Abstract
To design effective eradication strategies, it may be necessary to target HIV reservoirs in anatomic compartments other than blood. This study examined HIV RNA rebound following interruption of antiretroviral therapy (ART) in blood and cerebrospinal fluid (CSF) to determine whether the central nervous system (CNS) might serve as an independent source of resurgent viral replication.
Paired blood and CSF samples were collected longitudinally from 14 chronically HIV-infected individuals undergoing ART interruption. HIV env (C2-V3), gag (p24) and pol (reverse transcriptase) were sequenced from cell-free HIV RNA and cell-associated HIV DNA in blood and CSF using the Roche 454 FLX Titanium platform. Comprehensive sequence and phylogenetic analyses were performed to search for evidence of unique or differentially represented viral subpopulations emerging in CSF supernatant as compared with blood plasma.
Using a conservative definition of compartmentalization based on four distinct statistical tests, nine participants presented a compartmentalized HIV RNA rebound within the CSF after interruption of ART, even when sampled within 2 weeks from viral rebound. The degree and duration of viral compartmentalization varied considerably between subjects and between time-points within a subject. In 10 cases, we identified viral populations within the CSF supernatant at the first sampled time-point after ART interruption, which were phylogenetically distinct from those present in the paired blood plasma and mostly persisted over time (when longitudinal time-points were available). Our data suggest that an independent source of HIV RNA contributes to viral rebound within the CSF after treatment interruption. The most likely source of compartmentalized HIV RNA is a CNS reservoir that would need to be targeted to achieve complete HIV eradication.
Keywords: central nervous system, HIV reservoir, ART interruption, viral rebound.
Introduction
Antiretroviral therapy (ART) suppresses viral replication to undetectable levels in most HIV-infected individuals (Palella et al. 2006). However, ART cannot eradicate latently infected cells (Wong et al. 1997; Finzi et al. 1999; Siliciano et al. 2003), and robust viral replication resumes following treatment interruption (Hoen et al. 2005). Discouragingly, this pattern holds in 80–90% of cases even when ART is initiated during the earliest phase of HIV infection (2–10 weeks), specifically to limit the size of the HIV reservoir and improve immune reconstitution (Hocqueloux et al. 2010; Saez-Cirion et al. 2013). Among individuals who initiate ART during chronic HIV infection (i.e. at least 3 months post infection) viral rebound is virtually universal (Palmisano et al. 2007; Saez-Cirion et al. 2013).
Rebounding virus might originate from a variety of sources: the DNA compartment in blood, namely peripheral blood mononuclear cells (PBMC), RNA or DNA compartments in lymphoid tissues, and possibly other anatomical compartments that harbor replication-competent HIV-infected cells or viral particles (Lewin et al. 2011; Gray et al. 2014; Margolis, 2014).
In most HIV-infected individuals, the establishment of viral reservoirs in multiple tissues and anatomic compartments, including the central nervous system (CNS), occurs within the first few weeks of HIV infection (Clements et al. 2005; Thompson et al. 2011; Gray et al. 2014; Salemi and Rife, 2015; Sturdevant et al. 2015). The HIV population during early HIV infection is typically homogenous (Keele et al. 2008), but compartmentalization can occur as a consequence of tissue-specific genetic differentiation and restricted viral migration between anatomic sites or tissues (Zarate et al. 2007; Blackard, 2012; Svicher et al. 2014). Compartmentalized viral evolution is frequently a consequence of discordant selective pressures (Pillai et al. 2006), and gives rise to tissue-adapted variants, which subsequently contribute to disease pathogenesis (Strain et al. 2005; Pillai et al. 2006). Phylogenetic methods needed to quantify this restriction of gene flow between compartments are numerous and well developed (Zarate et al. 2007). Our group and others have used similar methods to describe the presence of compartmentalized HIV RNA populations in the cerebrospinal fluid (CSF) supernatant, mostly using samples from ART naïve individuals (Harrington et al. 2009; Smith et al. 2009; Choi et al. 2012).
While direct assessment of viral variants sampled from brain parenchyma in living subjects is rarely feasible, HIV RNA collected from the CSF can be an informative surrogate. Interestingly, viral rebound in CSF supernatant almost always follows the rebound in blood plasma after ART interruption (Monteiro de Almeida et al. 2005). This lag might indicate that rebounding virus in CSF is simply imported from blood, in which case, the sequences from the two compartments should be genetically indistinguishable early after viral rebound. In contrast, if sequences of the rebounding CSF viral population are demonstrably distinct from blood, they may be derived from separate CNS sources. Additionally, the degree of compartmentalization is expected to vary over time and can be influenced by pleocytosis and other factors (Smith et al. 2009), which are proxies for the extent of cellular trafficking between CNS and blood.
In this study, we quantified the reservoir size and characterized the complex dynamics of viral subpopulations in a unique cohort of 14 HIV-infected individuals who had been serially sampled in CSF and blood plasma before and after interruption of ART. Comprehensive sequence and phylogenetic analyses were performed to explore the relative contribution of viral reservoirs within the CNS to the HIV RNA rebound following ART interruption. This is important since compartmentalized HIV reservoirs within the CNS will be particularly difficult to target as part of viral eradication strategies, as a consequence of limited drug penetration, compartmentalization, tissue-specific viral adaptation and the presence of unique cellular targets (Gray et al. 2014).
Methods
Participants, samples and clinical laboratory tests
All subjects were prospectively enrolled in protocols that collected serial paired blood and CSF samples from chronically HIV-infected subjects undergoing ART interruption (Monteiro de Almeida et al. 2006). A group of five participants was prospectively enrolled in a structured ART interruption study (Group 1, suppressed >2 years) (Monteiro de Almeida et al. 2006). A second group of five participants was retrospectively determined to have undergone treatment interruption (self- reported) for clinical or personal reasons other than viral failure (Group 2, suppressed >6 months). A third group of four participants was retrospectively determined to have undergone treatment interruption (self-reported) for clinical or personal reasons (Group 3, uncertain or no documented suppression). Detailed inclusion criteria (for each group) and clinical information for each participant are provided as Supplementary material S1. Blood and CSF cellular pellets and supernatant were collected and processed as previously described (Smith et al. 2009; Oliveira et al. 2015). Blood CD4+ T-lymphocyte absolute counts were measured by flow cytometry, and HIV RNA levels in blood plasma and CSF supernatant were quantified by the Amplicor HIV Monitor Test (Roche Molecular Systems Inc.). The study was conducted with appropriate written subject informed consent and was approved by the Human Research Protections Program at the University of California San Diego.
HIV RNA extraction and next-generation sequencing from blood plasma and CSF supernatant
HIV RNA was isolated from blood plasma (QIAamp viral RNA minikit; Qiagen, Hilden, Germany), and cDNA was produced (RETROscript kit; Applied Biosystems/Ambion, Austin, TX) according to the instructions of the manufacturer (Gianella et al. 2011). For blood plasma, if HIV-1 RNA levels (i.e. viral load) exceeded 20,000 HIV-1 RNA copies/ml, 500 μl of blood plasma was used; otherwise, 1 ml of blood plasma was used. For CSF, viral population was concentrated from 1 or 2ml of supernatant in order to maximize template input in each 454 reaction (depending on viral load and sample availability). Three coding regions—gag p24 (HXB2 coordinates 1366–1619), pol RT (2708-3242) and env C2-V3 (6928-7344)—were amplified by PCR with region-specific primers, as previously described (Gianella et al. 2011). Rubber gaskets were used to physically separate 16 samples on a single 454 GS FLX titanium picoliter plate (454 Life Sciences/Roche, Branford, CT) during sequencing.
