Abstract
Objectives
Recent studies have found that cases with oropharyngeal squamous cell carcinoma (OPSCC) positive for HPV16 genotype have better overall survival compared with cases positive for other HPV genotypes. We sought to further replicate these studies and determine if this relationship is modified by expression of p16 tumor suppressor protein.
Material and Methods
We identified 238 OPSCC cases from the Carolina Head and Neck Cancer Study (CHANCE) study, a population based case-control study. Tumors that tested positive solely for HPV16 genotype and no other genotypes with PCR were classified as HPV16-positive. Tumors positive for any other high-risk HPV genotype were classified as non-HPV16-positive. Expression of p16 in the tumor was determined with immunohistochemistry. Follow-up time was calculated from the date of diagnosis to date of death or December 31, 2013. Overall survival was compared with the Kaplan-Meier curves and log-rank test. Hazard ratios (HR) adjusted for smoking, alcohol use, sex, race, and age was calculated with the Cox proportional hazard regression.
Results
Cases with HPV16-positive OPSCC had better overall survival than cases with non-HPV16-positive OPSCC (log-rank p-value: 0.010). When restricted to OPSCC cases positive for p16 expression, the same trend continued (log-rank p-value: 0.002). In the adjusted model, cases with non-HPV16-positive OPSCC had greater risk of death compared to cases with HPV16-positive tumors (HR: 1.92; 95% CI: 1.03, 3.60).
Conclusions
This finding indicates that HPV genotyping carries valuable prognostic significance in addition to p16 status and future survival studies of OPSCC should take into account differing HPV genotypes.
Keywords: Head and Neck Cancer, Oropharyngeal Cancer, HPV, HPV16, p16
Introduction
Over the last twenty years, the epidemiology of head and neck squamous cell carcinoma (HNSCC) has dramatically changed as a result of human papillomavirus (HPV). While head and neck cancers were traditionally associated with tobacco and alcohol use, approximately 60–70% of oropharyngeal squamous cell carcinoma (OPSCC) cases are now driven by HPV [1, 2]. Additionally, HPV is an independent determinant of recurrence and survival in OPSCC, in which HPV-positive cancer cases have better outcomes than HPV-negative cancer cases [1, 3, 4].
HPV16 is the most common genotype associated with OPSCC [5, 6]. Other HPV genotypes such as HPV33, HPV18 and HPV35, have been associated with OPSCC but their relation with clinical outcomes are less well known [5]. Previous studies have suggested patients with HNSCC positive for HPV16 genotype have better survival compared with patients with tumors that are positive with other HPV genotypes [6–9].
Immunohistochemical expression of p16, a tumor suppressor protein, is a widely accepted biomarker for assessing HPV status and is considered the standard for treatment stratification in clinical trials [10]. Following mucosal infection, two HPV-encoded oncoproteins, E6 and E7, inactivate tumor suppressor genes TP53 and pRb, respectively. This induces downstream overexpression of the p16 protein [11]. Polymerase chain reaction (PCR) can also be used for HPV identification in tumor cells. PCR is more sensitive than p16 immunohistochemistry and allows for identification of specific HPV genotypes, thus adding another dimension to the assessment of HPV status [12].
We sought to assess if survival differs by HPV genotype determined by PCR in the cases from a large population-based study, the Carolina Head and Neck Cancer Study (CHANCE). We hypothesize OPSCC cases positive for genotypes other than HPV16 will have worse survival than those with HPV16-positive tumors and this trend will hold among OPSCC positive for p16 expression.
Material and Methods
Study Population
Subjects for this longitudinal survival analysis were cases from CHANCE, the population-based case-control study. Cases were diagnosed with first primary squamous cell carcinoma of the oral cavity, pharynx, and larynx from January 1, 2002 to February 28, 2006, aged 20 to 80 years at diagnosis, and resided in a 46-county region in central North Carolina. Benign tumors, carcinomas in situ, papillary thyroid carcinomas, and adenoid carcinomas were excluded. All available cases of oropharyngeal tumors and a sample of non-oropharyngeal tumors were selected for p16 immunohistochemistry and HPV detection by PCR (N = 492). Cases with lip and hypopharynx cancers, cases for whom the hospital would not release tumor blocks, and cases in which interviews were completed by a proxy were also excluded from p16 immunohistochemistry and HPV typing. However, since HPV has only been consistently associated with oropharyngeal cancer, all other sites were excluded (N = 254), resulting 238 OPSCC cases for analysis. This research was approved by the institutional review board at the University of North Carolina at Chapel Hill.
