Abstract
As one of the major pathogens, bovine viral diarrhea virus caused a significant economic loss to the livestock industry worldwide. Although BVDV infections have increasingly been reported in China in recent years, the molecular aspects of those BVDV strains were barely characterized. In this study, we reported the identification and characterization of a novel BVDV isolate designated as SD-15 from cattle, which is associated with an outbreak characterized by severe hemorrhagic and mucous diarrhea with high morbidity and mortality in Shandong, China. SD-15 was revealed to be a noncytopathic BVDV, and has a complete genomic sequence of 12,285 nucleotides that contains a large open reading frame encoding 3900 amino acids. Alignment analysis showed that SD-15 has 93.8% nucleotide sequence identity with BVDV ZM-95 isolate, a previous BVDV strain isolated from pigs manifesting clinical signs and lesions resembling to classical swine fever. Phylogenetic analysis clustered SD-15 to a BVDV-1m subgenotype. Analysis of the deduced amino acid sequence of glycoproteins revealed that E2 has several highly conserved and variable regions within BVDV-1 genotypes. An additional N-glycosylation site (240NTT) was revealed exclusively in SD-15-encoded E2 in addition to four potential glycosylation sites (Asn-X-Ser/Thr) shared by all BVDV-1 genotypes. Furthermore, unique amino acid and linear epitope mutations were revealed in SD-15-encoded Erns glycoprotein compared with known BVDV-1 genotype. In conclusion, we have isolated a noncytopathic BVDV-1m strain that is associated with a disease characterized by high morbidity and mortality, revealed the complete genome sequence of the first BVDV-1m virus originated from cattle, and found a unique glycosylation site in E2 and a linear epitope mutation in Erns encoded by SD-15 strain. Those results will broaden the current understanding of BVDV infection and lay a basis for future investigation on SD-15-related pathogenesis.
Introduction
Bovine viral diarrhea virus (BVDV) is a small, enveloped virus with a single-stranded, positive-sense RNA genome. Together with classical swine fever virus (CSFV) and border disease virus (BDV), BVDV belongs to the genus of pestivirus within the family of Flaviviridae [1]. As one of the most important viral pathogens, BVDV causes significant economic losses to cattle industry worldwide [2–4]. In addition to cattle, BVDV also infects pigs, deer, sheep and other wild animals [5–8]. Based on the cytopathic effect (CPE) on cell culture, BVDV is divided into two biotypes, the cytopathic (CP) and noncytopathic (NCP) biotypes where CP or NCP isolates are divided into BVDV-1, BVDV-2, and atypical BVDV-3 genotypes based on viral sequence variations [9, 10]. While the epidemic isolates for BVDV mainly belong to BVDV-1, the more recent hypervirulent BVDV-2 strains have been isolated from cattle with acute diarrhea and fatal thrombocytopenia [11–13]. Genomic sequence comparisons revealed the diversity and genetic variability of BVDV strains isolated from different herds or even in the same herd [13]. Based on the genetic variability, seventeen BVDV-1 subgenotypes and four BVDV-2 subgenotypes have been reported so far [14–18].
The genome of BVDV is approximately 12.5 kb in length containing a single open reading frame (ORF) flanked by 5’-UTR and 3’-UTR [19–21]. The ORF encodes a precursor polyprotein of about 3,900 amino acids, which is subsequently processed by viral or cellular proteases into 11 or 12 individual proteins including Npro, C, Erns, E1, E2, p7, NS2/3, NS4A, NS4B, NS5A and NS5B from the N terminus to the C terminus [20, 22, 23]. The C, Erns, E1 and E2 are four structural proteins, and the remains are nonstructural viral proteins [23, 24]. Out of four structural proteins, E2 has a mass of 55 KDa and is classified as type I transmembrane protein, which is associated with virus entry, viral pathogenicity and immunity. Erns is structural glycoprotein that possess the intrinsic ribonuclease activity involved in virus attachment and entry into target cells. Study has demonstrated that envelope proteins are involved in several biological activities through participating host–cell interactions such as receptor binding, internalization and posttranslational modifications, in most viruses, the glycosylation [25]. Glycosylation has been demonstrated to play a crucial role in biogenesis, stability, antigenicity and infectivity. Many viruses are dependent on N-linked glycosylation for vital biological functions via promoting proper folding and subsequent trafficking using host cellular chaperones and folding factors [25]. It is well-recognized that glycosylation in many enveloped viruses is important to viral infection, and alteration of glycosylation sites affects the pathogenicity and antigenicity of the viruses [26].