Quantification and next-generation sequencing of HIV DNA in PBMC and CSF cellular pellets
DNA was extracted from 5 million PBMC for each participant (QIAamp DNA Mini Kit, Qiagen, CA). For CSF cellular pellets, DNA was extracted from all available cells (median: 10,000 cells/pellet, range: 514–80,800 cells) using lysis buffer, as previously described (Christopherson et al. 2000; Oliveira et al. 2015). For PBMC, total HIV DNA (pol) was quantified by droplet digital PCR (ddPCR) from extracted DNA (Strain et al. 2013). Briefly, approximately 1,000 ng of DNA per replicate was digested with BSAJ1 enzyme (New England Biolabs) prior to ddPCR. Total HIV DNA (Pol) was measured with VIC probe using the following cycling conditions: 10 min at 95 °C, 40 cycles consisting of a 30 s denaturation at 94 °C followed by a 60 °C extension for 60 s and a final 10 min at 98 °C. A 1:10 dilution of the digested DNA was used for host cell RPP30 (Ribonuclease P/MRP 30kDa Subunit) PCR (probe VIC) and cycled with the same parameters as Pol. Copy numbers were calculated as the mean of replicate PCR measurements and normalized to one million cells as determined by RPP30 (total cell count). Sequencing was performed as described above.
Sequence processing and bioinformatics analysis
Read and quality score files produced by the 454 instruments were further analyzed using a purpose-built bioinformatics pipeline, used by our group in numerous studies (Gianella et al. 2011; Wagner et al. 2013; Carter et al. 2015; Fisher et al. 2015). The pipeline is available at https://github.com/veg/HIV-NGS and consists of the following key steps:
Baseline quality control: Reads that are too short (≤50 base-pairs [bp]) or have too many low quality bases (≥8 bases with quality scores q ≤ 15) are removed, and all remaining bases with low scores are masked with the N character [missing data].
Error correction: Reads obtained from step 1 are aligned to the corresponding amplicon reference sequence (HXB2) using a codon-level extension to the Smith–Waterman local alignment algorithm, which directly accounts for frame-shift errors caused by homopolymer length miscalls—the most common error modality for the 454 platform. Pairwise alignments are merged into a global alignment of all reads, so that each base is mapped to a consistent system of genomic coordinates. Remaining sequencing errors are modeled as a mixture of multinomial distributions, and all individual single nucleotide variants, which cannot be reliably assigned to non-error multinomial components (posterior probability > 0.999, assumed error rate ≤0.5%) are reverted to position consensus.
Extraction of common and representative reads: Because the median read length is > 50% of the length of sequenced amplicons, we performed simple clustering of error-corrected reads, where two reads are merged in the same cluster if (a) they do not differ except in positions where one of the sequences had an error-induced ‘indel’; (b) they match at X or more nucleotide positions, where X is the maximum of 100 or half of the median read length. Condition (b) allows clusters to absorb shorter reads that match the longer sequences exactly. Each cluster is represented by its consensus sequence, and by the number of reads assigned to the cluster. Note that by construction each read in the cluster matches the consensus sequence in all positions where the read has resolved bases, so the pairwise distance between any two reads in the same cluster is 0 as defined in the next step. Sequences obtained from this step can be downloaded from https://goo.gl/DZhzh1 [see README.md for format explanations]
-
Compartmentalization analysis: We compute the fixation index (Hudson et al. 1992), defined as where πI is the estimate of mean pairwise intra-compartment genetic distance (TN93) (Tamura and Nei, 1993), and πD is its inter-compartment counterpart. Both quantities are computed by comparing all reads from two different compartments at a particular time-point (i.e. only contemporaneous sequences are compared), subject to the requirement that they share at least 150 aligned nucleotide positions. The large number of pairwise comparison (107–109) can be handled computationally using an efficient implementation of the TN93 distance calculator (github.com/veg/tn93), which achieves a throughput of 107 distances/second on a modern multi-core desktop. Note that as defined, could assume negative values (see an example as Supplementary material S2), but none of the samples with negative could be called compartmentalized (see [4c] below). Significance testing is carried out four different ways to improve robustness and to err on the conservative side. One specific patient/time-point is defined as compartmentalized if only if all four methods described below result significant.
We generate the null distribution of from 100 Monte-Carlo replicates, where reads are re-assigned to a random compartment, maintaining the relative sizes of the compartments. The P value of the permutation test must be ≤ 0.05 to establish significance.
We examine the robustness of estimates to extreme errors in haplotype frequencies. To do so, we discard all the frequency information (cluster sizes from step 3), and repeat the estimation and re-sampling as in (a) above [note that the typical range of cluster sizes is from 1 to ∼103–104, hence discarding frequency information is not a trivial modification]. The P value of the permutation test must be ≤ 0.05 to confirm significance.
We estimate the effects of sampling errors on bounds on estimates. To do so, we tabulate values from 100 replicates of data, where alignment sites are resampled with replacement (standard phylogenetic bootstrap) and where the estimated frequencies of haplotypes are perturbed by multiplying each with a uniformly distributed random number between 0.05 and 10, and rounding the resulting value down. The lower bound of the estimated 95% confidence interval must be strictly greater than zero as the third requirement for significance.
We extract 100 subsets of 100 sequences from each compartment (randomly, with larger clusters having proportionally more sampling probabilities), reconstruct ML phylogenies (see point 6 below for details) and apply the Slatkin–Maddison test (Slatkin and Maddison, 1989) for compartmentalization. All 100 replicates must have Slatkin–Maddison P≤0.01 to confirm significance.
Recombination screening: We screen all sets of representative reads from step 3 for evidence of recombination using GARD (Pond et al. 2006). Note that because step 4 does not rely on the assumption that all reads are related by a single phylogeny, it will not be adversely affected by the presence of recombinants.
Phylogenetic analysis: We further filtered the clusters from step 3 to include only those which comprised at least 10 reads, or 0.1% of the total reads (whichever was greater). The resulting collections of sequences are realigned using MUSCLE (Edgar, 2004), piped to FastTree 2 (Price et al. 2010) for maximum likelihood trees reconstruction, and subjected to codon-based (MG94) phylogenetic analyses in HyPhy (Pond et al. 2005). The output of the analyses includes divergence and diversity estimates, both at a nucleotide level, and at the levels of dS (synonymous) and dN (non-synonymous) substitutions.
Statistical analysis
Statistical analyses were performed using SAS (versions 9.2 and 9.4). Qualitative comparisons were performed using the Fisher exact text (for discrete data) or Mann–Whitney U test (for continuous variables). P values were adjusted for multiple comparisons using the Bonferroni methods.
For each compartment (blood, CSF), the time of viral rebound was estimated as the midpoint between the last undetectable HIV RNA viral load (or date of ART interruption if participant interrupted ART before the first study visit), and the first detectable HIV RNA viral load in blood or CSF supernatant respectively.
Results
Participants, samples and clinical laboratory tests
Study participants (N = 14) were HIV-infected individuals with a median age of 41 years (interquartile range [IQR]: [37–45]), who had been receiving ART for a median of 4 years (IQR: 1.7–8.5 years) and voluntarily interrupted therapy between February 1999 and December 2005. Characteristics of the study participants are summarized in Table 1.
Table 1.
Characteristics at baseline (14 individuals).
| Median or count | IQR or % | ||
|---|---|---|---|
| Age; years | 41 | 37–45 | |
| Gender | Male | 14 | 100 |
| Race/ethnicity | Caucasian | 10 | 71 |
| Black | 4 | 29 | |
| Time on ART, years | 4.0 | 1.7–8.5 | |
| CD4+ T-cell after ART interruption, counts/μl | 412 | 397–476 | |
| CD8+ T-cell after ART interruption, counts/μl | 1020 | 651–1449 | |
| Nadir CD4+ T-cell, counts/μl | 293 | 71–388 | |
| HIV RNA in CSF after ART interruptiona, copies/ml (log10) | 3.3 | 2.7–3.8 | |
| HIV RNA in blood after ART interruptiona, copies/ml (log10) | 4.7 | 4.2–5.1 | |
| Time to HIV rebound in blood, days | 19 | 8–31 | |
| Time to HIV rebound in CSF, days | 30 | 16–44 | |
| Time from rebound to sampling in blood, days | 24 | 20–87 | |
| Time from rebound to sampling in CSF, days | 14 | 4–87 | |
| Delay in rebound between blood and CSF, days | 10 | 0–15 |
HIV, human immunodeficiency virus; ART, antiretroviral therapy; IQR, interquartile range; CSF, cerebrospinal fluid.
aHIV RNA levels at the time of sampling for time point 1.