Outcome assessment
CHANCE data were linked to the National Death Index based on name, social security number, date of birth, sex, race, and state of residence to identify deaths through December 31, 2013. The National Death Index is a national file of identified death record information, including cause of death compiled from computer files submitted by State Vital Statistics offices. Greater than 75% of the cases in CHANCE were perfect or near-perfect matches on social security number, date of birth, and sex. The remaining near-matches were confirmed by examining the United States Social Security Death Index and obituaries on newspaper websites.
Questionnaire and Clinical Assessment
Demographic and risk factor information were collected using a structured questionnaire during an in-home visit conducted by trained nurse-interviewers. Demographic variables of interest include: age, race, sex, smoking (<10 pack-years and ≥10 pack-years) and alcohol (<1 drink/week and ≥1 drinks/week).
Clinical information such as TNM classification and cancer treatments were abstracted from the subjects’ medical records and reviewed independently by a head and neck surgeon and a pathologist. Only oropharynx cancers (C01.9, C02.4, C05.1, C05.2, C09.0, C09.1, C09.8, C09.9, C10.0-C10.4, C10.8, and C10.9) were included in this analysis.
HPV and p16 Typing
The International Agency for Research on Cancer performed HPV typing and centralized pathology review plus evaluation of p16 expression by immunohistochemistry. HPV infection was determined using a HPV-type-specific E7 PCR bead-based multiplex genotyping assay (E7-MPG).[13] We included the high-risk genotypes (HPV16, HPV18, HPV26, HPV31, HPV33, HPV35, HPV39, HPV45, HPV51, HPV52, HPV53, HPV56, HPV58, HPV59, HPV66, HPV68, HPV70, HPV73 and HPV82) as well as two low-risk genotypes (for the following genotypes: HPV6 and HPV11) for PCR. HPV genotype was classified into two categories: HPV16-positive and non-HPV16 positive. Tumors were classified as HPV16-positive if they were positive for HPV16 genotype only. Tumors positive for any high-risk genotype were classified as non-HPV16-positive. Immunocytochemistry for p16 was performed according to the protocol provided with the CINtec Histology p16INK4a Kit (9511, mtmlabs) for the detection of the p16 antigen on slides prepared from formalin-fixed, paraffin-embedded resections. The expression was scored through evaluation and semi-quantitative scoring of two variables: the percentage of stained cells (0% = 0, 1–10% = 1, 11–50% = 2, 51–80% = 3, 81–100% = 4) and the intensity of the nucleic or cytoplasmic staining (none = 0, weak = 1, moderate = 2, strong = 3). The two scores were then multiplied to yield the composite score, ranging from 0–12. Composite scores of 4 or greater were considered positive for p16 expression and scores of 0 to 3 were considered negative for p16 expression.
Statistical Analysis
We calculated overall survival as time from diagnosis to either date of death due to any cause or censoring on December 31, 2013, whichever came first. Differences in covariate distributions by HPV or p16 status were tested by two-sided Pearson chi-square tests or by Fisher exact tests when cells are sparse. Kaplan-Meier overall survival plots were constructed and log-rank p-values were calculated. Hazard ratios (HRs) for the independent effects HPV genotype with overall survival among all OPSCC cases were estimated by Cox proportional hazards regression adjusted for sex, age, race, smoking, and alcohol. These potential confounders were selected with a causal diagram or due to its relationship with head and neck cancer. We estimated HRs with a Cox proportional hazard regression that adjusted for treatment and N and T classification in addition to the previous variables. The other race classification and cases that are M1 were excluded from this multivariable analysis due to small numbers. The proportional hazards assumption for HPV genotype was tested and satisfied. Alpha of 0.05 was used for all statistical testing and confidence interval (CI) calculations. All statistical analyses were implemented using SAS 9.4 (SAS Institute, Cary, NC).
