT1, demonstrating the intersection of Golgi-ER and autophagic trafficking mechanisms.13
RINT1 was identified as a Rad50-interacting protein involved in G2/M checkpoint14 and was shown to be involved in telomerase independent maintenance of telomeres via the interaction with p130.15 The importance of RINT1 in Golgi-ER trafficking and thus in homeostasis of Cis- and Trans-Golgi was demonstrated in vitro16, 17, 18, 19, 20, 21 where in vivo studies were limited by the early embryonic lethality (E5.5) associated with Rint1 inactivation.19 Rint1 was proposed to be a tumor suppressor gene since its heterozygous mutation predisposed to tumor,19 but it was also reported to act like an oncogene in glioblastomas.22 The tumor suppression function of RINT1 was recently strengthened by the identification of rare RINT1 mutations predisposing to breast cancer and Lynch syndrome cancers.23 These findings suggested that RINT1 is essential for cell homeostasis and prevention of tumorigenesis. Nevertheless, the lack of mouse models prevented further understanding of its in vivo functions. Therefore, it motivated us to analyze a mouse model with a conditional Rint1 inactivation in the central nervous system (CNS). Here, we identify new Rint1 functions in maintenance of genomic stability and autophagy together with its role in ER-Golgi homeostasis in developing brain. We demonstrate that Rint1 is essential for CNS development and prevention of neurodegeneration.
Results
Specific CNS inactivation of Rint1 is lethal at birth
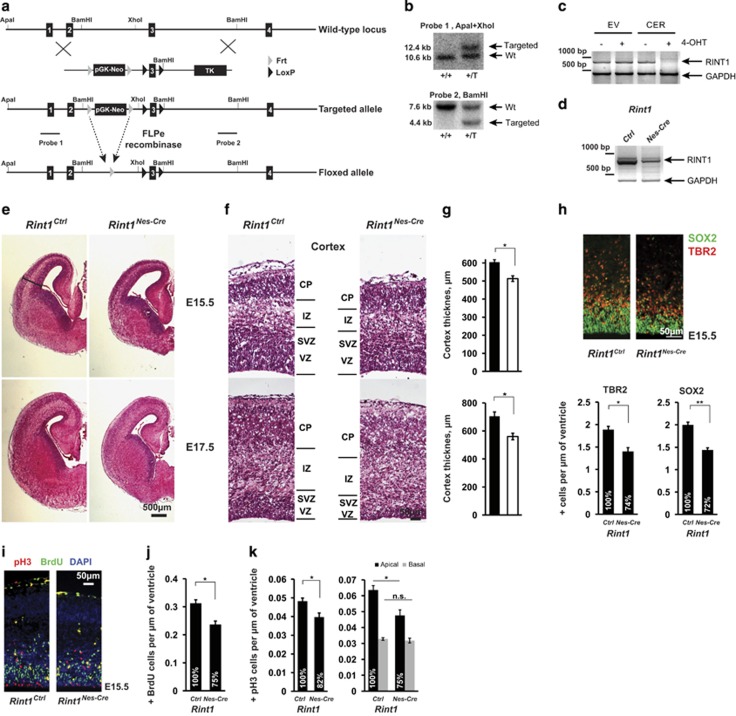
To overcome the early embryonic lethality associated with Rint1 inactivation in mouse,19 we generated a conditional Rint1 allele (Figures 1a and b). Deletion of exon 3 is predicted to generate an out of frame mutation. The pGK-Neo resistance cassette was excised in vivo using FLPe recombinase to create the ‘Floxed' allele. The ‘Floxed' mice were born at a normal Mendelian ratio (data not shown). Finally, the Nestin-Cre mouse was used to specifically inactivate Rint1 in neural progenitors (Rint1Nes-Cre). We confirmed the complete deletion of the Rint1 exon 3 by RT-PCR in embryonic E15.5 cortices and in immortalized mouse embryonic fibroblasts (MEFs) with inducible Rint1 inactivation (Figures 1c and d). Rint1Nes-Cre mice died at birth (Supplementary Table S1), suggesting an important role of Rint1 during embryonic CNS development.
Figure 1.
Rint1 deletion in the brain leads to developmental defects in cortex. (a) Targeting strategy for conditional inactivation of Rint1. Floxed allele has loxP sites flanking exon 3 that can be recombined by the Cre protein leading to exon 3 deletion and thus to the out of frame mutant. (b) Southern blot analysis with two different probes indicates ES clones with correctly integrated targeting vector. (c) mRNA expression analysis shows loss of Rint1 mRNA in immortalized MEFs after the 4-OHT mediated Rint1 inactivation. (d) mRNA analysis of cortices from E15.5 Rint1Nes-Cre and Rint1Ctrl embryos with indicated genotypes. (e) Comparison of Rint1Ctrl and Rint1Nes-Cre brains at E15.5 and E17.5 by H&E staining of coronal sections. (f) Defects in Rint1Nes-Cre cortex indicating reduction of ventricular (VZ) and sub-ventricular (SVZ) zones at E15.5 and E17.5. IZ, intermediate zone; CP, cortical plate. The quantification of cortex thickness is shown in (g). (h) Neuronal progenitors labeled by Sox2 and TBR2 are significantly reduced in Rint1Nes-Cre cortex at E15.5. Lower panel shows corresponding quantifications. (i) Comparison of mitotic pH3+ and proliferating cells (BrdU+ cells) in wild-type and Rint1Nes-Cre cortex from E15.5 embryos. Quantification for BrdU+ in (j) and for pH3+ in (k) shows significant reduction of both cell types in cortex (n=4). (g–j) Quantifications show average values with the standard error measured in four embryos. Stars indicate significance in two-tailed Student's t-test *P<0.05, **P<0.005, ***P<0.0005
Rint1 depletion induces apoptosis in the CNS
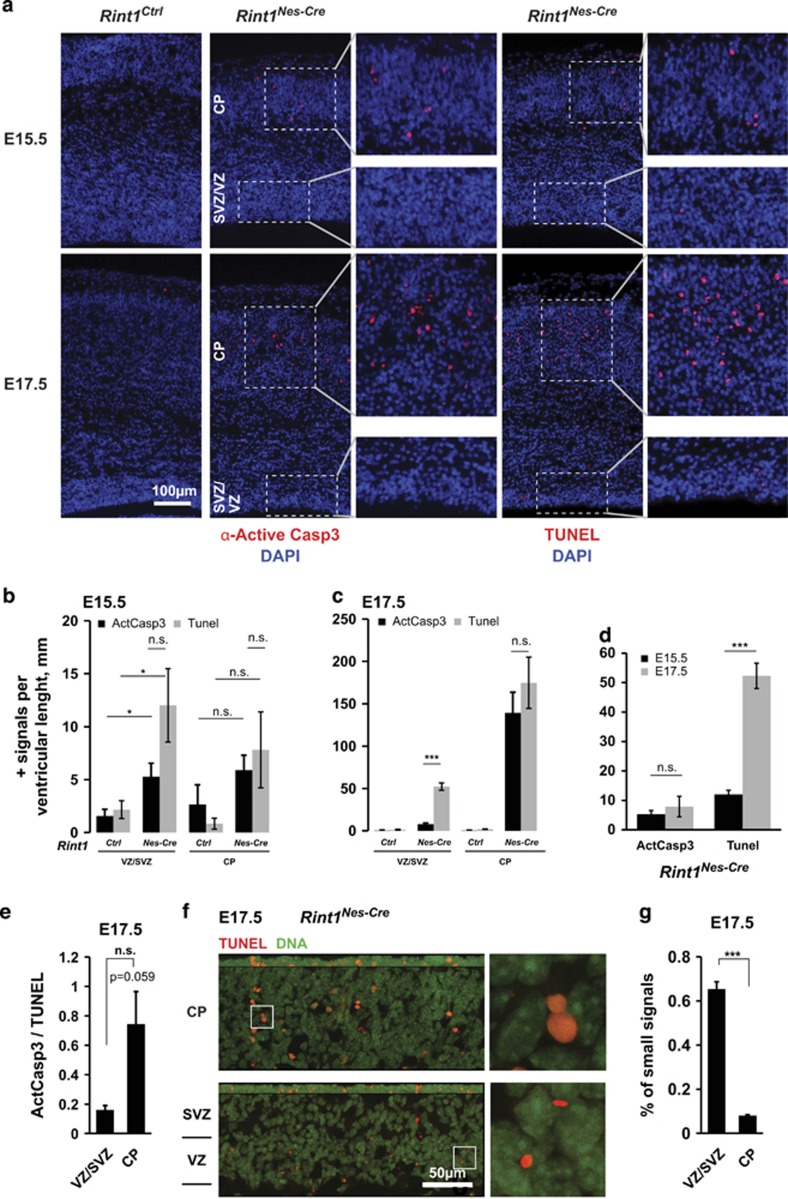
To understand the consequences of Rint1 inactivation in the CNS, we analyzed the Rint1Nes-Cre brains at embryonic stages E15.5 and E17.5. Coronal sections showed that the cortical proliferative areas composed of the ventricular (VZ) and the sub-ventricular zones (SVZ) were substantially reduced in Rint1Nes-Cre cortex (Figures 1e and g). Consistently, immunostaining of neuroprogenitors revealed a significant reduction of Sox2- and Trb2-positive cells at E15.5 (Figure 1h). To identify whether the reduction of neuroprogenitors in Rint1Nes-Cre cortex was due to proliferation defects or increased apoptosis, we first measured the number of BrdU-positive cells after a 1-h pulse and the number of phospho-Histone-3 (pH3)-positive mitotic cells. We found a significant reduction of both BrdU- and apical pH3-positive cells (Figures 1i–k). We then analyzed apoptosis in cortex using active-Caspase3 (ActCasp3) immunostaining and DNA fragmented nuclei by TUNEL assay (Figure 2a). In Rint1Nes-Cre cortices, a significant increase in both apoptotic signals was found in VZ/SVZ at E15.5 and interestingly a dramatic increase in cortical plate (CP) from E17.5 (Figures 2a and c). It indicates that reduction of neuroprogenitors pool is due to apoptosis rather than proliferative defects. Notably, the correlation between the TUNEL and ActCasp3 staining detected in CP was missing in VZ/SVZ regions at E17.5 (Figures 2d and e). Indeed, while the number of TUNEL-positive cells increased from E15.5 to E17.5, cells with ActCasp3 immunostaining remained unchanged (Figure 2d). Detailed analysis showed that the majority of TUNEL-positive signals in VZ/SVZ were smaller than the ones in CP (Figures 2f and g) indicating different origin of these signals. These small foci in proliferative area were reminiscent to micronuclei, appearing during mitosis upon defective DNA repair. Altogether we showed that Rint1 deficiency causes neuronal apoptosis in progenitor and post-mitotic neurons and that Rint1 is essential for maintenance of genomic stability in neuroprogenitors by preventing micronuclei formation. Finally, increased cell death was also detected in other parts of brain such as ganglionic eminence (Supplementary Figures S1a and b) indicating that Rint1 deficiency affects all the brain structures. It is expected that this massive cell death would impair brain function and trigger premature death.