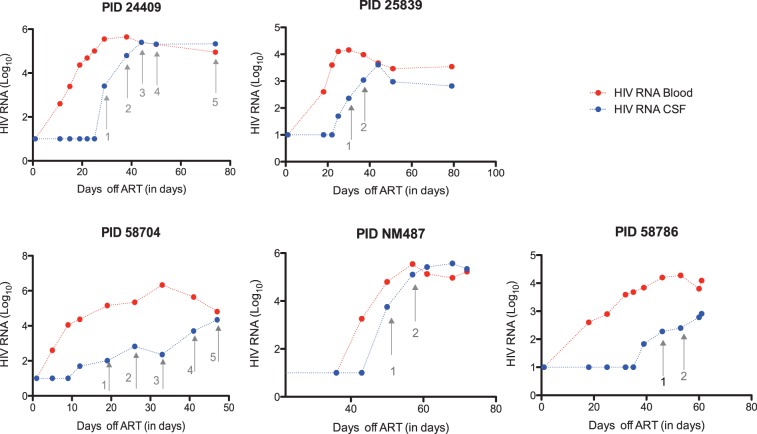
Ten participants had documented undetectable HIV RNA levels for at least 2 years (prospective Group 1) or 6 months (retrospective Group 2) before ART interruption. For the remaining four participants (convenience cohort), duration of HIV RNA suppression before treatment interruption was uncertain. Rebound dynamics of HIV RNA in blood plasma and CSF supernatant for five participants belonging to Group 1 are showed in Fig. 1.
Figure 1.
Dynamics of HIV RNA rebound in CSF supernatant (blue) and blood plasma (red) for five participants belonging to group 1 (prospective cohort). Each dot represents a study visit with blood and CSF collection. Time points used for sequence analysis are indicated with arrows. X axis shows time after interruption of ART (in days), Y axis shows HIV RNA viral load (log10).
The median time from ART interruption to viral rebound in blood plasma was 19 days (IQR: 8–31 days), while the median time from ART interruption to viral rebound in CSF supernatant was 30 days (IQR: 16–44 days). The median time from viral rebound to HIV RNA sampling in blood plasma (referred as Time point 1) was 24 days (IQR: 20–87 days), while the median time from viral rebound (within the CSF compartment) to HIV RNA sampling in CSF supernatant (Time point 1) was 14 days (IQR: 4–87 days). Since our study was primarily interested in analyzing viral rebound in CSF, the earliest available time points at which HIV RNA levels were sufficient for sequence analysis were included as Time point 1. Because of the characteristic delay between rebound in blood and CSF, HIV RNA levels in blood had plateaued and were no longer substantially increasing during this time period (see Fig. 1). At Time point 1, median CD4 T cell count was 412 cells/μl [IQR: 397–476] and HIV RNA levels in blood and CSF supernatant were 4.7 log10 (copies/ml) [IQR: 4.2–5.1] and 3.3 log10 (copies/ml) [IQR: 2.7–3.8], respectively. With the exception of participant 34535, we sampled HIV RNA from blood and CSF supernatant for one or more additional time points (referred as Time points 2–5) after a median of 55 days from Time point 1 (IQR: 9–364 days). For participants 34535, 26919 and 25839, plasma and CSF supernatant were available from a time point preceding the initiation of ART.
Sequencing metrics
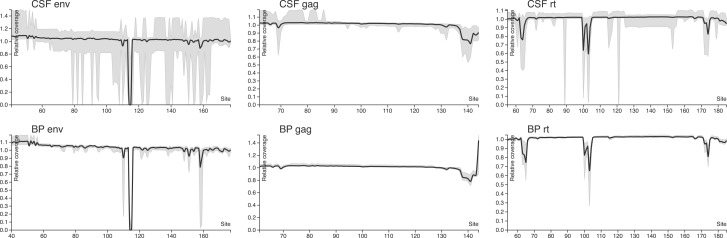
We generated 1.04 gigabases of mapped reads from 71 successfully sequenced time point amplicon (env, gag and RT mixed together) libraries, with a median of 11.76 megabases (IQR over runs 7.95–16.33) per sequencing run (recall that each run represents a 16-way physically multiplexed reaction). The original FASTQ files can be retrieved via study accession number PRJEB14227 at the European Nuelcotide Archive (www.ebi.ac.uk/ena). The median read length was 255 bp (IQR 254–393). The gag amplicon was represented by the largest number of mapped reads per sample, while RT and env had 40–60% yields relative to gag. Median coverage per amplicon position was >3,000 for all regions and compartments, ensuring sufficient resolution to detect sequenced minority variants < 1% (Gianella et al. 2011). The median number of reads assigned to a cluster (inversely correlated with sample diversity) was the highest in gag, intermediate in RT and the lowest in env, consistent with the relative rates of evolution of different viral genes (Alizon and Fraser, 2013). With the exception of two difficult to resolve homopolymeric regions, sequencing coverage was uniform across amplicons, with CSF samples showing more between-sample variation in env and RT (Fig. 2 and Table 2). Detailed characteristics of the viral population are given in Supplementary Tables 1 (blood plasma RNA), 2 (CSF supernatant RNA), 3 (PBMC DNA) and 4 (cellular pellet DNA).
Figure 2.
Coverage plots normalized to within-sample median (see text), used to illustrate the uniformity of coverage across each amplicon and compartment. Solid lines are median values computed over all 14 individuals, and grey areas show the 2.5–97.5 percentile range. Systematic dips in env coverage coincide with the region AAGAAAAA, where length miscalls of the A homopolymer are repaired by codon-aware alignment and partial codons (e.g. –AA) are not counted toward position coverage. In RT, the dip in coverage corresponds to a similar homopolymer context (AAAAAAGAAAAAA).
Table 2.
Sequencing metrics for HIV RNA, stratified by compartment and amplicon.
| Comp | Region | N | Mapped reads [IQR] | Clusters [IQR] | Reads/cluster [IQR] | Median coverage per amino-acid position [IQR] |
|---|---|---|---|---|---|---|
| BP | RT | 35 | 3,544 [1,390–7,404] | 868 [558–1,295] | 3.70 [2.61–5.59] | 3,464 [1,356–7,105] |
| CSF | RT | 32 | 5,352 [3,006–8,045] | 1,101 [706–1,495] | 4.15 [3.05–5.28] | 5,253 [2,932–7,947] |
| BP | GAG | 35 | 9,049 [7,389–13,246] | 1,324 [1,000–2,001] | 7.19 [5.60–8.71] | 8,067 [5,564–12,900] |
| CSF | GAG | 35 | 9,587 [6,821–14,753] | 1,316 [985–1,637] | 7.55 [5.60–11.21] | 7,587 [4,059–14,418] |
| BP | ENV | 35 | 4,913 [3,210–8,458] | 1,918 [1,254–3,122] | 2.29 [1.97–3.21] | 4,701 [2,851–7,159] |
| CSF | ENV | 35 | 5,891 [4,109–11,275] | 1,724 [1,318–3,270] | 3.32 [2.72–3.96] | 5,848 [3,566–10,872] |
BP, blood plasma; CSF, cerebrospinal fluid; N is the number of successful sequencing runs for the compartment/amplicon pair. ENV, envelope; RT, reverse transcriptase; medians and interquartile ranges [IQR] are computed over individual sequenced time points. For an individual sample, median coverage is computed over the entire amplicon.
Recombination screening
We screened alignments composed of five or more common sequence variants per time point: these analyses combined data from blood plasma and CSF (RNA only) and all available time points for a single amplicon for a given individual. With the exception of env in one individual (26618), GARD found no evidence of significant phylogenetic incongruence. Subsequent Shimodaira–Hasegawa testing confirmed one breakpoint in env for individual 26618 (P = 0.0002). Thus, except for one individual-gene pair, viral populations appear to not have experienced detectable recombination events.