Results
The demographic and clinical variables were not significantly different by HPV genotype in OPSCC (Table 1). However, several key clinical variables varied by HPV genotype. HPV16-positive cases were more likely to have smoked more than 10 pack-years (61.9%) compared with non-HPV16-positive case (42.9%). HPV16-positive OPSCCs were also more likely to have p16 expression than non-HPV16-positive OPSCCs (p-value: 0.177).
Table 1.
Descriptive statistics for OPSCC cases by HPV status. P-value comparing HPV16-positive with non-HPV16-positive
| HPV-negative N = 66 |
HPV16-positive N = 151 |
Non-HPV16-positive N = 21 |
|||
|---|---|---|---|---|---|
| N (%) | N (%) | N (%) | p-value | ||
| p16 | |||||
| Negative | 59 (89.4) | 20 (12.5) | 5 (23.8) | 0.177 | |
| Positive | 7 (10.6) | 140 (87.5) | 16 (76.2) | ||
| Missing | -- | 1 | -- | ||
| Smoking | |||||
| < 10 pack-years | 6 (9.1) | 61 (38.1) | 12 (57.1) | 0.095 | |
| ≥ 10 pack-years | 60 (90.9) | 99 (61.9) | 9 (42.9) | ||
| Missing | -- | 1 | -- | ||
| Alcohol Consumption | |||||
| < 1 drink/week | 26 (16.4) | 4 (19.0) | 0.757 | ||
| ≥ 1 drink/week | 64 (100.0) | 133 (83.6) | 17 (81.0) | ||
| Missing | 2 | 2 | -- | ||
| Race | |||||
| White | 24 (36.4) | 140 (87.0) | 18 (85.7) | 0.719 | |
| Black | 40 (60.6) | 16 (9.9) | 3 (14.3) | ||
| Other | 2 (3.0) | 5 (3.1) | 0 | ||
| Age (years) | |||||
| ≤50 | 20 (30.3) | 54 (33.5) | 6 (28.6) | 0.109 | |
| 51 – 65 | 35 (53.0) | 88 (54.7) | 9 (42.9) | ||
| 66≤ | 11 (16.7) | 19 (11.8) | 6 (28.6) | ||
| Sex | |||||
| Male | 52 (78.8) | 136 (84.5) | 16 (76.2) | 0.351 | |
| Female | 14 (21.2) | 25 (15.5) | 5 (23.8) | ||
| Subsite | |||||
| Base of Tongue | 23 (34.8) | 42 (26.1) | 8 (38.1) | 0.085 | |
| Tonsil | 33 (50.0) | 106 (65.8) | 9 (42.9) | ||
| Other | 10 (15.2) | 13 (8.1) | 4 (19.0) | ||
| T Stage | |||||
| T1 | 10 (15.2) | 47 (29.2) | 6 (28.6) | 0.423 | |
| T2 | 26 (39.4) | 70 (43.5) | 7 (33.3) | ||
| T3 | 15 (22.7) | 23 (14.3) | 6 (28.6) | ||
| T4 | 15 (22.7) | 21 (13.0) | 2 (9.5) | ||
| N Stage | |||||
| N0 | 23 (34.8) | 26 (16.1) | 6 (28.6) | 0.316 | |
| N1 | 9 (13.6) | 27 (16.8) | 3 (14.3) | ||
| N2 | 27 (40.9) | 94 (58.4) | 9 (42.9) | ||
| N3 | 7 (10.6) | 14 (8.7) | 3 (14.3) | ||
| Radiation | |||||
| No | 8(12.1%) | 7(4.3%) | 3(14.3%) | 0.093 | |
| Yes | 58(87.9%) | 154(95.7%) | 18(85.7%) | ||
| Chemotherapy | |||||
| No | 27(40.9%) | 56(34.8%) | 7(33.3%) | 0.896 | |
| Yes | 39(59.1%) | 105(65.2%) | 14(66.7%) | ||
| Surgery | |||||
| No | 42(63.6%) | 66(41.0%) | 9(42.9%) | 0.870 | |
| Yes | 24(36.4%) | 95(59.0%) | 12(57.1%) | ||
p-value comparing HPV16-positive and non-HPV16-positive
Table 2 displays the descriptive results for HPV genotype by p16 expression in OPSCC. As expected, the OPSCCs were mostly HPV-positive (73.4%). About 12.5% of HPV-positive oropharynx tumors were non-HPV16-positive. Most of the oropharynx tumors that are HPV16-positive also have p16 expression (88.3%). However, about 30% of the OPSCC negative for p16 expression were positive for HPV with PCR.