Figure 2.
Rint1 deficiency promotes apoptosis in the cortex. (a) Detection of apoptotic cells by labeling active Caspase3 (ActCasp3) (left panel—red cells) and fragmented DNA by TUNEL staining (right panel—red cells) after Rint1 inactivation in the brain at indicated embryonic stages. Inserts show magnification of VZ/SVZ and CP layers. (b) Increase in ActCasp3 (black columns) and TUNEL (gray columns) signals in cortical progenitors (VZ/SVZ) and neurons (CP) of Rint1Nes-Cre embryos at E15.5 is drastically enhanced at later stages E17.5 (c) with an exception for ActCasp3 signals in VZ/SVZ (d). Indeed the ratio between ActCasp3 and TUNEL signals at E17.5 (e) shows that cortical progenitors are much less affected by Caspase3-mediated apoptosis than the neurons in CP. (f) Confocal microscopy depicts differences in morphology of TUNEL-positive signals (Red) in different cortical layers. Magnified images on the right panel are taken from the areas indicated by white frames. (g) Majority of TUNEL signals in cortical progenitors are much smaller than in differentiated neurons. Stars indicate significance in two tailed Student's t-test *P<0.05, ***P<0.0005
Rint1 deficiency promotes mitotic defects due to genomic instability in neuroprogenitors
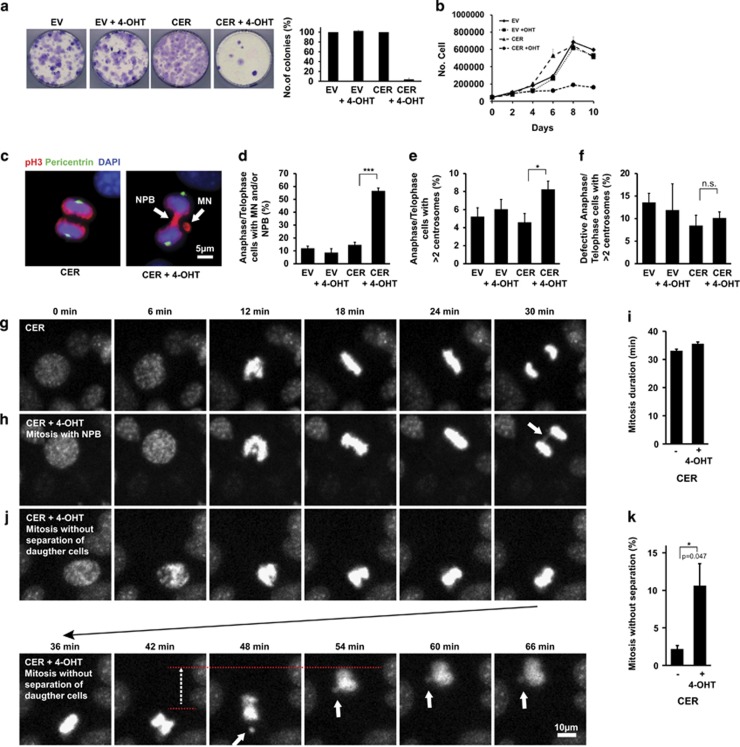
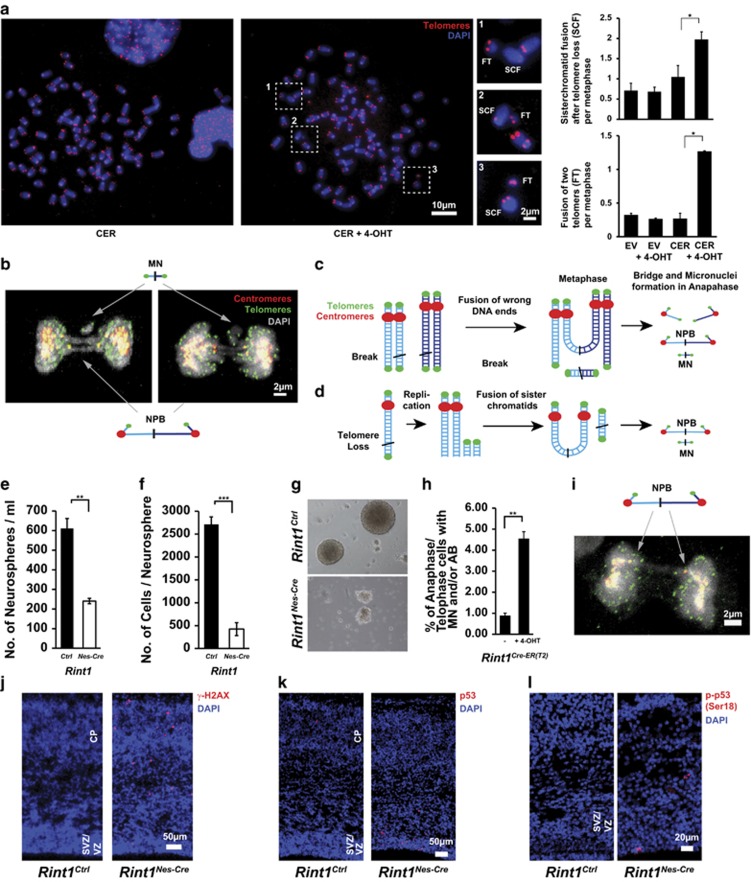
To elucidate the mechanisms underlying the putative genomic instability, we generated immortalized Rint1 inducible MEFs. Similar to in vivo findings, Rint1-deficient MEFs exhibited proliferation defects and defective colony formation (Figures 3a and b; Supplementary Figures S2a and b). Two days after Rint1 deletion, 50% of the cells exhibited micronuclei (MN) and nucleoplasmic bridges (NPB) (Figures 3c and d; Supplementary Figure 2c). Since it was demonstrated in RINT1-depleted Hela cells that those defects were due to centrosome amplification associated with RINT1 malfunction in Golgi,19 we investigated the centrosome amplification status in our MEFs. Although, we found a significant increase in cells with more than two centrosomes, we could not find a significant correlation between the centrosome amplification and the occurrence of mitotic defects (Figures 3e and f; Supplementary Figures S2d and e). This suggests that centrosome amplification is not a cause of MN and NPB formation in Rint1-deficient cells. Live-cell imaging did not show significant differences in mitosis duration between Rint1-proficient and -deficient cells (Figures 3g and i; Supplementary Figure S2f). However, we detected that 11% of the Rint1-deficient MEFs were not able to complete mitosis due to massive NPBs and thus impaired chromosome segregation (Figures 3j and k; Supplementary Fig. S2g). These cells were exiting mitosis without achieving cytokinesis, and sometimes generating MN (Figure 3j). Metaphase spread analysis showed a significant increase in chromosomal aberrations such as sister chromatid fusion (SCF) and fusion of two telomers (FT) (Figure 4a; Supplementary Figures S2h and i). In cells exhibiting MN and NPBs, we visualized telomeres and centromeres using immunofluorescent staining (Figure 4b). We discovered that DNA bridges between two separating daughter cells connected two centromeres and acentric DNA fragments around bridges had two telomeres but no centromere. This was obviously a result of the SCFs and FTs observed in metaphases and generated by improper joining of DNA ends (Figures 4c and d). To comprehend whether this mechanism can be generalized to neural stem cells (NSCs), we isolated NSCs from E13.5 Rint1Nes-Cre brains. After 7 days in culture, the total number of formed neurospheres and the number of cells per neurosphere were significantly reduced in NSCs from Rint1Nes-Cre compared with Rint1Ctrl brains, indicating that the pool of neural stem cells is dramatically reduced after Rint1 inactivation and that they have reduced proliferation capacity (Figures 4e and g). In addition, inducible NSCs, 2 days after 4-OHT-mediated Rint1 deletion exhibited a 4.5-fold increase in mitotic defects compared with control (Figure 4h). Confocal imaging confirmed that mitotic defects were DNA bridges identical to those found in MEFs (Figure 4i). Consistent to MEF findings, no centrosome amplification was found (data not shown). Finally, cells exhibiting positive staining for γ-H2AX were detected in E17.5 embryo cortices (Figure 4j). Additionally, p53 stabilization and its phosphorylation at serine18 were observed (Figures 4i and k). These results suggest that Rint1 is essential for prevention of chromosome fusions independently of centrosome amplification and thus is an important player in prevention of genomic instability and apoptosis in progenitor cells.
