Abstract
The tumor-suppressor p53 is a transcription factor that prevents cancer development and is involved in regulation of various physiological processes. This is mediated both by induction of cell cycle arrest and apoptosis and by controlling the expression of a plethora of target genes, including secreted proteins. It has been demonstrated that p53 may exert its effect in non-cell-autonomous manner by modulating the expression of genes that encode for secreted factors. In this study, we utilized our microarray data to identify and characterize novel p53 target genes expressed in human liver cells and associated with steroid hormones processing and transfer. We identified the steroid hormones binding factors, sex hormone-binding globulin (SHBG), corticosteroid-binding globulin (CBG) and cytochrome P450 family 21 subfamily A polypeptide 2, as novel p53 target genes. Their expression and secretion was increased following p53 activation in various hepatic cells. We observed that p53 wild-type mice exhibited higher levels of CBG compared with their p53 null counterparts. We demonstrated that the induction of the steroid hormones binding factors can be mediated by binding to specific p53 responsive elements within their promoters. In addition, utilizing conditioned medium experiments we have shown that p53-dependent induction of SHBG secretion from liver cells enhances apoptosis of breast cancer cells. Moreover, depletion of SHBG abolished the induction of breast cancer cells death. The newly identified p53 target genes suggest a novel non-cell-autonomous tumor-suppressive regulation mediated by p53 that is central for maintaining organism homeostasis.
The transcription factor p53 is a crucial tumor suppressor that functions to prevent cancer development.1 Under normal conditions, p53 protein is maintained in low levels because of the rapid degradation mediated by its main negative regulator, mouse double minute 2 homolog, MDM2. Following different insults, p53 becomes activated and elicits a variety of activities that include cell growth arrest, apoptosis or senescence to prevent proliferation of aberrant cells.1, 2 In addition to its classical tumor-suppressor activity, p53 was suggested to function as a homeostatic gene that coordinates a wide variety of cellular processes.3, 4, 5 Notably, it has been demonstrated that p53 activation within a cell affects not only that cell, but also its surroundings, by modulating the expression of genes that encode for secreted factors.6, 7 Recently, it was demonstrated that in normal tissue the non-cell-autonomous function of p53 can facilitate liver homeostasis following damage. This was shown to be mediated by induction of senescence-associated secretory phenotype (SASP) in hepatic stellate cells, which in turn reduces the accumulation of fibrotic tissue.8, 9 Moreover, a recent study by Lujambio et al.6 has revealed that SASP produced by hepatic stellate cells following p53 activation stimulates immune surveillance to maintain tissue homeostasis and suppress cancer development.9
In our previous study, we attempted to identify p53 transcriptome in liver cells. In our search for specific p53 target genes in hepatic cells, we used the human hepatoma-derived cell line, HepG2. p53 in HepG2 cells was either downregulated by short hairpin (sh) RNA or activated by Nutlin-3a treatment, which inhibits p53 degradation mediated by MDM2.10 Gene expression patterns of the different HepG2 cells were obtained following RNA profiling by microarray. The obtained data provided insights into novel roles of p53 in the regulation of various liver functions. So far, we have characterized the interaction of p53 and groups of genes involved in lipid homeostasis,11, 12 cytochrome P450 enzymes,13 as well as genes related to hepatic glucose production.14 Collectively, these findings have placed p53 as a regulator of diverse metabolic pathways and put forward the notion that p53 has a role in maintenance of systemic homeostasis.
In this study, we report that the aforementioned microarray analysis has revealed yet additional novel group of p53 target genes that are expressed in liver cells and are associated with steroid hormone processing and transfer. This group includes the sex hormone-binding globulin (SHBG), corticosteroid-binding globulin (CBG) and cytochrome P450 family 21 subfamily A polypeptide 2 (CYP21A2).
Steroid hormones influence a variety of vital processes including metabolism, salt and water balance, development of sexual characteristics. These lipophilic molecules derived from cholesterol are secreted from endocrine glands and transported through the bloodstream to the cells of various target organs.15 Within the target cells, steroid hormones bind to their specific receptors that allow the regulation of a wide range of physiological functions. Steroid hormones are typically classified into five major groups: androgens, estrogens, progestogens, glucocorticoids and mineralocorticoids.16
Two major types of enzymes are involved in the biosynthesis of steroid hormones from cholesterol: cytochromes P450 and other steroid oxidoreductases.17 The cytochrome P450 enzymes catalyze the hydroxylation and cleavage of the steroid substrate.18 The CYP21A2 is a member of cytochrome P450 enzymes that catalyzes the conversion of 17 alpha-hydroxyprogesterone to 11-deoxycortisol, the immediate precursor of cortisol.19
Steroid hormones are generally transported in the bloodstream while being bound to specific carrier proteins such as CBG or SHBG. Both CBG and SHBG are produced primarily by the liver and secreted to human plasma.20 CBG is a plasma transport glycoprotein, which binds cortisol with high affinity and facilitates its transport in the blood.21 SHBG binds testosterone and estradiol with high affinity and serves as the principal transporter of estrogens and androgens in the blood.22 According to the 'free hormone hypothesis' circulating steroid hormones that are bound to their carrier proteins considered biologically inactive. Whereas only unbound hormones exhibit biological activity and are capable to access their target tissues via diffusion.23 Accordingly, the hormone carrier proteins, such as CBG and SHBG, provide a mechanism to maintain a reservoir of inactive circulating hormones.24, 25
Nevertheless, it was suggested that both CBG and SHBG are able to bind their specific receptor (that is still unknown) on the membrane of target cells and thus function as protein hormones.20 CBG is able to influence the transport and disposal parameters of cortisol from plasma to tissue.26 It functions as a protein thermocouple that is able to release cortisol in response to increasing temperatures within the physiological rate.24, 27 It has been documented that steroid-free SHBG may bind to membranes of cells in several tissues such as prostate, testis, epididymis and endometrium and to established cell lines such as the estrogen-dependent breast cancer cell line, MCF7.28, 29, 30 Interestingly, SHBG was shown to counteract the estradiol-induced proliferation of breast cancer cells by regulating the expression of genes involved in cell growth and apoptosis.31, 32 Accordingly, numerous studies have reported that low levels of SHBG have been measured in breast cancer patients compared with healthy controls (CONs).32
In this study, we identified the steroid hormones binding factors, CYP21A2, CBG and SHBG, as novel p53 target genes in several hepatic-derived cell lines. In vivo analysis indicated that p53-deficient mice express lower levels of CBG compared with their p53-expressing counterparts. Activation of p53 by different agents led to elevation of CYP21A2, CBG and SHBG gene expression and protein secretion. This is mediated by direct binding of p53 to its regulatory elements at these loci. Furthermore, we show that p53-dependent induction of SHBG secretion in liver cells leads to increased apoptosis of breast cancer cells. Thus, demonstrating a novel aspect of non-cell-autonomous tumor-suppressive function of p53 and suggesting that p53 acts as a systemic homeostatic regulator.