Table 2.
Distribution of HPV genotype in OPSCC
| p16-positive N = 163 |
p16-negative N = 84 |
||
|---|---|---|---|
| N (%) | N (%) | ||
| HPV-positive | |||
| 6 | 0 | 1 (1.2) | |
| 11 | 0 | 0 | |
| 16 | 144 (88.3) | 22 (26.2) | |
| 18 | 4 (2.5) | 0 | |
| 26 | 1 (0.6) | 0 | |
| 31 | 0 | 0 | |
| 33 | 4 (2.5) | 0 | |
| 35 | 5 (3.1) | 3 (3.6) | |
| 39 | 0 | 1 (1.2) | |
| 58 | 1 (0.6) | 0 | |
| 59 | 2 (1.2) | 0 | |
| 82 | 0 | 0 | |
| HPV-negative | 7 (4.3) | 59 (70.2) | |
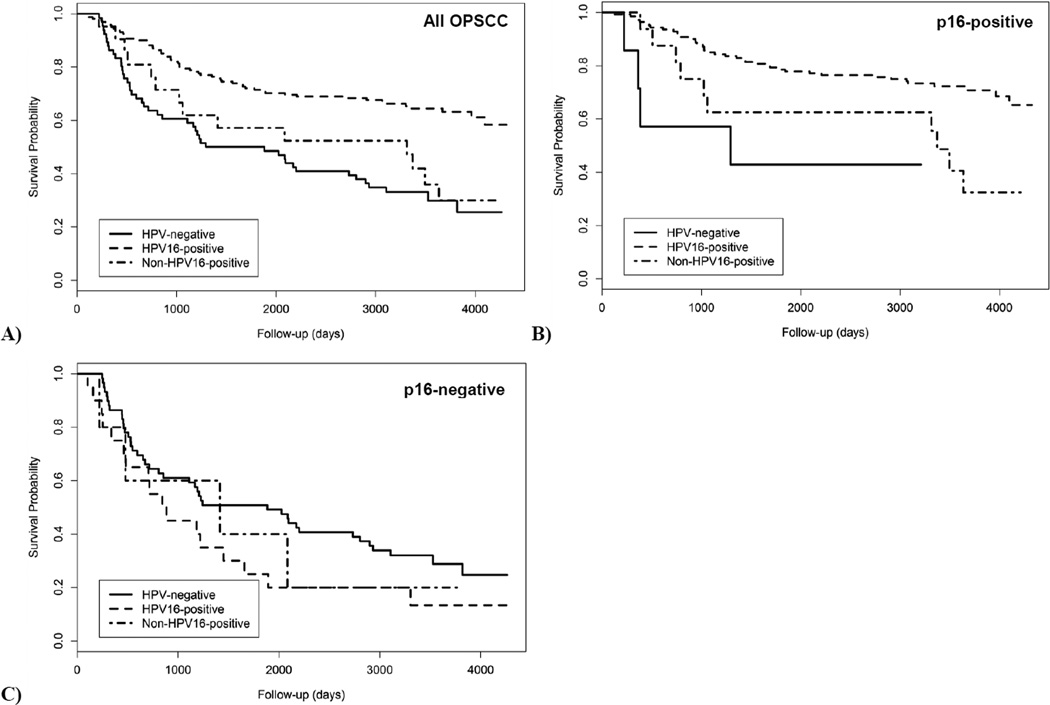
As seen in the Kaplan-Meier curve (Figure 1), cases with HPV16-positive OPSCC had better overall survival than cases with non-HPV16-positive OPSCCs (log-rank p-value: 0.010). Five-year survival for cases with HPV16-positive OPSCCs was 71.4% while cases with HPV-positive-other OPSCCs (57.1%) and HPV-negative OPSCCs (50.0%) had similar 5-year survival. When restricted to OPSCCs with positive expression of p16, cases with non-HPV16-positive OPSCCs had worse survival (62.5%) than cases with HPV16-positive OPSCCs (78.6%; Log-rank p-value: 0.002). Among cases with p16-negative OPSCCs, survival did not differ by HPV genotype (log-rank p-value: 0.751).