In China, bovine viral diarrhea-mucosal disease (BVD-MD) was first reported in 1980 on a farm where cattle were imported from Europe. The first BVDV strain named Changchun-184 (CC-184) was isolated from the same farm and classified to BVDV-1b subgenotype based on the sequence similarity [27, 28]. In 1995, a BVDV strain named ZM-95 was isolated from pigs in the Inner Mongolia autonomous region, which showed clinical signs and gross lesions similar to classical swine fever [7], thus discovering the BVDV infection in pigs in China. Sequence analysis revealed that ZM-95 belongs to BVDV-1m subgenotype [29]. During late 1990’s and early 2000’s, BVD occurred in many regions mainly due to the booming cattle industry and the circulation of live cattle across China. Analysis of the 5’-UTR sequence of BVDV strains isolated from 2005 to 2013 revealed that majority of BVDV strains belongs to BVDV-1, and BVDV-1b and BVDV-1m were the predominant subgenotype [30–35]. Recently, BVDV-2 infections have been reported in cattle and pig populations in China [36, 37]. Although BVDV infections has increasingly been reported in China recently, the majority of those reports were only focus on the genotyping using 5’-UTR sequences, thus resulting in the molecular aspects of BVDV strains largely unexplored, especially the genomic sequence of the predominated subgenotype BVDV-1m strains. To understand the origin and evolution of BVDV strain and determine the molecular characters of the BVDV strains predominantly spread in China, we collected and characterized the BVDV strains associated with severe diarrhea and mucosal diseases, and reported here the characterization of a novel BVDV genotype/subgenotype strain SD-15 that is associated with an unusual high morbidity and mortality on a cattle farm in Shandong province, China.
Materials and Methods
Ethics statement
All sample collection and processing from diseased or dead cattle was performed following the protocol approved by the animal ethics committee of Jilin University.
Outbreak investigation
A disease characterized by severe diarrhea occurred on a cattle farm with 320 cattle in Shandong province after 40 cattle purchased from Inner Mongolia autonomous region were newly introduced to the original herd in early 2015. The sick cattle were observed to manifest pyrexia, anorexia in early days, and later oral mucous ulcer and severe diarrhea containing mucous and hemorrhage excretions were found. Approximately 67% (214/320) of cattle showed clinical signs and 60% (128/214) of the sick cattle succumbed to death within 10–15 days. Treatment of the sick cattle with gentamycin or tobramycin yielded no effects on relieving the clinical signs. Spleen samples were collected from four cattle for virus isolation after postmortem examination following the protocol approved by the animal ethics committee of Jilin University, and processed for electron microscope examination and virus detection.
Cell culture and virus isolation
Madin-Darby Bovine Kidney (MDBK) cells were grown in Dulbecco’s modified Eagle medium (DEME) containing 10% fetal calf serum (FBS) (HyClone, Logan, UT) and maintained in DMEM containing 2% FBS. For virus isolation, spleen samples were homogenized in a dilution of 1:10 (W/V) with 10 mM phosphate buffered saline (PBS), centrifuged at 10,000 × g at 4°C for 10 min, then passed through 0.45 nm filter before infecting cells. After infection, cell cultures were examined for the presence or absence of cytopathic effect (CPE) before they were frozen and thawed. The 5th passages of infected cells were used as stock for further characterization.
Indirect immunofluorescence assay (IFA)
MDBK cells were seeded into 24-well plates and infected with the 5th passage of isolated virus. The uninfected MDBK cells were used as the negative controls. 24–48 h postinfection, cells were fixed with methanol/acetone (1:1) for 30 min at -20°C, blocked using 1% bovine serum albumin, and incubated with polyclonal antibody against BVDV E2 protein for 1h at 37°C before the addition of Rhodamine-conjugated goat anti-rabbit antibody (1:500 dilution). After incubation for 45 min at 37°C, the cells were washed 3 times with PBS, sealed with glycerol, and examined using the fluorescence microscope.
RNA isolation and RT-PCR
Total RNAs were extracted from either the spleen samples or infected cell cultures using TRNzol kit (Tiangen, Beijing) following manufacturer`s instructions. Briefly, the samples were lysed in TRNzol® reagent, mixed with 0.2 volume of chloroform and shaken vigorously for 30 sec. After centrifugation at 12,000 x g for 20 min at 4°C, the aqueous phase was mixed with equal volume of isopropanol and centrifuged at 12,000 x g for 20 min at 4°C. The pellets were washed with 70% ethanol and dissolved in DEPC-treated H2O. The resultant RNAs were kept at –80°C for further analysis.