Results
Activation of p53 augments the expression levels of steroid hormones binding factors
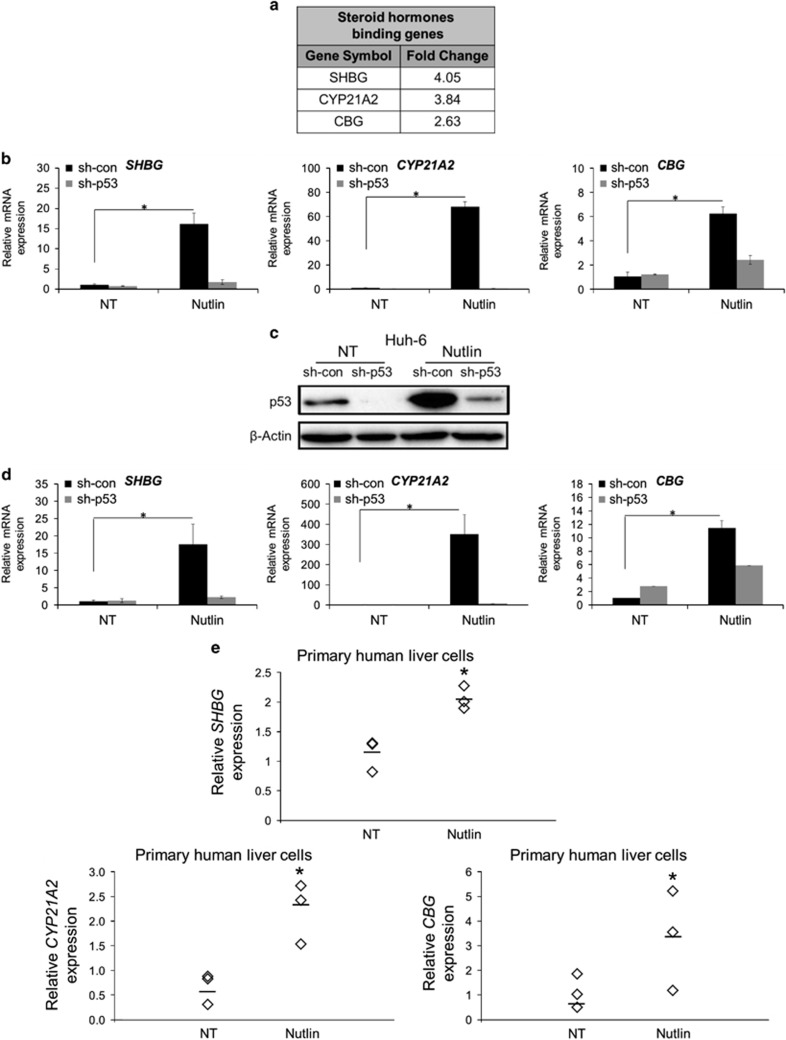
In our previous studies, we aimed to identify p53-dependent gene expression patterns in HepG2 cells. Utilizing global transcriptional expression analysis, we compared the expression patterns of cells in which p53 expression was downregulated by sh-RNA against p53 (HepG2 sh-p53) and non-relevant sh-RNA as CON (HepG2 sh-con). Furthermore, we analyzed the specific expression patterns of cells treated with Nutlin-3a.12, 13, 14 Analysis using the DAVID algorithm33 revealed a new group of p53 target genes that includes SHBG, CYP21A2 and CBG. These genes, implicated in steroid hormones binding processes, were significantly elevated in a p53-dependent manner (>2 fold, P<0.05; Figure 1a). To validate the actual expression of these potential novel targets, we measured the mRNA expression levels of SHBG, CYP21A2 and CBG by quantitative real-time PCR (qRT-PCR). Indeed, their expression levels were higher in HepG2 sh-con cells compared with HepG2 sh-p53 both in basal conditions and following Nutlin-3a treatment (Figure 1b).
Figure 1.
Activation of p53 by Nutlin-3a induces the mRNA expression of SHBG, CYP21A2 and CBG in several hepatic-derived cells. (a) HepG2 sh-con/sh-p53 cells treated with 10 μM Nutlin-3a for 24 h or maintained in CON medium were subjected to microarray analysis. Functional analysis of p53-induced genes using DAVID algorithm revealed list of genes related to steroid hormones binding factors. The fold change indicates the increase in gene expression in HepG2 sh-con cells treated with Nutlin-3a compared with the rest of the samples. (b) qRT-PCR analysis of SHBG, CYP21A2 and CBG mRNA expression in HepG2 sh-con/sh-p53 treated either with 10 μM Nutlin-3a for 24 h (Nutlin) or NT. Results are of a representative experiment from at least three experiments. (c) Protein levels of p53 were measured by western blot in Huh-6 sh-con/sh-p53 cells treated with 10 μM Nutlin-3a for 24 h (Nutlin) or maintained in CON medium, NT. β-Actin was used as loading CON. Results are of a representative experiment from at least three experiments. (d) qRT-PCR analysis of SHBG, CYP21A2 and CBG mRNA expression in Huh-6 sh-con/sh-p53 treated either with 10 μM Nutlin-3a for 24 h (Nutlin) or maintained in CON medium, NT. Results are of a representative experiment from at least three experiments. (e) qRT-PCR analysis of SHBG, CYP21A2 and CBG expression in human primary liver cells treated with 10 μM Nutlin-3a for 24 h (Nutlin) or maintained in CON medium, NT. Each rhombus represents the expression level in a distinct experiment. The horizontal bar represents the median of three individual experiments. *P<0.05, determined by Student's t-test analysis
To corroborate the effect of p53-dependent induction of steroid hormones binding factors, we knocked-down p53 in Huh-6 cells and applied treatment with Nutlin-3a (Figure 1c). Consistent with the results observed in HepG2 cells, qRT-PCR analysis of SHBG, CYP21A2 and CBG mRNA levels in Huh-6 cells indicated that steroid hormones binding factors expression was markedly increased in sh-con cells compared with sh-p53 cells following Nutlin-3a treatment (Figure 1d).
Next, we examined whether p53-dependent induction of steroid hormones binding factors is also applicable in human primary cells of hepatic origin. Concomitantly with the results obtained in hepatic cancer cell lines, primary human liver cells exhibited elevated levels of SHBG, CYP21A2 and CBG following Nutlin-3a treatment (Figure 1e). These data suggest that p53-dependent transcriptional activation of steroid hormones binding factors is a general phenomenon observed both in established hepatic cancer cell lines, as well as in physiological primary liver cells.
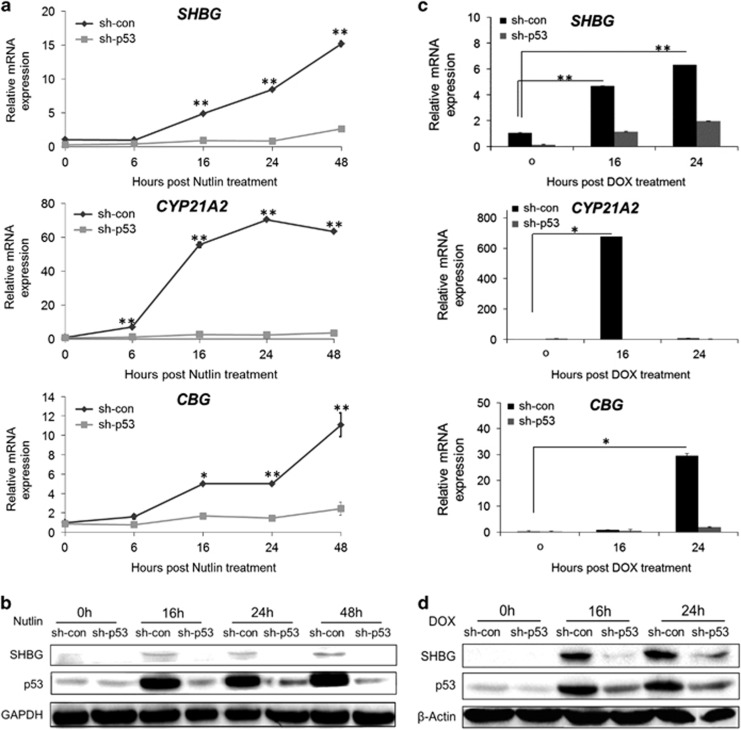
To evaluate the kinetics of SHBG, CYP21A2 and CBG expression pattern, as a result of p53 stabilization, their mRNA and protein levels were analyzed at different time points following Nutlin-3a treatment. The mRNA levels of SHBG, CYP21A2 and CBG in HepG2 sh-con cells increased as early as 6 h following exposure to Nutlin-3a. Although the mRNA levels of SHBG and CBG continued to increase gradually through the 48 h of the treatment, the mRNA expression of CYP21A2 peaked at 24 h following Nutlin-3a addition and thereafter began to decline (Figure 2a). As depicted in Figure 2b, Nutlin-3a treatment resulted in a pronounced accumulation of p53 protein in the HepG2 sh-con cells compared with the HepG2 sh-p53 cells that was accompanied by similar pattern of SHBG protein expression (Figure 2b).
Figure 2.