Characteristics of rebounding virus in blood and CSF
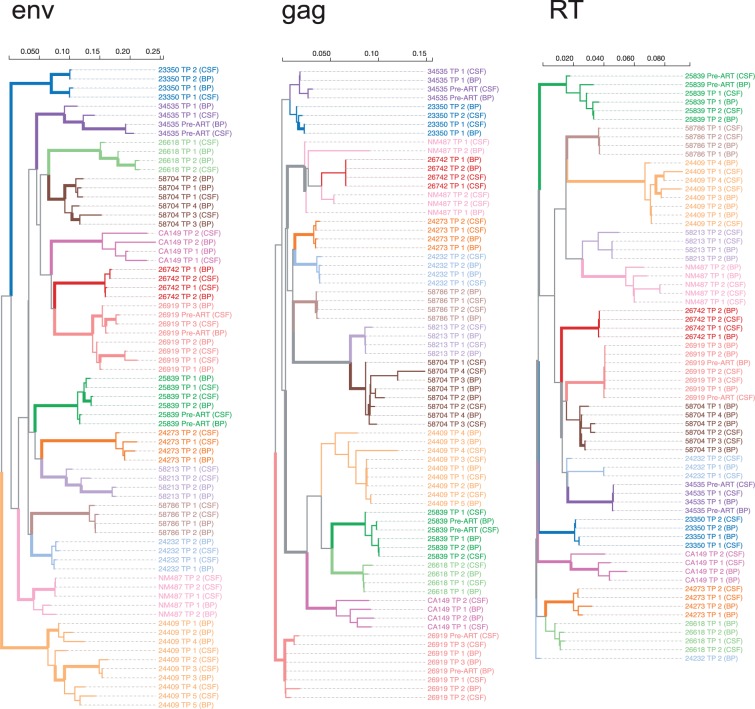
As expected for mono-infected individuals, time point consensus sequences from individual participants formed monophyletic (aLRT > 0.9) clades in the joint phylogenies in env (Fig. 3). This pattern was largely recapitulated in gag and RT, although not all individual clades had the same level of support due to lower sequence divergence. For the primary statistical analysis, we concentrated on the env amplicon and Time points 1 and 2 (which were available for 13/14 participants). Overall, the median nucleotide diversity for HIV RNA env sampled at Time point 1 was not statistically different (two-sided Mann–Whitney test, P = 0.65) between blood and CSF supernatant: median 2.48% (IQR 1.4–5.6%) in blood plasma, versus median 3.43% in CSF supernatant (IQR 1.35–4.08%). Similarly, the median diversity was not significantly different between the first and the second sampled time points (P = 0.78 for blood, P = 0.81 for CSF). Similar results were obtained for pol and gag.
Figure 3.
Approximate maximum-likelihood trees for each sequenced genomic region, based on the consensus sequences for every time point and source compartments. The trees are (arbitrarily) rooted on the reference (HXB2) sequence [not shown], and color-coded by individual. Interior branches are assigned a (non-gray) color if all of their descendants have the same color, i.e. they are assumed to represent evolution within the corresponding host, and are drawn as thick lines if they have 0.9 or greater aLRT support. Branch lengths are scaled in expected substitutions per nucleotide site.
To explore possible predictors of time to viral rebound, we divided our cohort into two groups using the median time from ART interruption to sampling (Time point 1) as a cut-off (43 days). None of the measured covariates (molecular diversity, levels of HIV DNA in PBMC or CSF pellet, time on ART, CD4 nadir, HIV RNA viral load in blood and CSF) were associated with shorter (or longer) time to rebound, after correcting for multiple testing. A negative finding is not surprising, given the relatively small sample size, the large number of tested variables and the uncertainty of the exact timing of rebound as a consequence of sparse sampling and retrospective study design.
Compartmentalization status at the first and second time-points following ART interruption
We detected a significant signal for viral compartmentalization based on four tests (i.e. FST > 0 and all permutation P ≤ 0.05, bootstrap confidence interval does not overlap zero, results are robust to ignoring haplotype frequencies, significant Slatkin–Maddison testing) at the first sampled time point in nine out of 14 participants for at least one gene, and in three out of 14 participants for all genes with available sequence data (Table 3). This included four out of five participants belonging to Group 1 (i.e. prospective group with documented viral suppression > 2 years before interruption of therapy).
Table 3.
Compartmentalization analysis for HIV RNA between blood plasma and CSF supernatant.
| PID | TP | Timea (days) | RT |
GAG |
ENV |
Compartmentalized? |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FST [CI] | P | FST [CI] | P | FST [CI] | P | RT | GAG | ENV | |||
| 24409 | 1 | 2 | 0.02 [−0.12:0.17] | 0.129 | −0.24 [−0.40:0.03] | 1.00 | 0.11 [0.05:0.18] | <0.01 | No | No | Yes |
| 24409 | 2 | 11 | 0.07 [−0.05:0.20] | <0.01 | −0.03 [−0.18:0.14] | 1.00 | 0.20 [0.14:0.29] | <0.01 | No | No | Yes |
| 24409 | 3 | 17 | −0.44 [−1.17: −0.00] | 1.00 | −0.15 [−0.95:0.14] | 1.00# | 0.07 [−0.58:0.24] | <0.01 | No | No | No |
| 24409 | 4 | 47 | 0.28 [0.10:0.40] | <0.01 | 0.22 [0.07:0.43] | <0.01 | 0.67 [0.58:0.78] | <0.01 | Yes | Yes | Yes |
| 24409 | 5 | 83 | NA | NA | NA | NA | 0.38 [0.19:0.57] | <0.01 | Yes | ||
| 25839 | Pre-ART | −2181 | 0.32 [0.07:0.57] | <0.01 | 0.18 [−0.10:0.71] | <0.01 | 0.18 [0.04:0.46] | <0.01§ | Yes | No | No |
| 25839 | 1 | 3.5 | −0.09 [−0.61:0.26] | 1.00# | 0.47 [−0.96:0.77] | <0.01 | −0.13 [−0.60:0.15] | 1.00 | No | No | No |
| 25839 | 2 | 10.5 | 0.08 [−0.05:0.24] | <0.01§ | −0.10 [−0.30:0.69] | 0.498 | 0.07 [−0.04:0.34] | <0.01 | No | No | No |
| 58704 | 1 | 3.5 | NA | NA | 0.87 [0.63:0.95] | <0.01 | 0.18 [0.00:0.44] | <0.01 | Yes | Yes | |
| 58704 | 2 | 10.5 | 0.17 [0.10:0.30] | <0.01 | 0.50 [0.13:0.67] | <0.01 | 0.02 [−0.02:0.09] | <0.01# | Yes | Yes | No |
| 58704 | 3 | 25.5 | −0.50 [−1.67: −0.07] | 1.00 | 0.23 [−0.12:0.45] | <0.01# | 0.23 [0.10:0.42] | <0.01 | No | No | Yes |
| 58704 | 4 | 31.5 | NA | NA | 0.32 [−0.02:0.67] | <0.01 | 0.10 [−0.51:0.46] | <0.01# | No | No | No |
| NM487 | 1 | 3.5 | 0.21 [0.15:0.30] | <0.01 | 0.24 [0.03:0.41] | <0.01 | 0.62 [0.24:0.86] | <0.01 | Yes | Yes | Yes |
| NM487 | 2 | 14.5 | 0.43 [0.28:0.61] | <0.01 | 0.60 [0.42:0.81] | <0.01 | 0.42 [0.30:0.61] | <0.01 | Yes | Yes | Yes |
| 58786 | 1 | 9 | 0.05 [−0.56:0.72] | 0.050 | 0.01 [−0.57:0.60] | <0.01§# | 0.36 [0.14:0.75] | <0.01 | No | No | Yes |
| 58786 | 2 | 23 | 0.03 [−0.27:0.45] | <0.01# | −0.01 [−0.37:0.43] | 1.00# | 0.08 [0.04:0.20] | <0.01§ | No | No | No |
| 24232 | 1 | 18 | 0.45 [0.23:0.65] | <0.01 | 0.36 [0.01:0.73] | <0.01§ | 0.41 [0.26:0.62] | <0.01 | Yes | No | Yes |
| 24232 | 2 | 375 | 0.65 [0.41:0.75] | <0.01 | 0.07 [−0.16:0.23] | <0.01# | −0.20 [−0.76:0.12] | 1.00 | Yes | No | No |