Figure 3.
Mitotic defects after Rint1 deletion in MEFs. (a) Colony formation assay after 4-OHT mediated Rint1 deletion in immortalized MEFs as well as cell counting (b) every 2 days demonstrate growth defects. (c) Two days after 4-OHT treatment MEFs were fixed and co-stained against phospho-Histone-3 (pH3) and Pericentrin. Arrows indicate formation of Nucleo-Plasmic Bridges (NBP) and Micronuclei (MN) independent of centrosome amplification in anaphases when Rint1 is deleted. (d) Percentage of Anaphase/Telophase cells with mitotic defects, (e) percentage of mitotic cells with amplified centrosomes and (f) percentage of the defective mitosis that have amplified centrosomes. (g, h and j) Live-cell imaging of immortalized MEFs mitosis. Nuclei of living cells were labeled with Hoechst33342 dye and imaged every 6 min. (g) Time course of wild-type cell mitosis compared with the mitosis of Rint1-deficient cells (h) arrow indicate NPB. (j) An example where Rint1-deficient daughter cells cannot separate due to anaphase bridges, performing tension-based movement indicated by red dashed lines and the dashed arrow. White arrow indicates Micronuclei formation. Duration of mitosis in MEFs does not change after Rint1 deletion (i), quantification was performed excluding mitosis without separation. Percentage of inhibited daughter cell separation is significantly increased in Rint1-deficient cells (k). Stars indicate significance in two tailed Student's t-test *P<0.05, ***P<0.0005
Figure 4.
Genetic instability induced by Rint1 deficiency. (a) Metaphase spreads show increased genomic instability after Rint1 inactivation in MEFs. Magnified regions show chromosomes without two telomeres (sister chromatid fusion, SCF) and nearby small DNA fragments with two telomeres (fused telomeres, FT). Evaluations in the right panel show a significant increase in these chromosomal aberrations after Rint1 inactivation. (b) Projection of confocal images of the defective mitosis with co-stained telomeres (green) and centromeres (red) demonstrating DNA bridges between two centromeres of daughter cells and micronuclei with two telomeres. (c) Mechanism of bridge and micronuclei formation in case of fusion of wrong DNA ends of DSBs in close proximity. (d) Mechanism of bridge and micronuclei formation in case of telomere loss. After replication the telomere-less chromatids are fused forming SCF and FT. Rint1Nes-Cre brains (n=5) have a reduced pool of neural stem cells (e) with altered capacity to proliferate in comparison with Rint1Ctrl mice (n=17) (f). (g) Representative images of neurospheres isolated from Rint1Ctrl and Rint1Nes-Cre mice brains. (h) A significant increase in mitotic defects in primary neuronal stem cells from Rint1Cre-ER(T2) mice 2 days after 4-OHT treatment. (i) Projection of confocal images of the defective mitosis in neuronal progenitor cell 2 days after 4-OHT treatment, co-stained against telomeres (green) and centromeres (red) showing the same bridge geometry as in MEFs. Stars indicate significance in two tailed Student's t-test *P<0.05, **P<0.005, ***P<0.0005
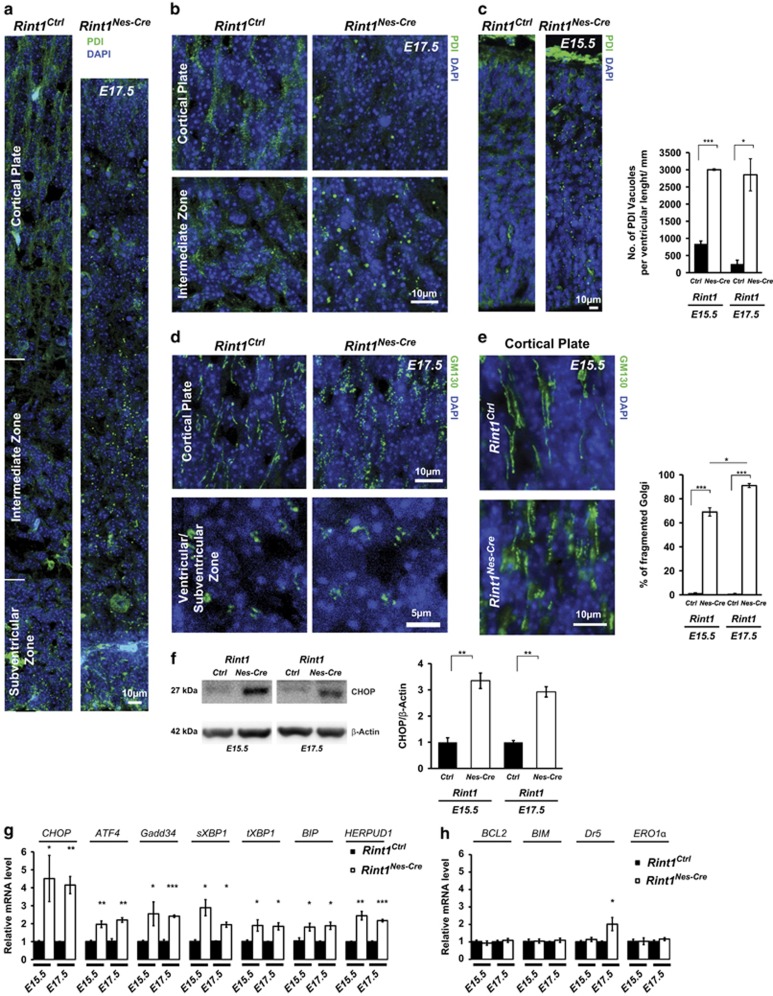
Rint1 depletion causes Cis/Trans-Golgi disruption and ER stress in cortical neurons
Several studies demonstrated that RINT1 is involved in ER-Golgi trafficking in vitro.16, 17, 19, 20, 21 Lin et al.19 associated the observed mitotic defects with Golgi dispersion and centrosome amplification. We demonstrated that centrosome amplification in MEFs is not correlated with mitotic defects and in NSC is not detected at all. To understand whether Golgi and ER alteration is associated with mitotic defects, we analyzed Golgi and ER morphology in inducible MEFs. Two days after Rint1 deletion, we could not observe any changes in Golgi labeled by GM130 and ER labeled by PDI (Supplementary Figure S3a). The morphology of these organelles started to change only 4 days after Rint1 inactivation, much later than the occurrence of genomic instability. Furthermore, we investigated whether ER-Golgi homeostasis was also disrupted in the cortex. We detected punctate staining of PDI indicating an ER vacuolization in Rint1Nes-Cre cortices starting from E15.5 (Figures 5a–c). GM130 staining revealed that ER vacuolization was associated with the disruption of Cis-Golgi structure in CP and VZ/SVZ (Figures 5d and e). These changes were accompanied by increased levels of the transcription factor CHOP (Figure 5f), that is a target of the integrated stress response including ER stress24 and DNA damage.25 qPCR analysis confirmed CHOP upregulation and also showed that mRNA levels of ATF4, Gadd34, spliced-XBP1, total-XBP1, BIP and HERPUD1 were significantly increased (Figure 5g). Upregulation of spliced-XBP1, BIP and HERPUD1 mRNA levels indicates that Rint1 deletion induces ER stress activating the Unfolded Protein Response.26 In order to understand whether CHOP increase mediated apoptosis, we measured mRNA levels of genes downstream CHOP (Figure 5h). We could detect neither downregulation of BCL2 nor upregulation of BIM (Figure 5h). mRNA levels of ERO1α (Figure 5h) involved in the ERO1-IP3R-Ca2+-CaMKII apoptotic pathway27 stayed unchanged. However, we detected upregulation of Dr5 (Figure 5h) that is known to be activated not only by ER stress28 via CHOP but also by DNA damage via p53.29 These findings suggest that the neuronal death in Rint1-deficient cortex is promoted by both ER stress and DNA damage.
Figure 5.
Rint1 deficiency destabilizes ER, Cis/Trans-Golgi homeostasis and cause ER stress in CNS. (a and b) Dot-like structure of ER indicates its vacuolization in Rint1Nes-Cre cortex at E17.5 visualized by immunofluorescent staining of ER luminal protein PDI. (c) ER vacuolization already at E15.5. (d) GM130 staining in indicated E17.5 cortex regions shows Cis-Golgi dispersion in CP as well as in VZ and SVZ of Rint1Nes-Cre cortex starting already at E15.5 (e). (f) Western blot analysis shows CHOP induction in Rint1Nes-Cre cortex at indicated developmental stages, indicating ER stress. (g) Relative mRNA expression of UPR genes in Rint1Nes-Cre vs Rint1Ctrl cortices quantified by qPCR analysis. (h) Relative mRNA expression of genes downstream of CHOP in Rint1Nes-Cre vs Rint1Ctrl cortices quantified by qPCR analysis. Stars indicate significance in two tailed or one tailed (g, h) Student's t-test *P<0.05, **P<0.005, ***P<0.0005
It was shown that RINT1 had a role in Trans-Golgi Network(TGN) homeostasis.17 To understand whether it is true in the cortical neurons, we analyzed TGN markers γ-Adaptin and Syntaxin 6. After 4 days in in vitro culture (DIV) Rint1Nes-Cre neurons revealed defective development (Supplementary Figure S4a) and Cis-Golgi disruption (Supplementary Figure S4b). Analysis of γ-Adaptin and Syntaxin 6 showed also Trans-Golgi disruption upon Rint1 inactivation (Supplementary Figure S4b). Surprisingly, despite RINT1 being linked to abnormal TGN distribution,17 we did not detect any alterations in the transport from endosomes to TGN. Monitoring the trafficking of Alexa488-conjugated Cholera toxin B (CTB) showed that after 45 and 90 min CTB was accumulated even in disrupted TGN in the neurons from Rint1Ctrl and Rint1Nes-Cre (Supplementary Figure S4c), suggesting that Rint1 does not have a role in transport from endosomes to TGN in neurons. Our findings show that Rint1 deficiency is leading first to mitotic defects and then to the disorganization of ER-Golgi network triggering ER stress and cell death both in progenitors and in post-mitotic neurons.