Steroid hormones binding factors expression is p53-dependent and augmented upon DNA damage. (a) HepG2 sh-con/sh-p53 cells were treated with 10 μM Nutlin-3a for 6, 16, 24 and 48 h. The mRNA levels of SHBG, CYP21A2 and CBG were measured by qRT-PCR. (b) Western blot analysis of SHBG and p53 protein levels in the same cells as in a. GAPDH was used as loading CON. (c) HepG2 sh-con/sh-p53 cells were treated with 0.5 μg/ml DOX for 16 and 24 h. mRNA levels of SHBG, CYP21A2 and CBG were measured by qRT-PCR analysis. (d) Western blot analysis of SHBG and p53 protein levels in the same cells as in c. β-Actin was used as loading CON. Results are of a representative experiment from at least three experiments. *P<0.05, **P<0.01, determined by Student's t-test analysis
Next, we examined whether expression of SHBG, CYP21A2 and CBG would be induced following p53 activation by the DNA damaging agent, doxorubicin (DOX). As demonstrated in Figure 2c, HepG2 sh-con cells exhibited considerable p53-dependent induction of SHBG mRNA both at 16 h and 24 h after treatment with DOX. The levels of CYP21A2 in HepG2 sh-con cells increased 16 h on DOX treatment; however, after 24 h this effect was abolished. The elevation of CBG mRNA level was observed 24 h following DOX treatment (Figure 2c). Figure 2d depicts significant stabilization of p53 protein following treatment with DOX. The accumulation pattern of SHBG protein was p53 dependent and analogous to the mRNA expression (Figure 2d).
In sum, the pattern of mRNA and protein expression of the steroid hormones binding factors correlates with the p53 protein expression in the cells.
The globulins, SHBG and CBG, are secreted in p53-dependent manner
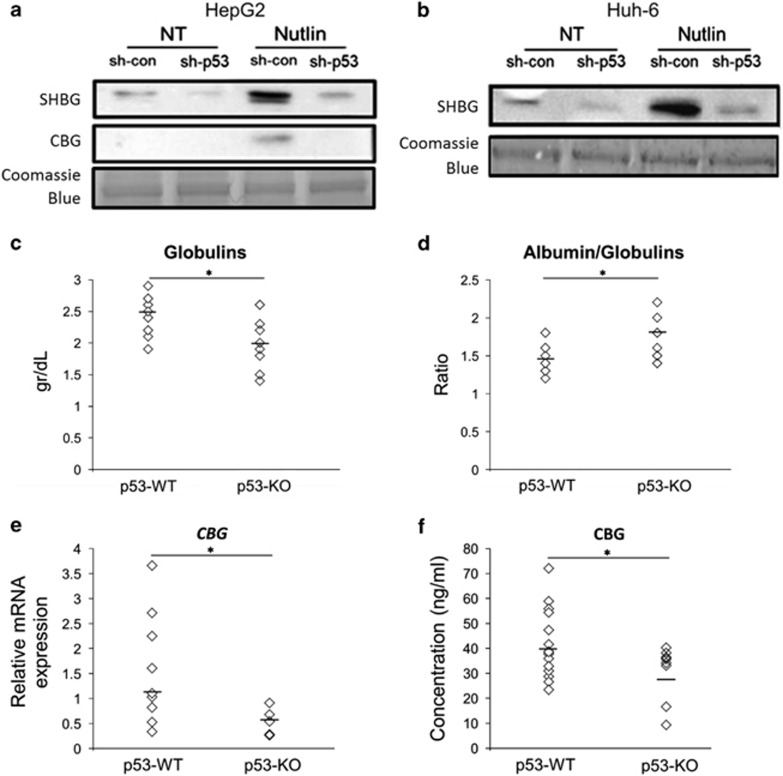
To execute their principal function as transport proteins for steroid hormones, SHBG and CBG are secreted by the liver to the vascular compartment.20 Thus, it was of interest to examine whether p53 stabilization in HepG2 and Huh-6 cells treated with Nutlin-3a would augment the secretion of the SHBG and CBG proteins. As seen in Figure 3, both cell lines downregulated for p53 expression exhibited a decrease in SHBG secretion compared with the corresponding CONs. However, upon Nutlin-3a treatment HepG2 sh-con cells exhibited an increase in SHBG and CBG secretion compared with their HepG2 sh-p53 counterparts (Figures 3a and b).
Figure 3.
SHBG and CBG proteins are secreted in p53-dependent manner. (a and b) HepG2 sh-con/sh-p53 cells and Huh-6 sh-con/sh-p53 cells were treated with 10 μM Nutlin-3a for 24 h (Nutlin) or maintained in CON medium, NT. Cell media were concentrated and the secreted protein levels of SHBG and CBG were measured by western blot. Coomassie blue staining is presented as loading CON. Results are of a representative experiment from at least three experiments. (c and d) Analysis of total globulins concentrations and albumin/globulins ratio in sera of p53-WT and p53-KO mice. (e) Total RNA was isolated from livers of p53-WT and p53-KO mice and CBG mRNA levels were determined using qRT-PCR. (f) CBG protein concentrations in the sera of p53-WT and p53-KO mice were assessed by ELISA assay. Each rhombus represents an individual mouse. The small horizontal bar in the columns represents the median. *P<0.05, determined by Student's t-test
Our in vitro studies have prompted us to examine whether the increased secretion of total sera globulins in p53 wild-type (WT) mice is retained in vivo. To this end, we analyzed sera from p53-WT and p53 knockout (p53-KO) mice34 for globulins and albumin concentrations. In accordance with our in vitro findings, we observed that p53-WT mice exhibited higher levels of total blood globulins compared with their p53-KO counterparts, and were also accompanied by lower albumin/globulins ratio (Figures 3c and d). As, postnatal, mice do not express SHBG in the liver,35, 36 we quantified the specific expression levels of CBG mRNA expression in murine livers and its protein concentration in the sera. The results showed that CBG mRNA expression is significantly reduced in the livers of p53-KO mice (Figure 3e). In addition, CBG protein concentration was higher in sera of p53-WT mice compared with p53 null mice (Figure 3f). Overall, these data suggest that p53 has an important role in regulation of SHBG and CBG expression and secretion both in vitro and in vivo.
p53 directly activates transcription of steroid hormones binding factors
The p53 transcription factor may regulate it's target genes either via direct or indirect mechanisms. It is well established that direct transcription of p53 target genes is mediated by p53 binding to its responsive elements (REs) entailed within the target gene promoter.37 When utilizing the indirect mechanism, p53 induces the expression of mediating factors that will be translated, and in turn will facilitate the transcription of a secondary, indirect p53 target genes.37, 38
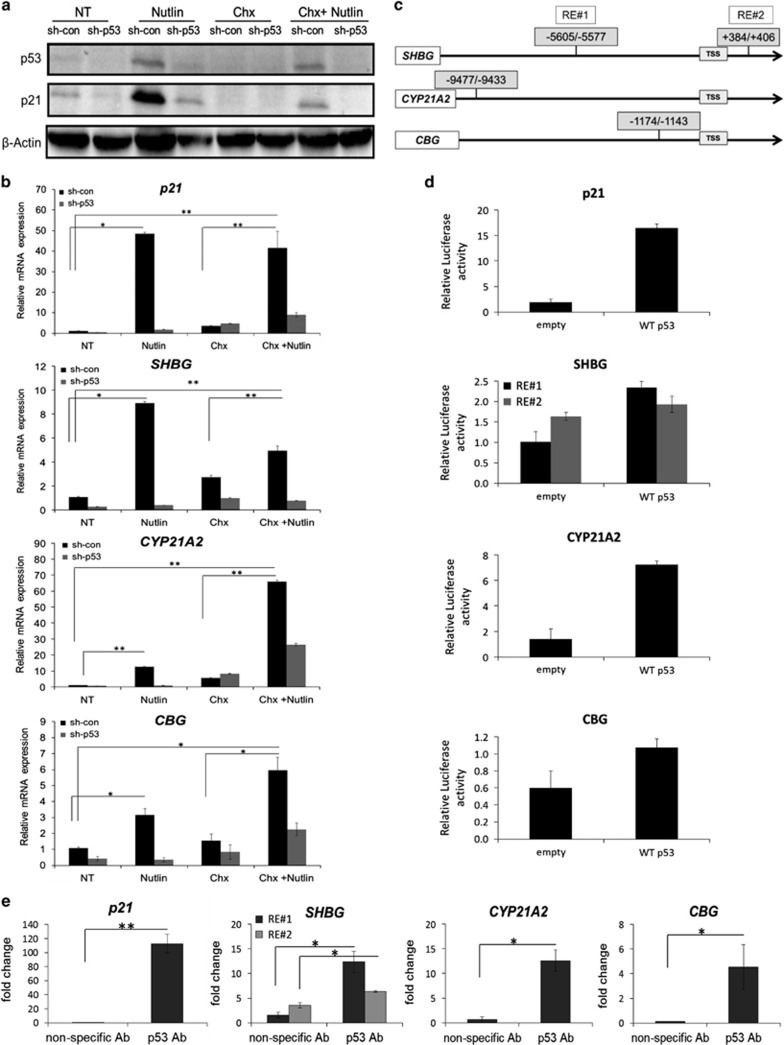
In order to elucidate the mechanism by which p53 induces the transcription of steroid hormones binding factors, we treated HepG2 cells with Nutlin-3a in combination with cycloheximide (Chx), protein biosynthesis inhibitor.39 We assumed that if steroid hormones binding factors are induced by p53 following Nutlin-3a treatment through an indirect mechanism, then Chx should prevent their induction. As a positive CON, we measured the expression of p21 that was induced in a p53-dependent manner (Figures 4a and b). Treatment with Chx alone resulted in a decrease in p21 protein, however, it led to consistent increase in the p21, SHBG, CYP21A2 mRNA expression. This upregulation of mRNA expression following Chx treatment could be attributed to one of the acknowledged characteristics of protein synthesis inhibitors – 'super-induction'.12, 40 Finally, treatment of the cells with Chx after their exposure to Nutlin-3a did not abolish p21 induction (Figures 4a and b); thus confirming that secondary mediator is not required. We observed a statistically significant induction of SHBG, CYP21A2 and CBG mRNA expression following Nutlin-3a and Chx combined treatment compared with their levels in non-treated (NT) CON and in Chx alone (Figure 4b), thus indicating that these genes, similarly to p21, are primary p53 targets.