| 24273 | 1 | 11 | 0.21 [0.06:0.36] | <0.01 | 0.22 [0.06:0.37] | <0.01 | 0.30 [0.16:0.51] | <0.01 | Yes | Yes | Yes |
| 24273 | 2 | 383 | 0.21 [0.01:0.35] | <0.01 | 0.14 [−0.02:0.32] | <0.01 | 0.61 [0.46:0.71] | <0.01 | Yes | No | Yes |
| 23350 | 1 | 19 | 0.04 [−0.06:0.33] | <0.01 | −0.24 [−0.47:0.36] | 1.00 | −0.01 [−0.22:0.45] | 0.703 | No | No | No |
| 23350 | 2 | 557 | 0.01 [−0.01:0.06] | <0.01§# | 0.06 [−0.02:0.19] | <0.01§ | −0.01 [−0.60:0.06] | 1.00 | No | No | No |
| 26618 | 1 | 150 | 0.13 [−0.04:0.29] | <0.01 | 0.15 [−0.06:0.54] | <0.01 | 0.15 [0.03:0.29] | <0.01 | No | No | Yes |
| 26618 | 2 | 514 | −0.04 [−0.29:0.16] | 1.00 | 0.26 [−0.09:0.48] | <0.01 | 0.04 [−0.04:0.12] | <0.01 | No | No | No |
| 34535 | Pre-ART | −732 | 0.12 [−0.12:0.47] | <0.01 | 0.82 [0.18:0.94] | <0.01 | 0.19 [0.03:0.43] | <0.01 | No | Yes | Yes |
| 34535 | 1 | 37.5 | 0.09 [−0.22:0.42] | <0.01 | 0.07 [−0.13:0.23] | <0.01§ | 0.33 [0.08:0.46] | <0.01 | No | No | Yes |
| 58213 | 1 | 175 | 0.14 [−0.02:0.30] | <0.01 | 0.11 [−0.09:0.37] | <0.01 | 0.40 [0.24:0.60] | <0.01 | No | No | Yes |
| 58213 | 2 | 319 | 0.03 [−0.92:0.35] | <0.01# | 0.32 [−0.28:0.65] | <0.01 | 0.59 [0.46:0.69] | <0.01 | No | No | Yes |
| 26742 | 1 | 104 | −0.60 [−0.83: −0.30] | 1.00 | 0.01 [−0.10:0.18] | 0.079 | 0.00 [−0.04:0.07] | 0.337 | No | No | No |
| 26742 | 2 | 159 | 0.39 [0.12:0.62] | <0.01 | 0.01 [−0.09:0.15] | 0.050 | 0.01 [−0.00:0.07] | <0.01§# | Yes | No | No |
| 26919 | Pre-ART | −21 | 0.25 [0.10:0.42] | <0.01 | 0.13 [0.02:0.32] | <0.01 | 0.20 [0.11:0.34] | <0.01 | Yes | Yes | Yes |
| 26919 | 1 | 7 | NA | NA | 0.18 [−0.02:0.34] | <0.01 | 0.08 [−0.02:0.18] | <0.01 | No | No | |
| 26919 | 2 | 16 | 0.06 [0.02:0.14] | <0.01 | 0.30 [−0.07:0.64] | <0.01 | 0.08 [0.01:0.14] | <0.01 | Yes | No | Yes |
| 26919 | 3 | 99 | −0.01 [−0.06:0.08] | 0.743 | 0.13 [0.01:0.26] | <0.01 | 0.19 [0.11:0.29] | <0.01 | No | Yes | Yes |
| CA149 | 1 | 137 | 0.08 [−0.28:0.28] | <0.01# | −0.06 [−0.83:0.37] | 1.00 | −0.45 [−0.65: −0.24] | 1.00 | No | No | No |
| CA149 | 2 | 503 | 0.60 [0.48:0.72] | <0.01 | 0.68 [0.50:0.83] | <0.01 | 0.71 [0.62:0.79] | <0.01 | Yes | Yes | Yes |
TP, time point collected sequentially after ART interruption, pre-ART time points were collected before starting antiretroviral therapy; ENV, envelope; RT, reverse transcriptase; BP, blood plasma; CSF, cerebrospinal fluid; confidence intervals are estimated by computing FST values for 100 replicates drawn by sampling alignment sites with replacement and adding noise to haplotype frequencies [see text]; P values <0.01 are derived from 100 bootstrap replicates, when 0/100 replicates had FST values equal to or greater than the estimate obtained from the data. Compartmentalization is called when four tests indicate compartmentalization [see text]
#Indicates tests which become significant (or not significant) if copy numbers are ignored during FST calculations.
§Indicates tests which become insignificant under the phylogeny-based SlatkinMaddison test [see text].
*time in days elapsed from viral rebound to sampling in CSF.
At subsequent time points (i.e. 2–5), 10 participants exhibited compartmentalization between blood and CSF in at least one gene. Overall, 58% of all samples were compartmentalized in env, 28% were compartmentalized in gag and 39% in RT. Seven time-points (19%) were found compartmentalized in all three regions. Interestingly, data collected pre-ART for a subset of three participants indicate that compartmentalization was present at the time of ART initiation but disappeared in two cases at the first samples time-point. Overall, compartmentalization pattern changed over time in most participants and across different coding regions.
Structure of the rebounding HIV RNA populations
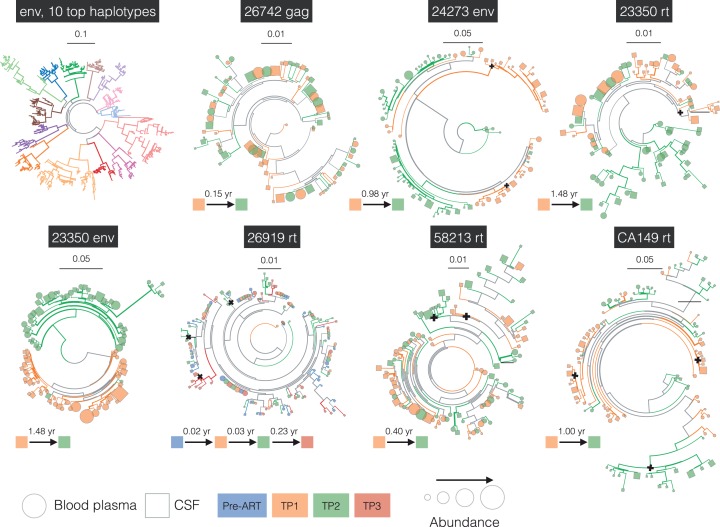
A complete set of interactive phylogenetic trees showing the relationships between the samples (distinguishing up to the 25 most frequent viral haplotypes per sample) are available at http://bit.ly/gianella-pond-ellis-trees. Phylogenies including DNA-based sequences are also available for viewing at the same address. Representative phylogenies illustrating different patterns of dynamics and evolution are shown in Fig. 4. As with the consensus phylogeny (shown in Fig. 3), individuals form monophyletic clades in the tree constructed from 10 most frequent haplotypes (env, 10 top haplotypes). In some cases (e.g. 26742 gag), the blood and CSF sequences are intermixed, which, coupled with low FST, suggest trafficking between compartments or independent rebound of near-identical archived viruses in both compartments. Remarkably, for 10 out 14 participants (for example, 24273 env showed in Fig. 4), the phylogenies contain unique clades in the CSF at the earliest time point after ART interruption, suggesting possible independent rebound events wholly within CSF (summarized in Table 4). Frequencies of CSF-only clades ranged from being a minority variant up to representing over half of the inferred population. These CSF-only clades persisted over time and were often visible over longitudinal time-points within one subject. In two individuals with available pre-ART samples (34535 and 26919), the same pre-ART population re-appeared in the CSF after treatment interruption. Similarly, some trees showed blood-only clades, but this observation should not be over-interpreted because lower viral loads in the CSF compartment necessarily make representative sampling more challenging.