Autophagy blockage in Rint1-deficient neurons causes cell death
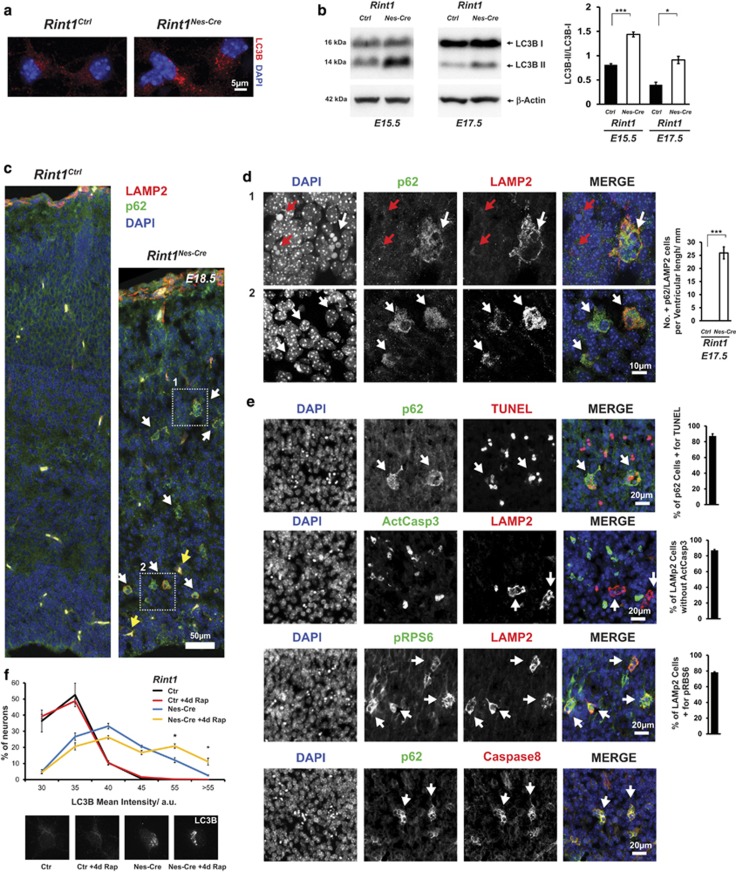
Studies reported that ER stress led to autophagy30, 31, 32 and that RINT1 was associated with autophagy.21 Therefore, we decided to monitor autophagy in Rint1-deficient cortices. We found that Rint1-deficient neurons exhibited autophagosome accumulation as indicated by the accumulation of LC3B punctate in cortical neurons and the increased conversion of the unconjugated LC3B-I to the phosphatidylethanolamine-conjugated form LC3B-II in E15.5 and E17.5 cortices (Figures 6a and b). This accumulation could be due to increased autophagic flux or autophagy inhibition in later stages.33 Therefore, we analyzed p62 that is known to be downregulated upon autophagy induction.34 In CP and in SVZ we detected p62 accumulation (Figure 6c). In the same neurons we also detected accumulation of LAMP2 that is required for autophago-lysosomes (Figure 6d). Interestingly in CP, p62 and LAMP2 accumulation was detected in pyknotic cells, which was confirmed by TUNEL staining to undergo apoptosis independent of caspase-3 pathway (Figure 6e). p62/LAMP2-positive cells were also positive for phosphorylated Ribosomal Protein S6 (p-RPS6) that is a target of RPS6 kinase and is downregulated during increased autophagy (Figure 6e). Although Caspase-8 was also found to be accumulated in these cells (Figure 6e), it was not activated (data not shown). To confirm the late autophagy inhibition, we induced autophagy in neuronal cultures using rapamycin and monitored LC3B levels. They were not changed in the neurons from Rint1Ctrl cortices, with or without rapamycin treatment underscoring the efficient autophagosome clearance (Figure 6f). However, Rint1Nes-Cre neurons exhibited higher percentage of cells with increased LC3B intensity upon rapamycin treatment (Figure 6f), confirming that autophagosomal clearance in Rint1-deficient neurons is in vivo and in vitro inhibited. It indicates that in Rint1Nes-Cre cortical neurons autophagy is inhibited in the late stage leading to accumulation of p62, LAMP2 and p-RPS6 and causes caspase-3-independent neuronal death (Figure 6e).
Figure 6.
Defective autophagy after Rint1 inactivation. (a) Accumulation of autophagosomes labeled by LC3B in cortical neurons. (b) Western blot analysis of protein extracts from cortex shows increased conversion from LC3B-I to LC3B-II. Right panel shows quantification of LC3B-II/LC3B-I ratio from three different embryos. (c) Confocal microscopy of p62- (green) and LAMP2- (red) positive neurons marked by white arrows in Rint1Nes-Cre cortex from E18.5. Yellow arrows indicate blood cells. Magnified images from frames 1 and 2 in (c) are depicted in (d). All p62- and LAMP2-positive cells in CP are with pyknotic nuclei (white arrow) representing dying cells (d1), however, not all cells with pyknotic nuclei show accumulation of p62 and LAMP2 (red arrows). In lower panel, confocal microscopy shows that in SVZ p62/LAMP2-positive neurons does not exhibit pyknotic nuclei (white arrows). Right panel shows quantification of p62- and LAMP2-positive cells in Rint1Nes-Cre cortices. p62- and LAMP2-positive cells (white arrows) co-localize with TUNEL, p-RPS6, Caspase8 but not with active Caspase3 (e) at E17.5. Right panel shows percentage for co-localization of indicated signals. (f) Percentage of neurons with given mean intensity of LC3B signals from Rint1Ctrl and Rint1Nes-Cre cortices after 4 days treatment with 100 nM rapamycin. Stars indicate significance in two tailed Student's t-test *P<0.05, ***P<0.0005
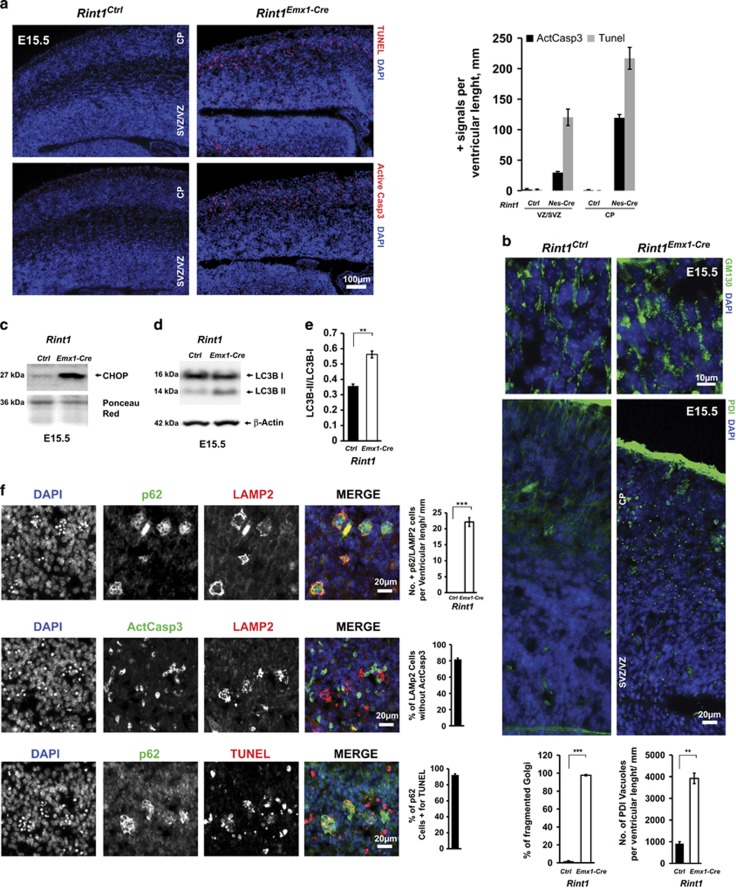
To make sure that observed phenotype is originating in the neurons generated in cortical VZ/SVZ, we performed the same analysis on Rint1Emx1-Cre mice in which deletion occurs only in the early progenitors of dorsal telencephalon.35 The results obtained in Rint1Emx1-Cre at E15.5 and Rint1Nestin-Cre at E17.5 were identical (Figures 7a and f), suggesting that the ER stress and autophagy blockage accompanied by neuronal death is affecting neurons that originate from the dorsal telencephalon. The stronger early phenotype in Rint1Emx1-Cre mice compared with Rint1Nes-Cre mice can be explained by lower recombination efficiency of Nestin-Cre,36 or increased susceptibility of earlier cortical neural progenitors.
Figure 7.