Figure 4.
p53 directly induces SHBG, CYP21A2 and CBG transcription by binding to specific p53 REs entailed in their loci. HepG2 sh-con/sh-p53 cells were maintained in CON medium, NT or treated with either 10 μM Nutlin-3a for 24 h (Nutlin) or with 10 μg/ml Chx for 6 h (Chx) alone or with a combination of Nutlin-3a for 18 h followed by Chx treatment for additional 6 h (Chx+Nutlin). (a) Protein levels of p53 and p21 were measured by western blot. β-Actin was used as loading CON. (b) qRT-PCR analysis of p21, SHBG, CYP21A2 and CBG genes. (c) SHBG, CYP21A2 and CBG genomic sequences were analyzed by p53MH algorithm. Representative scheme of the potential p53 REs locations relative to TSS are indicated. (d) Reporter assay analysis of p53 REs in SHBG, CYP21A2 and CBG promoters. H1299 cells were transiently co-transfected with Luciferase reporter construct harboring distinct p53 REs of the steroid hormone binding factors loci as well as with WT p53 expression vector or corresponding empty vector as CON. Luminescence values were estimated 48 h after transfection. Transfection efficacy was normalized on the basis of the activity of the co-transfected Gaussia luciferase. (e) ChIP analysis of HepG2 cells treated with 10 μM Nutlin-3a for 24 h. p53 protein was immunoprecipitated using p53-specific H47 polyclonal antibody (p53 Ab). Nonspecific antibody-IgG was used as a CON (nonspecific Ab). qRT-PCR was performed using specific primers against the p53 REs indicated in (c). Values were normalized to 1% input of the corresponding sample. Results are of a representative experiment from at least three experiments. *P<0.05, **P<0.01, determined by Student's t-test analysis
All in all, these data indicate that p53 may activate the transcription of steroid hormones binding factors via direct mechanism that does not require the synthesis of intermediating proteins.
Next, we used the p53MH algorithm,41 which searches for the potential p53 REs in the steroid hormones binding factors promoters and intronic sequences, defined by us as 10 000 base pairs upstream and downstream from the transcription start site (TSS), respectively. In the SHBG locus, we found two potential p53 REs with high similarity to p53 consensus sequences; one in SHBG promoter (RE #1) and another in SHBG second intron (RE #2). In addition, putative p53 binding sites were found in the promoters of CYP21A2 and CBG (Figure 4c).
In order to examine whether the p53-mediated upregulation of SHBG, CYP21A2 and CBG mRNA results from transcriptional activation of the potential p53 REs in their loci, DNA fragments with the potential p53 REs were fused to luciferase reporter. As depicted in Figure 4d, ectopic expression of p53 resulted in induction of the luciferase expression in the construct harboring p53 RE of the p21 gene, serving as the positive CON, as well as in the constructs harboring the promoters of steroid hormone binding factors.
To determine whether p53 indeed binds to these potential REs, we performed a chromatin immunoprecipitation (ChIP) analysis of HepG2 cells treated with Nutlin-3a. The chromatin was precipitated using p53-polyclonal antibodies or nonspecific CON antibodies. Next, the enrichment of p53 binding was measured by qRT-PCR using specific primers targeting the p53 REs in steroid hormones binding factors loci. In the precipitated DNA of HepG2 Nutlin-3a-treated cells, there was an enrichment for the p53 REs entailed in all steroid hormones binding factors and p21 (Figure 4e). Taken together, these results suggest that p53 trans-activated the steroid hormones binding factors transcription by direct binding to p53 REs in the SHBG, CYP21A2 and CBG loci.
p53-dependent non-cell-autonomous induction of apoptosis involves SHBG secretion
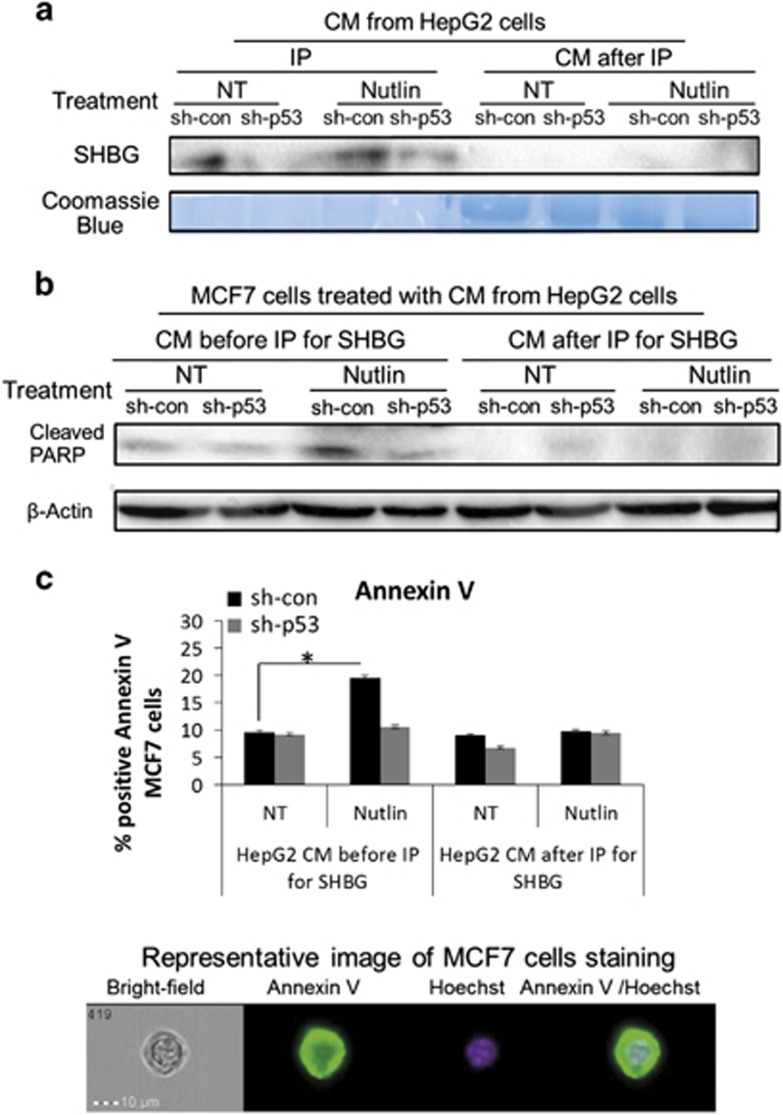
One of the prime p53 activities is the ability to induce growth arrest and apoptosis.42 Interestingly, numerous studies have demonstrated that SHBG is able to inhibit estrogen-dependent cell growth.32 For example, Catalano et al.31 have shown that pre-incubation of breast cancer cell line, MCF7, with recombinant human SHBG protein, before treatment with estrogen, antagonizes the anti-apoptotic effect of estrogen on MCF7 cells. Notably, a recent study has revealed that in hepatic cells p53 acts in a non-cell-autonomous manner to suppress tumor development through secretion of factors that modulate tissue microenvironment.6 With these observations in mind, we examined whether authentic hepatic-derived SHBG protein secreted upon p53 activation in HepG2 cells, will indeed promote apoptosis of MCF7 cells. To this end, p53-dependent SHBG secretion was achieved by treatment of HepG2 cells with Nutlin-3a. Then, the medium containing Nutlin-3a was removed and cells were further grown in fresh medium. The latter's conditioned medium (CM) was collected and incubated with MCF7 cells. This was followed by the addition of 17β-estradiol, which is necessary for SHBG function.31 To assess the specific effect of hepatic-derived SHBG on MCF7 apoptosis, we depleted SHBG from the HepG2 CM using specific antibody (Figure 5a). To detect the apoptotic effect of SHBG on MCF7 cells following incubation with CM from HepG2 prior and after SHBG depletion, we measured both cleaved Poly (ADP-ribose) polymerase (PARP) and positive Annexin V staining. As demonstrated in Figure 5, HepG2 sh-con cells displayed higher SHBG secretion compared with their HepG2 sh-p53 counterparts, which was further augmented upon Nutlin-3a treatment. Following immunoprecipitation (IP) with specific antibody, SHBG was not detected in the HepG2 CM (Figure 5a). Incubation of MCF7 cells with the CM from the Nutlin-3a-treated HepG2 sh-con cells, followed by 17β-estradiol treatment, resulted in high levels of cleaved PARP and increased percentage of Annexin V-positive cells, which was abolished by SHBG depletion (Figures 5b and c). This suggests that activated p53 in hepatic cells induced the secretion of SHBG, which in turn promoted apoptosis of breast cancer cells, counteracting the proliferative effect of estrogen.