Figure 4.
Examples of intra-host population dynamics, and the phylogenetic relationship between different individuals. For individual subjects, the tree is restricted to represent up to 25 most common haplotypes per time point (see text), whereas the joint env tree uses up to 10 most common haplotypes. Branch coloring and thickness conventions are the same in Fig. 2, except that black crosses mark the most recent common ancestors for CSF-only clades. For an alternative rendering of the trees as cladograms, refer to Supplementary materials.
Table 4.
The distribution of unique clades within the CSF (not observed in paired blood plasma samples).
| PID | RT | GAG | ENV |
|---|---|---|---|
| 24409 | None | TP2: 0.5% | TP4: 8.1% |
| TP3: 26.5% | |||
| 25839 | None | Pre-ART: 17.8% | Pre-ART: 6.5% |
| 58704 | None | None | TP1: 50.2% |
| TP2: 5.6% | |||
| NM487 | TP1: 4.1% (2) | TP2: 37.2% (1), 21.8% (2) | TP1: 2.8% (1), 1.2% (2), 1.1% (3), 3.8% (4) |
| TP2: 9.1% (1), 81.7% (2) | TP2: 24.6% (1), 16.0% (2), 33.7% (3), 3.4% (4), 6.0% (5) | ||
| 58786 | TP2: 7.0% | TP2: 15.3% | None |
| 24232 | TP1: 27.3% (1) | None | None |
| TP2: 10.1% (2) | |||
| 24273 | TP2: 5.4% | None | TP1: 37.9% (1), 13.6% (2) |
| 23350 | TP1: 11.3% | TP1: 14.4% | None |
| TP2: 0.7% | |||
| 26618 | TP1: 4.7% (1), | TP1: 15.5% | TP2: 22.7% (1), 14.6% (2) |
| TP2: 7.8% (1), 13.4% (2) | TP2: 1.2% | ||
| 34535 | None | None | Pre-ART: 10.6% (1) |
| TP1: 5.1% | |||
| 58213 | TP1: 38.0% (1) | TP1: 13.1% | TP1: 0.9% (1) |
| TP2: 26.3% (1), 24.9% (2) | TP2: 59.6% | TP2: 50.6% (1) | |
| 26742 | None | None | None |
| 26919 | Pre-ART: 2.0% (1) | None | TP1: 38.6% (1) |
| TP2: 9.2% (1), 14.7% (2) | TP2: 15.3% (2) | ||
| TP3: 14.3% (3) | TP2: 12.6% (3) | ||
| CA149 | TP1: 10.7% (1), 41.7% (2) | TP2: 40.3% | TP1: 10.8% (1) |
| TP2: 61.4% (3) | TP2: 60.5% (2) |
Unique clusters observed within the CSF are defined as phylogenetically supported (aLRT≥0.9) clades with CSF only sequences, including three or more haplotypes with total frequency of constituent reads at 5% or greater. For each PID/genetic region, clusters are broken down by the time of sampling of contributing reads. For example, NM487 RT has two CSF-only clusters: Cluster 1 has 9.0% sequences from Time point 2, and Cluster 2 has 81.7% of sequences from Time point 2 and 4.1% of sequences from Time point 1.
The phylogenies for different viral genes sampled from the same individuals do not necessarily show identical dynamics: for example, Fig. 4 shows the env and rt genes for the same individual (23350) at Time point 1, with no evidence for CSF-only clades in env, but containing a well-separated and statistically supported CSF-only clade at the first time point in rt. This observation could result from an independent rebound in CSF, for which the archived viruses in blood and CSF had homogenized env but different rt genes, for example, due to a past recombination event. Previous studies have suggested complex intra-host recombination dynamics (Brown et al. 2011; Pacold et al. 2012), and demonstrated how selection can affect HIV sequences in different compartments (Hightower et al. 2009; Neher and Leitner, 2010; Holman and Gabuzda, 2012), indicating that such eventualities may be common. The phylogenies for both these genes (i.e. env and RT for participants 23350) suggest that by the second time point (about seven months later), populations in both compartments have diverged from the population at the first time point and have become intermixed.
In contrast, 26919 RT shows a case where there is no noticeable divergence across four time points spanning a 3-month period, and in which, some separation by compartment appears to persist. Similarly, 58213 RT shows a case where a large CSF-only clade persists across two time points spanning a 4-month period, with no noticeable divergence between the time points.
Finally, CA149 RT shows a case where the blood and CSF populations appear to have been mixed at the first time point, but became segregated by the second time point about 1 year later. Table 4 provides an overview about the distribution of the unique clades within the CSF across 14 subjects.
This panoply of apparent viral dynamics is not surprising, because there is no compelling reason to expect any particular evolutionary scenario to be common or dominant (Lemey et al. 2006).
Comparative analysis of HIV RNA and DNA populations within each anatomic compartment
For a subset of participants, we were able to generate sequences from HIV DNA in peripheral blood mononuclear cells (N = 10 participants at 15 individual time-points), and CSF cellular pellet (N = 9 participants at 10 individual time-points, see Supplementary Tables 3 and 4). Eight time points had paired data from HIV RNA and HIV DNA sequenced from both blood and CSF. Interestingly, HIV RNA virus rebounding in blood plasma appeared genetically segregated from paired PBMC HIV DNA in eight out of 13 participants. HIV DNA samples from CSF cellular pellet were also genetically segregated from the HIV RNA population in the CSF in 9/10 of analyzed cases. PBMC and CSF cellular pellets were genetically segregated in 5/6 (26618, CA149, 58786, 58704 and 24232, but not 23350) analyzed cases (based on the FST permutation testing only). These statistical analyses could be made unreliable by the low template input, especially for the CSF cell pellet, which might make a population look more homogenous than it really is due to template resampling, or simply miss large swaths of viral genetic diversity in the DNA reservoir.
Discussion
A successful HIV eradication strategy must block or cripple all known mechanisms of viral persistence and recovery (Richman et al. 2009). One such mechanism is the establishment of genetically distinct viral populations in different tissues and anatomical compartments (Svicher et al. 2014). Completely eradicating viral replication and reservoirs in blood might be insufficient to achieve a cure if the virus is able to reconstitute its population within the CNS compartment. Numerous previous studies estimated that 30–60% of HIV-infected individuals harbor viral populations that are compartmentalized between blood and CNS (Blower et al. 2005; Pillai et al. 2006; Harrington et al. 2009; Sturdevant et al. 2015). Through the use of more sensitive sequencing techniques and high-throughput bioinformatics methods, our study suggests that an even larger proportion of individuals may have some degree of compartmentalization in rebounding HIV RNA populations following treatment interruption. Most of our participants (nine out of 14) presented a compartmentalized HIV RNA rebound within the CNS after ART interruption (conservatively based on 4 different tests), even when sampled a few days after viral rebound.
Two processes can affect the extent of differences between viral populations between blood and CSF. The first such process is genetic divergence: standard population genetics results imply that in the absence of mixing, two anatomically separate viral populations will diverge genotypically via neutral drift and/or adaptation. It is possible that a considerable amount of divergence happens before viral suppression, particularly in patients starting ART during chronic infection, as most of treated patients in the USA. Such pre-suppression divergence in turn would lead to distinct populations rebounding in the two compartments, if the rebounding populations arise independently from compartment-specific sources. The second process is trafficking between the CNS and other compartments, most importantly blood, which reduces differences between compartments by mixing and/or recombination between the viral populations (this process has been called “a fundamental evolutionary mechanism that helps to shape HIV-1 env intrahost diversity in natural infection” (Brown et al. 2011)). When significant compartmentalization is observed, as we show here in most cases of viral rebound after treatment interruption, this might suggest that the rate of trafficking is not sufficiently high to completely homogenize the populations. In contrast, the absence of compartmentalization (i.e. high population inter-mixing) could reflect either high trafficking or that the populations were not distinguishable to begin with (due to low divergence rates). We remark that the finding of viral compartmentalization does not imply completely segregated viral populations, but that there is a detectable divergence between some subpopulations in each compartment. Nevertheless, in 12 cases out of 14, we identified unique viral populations within the CSF after ART interruption, which were phylogenetically distinct from those present in the paired blood plasma. These CSF-only clades persist over time and, in two cases (of whom one was suppressed for >1.2 years before interrupting ART), the same pre-ART population reappeared in the CSF (but not in blood) after interrupting therapy. This pattern implies that at least some of the rebounding virus within the CNS originates from a CNS source and might represent a notable barrier to sterilizing cure.