Deletion of Rint1 in progenitors of dorsal telencephalon cause severe defects already at E15.5. (a) TUNEL and Activated Caspase 3 staining of Rint1Ctrl and Rint1Emx1Cre cortex at 15.5 exhibits extensive apoptosis not only in CP but also in VZ/SVZ as shown in the quantification in the right panel. (b) Destabilization of Golgi (GM130) and ER (PDI) homeostasis in Rint1Emx1Cre cortices with quantification of Golgi fragmentation and ER vacuolization in the bottom panel. Levels of ER stress marker CHOP (c) as well as autophagosome marker LC3B-II (d) are increased in Rint1Emx1Cre cortex. (e) Quantification of LC3B-II/LC3B-I and LC3B-II/Actin ratios in cortex from three different embryos. (f) p62 and LAMP2 accumulation in apoptotic active Caspase3-negative cells in Rint1Emx1Cre cortex at E15.5 with statistic evaluation in the right panel. Stars indicate significance in two tailed Student's t-test **P<0.005, ***P<0.0005
Rint1 deletion causes neurodegeneration in Purkinje cells
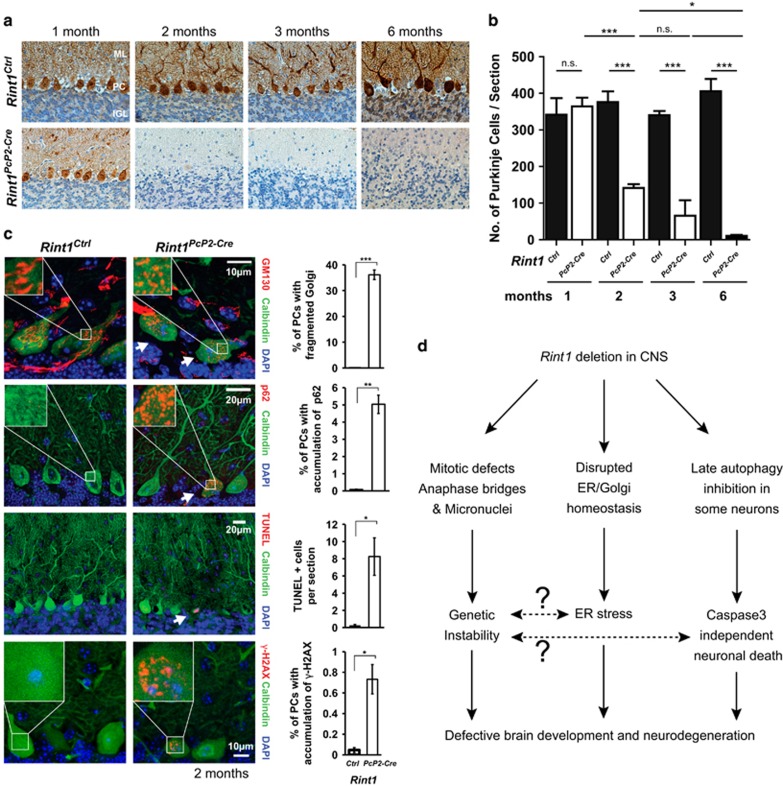
We showed cell death in the brain initiated by Rint1 deletion in neural progenitors. Since we also detected genomic instability in dividing cells, we asked whether genomic instability triggered in progenitor cells could be responsible for ER stress and defective autophagy-driven neurodegeneration as a secondary result of chromosome fusions. To address this question, we generated Rint1Pcp2-Cre mice that enabled inactivation of targeted gene in non-dividing Purkinje cells (PC) around 2 weeks after birth.37 One month after the birth, we did not observe any change in PC distribution; however, between 1 and 5 months old, we detected loss of PC in Rint1Pcp2-Cre cerebella (Figures 8a and b). Microscopy analysis revealed that PC showed the same phenotype as the Rint1Nes-Cre neurons. Indeed, 2-month-old Rint1Pcp2-Cre mice exhibited Golgi fragmentation in 35% of PCs. p62 accumulation was found in 5% of PC (Figure 8c). Surprisingly, we detected few PCs exhibiting accumulation of γ-H2AX, indicating that non-dividing Rint1-deficient cells could accumulate DNA damage (Figure 8c). We observed a significant increase in PC death using TUNEL assay. However, ActCasp3 was negative indicating that the cell death is caspase-3 independent. Given that inactivation of DSBs repair genes in PC does not cause any defects,38 and the fact that γ-H2AX-positive PC are few compared with those exhibiting Golgi fragmentation and p62 accumulation, we suggest that Rint1 deficiency triggered neurodegeneration in vivo is not a secondary result of genomic instability. It supports a direct role for Rint1 in ER-Golgi homeostasis and autophagy, and highlights the complexity and the variety of pathways in which Rint1 is involved.
Figure 8.
Neurodegeneration triggered by Rint1 inactivation in Purkinje cells. (a) Calbindin D-28K (brown) staining of Purkinje cells on paraffin sections of cerebellum. (b) Quantification of Purkinje cell numbers per section in postnatal animals. (c) Fragmentation of Golgi (GM130), accumulation of p62, γ-H2AX and TUNEL signals in Purkinje cells induced by Rint1 inactivation, visualized by confocal (GM130, p62, γ-H2AX) and wide field (TUNEL) microscopy, with the quantification in the right panel. (d) Scheme of Rint1 role in CNS. Stars indicate significance in two tailed Student's t-test *P<0.05, **P<0.005, ***P<0.0005
Discussion
We demonstrate that Rint1 is essential for the prevention of genetic instability by preventing anaphase bridges. In contrast to Lin et al.,19 we rule out that mitotic defects are due to centrosome amplification. Anaphase bridges are found among cells exhibiting impaired telomere homeostasis39, 40, 41, 42 and defective DNA DSB repair.36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49 RINT1 is known to be involved in both pathways through interaction with p13015 or Rad50.14 The structure of chromosome fusion in the Rint1-deficient cells supports the hypothesis of defective DSB repair (Figures 4c and d). Notably, the frequency of mitotic defects in Rint1-deficient NSCs was significantly increased. It indicates that primary cells do not tolerate chromosomal fusions and such cells are eliminated quickly. The role of RINT1 in DNA repair and maintenance of genomic stability was emphasized by the finding that RINT1 is mutated in a subset of Lynch syndrome patients.23 Lynch syndrome is a cancer predisposition disease associated with mutations of mismatch repair genes. DSB response was shown to be essential in developing CNS but CNS-specific DSB repair mutants obtained with Nestin-Cre such as Nbn,50 Brca251 and Atr52 are viable at birth and did not exhibit any apoptosis in mature neurons.53 In contrast, Rint1 inactivation in NSCs prevents postnatal survival, suggesting that the role of Rint1 in maintaining genomic stability is not the only cause of the premature death.
In fact, we demonstrate that Rint1 inactivation in brain leads to ER stress due to altered ER and Cis/Trans-Golgi homeostasis and impaired clearance of autophagosomes that were shown to be involved in neurodegenerative disease including Alzheimer, Parkinson and Huntington.1, 2, 54 The phenotype of Rint1-deficient brain with the exception of autophagy defects is reminiscent of the SNARE p31-deficient mice in which ER stress was claimed to be responsible of neuronal cell death.55 Interestingly, p31 and RINT1 are forming the ‘Syntaxin 18 SNARE' complex together with Syntaxin 18, BNIP1, Sec22B, Sly1p, NAG and ZW10.10, 11 ‘Syntaxin 18 SNARE' complex was found to be implicated in membrane fusion of retrograde transport10, 56, 57 while its members Sec22B and ZW10/RINT1 are also involved in the anterograde transport.16, 58 RINT1 was shown to be also involved in the more specific Rab6-dependent recycling pathway from the Golgi to the ER.20 Disruption of the balance of the bi-directional ER/Golgi transport was shown to trigger Golgi fragmentation followed by ER stress.1, 55 Complementary to former RINT1 studies where ER stress was not reported16, 17, 19 here we postulate that in our model, Rint1 deficiency generates ER/Golgi imbalance leading to Golgi apparatus fragmentation, induction of ER stress and ultimately cell death. Since CHOP, GADD34 and Dr5 can also be activated by DNA damage, we cannot exclude the role of the DNA damage response in Rint1-deficient phenotype. We also observe that some of the Rint1-deficient neurons exhibit autophagy inhibition. It was shown that UPR resulting from ER homeostasis disruption led preferentially to activation of autophagy rather than proteasome degradation in the neuronal cells.32 Although autophagic activity is hardly detectable in healthy brains compared with other tissues,59 it was shown that basal autophagy was essential for the development and homeostasis of the brain.60, 61, 62 Neurons exhibit heterogeneous sensitivity to autophagy inhibition or induction depending of their type and developmental stage.60 It is often considered that autophagy is associated with survival but in certain situation when it is inhibited at late stages, it could lead to neurodegeneration.3, 63, 64 This is consistent with the phenotype in Rint1Nes-Cre and Rint1Emx1-Cre cortices, where we observed accumulation of p62, LAMP2 and p-RPS6 as an evidence for autophagy inhibition in late stage. These cells were also positive for Caspase 8 that is known to bind p62 on autophagosomes after inhibition of proteasome. Although caspase-8 was accumulated, it was not activated, leading to caspase-3-independent neuronal death.