Figure 5.
Activation of p53 in liver cells results in elevated SHBG secretion, which enhances apoptosis in breast cancer cells. (a) CM from HepG2 sh-con/sh-p53 cells was collected after 24 h during which cells were treated with 10 μM Nutlin-3a for 18 h followed by maintenance in fresh medium for additional 6 h (Nutlin). As CON, cells were grown in normal medium and were left NT. For depletion of SHBG from CM, IP with beads conjugated to SHBG antibody was performed (IP). Western blot analysis was done to detect both the precipitated SHBG and the possible remaining SHBG in the CM (CM after IP). Coomassie blue staining is presented as loading CON. (b and c) MCF7 cells were treated with the CMs from HepG2 sh-con/sh-p53 cells that were collected either before or after the IP assay of SHBG as described in a, and followed by the addition of 17β-estradiol. (b) Cleaved PARP protein levels were measured by western blot analysis. β-Actin was used as loading CON. (c) Annexin V-positive staining was determined by Imaging Flow Cytometry. The upper panel depicts the quantification of the percentage of MCF7 Annexin V-positive cells. Representative image of Annexin V staining as visualized by imaging flow cytometry is demonstrated in the lower panel. Results are of a representative experiment from at least three experiments. *P<0.05, determined by Student's t-test analysis
Discussion
It is well known that the p53 transcription factor prevents tumor development.1 Ample data support the notion that p53 is a multitask protein, which is involved in regulation of additional diverse cellular processes. Interestingly, accumulated data suggest that p53 may act in non-cell-autonomous manner and influence other cells via paracrine and endocrine mechanisms.6, 7 Our present data support the growing body of evidence suggesting a crosstalk between p53 and the endocrine system.43, 44, 45 Here we report on the identification of novel p53 target genes, CYP21A2, CBG and SHBG, that are associated with steroid hormones processing and transfer. We showed that activation of p53 by either Nutlin-3a or DOX induced the expression and the secretion of the steroid hormones binding factors in distinct liver-derived cell lines. In addition, we observed that p53-WT mice exhibit higher CBG expression compared with p53-KO counterparts. Our data indicated that p53 induces the expression of CYP21A2, CBG and SHBG through direct binding to the p53 REs contained within these genes. Moreover, we suggest here an intriguing aspect of non-cell-autonomous action of the p53 tumor suppressor, showing that activated p53 in liver cells leads to secretion of SHBG, which in turn is able to increase apoptosis of breast cancer cells. Taken together, these results suggest that p53 functions as a systemic regulator with comprehensive impacts on different systems, combined together to maintain human body homeostasis.
Deficiency in the CYP21A2 enzyme causes the congenital adrenal hyperplasia disorder.46 Adrenal hyperplasia serves as one of the origins for the development of adrenocortical tumors. Interestingly, it has been reported that adrenal hyperplasia is also associated with the Li-Fraumeni syndrome. The Li-Fraumeni patients that carry a germline mutation in the p53 allele display high incidence of various tumors in early age, including adrenocortical carcinoma, as one of the most typical cancers.47, 48 Mutant p53 proteins lose the WT p53 transactivation and tumor suppression functions.49 Therefore, it might be suggested that adrenal hyperplasia associated with Li-Fraumeni syndrome patients may develop because of low levels of CYP21A2, which is not trans-activated because of lack of functional WT p53.
In the presence of liver damage, functional p53 eventually assures liver homeostasis by inducing senescence of hepatic stellate cells, which is associated with secretion of factors leading to clearance of senescent cells.6 Interestingly, increased CBG levels have been found in sera of patients with chronic active hepatitis or alcoholic cirrhosis.50 Taken together, a possible mechanism underlying the physiological elevation of secreted CBG in plasma of patients with liver damage might be due to its induction by activated p53 in liver cells.
In this study, we identified SHBG as a novel p53 target gene, which is involved in the p53-mediated counteraction of proliferative effects of estrogen. Our results in the human hepatocyte-derived cell lines, as well as in primary human liver cells revealed that SHBG transcription levels were induced by p53 via a direct mechanism.
Accumulating data support the notion that SHBG functions as a negative modulator of estrogen action in breast cancer cells, reducing breast cancer growth and proliferation. In particular, it was previously demonstrated that administration of human purified SHBG to MCF7, breast cancer cells, attenuates the anti-apoptotic effect of estrogen on these cells.31 Similarly, we demonstrate that activation of p53 in liver cells results in p53-dependent induction of authentic, hepatic-derived SHBG protein secretion. Further administration of medium containing this SHBG protein to breast cancer cells, promotes elevated PARP cleavage and positive Annexin V staining, which is completely abolished following IP of SHBG by a specific antibody. Hence, indicating increased apoptosis and counteraction of the proliferative effect expected after estrogen treatment. These data suggest a novel function for the tumor-suppressor p53 that is associated with the regulation of the endocrine system, whereby p53 may mediate cancer cell death through SHBG induction. This highlights the possible systemic role of p53 in tumor suppression.
In summary, our data reveal novel p53 target genes CYP21A2, CBG and SHBG that are implicated in several aspects of steroid hormones production, bioavailability and signaling. Thus, positioning p53 as a systemic regulator that operates to maintain homeostasis and prevent malignant transformation not only in single cells rather in the organism, at large.
Materials and Methods
Cell culture
HepG2 cells, were kindly provided by Prof. Yehiel Zick, Huh-6 cells were kindly provided by Prof. Yosef Shaul (both from the Weizmann Institute of Science, Rehovot, Israel). Primary human liver cells were kindly provided by Prof. Sara Ferber (Sheba Medical Center, Tel Hashomer, Israel) and isolated as previously described.51, 52 Informed consent was obtained from each donor and the study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki, as reflected in a priori approval by the Committee on Clinical Investigations (Institutional Review Boards of Sheba Medical Center and Rabin Medical Center, Petah Tikva, Israel). Non-small cell lung carcinoma (H1299) cells were obtained from the ATCC (Manassas, VA, USA) and MCF7 cells were provided by NCI-DCTD Repository (Bethesda, MD, USA). All cell lines were cultured in humidified incubator at 37 °C and 5% CO2. Primary human liver cells, HepG2 and Huh-6 cell lines were cultured in DMEM supplemented with 10% FCS, 2 mM l-glutamine, 1 mM sodium pyruvate and 100 mg/ml penicillin–streptomycin (Biological Industries, Beit-Haemek, Israel). H1299 and MCF7 cell lines were maintained in RPMI-1640 supplemented with 10% FCS, 2 mM l-glutamine, 1 mM sodium pyruvate and 100 mg/ml penicillin–streptomycin (Biological Industries).