Figure 1.
Kaplan-Meier curve for HPV16-positive, non-HPV16-positive and HPV-negative OPSCC for A) all OPSCC B) OPSCC positive for p16 expression and C) OPSCC negative for p16 expression
In the multivariable Cox regression analysis (Table 3) adjusting age, race, sex, smoking and alcohol, both cases with non-HPV16-positive OPSCCs (Hazard ratio (HR): 1.92; 95% CI: 1.03, 3.60). When including T classification, N classification and treatment in the adjustment, there is little difference in the HR (Table 3).
Table 3.
Mutually adjusted hazard ratios from the Cox proportional hazard regression
| With treatment adjustment | Without treatment adjustment | ||||
|---|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | ||
| HPV Status | |||||
| HPV16-positive | Ref | Ref | |||
| Non-HPV16-positive | 1.75(0.91, 3.37) | 0.096 | 1.92(1.03, 3.60) | 0.041 | |
| HPV-negative | 1.44(0.85, 2.46) | 0.177 | 1.38(0.81, 2.34) | 0.236 | |
| Sex | |||||
| Male | Ref | Ref | |||
| Female | 0.80(0.47, 1.37) | 0.413 | 0.78(0.45, 1.35) | 0.376 | |
| Race | |||||
| White | Ref | Ref | |||
| Black | 1.65(1.00, 2.72) | 0.051 | 1.87(1.13, 3.09) | 0.015 | |
| Smoking | |||||
| <10 Pack-years | Ref | Ref | |||
| ≥10 Pack-years | 1.75(1.02, 3.02) | 0.043 | 1.82(1.07, 3.10) | 0.028 | |
| Alcohol | |||||
| <1 drink/week | Ref | Ref | |||
| ≥1 drink/week | 1.04(0.49, 2.22) | 0.911 | 1.11(0.52, 2.38) | 0.787 | |
| Age | 1.02(1.00, 1.04) | 0.114 | 1.02(1.00, 1.04) | 0.100 | |
| T Classification | |||||
| T1–T2 | Ref | ||||
| T3–T4 | 1.85(1.24, 2.75) | 0.002 | |||
| N Classification | |||||
| N0 | Ref | ||||
| N+ | 1.08(0.64, 1.83) | 0.765 | |||
| Treatment | |||||
| Single modality | Ref | ||||
| Multiple modality | 1.00(0.53, 1.90) | 0.99 | |||
Although not significant, among OPSCC that tested positive for p16 expression, cases with non-HPV16-positive (n = 16) tumors had greater risk of death compared with HPV16-positve (n = 135) tumors (HR: 1.82; 95% CI: 0.82, 4.02). The HR remains similar after adjustment for T classification, N classification and treatment. Among OPSCC negative for p16 expression, non-HPV16-positive OPSCC may have a slightly increased risk of death compared with HPV16-positive (n = 20) OPSCC (HR: 1.40; 95% CI: 0.39, 4.99). However, when including T classification, N classification and treatment into the model, the HRs are attenuated (HR: 1.28; 95% CI: 0.33, 5.05).
Discussion
The purpose of this study is to determine if HPV genotype is associated with differences in survival for patients with OPSCC and if this trend holds for oropharyngeal tumors positive for p16 expression. Our results replicated previous findings that cases with HPV16 genotype have improved survival, even among cases with OPSCC positive for p16 expression. However, among OPSCC negative for p16 expression HPV genotype may not be associated with overall survival. This survival benefit for the HPV16-genotype holds true in the Cox proportional hazard regression adjusting for stage at presentation and treatment as well as other confounders. These results suggest that the survival benefit of HPV-positivity may be restricted with HPV16 genotype only and HPV genotype may provide valuable prognostic information and help guide optimal treatment.