Little is known about RINT1 function in autophagy. RINT1 was shown to interact with UVRAG, a protein essential for Atg9-vesicle formation, but this interaction only occurs in the absence of autophagy induction.21 Interestingly, RINT1 interaction partner BNIP1 was reported to be involved in mitochondrial autophagy.65 Sec22B was shown to regulate clearance of autophagosomes indirectly by assuring the anterograde transport of lysosomal proteases from ER via Golgi to lysosomes.58 In this context, Rint1 could be involved in the clearance of autophagosomes by a mechanism identical to Sec22B and its function in anterograde transport. An alternative mechanism that could explain the Rint1-deficient autophagic defects in neurons is the disruption of autophagosome transport. In the neurons, autophagosomes formed in distal axons mature and fuse with late endosomes and/or lysosomes during transport towards the cell soma.66 It was shown that disruption of autophagosome transport on microtubules by vinblastin led to the inhibition of autophagosome and lysosome fusion and to accumulation of these vesicles.3, 67 Dynein inhibition by EHNA suppressed fusion of autophagosomes and lysosomes.68 Dynactin 1, a member of the Dynactin complex, which is essential for dynein function in motility was also linked to autophagosomal transport.69 RINT1 and Dynactin1 interact with ZW10 in a competitive manner70 and it was suggested that the RINT1/ZW10 complex may play as an ER anchor for the Dynein–Dynactin complex carried vesicles.16 It is conceivable that the absence of RINT1 causes defects in transport of vesicles and prevents clearance of autophagosomes.
In conclusion, our results reveal novel in vivo functions of Rint1. For the first time, we report the involvement of Rint1 in prevention of genomic instability, ER stress and in clearance of autophagosomes (Figure 8c). These in vivo findings illustrate the essential requirement of Rint1 for brain development and ageing.
Materials and Methods
Ethics statement
All animal care and procedures followed German legal regulations and were previously approved by the governmental review board of the state of Baden-Württemberg (Regierungspräsidium Karlsruhe-Abteilung 3-Landwirtschaft, Ländlicher Raum, Veterinär-und Lebensmittelwesen). All the aspects of the mouse work were carried out following strict guidelines to insure careful, consistent and ethical handling of mice.
Mice
The Rint1 floxed mice were generated as followed. The targeting vector was created using the recombineering method developed by the National Cancer Institute at Frederick (USA). The BAC containing the Rint1 gene was obtained from the Sv129/J BAC library from Invitrogen (Darmstadt, Germany). The sequence of the mouse BAC containing the Rint1 gene was obtained from the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk). The pGK-neo cassette flanked by two Frt sites was inserted upstream the exon 3 and two loxP sites were inserted upstream and downstream the exon 3. The targeting vector was linearized using the enzyme NotI, and was electroporated into E14.1 ES cells. Southern-blot analysis was performed using the genomic probe 1 (generated by PCR amplification using the primer RINT109F: 5′-AGCCATTTTGAGATAGGTGTT-3′) and RINT901R: (5′-CTTCTGGGACATCGGACATT-3′) and the ApaI+XhoI digestion. The presence of the loxP downstream the exon 3 was also analyzed both by Southern blot using the genomic probe 2 (generated by PCR amplification using the primer RINT10738F: 5′-GATGGAAACAGAATGCTTGGA-3′ and RINT11444R: 5′-CATACATACATAGAGAGA-3′) and by BamHI digestion. Two independent clones 1G7 and 1G9 were injected and gave rise to germline transmission. The neomycine cassette was removed in vivo using a ubiquitous FLPe-recombinase mice (B6;SJL-Tg(ACTFLPe)9205Dym/J). The mice were routinely genotyped using the following primers RINT7512F (5′-TTCCTACTGACTTGCTGTGAT-3′) and RINT8345R (5′-ACTTCTGGATGACTGAGGAC-3′). The Δ allele was amplified using the primers: RINT6542F (5′-TAACCCCTGACCCATCTCTC-3′) and RINT8345R. The following used Cre-recombinase mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA): Nestin-Cre (B6.Cg(SJL)-Tg(Nes-cre)1Kln/J), Emx1-Cre (B6.129-Emx1tm1(cre)Krj/J) and PcP2-Cre (B6.129-Tg(Pcp2-cre)2Mpin/J) mice. The control group (Rint1Ctrl) depending on the mouse Cre line was composed of Rint1+/F; Rint1F/F; Rint1+/F, Cre+. Mice of other experimental groups were identified as follows: Rint1Nes-Cre= Rint1F/F, Nestin-Cre+; Rint1Emx1-Cre= Rint1F/F, Emx1-Cre+; Rint1PcP2-Cre=Rint1F/F, PcP2-Cre+.
Histology and immunohistochemistry
Brains were collected in 4% (w/v) phosphate-buffered saline (PBS)-buffered paraformaldhehyde (PFA) for 24 h at 4 °C then cryopreserved overnight in PBS-buffered 25% sucrose solution. In all, 10 μm cryosections were performed. The following antibodies were used after 40 min citrate buffer antigen retrieval at 95 °C: anti-active Caspase3 (1 : 400, BD Bioscience, Heidelberg, Germany, 559565), anti-BrdU (1 : 200, Linaris, Dossenheim, Germany, LOB0030G), anti-Phospho-H3 (1 : 400, Cell Signaling, Danvers, MA, USA, 9701), anti-TBR2 (1 : 200, Millipore, Billerica, MA, USA, ab15894), anti-Sox2 (1 : 200, Abcam, Cambridge, UK, ab97959), anti-γ-H2AX (1 : 200, Millipore, 05-636 and Cell Signaling 2577). Those ones required 60 min citrate buffer antigen retrieval at 60 °C: anti-GM130 (1 : 400, BD Bioscience, 610822), anti-PDI (1 : 400, Enzo-Life, Farmingdale, NY, USA, ADI-SPA-891), anti-p62 (1 : 500, MBL, Woburn, MA, USA, PM045), anti-LAMP2 (1 : 100, Santa Cruz, Heidelberg, Germany, sc20004), anti-Phospho-S6 Ribosomal Protein (1 : 200, Cell Signaling, 2211), anti-Caspase8 (1 : 100, Cell Signaling, 4927), anti-p53 CM5 (Vector Laboratories, Peterborough, UK, VP-P956) and anti-pospho Ser15-p53 (1 : 100, Cell Signaling, 9284 S). Immunoreactivity was visualized using secondary donkey antibodies conjugated with FITC, Cy3 or Alexa 647 or biotin (Jackson Immunoresearch, Newmarket, UK) followed by the incubation with Cy3-conjugated Streptavidin (Sigma-Aldrich, Taufkirchen, Germany, GEPA43001). Nuclei were counterstained with Hoechst 33342 (1 : 10 000, Life Technologies, Darmstadt, Germany) and mounted in ProLong Gold Antifade reagent (Life Technologies). For the in vivo proliferation study, 50 μg/g body weight BrdU (Sigma-Aldrich, B9285) was injected into mice intraperitonally. Brains were collected 1 h after injection. TUNEL analysis was performed on cryosections as described.71 Images were captured with Zeiss Axiovert fluorescence microscope or Zeiss LSM700 confocal microscope (Oberkochen, Germany).
Rint1 RT-PCR and quantitative RT-PCR
Cortex tissue was obtained from E15.5 and E17.5 embryos mice. RNA was extracted using TRIzol reagent according to the manufacturer's instructions and genomic DNA was removed using RNase-Free DNase Set from Qiagen. RNA was reverse transcribed using the SuperScript II cDNA synthesis kit (Life Technologies). The PCR was done using the following primers 5′-GCGCTCCTTTCCTATGTGTCTG-3′ and 5′-AGCCCTGGATGGATGACCTTGG-3′ for Rint1 and 5′-gtcatcatctccgccccttctgc-3′ and 5′-gatgcctgcttcaccaccttcttg-3′ for GAPDH. The following PCR parameters were used: incubation at 94 °C for 2 min, followed by 44 cycles of 95 °C for 30 s and 60 °C for 1 min. cDNA was used also for quantitative real-time PCR analysis using Power Sybr Green Master Mix and Applied Biosystems 7500 according to the manufacturer's instructions. Following primers has been used: CHOP-fw: CCTGAGGAGAGAGTGTTCCA; CHOP-rv: ATCCTCATACCAGGCTTCCA; BIP-fw: CAAGGTCTATGAAGGTGAACGA; BIP-rv: CAGCTGTCACTCGGAGAATAC; HERPUD1-fw: GATGGTTTACGGCAAAGAGAAG; HERPUD1-rv: GGCAGCTAAGTATTGCATGTAG; GADD34-fw: GAAGTGTACCTTCCGAGCTTT; GADD34-rv: CTTCCAGGTGGCCTTCTATTT; ATF4-fw: CCTGAACAGCGAAGTGTTGG; ATF4-rv: GAAAAGGCATCCTCCTTGCC; tXBP1-fw: AGCAGCAAGTGGTGGATTTG; tXBP1-rv: GAGTTTTCTCCCGTAAAAGCTGA; sXBP1-fw: TGAGTCCGCAGCAGGTGCA; sXBP1-rv: GTCCATGGGAAGATGTTCTGG; ACTIN-fw: TCACCCACACTGTGCCGATCTACGA; ACTIN-rv: GGATGCCACAGGATTCCATACCCA; BCL2-fw: TGAGTACCTGAACCGGCATCT; BCL2-rv: GGTATGCACCCAGAGTGATGC; BIM-fw: CCCGGAGATACGGATTGCAC; Bim-rv: GCCTCGCGGTAATCATTTGC; Dr5-fw: AGTAGTGCTGCTGATTGGAG; Dr5-rv: CCTGTTTTCTGAGTCTTGCC; ERO1-fw: TTCTGCCAGGTTAGTGGTTACC; ERO1-rv: GTTTGACGGCACAGTCTCTTC. The relative mRNA expression levels were calculated by Pfaffl method using Actin as a house keeping gene.