Mice
C57BL/6 p53-WT and p53-KO mice34 were kindly provided by G. Lozano (University of Texas M D Anderson Cancer Center, Houston, TX, USA) and maintained inside a barrier facility. Experiments were performed in accordance with Weizmann Institute of Science regulations for institutional animal care and use committees. For sera analysis, blood was collected from 8-week-old male p53-WT and p53-KO mice, centrifuged at 14 000 r.p.m. for 15 min. The obtained sera was collected and analyzed for albumin and total globulins by diagnostic veterinary pathology services, PathoVet Ltd (Kfar Bilu B, Israel).
For isolation of liver RNA, murine liver tissues were rapidly removed from killed animals and kept on ice for several minutes before initiation of RNA purification as described below.
For CBG protein concentration measurement, mice sera was diluted with phosphate-buffered saline (PBS) 1 : 2000 and CBG enzyme-linked immunosorbent assay (ELISA) kit (USCN Life Science Inc., Houston, TX, USA) was used according to the manufacturer's recommendation.
CM assay
MCF7 and HepG2 cells were grown in RPMI-1640 without phenol red supplemented with 10% charcoal stripped serum, 2 mM l-glutamine and 1 mM sodium pyruvate (Biological Industries). HepG2 cells were grown for 18 h in RPMI medium with 10 μM Nutlin-3a. Then, the medium was aspirated, cells were washed with PBS, following by replacement with fresh medium for additional 6 h. The HepG2 CM was collected and applied on MCF7 cells for 40 min. Then, 10 nM 17β-estradiol was added to MCF7 medium and the cells were collected after 24 h and analyzed.
Chemicals compounds
Chx, DOX and 17β-estradiol were purchased from Sigma-Aldrich (Rehovot, Israel). Nutlin-3a was purchased from Alexis-Biochemical (San Diego, CA, USA).
Retroviral constructs and infections
Constructs
pRetroSuper vectors harboring sh-RNA against human p53 (sh-p53) or mouse noxa (sh-con) were kindly provided by Dr. Doron Ginsburg (Bar-Ilan University, Israel).
Retroviral infection procedure
Ecotropic Phoenix-packaging cells were transfected with 10 μg of DNA per 10 cm plate of the appropriate retroviral construct by a standard calcium–phosphate procedure. Supernatants were collected 48 h after transfection and filtered. Recipient cells were infected with the filtered viral supernatants in the presence of 4 μg/ml polybrene (Sigma-Aldrich) for 12 h, after which fresh viral suspensions were added for an additional 12 h. Infected cells were selected for 20 days with 10 μg/ml Blasticidin.
Apoptosis assay
MCF7 cells were analyzed for apoptosis using the Annexin V-FLUOS Staining Kit (Roche, Penzberg, Germany). Apoptotic cells exposing phosphatidylserine on their surface were stained and subsequently detected by Imaging Flow Cytometry analysis. Cells were imaged by ImageStreamX mark II (Amnis Corp., part of EMD Millipore, Seattle, WA, USA) using bright field and the 488 and 405 nm lasers. Cells were gated for single cells using the area and aspect ratio features, and for focused cells using the Gradient RMS feature, as previously described.53 Data were analyzed using the IDEAS 6.1; Amnis Corp. software. Cells were then gated for Hoechst-positive cells (at least 10 000 cells in each population). Annexin V-positive cells were gated using the intensity (sum of the background-subtracted pixel values) and the Max Pixel (the largest value of the background-subtracted pixels) features. In addition, only cells with membrane staining were further gated using the Max Contour Position (the location of the contour in the cell that has the highest intensity concentration mapped to a number between 0 and 1, with 0 being the object center and 1 being the object perimeter) and their percentage of the single, focused cells was calculated. Examples of cell images and gating are shown in Supplementary Figure S1.
Western blot analysis
Cells pellets were lysed in TLB buffer (50 mM Tris-HCl, 100 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with Protease Inhibitor Cocktail (Sigma-Aldrich) for 15 min on ice and centrifuged for 15 min. Supernatants were analyzed for protein concentration using BCA reagent (Thermo-Scientific, Grand Island, NY, USA). In all, 50 μg of protein extracts were boiled and loaded on 10–12% SDS-polyacrylamide gel. Proteins were transferred to a nitrocellulose membrane at semi-dry conditions. Membranes were blocked using 5% dry milk in PBST.
Coomassie blue staining was performed using GelCode Blue stain reagent (Thermo-Scientific) according to the manufacturer's protocol.
The following primary antibodies were used: α-SHBG (sc-377031), α-CBG (SERPINA6) (sc-100851), α-p21 (sc-397) and α-β actin (sc-47778) were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). α-PARP1 (C-2-10) was purchased from Enzo Life Sciences (Farmingdale, NY, USA). α-GAPDH (mab374) was purchased from EMD Milipore (Billerica, MA, USA). α-p53 (DO-1) was kindly provided by Prof. David Lane. The protein–antibody complexes were detected using goat anti-mouse or goat anti-rabbit horseradish peroxidase-conjugated secondary antibodies (Jackson Immunoresearch Laboratories, West Grove, PA, USA) and the ECL western blotting detection reagents (Thermo Scientific), followed by analysis in ChemiDoc MP (Bio-Rad, Hercules, CA, USA).
RNA isolation and qRT-PCR
Total RNA from cells was isolated using the NucleoSpin RNA II kit (Macherey-Nagel, Duren, Germany) and total RNA from murine liver tissues was isolated using the Direct-zol RNA MiniPrep kit (Zymo Research, Irvine, CA, USA). A 2 μg aliquot of the total RNA was reverse transcribed into cDNA using Bio-RT (Bio-Lab, Jerusalem, Israel), dNTPs and random hexamer primers. qRT-PCR was performed on Step One Plus, ABI instrument (Applied Biosystems, Grand Island, NY, USA) using SYBR Green PCR Master Mix (Quanta BioSciences, Gaithersburg, MD, USA). The values for the specific genes were normalized to GAPDH (human) or to B2M (mouse) as housekeeping CON and the data are described in arbitrary units. PCR reactions were performed in duplicate. The specific primers used for qRT-PCR are listed in Table 1.
Table 1. List of specific primers that were used for qRT-PCR.
| Gene | 5′ Primer | 3′ Primer |
|---|---|---|
| GAPDH | ACCCACTCCTCCACCTTTGA | CTGTTGCTGTAGCCAAATTCGT |
| p53 | CCCAAGCAATGGATGATTTGA | GGCATTCTGGGAGCTTCATCT |
| p21 | GGCAGACCAGCATGACAGATT | GCGGATTAGGGCTTCCTCTT |
| SHBG | GCCCAGGACAAGAGCCTATC | CCTTAGGGTTGGTATCCCCATAA |
| CYP21A2 | CAAGCTGGTGTCTAGGAACTACC | TCTCATGCGCTCACAGAACTC |
| h-CBG | GTGAACATGAGTAACCATCACCG | CCTGGTGGATCTCAGTCTCAG |
| m-CBG | GATGTCTGAAGCTGAGATCCAC | CCGGCTTTAGTCCAGTCCT |
| m-B2M | CTCGGTGACCCTGGTCTTTC | TTGAGGGGTTTTCTGGATAGCA |
Medium concentration
Culture medium obtained from HepG2 or Huh-6 growing cells, was concentrated by centrifugation (3000 r.p.m. 4 °C) using the VIVAspin 20 columns (Sartorius Stedim Biotech, Goettingen, Germany) for 3 h.
Immunoprecipitation
Cells media was precipitated by α-SHBG (sc-377031, Santa Cruz Biotechnology), which was conjugated to protein A beads (Santa Cruz Biotechnology). The immunoprecipitated material was washed with 150 mM NaCl, 50 mM Tris-HCl (pH 7.5). For immunoblotting, it was re-suspended in SDS sample buffer, and subjected to western blot analysis.