Increased survival for HPV-positive OPSCC, regardless of HPV genotype, is well documented. However, few studies have examined survival trends in cases stratified by HPV genotype [7–9]. Bratman et al. demonstrated a significant survival benefit for HPV16-positive cases (N = 61) compared with non-HPV16-positive (N = 12) and HPV-negative (N = 442) among all HNSCC (log-rank: <0.001) with cases from The Cancer Genome Atlas [7]. The authors concluded that validation of these results specifically in OPSCC patients is warranted to inform treatment decisions. Although we also see a significant association by HPV genotype across all sites (results not shown), we concentrated on OPSCC since the prognostic value of HPV status in non-oropharynx tumors has yet to be established [14, 15]. Two other groups in the United States conducted studies comparing HPV genotypes in OPSCC. A study by Varier et al. found HPV16-positive OPSCC from the Icahn School of Medicine at Mount Sinai had non-significantly better survival than patients with non-HPV16-positive OPSCC (n=27) [9]. Another study from Centers for Disease Control Cancer Registry Sentinel Surveillance, a cancer registry-based residual tissue, also found decreased survival for cases with non-HPV16-positive OPSCC compared to HPV16-positive OPSCC even after adjustment for covariates [6]. However these studies did not examine HPV genotype among cases that were p16-positive, the most clinically used indicator of HPV driven disease due to its downstream position in the HPV tumorigenesis pathway.
The exact mechanisms underlying the decreased survival of cases with non-HPV16-positive OPSSC have yet to be fully elucidated. A recent study found the HPV18 genotype induced colony growth primary human keratinocytes more effectively than HPV16 or HPV31 [16]. This could be due to decreased apoptosis and thus increased carcinogenicity of HPV18. Rat fibrosarcomas transfected with HPV18 had significantly less apoptosis than fibroscaromas transfected with HPV16 [17]. Similarly, the HPV18 LCR-E6-E7 region was approximately 10- to 50-fold more active than that of HPV16 [18].
Examining the association between HPV genotype and survival is an important step in the development of individualized oncologic treatment plans for patients with OPSCC. Recently, several clinical trials have been developed to enroll HPV-positive OPSCC patients in trials with de-intensified therapy [19]. Improved survival and oncologic outcomes with HPV-positive OPSCCs are the principal factors underlying the development of these trials. Current treatment regimens used in clinical practice are associated with significant toxicities and treatment related morbidity [20]. Further, since patients with HPV-positive tumors have improved oncologic outcomes and are living longer after diagnosis and treatment, patients must live with these adverse treatment-related effects for longer periods of time. Thus, identifying HPV-positive low-risk patients who are candidates for trials with de-intensified treatment in which toxicity is minimized while clinical outcomes are maximized has important clinical implications.
Currently, many of the clinical trials that de-intensify treatment for patients with HPV-positive tumors use p16 expression alone to stratify patients [19]. Previous studies have suggested that p16 immunohistochemistry is the best method for patient risk-stratification since it is widely available [21, 22]. However, based upon our results, about 12% of cases have non-HPV16-positive and HPV-negative tumors. Treatment de-intensification based on p16 immunohistochemistry alone could adversely affect this small proportion of patients who have worse outcomes and may not receive appropriate treatment consistent with standard of care if included in trials with de-intensified treatment. Our study further emphasizes a differential survival based upon HPV genotype and suggests a clinical and biological difference by HPV genotype may exist. Additional studies should be conducted to determine if this discrepancy is due to other characteristics associated with HPV status or possible false positives due to the high sensitivity of PCR and further clarify if this association is demonstrated in populations with larger sample sizes of OPSCC p16-positive patients.
Our study has some limitations and strengths. First, similar to previous studies, there are relatively few cases of OPSCC that are non-HPV16-positive. Due to this limitation, we are unable to further explore the relationship between other HPV genotypes specifically and survival and their relationship with p16 expression. It is also possible that the non-HPV16 genotypes are false-positives. However, we are the first study to have both HPV genotype and p16 from a large population-based cohort of cases and the results were similar when restricted to p16-positive OPSCC. Additionally, because this study was conducted prior to HPV becoming widely accepted as a risk-factor for OPSCC, this HPV status should not affect treatment of these cases. All cases were recruited from North Carolina, which would reduce the variability in the treatment and thus impacting survival. Lastly, we are only able to study overall survival as a proxy for disease-specific survival. However, since this case-control study was linked to the National Death Index, we have reasonably good measure of the outcome for all the cases from 7 to 12 years after diagnosis.