Neurosphere culture
Neurosphere cultures were isolated and cultured as described.50 For immunofluorescent staining, neuronal stem cells were trypsinised and seeded on 12 mm diameter cover glasses coated with with 50 μg/ml Poly-L-Lysine (Sigma-Aldrich, P2636) for 30 min and then with 50 μg/ml laminin (Sigma-Aldrich, L2020) for 30 min at 37 °C.
Primary cortical neuron culture
Cortical neurons from E15.5 embryos were dissociated and plated at a density of 75 000 cells/cm2 on 12 mm coverslips coated with 50 μg/ml with Poly-L-lysine (Sigma-Aldrich, P2636) for 30 min. Neuron plating medium (0.5% Glutamax, 2% B-27 supplement, 100 μ/ml Penicilin, 100 μg/ml Streptomycin, 1 mM Na-Pyruvate and 10% FCS in DMEM) was changed after 12 h with neuronal maintenance medium (0.5% Glutamax, 2% B-27 supplement, 100 u/ml Penicilin, 100 μg/ml Streptomycin, 10 mM Hepes in Neurobasal-R medium). Cholera Toxin B (CTB) transport assay was performed as described.20 Autophagy induction was performed by treating neurons 4 days with 100 nM Rapamycin from the day of their isolation.
Cell culture
MEFs were isolated from E13.5 Rint1F/F embryos and immortalized following 3T3 protocol. Immortalized cells were infected with pMSCVpuro (EV) for control and pMSCVpuro-Cre-ER(T2) (CER) retroviruses produced in Phoenix-Eco cells. Rint1 deletion was obtained by adding to the medium 1 μM 4-hydroxytamoxifen (4-OHT) (Sigma-Aldrich, H7904) for 48 h. Metaphase spreads and telomere staining were performed as described.57
Live-cell imaging
MEFs were seeded into 8-well μ-Slides (Ibidi) 2 days before the experiment and treated with 4-OHT. Before imaging, cells were incubated at 37 °C for 30 min in medium containing 0.1 ng/μl Hoechst 33342. Time series images were recorded every 6 min for 16 h using Cell Observer Microscope (Zeiss).
Immunofluorescence staining
Cover glasses with cells (MEFs, cortical neurons or NSCs) were washed once in PBS and fixed for 10 min in 4% PFA, permeabilized with 0.7% Triton X-100 in PBS, washed again and incubated with primary antibody in blocking buffer (5% Normal Donkey Serum, 1% BSA and 0.1% Triton in PBS) overnight at 4 °C. Following antibodies were used against: Centomeres (CREST) (1 : 400, Antibodies Incorporated, Davis, CA, USA, 15–235), Telomeres (1 : 800, a kind gift from Maria Blasco, Madrid, Spain), LC3B (1 : 100, Cell Signaling, 2775), Pericentrin (1 : 400, BD Bioscience, 611815), Phospho-H3 (1 : 400, Cell Signaling, 9701), GM130 (1 : 200, BD Bioscience, 610822), PDI (1 : 200, Enzo-Life, ADI-SPA-891), β-Tubulin III (TUJ1) (1 : 200, Covance, Munich, Germany, MMS-435 P), γ-Adaptin (1 : 100, BD Bioscience, 610385), Syntaxin6 (1 : 100, BD Bioscience, 610635). After incubation with primary antibody, the slides were washed in PBS, PBS/0.05% Tween-20 and again in PBS for 10 min each at room temperature. For indirect immunochemical detection, the following secondary antibodies diluted in blocking buffer have been used: anti-mouse-FITC/Cy3/Alexa647, anti-rabbit-FITC/Cy3/Alexa647, anti-human-Cy3 (Jackson Immunoresearch) all at 1 : 400 incubated at room temperature for 1 h. After washing, the specimens were counterstained with 2 min pulse of Hoechst 33342 (1 : 10 000, Life Technologies) and mounted in ProLong Gold Antifade reagent (Life Technologies).
Mean intensity of LC3B in neurons was measured using Image J software. For this purpose threshold at value 20 was set up for more than 20 (8-bit) images and the mean intensity of the selected area by the Wand (Tracing) Tool was measured for more than 100 cells for each condition.
Western blot analysis
Proteins were extracted and separated as described.50 The following antibodies were used: anti-CHOP (1 : 1000, Cell Signaling, 5554), anti-Gadd153 (1 : 1000, Santa Cruz, sc575), anti-LC3B (1 : 1000, Cell Signaling, 2775), anti-Caspase12 (1 : 1000, Cell Signaling, 2202), anti-ß-Actin (1 : 300000, Sigma-Aldrich, A5441).
Statistical analysis
All experiments were independently repeated at least three times and error bars in the graphs show calculated standard error. Statistical significance was calculated using Student's t-test unless otherwise stated. Statistically significant differences were considered by the P-values<0.05. Microsoft Excel and GraphPad Prims6 (GraphPad Software, La Jolla, CA, USA), were used for statistical and graphical data evaluations.
Acknowledgments
We thank the DKFZ animal facility for excellent husbandry. We also thank Alexander Ernst and Lukas Leib for great technical assistance as well as Lionel Larribere and Johannes Ridinger for their advices on qPCR experiments. This work was supported by the Deutsche Forschungsgemeinschaft (DFG): FR 2704/1-1 to POF ‘Emmy Noether program'.
Author contributions
PG performed all the experiments for the characterization of Rint1-deficient mouse. CH and PJM contributed to the creation of the Rint1 conditional mouse. AvD provided histology and pathology expertise. PJM and AvD contributed also to the final version of the manuscript. POF was project leader and produced the final version of the manuscript.
Glossary
- 4-OHT
4-hydroxytamoxifen
- ActCasp3
active caspase 3
- CNS
central nervous system
- CP
cortical plate
- CTB
cholera toxin B
- DSB
double-stranded breaks
- ER
endoplasmic reticulum
- FT
fusion of two telomers
- MEFs
mouse embryonic fibroblasts
- MN
micronuclei
- NPB
nucleoplasmic bridge
- NSCs
neural stem cells
- PC
Purkinje cells
- pH3
phospho-histone-3
- SCF
sister chromatid fusion
- SNARES
soluble N-ethylmaleimidine-sensitive factor attachment protein receptors
- SVZ
sub–ventricular zone
- TGN
trans-Golgi network
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- VZ
ventricular zone
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by D Rubinsztein
Supplementary Material
References
- Atkin JD, Farg MA, Soo KY, Walker AK, Halloran M, Turner BJ et al. Mutant SOD1 inhibits ER-Golgi transport in amyotrophic lateral sclerosis. J Neurochem 2014; 129: 190–204. [DOI] [PubMed] [Google Scholar]
- Jellinger KA. Basic mechanisms of neurodegeneration: a critical update. J Cell Mol Med 2010; 14: 457–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J Neurosci 2008; 28: 6926–6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci 2010; 13: 805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Fujita N, Yoshimori T. The late stages of autophagy: how does the end begin? Cell Death Differ 2009; 16: 984–990. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009; 43: 67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K, Renna M, Rubinsztein DC. Connections between SNAREs and autophagy. Trends Biochem Sci 2013; 38: 57–63. [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Barlowe C. Organization of the ER-Golgi interface for membrane traffic control. Nat Rev Mol Cell Biol 2013; 14: 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MCS, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol 2004; 20: 87–123. [DOI] [PubMed] [Google Scholar]
- Aoki T, Ichimura S, Itoh A, Kuramoto M, Shinkawa T, Isobe T et al. Identification of the Neuroblastoma-amplified Gene Product as a Component of the Syntaxin 18 Complex Implicated in Golgi-to-Endoplasmic Reticulum Retrograde Transport. Mol Biol Cell 2009; 20: 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose H, Arasaki K, Dohmae N, Takio K, Hatsuzawa K, Nagahama M et al. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J 2004; 23: 1267–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt HD. Dsl1p/Zw10: common mechanisms behind tethering vesicles and microtubules. Trends Cell Biol 2010; 20: 257–268. [DOI] [PubMed] [Google Scholar]
- He S, O'Connell D, Zhang X, Yang Y, Liang C. The intersection of Golgi-ER retrograde and autophagic trafficking. Autophagy 2014; 10: 180–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Liu CC, Chen PL, Lee WH. RINT-1, a novel Rad50-interacting protein, participates in radiation-induced G(2)/M checkpoint control. J Biol Chem 2001; 276: 6105–6111. [DOI] [PubMed] [Google Scholar]
- Kong L-J, Meloni AR, Nevins JR. The Rb-related p130 protein controls telomere lengthening through an interaction with a Rad50-interacting protein, RINT-1. Mol Cell 2006; 22: 63–71. [DOI] [PubMed] [Google Scholar]
- Arasaki K, Taniguchi M, Tani K, Tagaya M. RINT-1 Regulates the Localization and Entry of ZW10 to the Syntaxin 18 Complex. Mol Biol Cell 2006; 17: 2780–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasaki K, Takagi D, Furuno A, Sohda M, Misumi Y, Wakana Y et al. A new role for RINT-1 in SNARE complex assembly at the trans-Golgi network in coordination with the COG complex. Mol Biol Cell 2013; 24: 2907–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Hirose H, Taniguchi M, Kurashina H, Arasaki K, Nagahama M et al. Involvement of BNIP1 in apoptosis and endoplasmic reticulum membrane fusion. EMBO J 2004; 23: 3216–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Liu C-C, Gao Q, Zhang X, Wu G, Lee W-H. RINT-1 serves as a tumor suppressor and maintains Golgi dynamics and centrosome integrity for cell survival. Mol Cell Biol 2007; 27: 4905–4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Shestakova A, Hunt L, Sehgal S, Lupashin V, Storrie B. Rab6 Regulates Both ZW10/RINT-1—and Conserved Oligomeric Golgi Complex-dependent Golgi Trafficking and Homeostasis. Mol Biol Cell 2007; 18: 4129–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Ni D, Ma B, Lee J-H, Zhang T, Ghozalli I et al. PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nat Cell Biol 2013; 15: 1206–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quayle SN, Chheda MG, Shukla S a, Wiedemeyer R, Tamayo P, Dewan RW et al. Integrative functional genomics identifies RINT1 as a novel GBM oncogene. Neuro Oncol 2012; 14: 1325–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DJ, Tao K, Le Calvez-Kelm F, Nguyen-Dumont T, Robinot N, Hammet F et al. Rare mutations in RINT1 predispose carriers to breast and Lynch syndrome-spectrum cancers. Cancer Discov 2014; 4: 804–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XZ, Lawson B, Brewer JW, Zinszner H, Sanjay A, Mi LJ et al. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153). Mol Cell Biol 1996; 16: 4273–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luethy JD, Holbrook NJ. Activation of the gadd153 promoter by genotoxic agents: a rapid and specific response to DNA damage. Cancer Res 1992; 52: 5–10. [PubMed] [Google Scholar]