Chromatin immunoprecipitation
Cell lysis
Cells underwent cross-linking (1% formaldehyde, room temperature for 10 min) followed by quenching (glycine 0.125 m). Cells were rinsed with cold PBS and incubated with 20% trypsin (Gibco Life Technologies, Grand Island, NY, USA) in PBS. Cells were removed by scraping, collected by centrifugation and washed in cold PBS. Cells were lysed (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP-40, 1% protease inhibitors) on ice for 10 min. Nuclei were collected by centrifugation (13 000 r.p.m.), re-suspended in nuclear lysis buffer (50 mM Tris-Cl, pH 8.0, 10 mM EDTA, 1% SDS, 1% protease inhibitors) and incubated on ice for 10 min. Samples were sonicated to obtain an average DNA fragment length of 500 bp and then centrifuged at 14 000 r.p.m. The chromatin solution was pre-cleared by incubating with blocked protein A beads (Santa Cruz Biotechnology) for 2 h at 4 °C.
IP of chromatin
the pre-cleared chromatin was mixed by rotation for 12 h at 4 °C with blocked protein A beads and 1 μg of polyclonal antibody. The following antibodies were used: polyclonal h47 α-p53 (produced in our laboratory), polyclonal IgG as a CON.
Immunoprecipitates were consecutively washed with dilution buffer (100 mM Tris-Cl, pH 9.0, 500 mM LiCl, 1% NP-40, 1% Deoxycholic acid, 1% protease inhibitors), TSE 150, TSE 500 and TE pH=8.
Elution and DNA analysis
Elution was carried out with elution buffer (Tris EDTA pH 6.8, 0.5% SDS). Samples were treated with 10 μg RNase A for 30 min at 37 °C, followed by 2 h treatment with 30 μg of proteinase K at 50 °C, and incubation at 65 °C overnight. DNA samples were extracted using QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). qRT-PCR was performed as described above with each sample containing 2 μl of immunoprecipitated DNA. Values were normalized for 1% input values.
The specific primers surrounding p53 RE used for qRT-PCR are listed in Table 2.
Table 2. List of specific primers surrounding the p53 REs that were used for qRT-PCR.
| Gene | 5′ Primer | 3′ Primer |
|---|---|---|
| p21 | AGCAGGCTGTGGCTCTGATT | CAAAATAGCCACCAGCCTCTTCT |
| SHBG RE #1 | CTCCACCCTCAACCTCTCAA | AGGCATGAGAATCGCTTGAT |
| SHBG RE #2 | GGGCAACATAGCAGAGACCT | GTCAGAGTGCAGTGGCACAT |
| CYP21A2 | TAGGAGGCTCGCCTGAGAAA | CCCTAGTAAGGGGAGAAAAGATGT |
| CBG | GGACTTATCAAAGGGAGAAAAGACA | TGTGAATGCCTCGCTAGGGT |
Reporter gene assay
The p53 construct pC53-SN3 was kindly provided by Prof. Bert Vogelstein (Johns Hopkins University School of Medicine, Baltimore, MD, USA). Reporter gene vectors were purchased from New England Biolabs (Ipswich, MA, USA).
Reporter assay was performed using the BioLox Gaussia and Cypridina Luciferase assay kit (NEB, Ipswich, MA, USA). H1299 cells were plated in 48-well plates in triplicate (40 000 cells per well) and 24 h later were transiently co-transfected with 480 ng per well of pCLuc-Basic2 (Cypridina luciferase) harboring the p53 REs, 10 ng per well of pCMV-GLuc (Gaussia luciferase) and 10 ng per well of pC53-SN3 or corresponding empty vector, using jetPEI (Polyplus-transfection, New York, NY, USA). Luminescence values were estimated after 48 h. In order to estimate transfection efficacy, values of the Cypridinia enzyme (encoded from pCLuc-Basic2) were normalized to values of Gaussia luciferase (encoded from pCMV-GLuc). Primers utilized for cloning of p53 REs entailed in steroid hormones binding factors loci into pCLuc-Basic2 vector are listed in Table 3.
Table 3. List of primers utilized for cloning of p53 REs entailed in steroid hormones binding factors loci into pCLuc-Basic2 vector.
| Gene | Forward | Reverse |
|---|---|---|
| CBG | 5'-GGGAGATCTTGGAATTCTGCAAACTTGCCTTATAGGATAGGAAACTTGAAACTCGGATCCTTCGCATATTA-3' | 5'-TAATATGCGAAGGATCCGAGTTTCAAGTTTCCTATCCTATAAGGCAAGTTTGCAGAATTCCAAGATCTCCC-3' |
| SHBG RE #1 | 5'-GGGAGATCTTGGAATTCTGCGGTCTTGCCCATGTTGCCTAGGCTGGTCTCTCGGATCCTTCGCATATTA-3' | 5'-TAATATGCGAAGGATCCGAGAGACCAGCCTAGGCAACATGGGCAAGACCGCAGAATTCCAAGATCTCCC-3' |
| SHBG RE #2 | 5'-GGGAGATCTTGGAATTCTGCGCACATGTCTGCTGCCCTAGCTACTCGGATCCTTCGCATATTA-3' | 5'-TAATATGCGAAGGATCCGAGTAGCTAGGGCAGCAGACATGTGCGCAGAATTCCAAGATCTCCC-3' |
| CYP21A2 | 5'-GGGAGATCTTGGAATTCTGCGAGCATGCAGGTGCGGGCATGCTGCTCGGATCCTTCGCATATTA-3' | 5'-GGGAGATCTTGGAATTCTGCGAGAATACAGGTGCGGGAATACTGCTCGGATCCTTCGCATATTA-3' |
Microarray analysis
Microarray data were analyzed as previously described.11
Statistical analysis
Statistical significance was evaluated using one-tailed unpaired Student t-test. *P<0.05, **P<0.01.
Acknowledgments
This research was supported by a Center of Excellence grant from Flight Attendant Medical Research Institute (FAMRI); Center of Excellence Grant 2084/15 from the Israel Science Foundation; Yad Abraham Center for Cancer Diagnosis and Therapy. This publication reflects the authors' views and not necessarily those of the European Community. VR is the incumbent of the Norman and Helen Asher Professorial Chair Cancer Research at the Weizmann institute.