In conclusion, we have demonstrated that among OPSCC positive for p16 expression, HPV16-positive tumors have greater survival compared with OPSCC that are non-HPV16-postive. This finding indicates that HPV genotyping carries valuable prognostic significance and is especially noteworthy given the recent clinical trials for de-intensification of treatment for HPV-positive OPSCC patients based upon p16 expression. Because both non-HPV16-positive and HPV-negative cases positive for p16 expression have worse survival, even with standard of care treatment, treatment de-intensification may not represent a viable option for these subsets of cases with p16 expression. Overall, due to these differences in survival, future studies should distinguish between HPV16-positive cancers and non-HPV16-positive cancers when evaluating treatment efficacy for OPSCC positive for p16 expression. However, due to our small sample size, additional studies should be conducted to replicate these results.
Highlights.
First to study the prognostic significance of non-HPV16 genotypes with p16 status.
HPV16-positve oropharynx tumors have better survival than non-HPV16-positive genotypes
The survival benefit for HPV16-positve genotypes holds for tumors positive for p16, but not for p16-negative tumors
Acknowledgments
This study was supported in part by the National Cancer Institute (R01-CA90731). HPV testing was partly supported by the Health General Directorate of the French Social Affairs and Health Ministry.
Abbreviations
- OPSCC
oropharyngeal squamous cell carcinoma
- HNSCC
Head and neck squamous cell carcinoma
- HPV
Human Papillomavirus
- CHANCE
Carolina Head and Neck Cancer Study
- HR
Hazard Ratio
- CI
Confidence Interval
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
There are no conflicts of interest to disclose.
References
- 1.Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaturvedi AK. Beyond cervical cancer: burden of other HPV-related cancers among men and women. J Adolesc Health. 2010;46:S20–S26. doi: 10.1016/j.jadohealth.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 3.Argiris A, Li S, Ghebremichael M, Egloff AM, Wang L, Forastiere AA, et al. Prognostic significance of human papillomavirus in recurrent or metastatic head and neck cancer: an analysis of Eastern Cooperative Oncology Group trials. Ann Oncol. 2014;25:1410–1416. doi: 10.1093/annonc/mdu167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fakhry C, Zhang Q, Nguyen-Tan PF, Rosenthal D, El-Naggar A, Garden AS, et al. Human papillomavirus and overall survival after progression of oropharyngeal squamous cell carcinoma. J Clin Oncol. 2014;32:3365–3373. doi: 10.1200/JCO.2014.55.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castellsague X, Alemany L, Quer M, Halec G, Quiros B, Tous S, et al. HPV Involvement in Head and Neck Cancers: Comprehensive Assessment of Biomarkers in 3680 Patients. J Natl Cancer Inst. 2016;108 doi: 10.1093/jnci/djv403. [DOI] [PubMed] [Google Scholar]
- 6.Goodman MT, Saraiya M, Thompson TD, Steinau M, Hernandez BY, Lynch CF, et al. Human papillomavirus genotype and oropharynx cancer survival in the United States of America. Eur J Cancer. 2015;51:2759–2767. doi: 10.1016/j.ejca.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bratman SV, Bruce JP, O'Sullivan B, Pugh TJ, Xu W, Yip KW, et al. Human Papillomavirus Genotype Association With Survival in Head and Neck Squamous Cell Carcinoma. JAMA oncology. 2016 doi: 10.1001/jamaoncol.2015.6587. [DOI] [PubMed] [Google Scholar]
- 8.No JH, Sung MW, Hah JH, Choi SH, Lee MC, Kim HS, et al. Prevalence and prognostic value of human papillomavirus genotypes in tonsillar squamous cell carcinoma: a Korean multicenter study. Cancer. 2015;121:535–544. doi: 10.1002/cncr.29086. [DOI] [PubMed] [Google Scholar]
- 9.Varier I, Keeley BR, Krupar R, Patsias A, Dong J, Gupta N, et al. Clinical characteristics and outcomes of oropharyngeal carcinoma related to high-risk non-human papillomavirus16 viral subtypes. Head Neck. 2016 doi: 10.1002/hed.24442. [DOI] [PubMed] [Google Scholar]