- Kadowaki H, Nishitoh H. Signaling pathways from the endoplasmic reticulum and their roles in disease. Genes (Basel) 2013; 4: 306–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol 2011; 13: 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Wang HG. CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J Biol Chem 2004; 279: 45495–45502. [DOI] [PubMed] [Google Scholar]
- Sheikh MS, Burns TF, Huang Y, Wu GS, Amundson S, Brooks KS et al. p53-dependent and -independent regulation of the death receptor KILLER/DR5 gene expression in response to genotoxic stress and tumor necrosis factor alpha. Cancer Res 1998; 58: 1593–1598. [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 2006; 26: 9220–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W-X, Ni H-M, Gao W, Hou Y-F, Melan MA, Chen X et al. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem 2007; 282: 4702–4710. [DOI] [PubMed] [Google Scholar]
- Nijholt D a T, de Graaf TR, van Haastert ES, Oliveira AO, Berkers CR, Zwart R et al. Endoplasmic reticulum stress activates autophagy but not the proteasome in neuronal cells: implications for Alzheimer's disease. Cell Death Differ 2011; 18: 1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010; 140: 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8: 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski J a, Talley T, Qiu M, Puelles L, Rubenstein JLR, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci 2002; 22: 6309–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Hippenmeyer S, Ghashghaei HT. A Nestin-cre transgenic mouse is insufficient for recombination in early embryonic neural progenitors. Biol Open 2012; 1: 1200–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski JJ, Dethleffsen K, Meyer M. Cre recombinase expression in cerebellar Purkinje cells. Genesis 2000; 28: 93–98. [PubMed] [Google Scholar]
- Rodrigues PMG, Grigaravicius P, Remus M, Cavalheiro GR, Gomes AL, Martins MR et al. Nbn and Atm Cooperate in a Tissue and Developmental Stage-Specific Manner to Prevent Double Strand Breaks and Apoptosis in Developing Brain and Eye. PLoS One 2013; 8: e69209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez P, Thanasoula M, Muñoz P, Liao C, Tejera A, McNees C et al. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev 2009; 23: 2060–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisaingo K, Uringa E-J, Lansdorp PM. Resolution of telomere associations by TRF1 cleavage in mouse embryonic stem cells. Mol Biol Cell 2014; 25: 1958–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell 1998; 92: 401–413. [DOI] [PubMed] [Google Scholar]
- Bailey SM, Bedford JS. Studies on chromosome aberration induction: what can they tell us about DNA repair? DNA Repair (Amst) 2006; 5: 1171–1181. [DOI] [PubMed] [Google Scholar]
- Laulier C, Cheng A, Stark JM. The relative efficiency of homology-directed repair has distinct effects on proper anaphase chromosome separation. Nucleic Acids Res 2011; 39: 5935–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Choi ES, Hwang K, Kim J, Sampath S, Venkitaraman AR et al. The breast cancer susceptibility gene BRCA2 is required for the maintenance of telomere homeostasis. J Biol Chem 2012; 287: 5091–5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue A, Coulombe Y, Jacquet K, Gagné J-P, Roques C, Gobeil S et al. The RAD51 paralogs ensure cellular protection against mitotic defects and aneuploidy. J Cell Sci 2013; 126: 348–359. [DOI] [PubMed] [Google Scholar]
- Goytisolo FA, Samper E, Edmonson S, Taccioli GE, Blasco MA, Goytisolo NA. The absence of the dna-dependent protein kinase catalytic subunit in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol Cell Biol 2001; 21: 3642–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhn C, Zhou Z-W, Ai H, Wang Z-Q. The essential function of the MRN complex in the resolution of endogenous replication intermediates. Cell Rep 2014; 6: 182–195. [DOI] [PubMed] [Google Scholar]
- Vannier J-B, Depeiges A, White C, Gallego ME. Two roles for Rad50 in telomere maintenance. EMBO J 2006; 25: 4577–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder-Heurich B, Wieland B, Lavin MF, Schindler D, Dörk T. Protective role of RAD50 on chromatin bridges during abnormal cytokinesis. FASEB J 2014; 28: 1331–1341. [DOI] [PubMed] [Google Scholar]
- Frappart P-O, Tong W-M, Demuth I, Radovanovic I, Herceg Z, Aguzzi A et al. An essential function for NBS1 in the prevention of ataxia and cerebellar defects. Nat Med 2005; 11: 538–544. [DOI] [PubMed] [Google Scholar]
- Frappart P-O, Lee Y, Lamont J, McKinnon PJ. BRCA2 is required for neurogenesis and suppression of medulloblastoma. EMBO J 2007; 26: 2732–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Shull ERP, Frappart P-O, Katyal S, Enriquez-Rios V, Zhao J et al. ATR maintains select progenitors during nervous system development. EMBO J 2012; 31: 1177–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon PJ. Maintaining genome stability in the nervous system. Nat Neurosci 2013; 16: 1523–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal R, Caballero B, Couve A, Hetz C. Converging pathways in the occurrence of endoplasmic reticulum (ER) stress in Huntington's disease. Curr Mol Med 2011; 11: 1–12. [DOI] [PubMed] [Google Scholar]
- Uemura T, Sato T, Aoki T, Yamamoto A, Okada T, Hirai R et al. P31 deficiency influences endoplasmic reticulum tubular morphology and cell survival. Mol Cell Biol 2009; 29: 1869–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Pelham HR. SNARE-mediated retrograde traffic from the Golgi complex to the endoplasmic reticulum. Cell 1996; 85: 205–215. [DOI] [PubMed] [Google Scholar]
- Patel SK, Indig FE, Olivieri N, Levine ND, Latterich M. Organelle membrane fusion: a novel function for the syntaxin homolog Ufe1p in ER membrane fusion. Cell 1998; 92: 611–620. [DOI] [PubMed] [Google Scholar]
- Renna M, Schaffner C, Winslow AR, Menzies FM, Peden A a, Floto RA et al. Autophagic substrate clearance requires activity of the syntaxin-5 SNARE complex. J Cell Sci 2011; 124: 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In Vivo Analysis of Autophagy in Response to Nutrient Starvation Using Transgenic Mice Expressing a Fluorescent Autophagosome Marker. Mol Biol Cell 2004; 15: 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Yue Z. Autophagy and Its Normal and Pathogenic States in the Brain. Annu Rev Neurosci 2014; 37: 55–78. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006; 441: 885–889. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006; 441: 880–884. [DOI] [PubMed] [Google Scholar]
- Alves S, Cormier-Dequaire F, Marinello M, Marais T, Muriel M-P, Beaumatin F et al. The autophagy/lysosome pathway is impaired in SCA7 patients and SCA7 knock-in mice. Acta Neuropathol 2014; 128: 705–722. [DOI] [PubMed] [Google Scholar]
- Pan J-A, Ullman E, Dou Z, Zong W-X. Inhibition of protein degradation induces apoptosis through a microtubule-associated protein 1 light chain 3-mediated activation of caspase-8 at intracellular membranes. Mol Cell Biol 2011; 31: 3158–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Wang B, Li N, Wu Y, Jia J, Suo T et al. RNF185, a novel mitochondrial ubiquitin E3 ligase, regulates autophagy through interaction with BNIP1. PLoS One 2011; 6: e24367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Wallace KE, Holzbaur ELF. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol 2012; 196: 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köchl R, Hu XW, Chan EYW, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic 2006; 7: 129–145. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Acevedo-Arozena A, Imarisio S, Berger Z, Vacher C, O'Kane CJ et al. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet 2005; 37: 771–776. [DOI] [PubMed] [Google Scholar]
- Ikenaka K, Kawai K, Katsuno M, Huang Z, Jiang Y-M, Iguchi Y et al. Dnc-1/Dynactin 1 knockdown disrupts transport of autophagosomes and induces motor neuron degeneration. PLoS One 2013; 8: e54511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Arasaki K, Ueda A, Aoki T, Tagaya M. N-terminal region of ZW10 serves not only as a determinant for localization but also as a link with dynein function. Genes Cells 2008; 13: 905–914. [DOI] [PubMed] [Google Scholar]
- Gruber R, Zhou Z, Sukchev M, Joerss T, Frappart PO, Wang ZQ. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nat Cell Biol 2011; 13: 1325–1334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.