Glossary
- CBG
corticosteroid-binding globulin
- ChIP
chromatin immunoprecipitation
- Chx
cycloheximide
- CM
conditioned medium
- CON
control
- CYP21A2
cytochrome P450 family 21 subfamily polypeptide 2
- DOX
doxorubicin
- ELISA
enzyme-linked immunosorbent assay
- IP
immunoprecipitation
- KO
knockout
- MDM2
mouse double minute 2 homolog
- NT
non-treated
- PARP
cleaved Poly (ADP-ribose) polymerase
- PBS
phosphate-buffered saline
- RE
responsive element
- SASP
senescence-associated secretory phenotype
- Sh
short hairpin
- SHBG
sex hormone-binding globulin
- TSS
transcription start site
- WT
wild type
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by E Gottlieb
Supplementary Material
References
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 2000; 408: 307–310. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer 2009; 9: 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer 2009; 9: 691–700. [DOI] [PubMed] [Google Scholar]
- Cooks T, Harris CC, Oren M. Caught in the cross fire: p53 in inflammation. Carcinogenesis 2014; 35: 1680–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molchadsky A, Rivlin N, Brosh R, Rotter V, Sarig R. p53 is balancing development, differentiation and de-differentiation to assure cancer prevention. Carcinogenesis 2010; 31: 1501–1508. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Akkari L, Simon J, Grace D, Tschaharganeh DF, Bolden JE et al. Non-cell-autonomous tumor suppression by p53. Cell 2013; 153: 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar J, Moskovits N, Oren M. Involvement of stromal p53 in tumor-stroma interactions. Semin Cell Dev Biol 2010; 21: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C et al. Senescence of activated stellate cells limits liver fibrosis. Cell 2008; 134: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj N, Attardi LD. Tumor suppression: p53 alters immune surveillance to restrain liver cancer. Curr Biol 2013; 23: R527–R530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004; 303: 844–848. [DOI] [PubMed] [Google Scholar]
- Goldstein I, Rotter V. Regulation of lipid metabolism by p53 - fighting two villains with one sword. Trends Endocrinol Metab 2012; 23: 567–575. [DOI] [PubMed] [Google Scholar]
- Goldstein I, Ezra O, Rivlin N, Molchadsky A, Madar S, Goldfinger N et al. p53, a novel regulator of lipid metabolism pathways. J Hepatol 2012; 56: 656–662. [DOI] [PubMed] [Google Scholar]
- Goldstein I, Rivlin N, Shoshana OY, Ezra O, Madar S, Goldfinger N et al. Chemotherapeutic agents induce the expression and activity of their clearing enzyme CYP3A4 by activating p53. Carcinogenesis 2013; 34: 190–198. [DOI] [PubMed] [Google Scholar]
- Goldstein I, Yizhak K, Madar S, Goldfinger N, Ruppin E, Rotter V. p53 promotes the expression of gluconeogenesis-related genes and enhances hepatic glucose production. Cancer Metab 2013; 1: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenstein E, Tillmann HC, Christ M, Feuring M, Wehling M. Multiple actions of steroid hormones—a focus on rapid, nongenomic effects. Pharmacol Rev 2000; 52: 513–556. [PubMed] [Google Scholar]
- Beato M, Klug J. Steroid hormone receptors: an update. Hum Reprod Update 2000; 6: 225–236. [DOI] [PubMed] [Google Scholar]
- Hanukoglu I. Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis. J Steroid Biochem Mol Biol 1992; 43: 779–804. [DOI] [PubMed] [Google Scholar]
- Payne AH, Hales DB. Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr Rev 2004; 25: 947–970. [DOI] [PubMed] [Google Scholar]
- Wedell A. Molecular genetics of 21-hydroxylase deficiency. Endocr Dev 2011; 20: 80–87. [DOI] [PubMed] [Google Scholar]
- Rosner W, Hryb DJ, Khan MS, Singer CJ, Nakhla AM. Are corticosteroid-binding globulin and sex hormone-binding globulin hormones? Ann N Y Acad Sci 1988; 538: 137–145. [DOI] [PubMed] [Google Scholar]
- Lin HY, Muller YA, Hammond GL. Molecular and structural basis of steroid hormone binding and release from corticosteroid-binding globulin. Mol Cell Endocrinol 2010; 316: 3–12. [DOI] [PubMed] [Google Scholar]
- Avvakumov GV, Cherkasov A, Muller YA, Hammond GL. Structural analyses of sex hormone-binding globulin reveal novel ligands and function. Mol Cell Endocrinol 2010; 316: 13–23. [DOI] [PubMed] [Google Scholar]
- Mendel CM. The free hormone hypothesis: a physiologically based mathematical model. Endocr Rev 1989; 10: 232–274. [DOI] [PubMed] [Google Scholar]
- Henley DE, Lightman SL. New insights into corticosteroid-binding globulin and glucocorticoid delivery. Neuroscience 2011; 180: 1–8. [DOI] [PubMed] [Google Scholar]
- Wallace IR, McKinley MC, Bell PM, Hunter SJ. Sex hormone binding globulin and insulin resistance. Clin Endocrinol (Oxf) 2013; 78: 321–329. [DOI] [PubMed] [Google Scholar]
- Bright GM. Corticosteroid-binding globulin influences kinetic parameters of plasma cortisol transport and clearance. J Clin Endocrinol Metab 1995; 80: 770–775. [DOI] [PubMed] [Google Scholar]
- Mickelson KE, Forsthoefel J, Westphal U. Steroid-protein interactions. Human corticosteroid binding globulin: some physicochemical properties and binding specificity. Biochemistry 1981; 20: 6211–6218. [DOI] [PubMed] [Google Scholar]
- Willnow TE, Nykjaer A. Cellular uptake of steroid carrier proteins—mechanisms and implications. Mol Cell Endocrinol 2010; 316: 93–102. [DOI] [PubMed] [Google Scholar]
- Rosner W, Hryb DJ, Kahn SM, Nakhla AM, Romas NA. Interactions of sex hormone-binding globulin with target cells. Mol Cell Endocrinol 2010; 316: 79–85. [DOI] [PubMed] [Google Scholar]
- Hryb DJ, Khan MS, Romas NA, Rosner W. The control of the interaction of sex hormone-binding globulin with its receptor by steroid hormones. J Biol Chem 1990; 265: 6048–6054. [PubMed] [Google Scholar]
- Catalano MG, Frairia R, Boccuzzi G, Fortunati N. Sex hormone-binding globulin antagonizes the anti-apoptotic effect of estradiol in breast cancer cells. Mol Cell Endocrinol 2005; 230: 31–37. [DOI] [PubMed] [Google Scholar]
- Fortunati N, Catalano MG, Boccuzzi G, Frairia R. Sex hormone-binding globulin (SHBG), estradiol and breast cancer. Mol Cell Endocrinol 2010; 316: 86–92. [DOI] [PubMed] [Google Scholar]
- Huang, da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr., Butel JS et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 1992; 356: 215–221. [DOI] [PubMed] [Google Scholar]
- Sullivan PM, Petrusz P, Szpirer C, Joseph DR. Alternative processing of androgen-binding protein RNA transcripts in fetal rat liver. Identification of a transcript formed by trans splicing. J Biol Chem 1991; 266: 143–154. [PubMed] [Google Scholar]
- Janne M, Hogeveen KN, Deol HK, Hammond GL. Expression and regulation of human sex hormone-binding globulin transgenes in mice during development. Endocrinology 1999; 140: 4166–4174. [DOI] [PubMed] [Google Scholar]
- Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer 2009; 9: 724–737. [DOI] [PubMed] [Google Scholar]
- Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol 2: a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga BS, Pronczuk AW, Munro HN. Mechanism of cycloheximide inhibition of protein synthesis in a cell-free system prepared from rat liver. J Biol Chem 1969; 244: 4480–4489. [PubMed] [Google Scholar]
- Hershko DD, Robb BW, Wray CJ, Luo GJ, Hasselgren PO. Superinduction of IL-6 by cycloheximide is associated with mRNA stabilization and sustained activation of p38 map kinase and NF-kappaB in cultured caco-2 cells. J Cell Biochem 2004; 91: 951–961. [DOI] [PubMed] [Google Scholar]
- Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci USA 2002; 99: 8467–8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer 2002; 2: 594–604. [DOI] [PubMed] [Google Scholar]
- Scrable H, Burns-Cusato M, Medrano S. Anxiety and the aging brain: stressed out over p53? Biochim Biophys Acta 2009; 1790: 1587–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C, Qian Y, Chen X. The p53-estrogen receptor loop in cancer. Curr Mol Med 2013; 13: 1229–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Wasylyk B. Physiological and pathological consequences of the interactions of the p53 tumor suppressor with the glucocorticoid, androgen, and estrogen receptors. Ann N Y Acad Sci 2004; 1024: 54–71. [DOI] [PubMed] [Google Scholar]
- Goncalves J, Friaes A, Moura L. Congenital adrenal hyperplasia: focus on the molecular basis of 21-hydroxylase deficiency. Expert Rev Mol Med 2007; 9: 1–23. [DOI] [PubMed] [Google Scholar]
- Malkin D, Li FP, Strong LC, Fraumeni JF Jr., Nelson CE, Kim DH et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 1990; 250: 1233–1238. [DOI] [PubMed] [Google Scholar]
- Mazzuco TL, Durand J, Chapman A, Crespigio J, Bourdeau I. Genetic aspects of adrenocortical tumours and hyperplasias. Clin Endocrinol (Oxf) 2012; 77: 1–10. [DOI] [PubMed] [Google Scholar]
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009; 9: 701–713. [DOI] [PubMed] [Google Scholar]
- Orbach O, Schussler GC. Increased serum cortisol binding in chronic active hepatitis. Am J Med 1989; 86: 39–42. [DOI] [PubMed] [Google Scholar]
- Kim HM, Han SB, Hyun BH, Ahn CJ, Cha YN, Jeong KS et al. Functional human hepatocytes: isolation from small liver biopsy samples and primary cultivation with liver-specific functions. J Toxicol Sci 1995; 20: 565–578. [DOI] [PubMed] [Google Scholar]
- Meivar-Levy I, Sapir T, Gefen-Halevi S, Aviv V, Barshack I, Onaca N et al. Pancreatic and duodenal homeobox gene 1 induces hepatic dedifferentiation by suppressing the expression of CCAAT/enhancer-binding protein beta. Hepatology 2007; 46: 898–905. [DOI] [PubMed] [Google Scholar]
- George TC, Fanning SL, Fitzgerald-Bocarsly P, Medeiros RB, Highfill S, Shimizu Y et al. Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. J Immunol Methods 2006; 311: 117–129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.