To investigate the possible source of rebounding viruses, we also sequenced HIV DNA collected from PBMC (N = 15) and CSF cellular pellets (N = 10) for a subset of individuals. While approximately 40% of the blood populations were not compartmentalized between rebounding HIV RNA and HIV DNA sampled from the corresponding PBMC (e.g., that would occur if viral rebound were seeded by cells circulating in the blood), all but one (80%) of the CSF populations were compartmentalized between HIV RNA and HIV DNA sampled from corresponding CSF cellular pellets. While this analysis is limited by sampling bias as a consequence of low cellular input especially from the CNS cellular pellets, it is also consistent with the hypothesis that HIV RNA rebound within the CNS compartment most likely originates from cells within the brain parenchyma, which cannot be directly sampled using our methods.
None of the tested variables were associated with time to viral rebound, which is likely a consequence of small sample size and retrospective study design with uncertain time to rebound for most included individuals.
This study has several limitations. First, it only includes 14 individuals (of whom only 10 were certainly suppressed at the time of treatment interruption), thereby limiting the power of subsequent statistical analysis; we note that despite the small sample size, our study delivers the most detailed characterization of a cohort of such size to date, highlighting the amount of clinical, laboratory and informatics effort involved. Second, this is a retrospective study taking advantage of samples collected from people interrupting ART, and the optimal samples (closest to ART interruption) were not always available (except for the five participants belonging to our prospective Group 1). Consequently, there are considerable differences in timing of sampling time-points after treatment interruption. While cohort characteristics are relatively heterogeneous, our results were robust to choosing individuals sampled early or late during rebound, and independent of HIV RNA levels, time on ART or any other clinical variables. Nevertheless, the HIV RNA populations were evaluated several days after viral rebound in most participants and we cannot exclude the possibility that HIV RNA did cross from the blood to the CNS very early and diversified fast enough to confound our analysis.
Third, low template input into the NGS reaction and sequencing errors could cause biases or skewing in the sampling of the original viral population, and negatively impact our ability to perform accurate analyses on these samples. We have, therefore, significantly elevated the threshold of compartmentalization detection and specifically included computational tests for robustness against significant errors in frequency estimation. Template input was particularly low in some (but not all) CSF samples, which could negatively impact our capacity to find unique clades within the CSF: assuming we are simply resampling the most common variants, we are more likely to find that CSF sequences fall within better sampled blood clades. In contrast, despite the possible sampling bias in CSF, we were still able to observe CSF-unique clades in all compartmentalized time points and some were repeated across longitudinal samples further confirming the validity of our methods.
Despite these limitations, our study suggests that the HIV RNA population often rebounds independently within the CSF and that the HIV DNA reservoirs in anatomic compartments might present additional obstacles to eradication and need to be actively targeted to achieve a complete cure. Our data suggest that failing to perform lumbar punctures during structured ART interruption in the setting of cure trials might overlook important events emanating from the CNS, which could be contributing to overall rebound. Future prospective studies with more frequent sampling should determine if the virus originated within the CNS meaningfully contributes to reconstituting the HIV RNA population in blood.
Supplementary data
Supplementary data are available at Virus Evolution online.
Conflict of interest: None declared
Acknowledgements
We would like to acknowledge all the study participants.
Author Contributions
SG participated in the study design, performed laboratory experiments, participated in the data analyses for this study and wrote the primary version of the manuscript; SKP designed and implemented the informatics tools for data analysis and visualization; wrote the primary version of the manuscript, MFO participated in design of laboratory experiments, preformed laboratory experiments, and revised the manuscript; DMS and SL participated in the study design, and helped with manuscript revisions; RJE, SKP, MS and KS participated in study design, performed statistical analysis and wrote the primary version of the manuscript; ADT participated in the study design, data analysis and revised the manuscript; RJE designed the clinical trial, enrolled participants and collected and archived the samples. All authors read and approved the final manuscript.
Competing Interests
DMS has received grant support from ViiV Pharmaceuticals and consultant fees from Gen-Probe and Testing Talent Services.
Funding
This work was supported by the Department of Veterans Affairs, the James Pendleton Charitable Trust; the US National Institutes of Health (NIH) awards MH101012-3, AI43638, AI100665, MH097520, DA034978, AI036214, AI007384, AI027763, AI106039, AI074621, AI110181, AI068636, AI106039, AI027763, GM093939, GM110749; University of California San Diego Center for AIDS Research (Translational Virology and Bioinformatics Cores), P30-AI027763; the California HIV/ AIDS Research Program RN07-SD-702, MC08-SD-700 and EI-11-SD-005; the Interdisciplicinary Research Fellowship in NeuroAIDS R25-MH081482. MFO was supported by the CNPq-Brazil. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Alizon S.,, Fraser C. (2013) ‘Within-host and Between-host Evolutionary Rates Across the HIV-1 Genome’, Retrovirology, 10: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackard J. T. (2012) HIV compartmentalization: A Review On A Clinically Important Phenomenon. Current HIV research. 2012. Epub 2012/02/15. PubMed PMID: 22329519. [DOI] [PubMed]
- Blower S., et al. (2005) ‘The Antiretroviral Rollout and Drug-resistant HIV in Africa: Insights from Empirical Data and Theoretical Models’, AIDS(Aids), 19/1: 1–14. [DOI] [PubMed] [Google Scholar]
- Brown R. J., et al. (2011) ‘Intercompartmental Recombination of HIV-1 Contributes to env Intrahost Diversity and Modulates Viral Tropism and Sensitivity to Entry Inhibitors’, Journal of Virology, 85/12: 6024–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C. C., et al. (2015) ‘HIV-1 Neutralizing Antibody Response and Viral Genetic Diversity Characterized with Next Generation Sequencing’, Virology, 474: 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. Y., et al. (2012) ‘Genetic Features of Cerebrospinal Fluid-derived Subtype B HIV-1 tat’, Journal of Neurovirology, 18/2: 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopherson C., et al. (2000) ‘PCR-Based Assay to Quantify Human Immunodeficiency Virus Type 1 DNA in Peripheral Blood Mononuclear Cells’, Journal of Clinical Microbiology, 38/2: 630–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements J. E., et al. (2005) ‘The Central Nervous System is a Viral Reservoir in Simian Immunodeficiency Virus-infected Macaques on Combined Antiretroviral Therapy: A Model for Human Immunodeficiency Virus Patients on Highly Active Antiretroviral Therapy’, Journal of Neurovirology, 11/2: 180–9. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004) ‘MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity’, BMC Bioinformatics, 5: 113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D., et al. (1999) ‘Latent Infection of CD4+ T Cells Provides a Mechanism for LifeLong Persistence of HIV-1, Even in Patients on Effective Combination Therapy’, Nature Medicine, 5/5: 512–7. [DOI] [PubMed] [Google Scholar]