- 10.Schache A, Croud J, Robinson M, Thavaraj S. Human papillomavirus testing in head and neck squamous cell carcinoma: best practice for diagnosis. Methods Mol Biol. 2014;1180:237–255. doi: 10.1007/978-1-4939-1050-2_13. [DOI] [PubMed] [Google Scholar]
- 11.Fotopoulos G, Pavlidis N. The role of human papilloma virus and p16 in occult primary of the head and neck: A comprehensive review of the literature. Oral Oncol. 2015;51:119–123. doi: 10.1016/j.oraloncology.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 12.Pannone G, Rodolico V, Santoro A, Lo Muzio L, Franco R, Botti G, et al. Evaluation of a combined triple method to detect causative HPV in oral and oropharyngeal squamous cell carcinomas: p16 Immunohistochemistry, Consensus PCR HPV-DNA, and In Situ Hybridization. Infect Agent Cancer. 2012;7:4. doi: 10.1186/1750-9378-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitt M, Dondog B, Waterboer T, Pawlita M, Tommasino M, Gheit T. Abundance of multiple high-risk human papillomavirus (HPV) infections found in cervical cells analyzed by use of an ultrasensitive HPV genotyping assay. J Clin Microbiol. 2010;48:143–149. doi: 10.1128/JCM.00991-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zafereo ME, Xu L, Dahlstrom KR, Viamonte CA, El-Naggar AK, Wei Q, et al. Squamous cell carcinoma of the oral cavity often overexpresses p16 but is rarely driven by human papillomavirus. Oral Oncol. 2016;56:47–53. doi: 10.1016/j.oraloncology.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young RJ, Urban D, Angel C, Corry J, Lyons B, Vallance N, et al. Frequency and prognostic significance of p16(INK4A) protein overexpression and transcriptionally active human papillomavirus infection in laryngeal squamous cell carcinoma. Br J Cancer. 2015;112:1098–1104. doi: 10.1038/bjc.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lace MJ, Anson JR, Klingelhutz AJ, Lee JH, Bossler AD, Haugen TH, et al. Human papillomavirus (HPV) type 18 induces extended growth in primary human cervical, tonsillar, or foreskin keratinocytes more effectively than other high-risk mucosal HPVs. J Virol. 2009;83:11784–11794. doi: 10.1128/JVI.01370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arends MJ, Wyllie AH, Bird CC. Human papillomavirus type 18 is associated with less apoptosis in fibroblast tumours than human papillomavirus type 16. Br J Cancer. 1995;72:646–649. doi: 10.1038/bjc.1995.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villa LL, Schlegel R. Differences in transformation activity between HPV-18 and HPV-16 map to the viral LCR-E6-E7 region. Virology. 1991;181:374–377. doi: 10.1016/0042-6822(91)90507-8. [DOI] [PubMed] [Google Scholar]
- 19.Psyrri A, Rampias T, Vermorken JB. The current and future impact of human papillomavirus on treatment of squamous cell carcinoma of the head and neck. Ann Oncol. 2014;25:2101–2115. doi: 10.1093/annonc/mdu265. [DOI] [PubMed] [Google Scholar]
- 20.Chan KK, Glenny AM, Weldon JC, Furness S, Worthington HV, Wakeford H. Interventions for the treatment of oral and oropharyngeal cancers: targeted therapy and immunotherapy. The Cochrane database of systematic reviews. 2015;12:Cd010341. doi: 10.1002/14651858.CD010341.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis JS, Jr, Thorstad WL, Chernock RD, Haughey BH, Yip JH, Zhang Q, et al. p16 positive oropharyngeal squamous cell carcinoma:an entity with a favorable prognosis regardless of tumor HPV status. Am J Surg Pathol. 2010;34:1088–1096. doi: 10.1097/PAS.0b013e3181e84652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oguejiofor KK, Hall JS, Mani N, Douglas C, Slevin NJ, Homer J, et al. The prognostic significance of the biomarker p16 in oropharyngeal squamous cell carcinoma. Clin Oncol (R Coll Radiol) 2013;25:630–638. doi: 10.1016/j.clon.2013.07.003. [DOI] [PubMed] [Google Scholar]