- Fisher R. G., et al. (2015) ‘Next Generation Sequencing Improves Detection of Drug Resistance Mutations in Infants After PMTCT Failure’, Journal of Clinical Virology, 62: 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianella S., et al. (2011) ‘Detection of Minority Resistance during Early HIV-1 Infection: Natural Variation and Spurious Detection Rather than Transmission and Evolution of Multiple Viral Variant’, Journal of Virology, 85/16: 8359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray L. R., et al. (2014) ‘Is the Central Nervous System a Reservoir of HIV-1?’, Current Opinion in HIV and AIDS, 9/6: 552–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington P. R., et al. (2009) ‘Cross-sectional Characterization of HIV-1 env Compartmentalization in Cerebrospinal Fluid Over the Full Disease Course’, AIDS, 23/8: 907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hightower G. K., et al. (2009) ‘Select Resistance-associated Mutations in Blood are Associated with Lower CSF Viral Loads and Better Neuropsychological Performance’, Virology, 394/2: 243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocqueloux L., et al. (2010) ‘Long-term Immunovirologic Control Following Antiretroviral Therapy Interruption in Patients Treated at the Time of Primary HIV-1 Infection’, AIDS, 24/10: 1598–601. [DOI] [PubMed] [Google Scholar]
- Hoen B., et al. (2005) ‘Structured Treatment Interruptions in Primary HIV-1 Infection: The ANRS 100 PRIMSTOP Trial’, Journal of Acquired Immune Deficiency Syndromes, 40/3: 307–16. PubMed PMID: 16249705. [DOI] [PubMed] [Google Scholar]
- Holman A. G.,, Gabuzda D. (2012) ‘A Machine Learning Approach for Identifying Amino Aacid Signatures in the HIV env Gene Predictive of Dementia’, PLoS One, 7/11: e49538.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Slatkin M., Maddison W. P. (1992) ‘Estimation of Levels of Gene Flow from DNA Sequence Data’, Genetics, 132/2: 583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele B. F., et al. (2008) ‘Identification and Characterization of Transmitted and Early Founder Virus Envelopes in Primary HIV-1 Infection’, Proceedings of the National Academy of Sciences of the United States of America, 105/21: 7552–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P., Rambaut A., Pybus O. G. (2006) ‘HIV Evolutionary Dynamics Within and Among Hosts’, AIDS Reviews, 8/3: 125–40. [PubMed] [Google Scholar]
- Lewin S. R., et al. (2011) ‘Finding a Cure for HIV: Will it Ever be Achievable?’, Journal of the International AIDS Society, 14: 4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis D. M. (2014) ‘How Might We Cure HIV?’, Current Infectious Disease Reports, 16/3: 392.. [DOI] [PubMed] [Google Scholar]
- Monteiro de Almeida S., et al. (2005) ‘Dynamics of Monocyte Chemoattractant Protein Type one (MCP-1) and HIV Viral Load in Human Cerebrospinal Fluid and Plasma’, Journal of Neuroimmunology, 169/1-2: 144–52. [DOI] [PubMed] [Google Scholar]
- Monteiro de Almeida S., et al. (2006) ‘Relationship of CSF Leukocytosis to Compartmentalized Changes in MCP-1/CCL2 in the CSF of HIV-Infected Patients Undergoing Interruption of Antiretroviral Therapy’, Journal of Neuroimmunology, 179/1-2: 180–5. [DOI] [PubMed] [Google Scholar]
- Neher R. A.,, Leitner T. (2010) ‘Recombination Rate and Selection Strength in HIV Intra-patient Evolution’, PLoS Computational Biology, 6/1: e1000660.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira M. F., et al. (2015) Comparative analysis of cell-associated HIV DNA levels in Cerebrospinal Fluid and Peripheral Blood by droplet digital PCR, PLoS One, 10/10:e0139510. [DOI] [PMC free article] [PubMed]
- Pacold M. E., et al. (2012) ‘Clinical, Virologic, and Immunologic Correlates of HIV-1 Intraclade B Dual Infection Among Men Who Have Sex with Men’, AIDS(Aids), 26/2: 157–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palella F. J., Jr., et al. (2006) ‘Mortality in the Highly Active Antiretroviral Therapy Era: Changing Causes of Death and Disease in the HIV Outpatient Study’, Journal of Acquired Immune Deficiency Syndromes, 43/1: 27–34. [DOI] [PubMed] [Google Scholar]
- Palmisano L., et al. (2007) ‘Determinants of Virologic and Immunologic Outcomes in Chronically HIV-Infected Subjects Undergoing Repeated Treatment Interruptions: The Istituto Superiore di Sanita-Pulsed Antiretroviral Therapy (ISS-PART) Study’, Journal of Acquired Immune Deficiency Syndromes, 46/1: 39–47. [PubMed] [Google Scholar]
- Pillai S. K., et al. (2006) ‘Genetic Attributes of Cerebrospinal Fluid-derived HIV-1 env’, Brain, 129/Pt 7: 1872–83. [DOI] [PubMed] [Google Scholar]
- Pond S. L., et al. (2006) ‘Automated Phylogenetic Detection of Recombination Using a Genetic Algorithm’, Molecular Biology Evolution, 23/10: 1891–901. [DOI] [PubMed] [Google Scholar]
- Pond S. L., Frost S. D., Muse S. V. (2005) ‘HyPhy: Hypothesis Testing Using Phylogenies’, Bioinformatics, 21/5: 676–9. [DOI] [PubMed] [Google Scholar]
- Price M. N., et al. (2010) ‘FastTree 2–Approximately Maximum-Likelihood Trees for Large Alignments’, PLoS ONE, 5/5: e9490. doi:10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman D. D., et al. (2009) ‘The Challenge of Finding a Cure for HIV Infection’, Science, 323/5919: 1304–7. [DOI] [PubMed] [Google Scholar]
- Saez-Cirion A., et al. (2013) ‘Post-treatment HIV-1 Controllers with a Long-term Virological Remission After the Interruption of Early Initiated Antiretroviral Therapy ANRS VISCONTI Study’, PLoS Pathogens, 9/3: e1003211.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemi M.,, Rife B. (2015) Phylogenetics and Phyloanatomy of HIV/SIV Intra-Host Compartments and Reservoirs: The Key Role of the Central Nervous System. Curr HIV Res. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano J. D., et al. (2003) ‘Long-term Follow-up Studies Confirm the Stability of the Latent Reservoir for HIV-1 in Resting CD4+ T Cells’, Nature Medicine, 9/6: 727–8. [DOI] [PubMed] [Google Scholar]
- Slatkin M., Maddison W. P. (1989) ‘A cladistic measure of gene flow inferred from the phylogenies of alleles’, Genetics, 123/3: 603–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. M., et al. (2009) ‘Pleocytosis is Associated with Disruption of HIV Compartmentalization Between Blood and Cerebral Spinal Fluid Viral Populations’, Virology, 385/1: 204–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strain M. C., et al. (2005) ‘Genetic Composition of Human Immunodeficiency Virus Type 1 in Cerebrospinal Fluid and Blood Without Treatment and During Failing Antiretroviral Therapy’, Journal of Virology, 79/3: 1772–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strain M. C., et al. (2013) ‘Highly Precise Measurement of HIV DNA by Droplet Digital PCR’, PLoS One, 8/4: e55943.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturdevant C. B., et al. (2015) ‘Compartmentalized Replication of R5 T Cell-tropic HIV-1 in the Central Nervous System Early in the Course of Infection’, PLoS Pathogens, 11/3: e1004720.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svicher V., et al. (2014) ‘Understanding HIV Compartments and Reservoirs’, Current HIV/AIDS Reports, 11/2: 186–94. [DOI] [PubMed] [Google Scholar]
- Tamura K., Nei S. M., et al. (1993) ‘Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees’, Molecular Biology Evolution, 10/3: 512–526. [DOI] [PubMed] [Google Scholar]
- Thompson K. A., et al. (2011) ‘Brain Cell Reservoirs of Latent Virus in Presymptomatic HIV-Infected Individuals’, The American Journal of Pathology, 179/4: 1623–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G. A., et al. (2013) ‘Incidence and Prevalence of Intrasubtype HIV-1 Dual Infection in At-Risk Men in the United States’, J Infect Dis, doi: 10.1093/infdis/jit633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J. K., et al. (1997) ‘Recovery of Replication-competent HIV Despite Prolonged Suppression of Plasma Viremia’, Science, 278/5341: 1291–5. [DOI] [PubMed] [Google Scholar]
- Zarate S., et al. (2007) ‘Comparative Study of Methods for Detecting Sequence Compartmentalization in Human Immunodeficiency Virus Type 1’, Journal of Virology, 81/12: 6643–51. [DOI] [PMC free article] [PubMed] [Google Scholar]