Reverse transcriptase reactions were performed using SuperScriptTM II Reverse Transcriptase (Invitrogen, Carlsbad, CA). Briefly, cDNA was synthesized in a volume of 20 μl containing 25 mM Tris-HCI, pH 8.3, 37.5 mM KCI, 1.5 mM MgCI2, 5mM DTT, 0.25 mM each of dATP, dCTP, dGTP and dTTP, 40 units of RNase inhibitor, 200 units of M-MLV reverse transcriptase, 2 μg of total RNA, and 2.5 μM random primers. The cDNA synthesis was performed at 42°C for 60 min. PCR amplification was done using Taq DNA polymerase (Takara, Dalian, Liaoning). The reaction was performed in a total volume of 50 μl containing 20 mM Tris-HCI, pH 8.4, 50 mM KCI, 3 mM MgCI2, 0.25 mM each of dATP, dCTP, dGTP and dTTP, 5 unit of Taq DNA polymerase, 1 μM of each primer, and 2 μl of the cDNA synthesized above. The amplification was done after the conditions were optimized. The primers used for detection of the potential pathogens were listed as Table 1. The primers used to amplify the complete genomic sequence for SD-15 were designed based on the nucleotide sequence alignment of the known BVDV strains in the GenBank and listed as Table 2.
Table 1. Primers used for detection of potential agents.
| Virus | Primer sequence (5’-3’) | Positions | |
|---|---|---|---|
| BMCFV | S | ATGACAGCAAGAGAATTAAACT | 49–70 |
| AS | ATGAATGACACCTCCAACAAGA | 479–458 | |
| RPV | GGGCTCTCATCAGCATCTTATC | 319–340 | |
| AS | GTATTTCACCCACCTCCGTAAC | 704–683 | |
| BTV | S | ATGAGATGTTTTTCATGTGTCT | 193–214 |
| AS | TTTAGTATTGCCGTTCTTAGTC | 877–856 | |
| BVDV | S | GAACCAGTTTATGACAAGGAAG | 454–475 |
| AS | GGGCAGTCTAGTCTGTTGTGGA | 875–854 | |
BMCFV: bovine malignant catarrhal fever virus; RPV: rinderpest virus; BTV: bluetongue virus; BVDV: bovine viral diarrhea virus; S: sense; AS: antisense.
Table 2. Primers used for amplification of the complete genome sequence.
| Primers | primer sequence(5’-3’) | Positions | |
|---|---|---|---|
| P1 | S | ACATGGGGTATACGAGATTTA | 1–21 |
| AS | CTCTTGTGTGGGAGCTTTA | 542–524 | |
| P2 | S | CCCACTGTGTTGCTACTAAAAAT | 350–372 |
| AS | CATCACTACCGGTAACTCTCCCAA | 705–682 | |
| P3 | S | TCCAGTTTACCACAGAGCCCCA | 675–696 |
| AS | GGTCTTCTCACTTGCATCCATCATA | 1337–1313 | |
| P4 | S | AGAACATAACACAATGGAAC | 1147–1166 |
| AS | AGTTTTCCAGTTTCTTTCCTAG | 1841–1821 | |
| P5 | S | ACTGCTCATTATCTAGTTGATGGAG | 1677–1701 |
| AS | ACCACCTTTACTAGACACACTAGGA | 2353–2329 | |
| P6 | S | TTTGAGGGCACTTAGAGACTTA | 2273–2294 |
| AS | GTCAGCAAGTTGCCCATCAT | 3583–3564 | |
| P7 | S | AACAGCGTGCACTATCAATTAC | 3290–3311 |
| AS | TACCTAAAGTAGTCTGTCACATAAC | 3979–3955 | |
| P8 | S | ATTGTAATAGGACTAATCGTGG | 3807–3828 |
| AS | TACCATGAGCATTTTTACTTTTG | 5051–5029 | |
| P9 | S | AGCCAGTACATTGAATAAAAACAGG | 4760–4784 |
| AS | ACTAGCATGTTACCCTTCATCTCC | 6313–6290 | |
| P10 | S | TTTATAGCCCCTGAAGTGATGAAAG | 6210–6234 |
| AS | ACTGCAACTTGTCTGATGTGGTC | 7678–7656 | |
| P11 | S | ACAGCACTCTACAAGAGCATAGC | 7557–7579 |
| AS | CTGTGTACCAGTTCAATCAACCT | 9040–9018 | |
| P12-Out | S | ACCAGGTTGGCTAAGAGATATACC | 8838–8861 |
| AS | ATCTATCTTGTCCCTGATTGCCTC | 10397–10374 | |
| P12-In | S | CAATCACCGTGATCTAGTAGAGAGG | 8900–8924 |
| AS | CTTTTCCAGTCCTATGCCTGC | 10331–10311 | |
| P13 | S | ATACTCAACCCTGGGAAGTTGTC | 10218–10240 |
| AS | CCCAAGTGTAGATAGGCTCAGGTTT | 11738–11714 | |
| P14 | S | GGACCCAATAGGGGCATA | 11630–11647 |
| AS | GGGGGCTGTTAAGGGTCTT | 12220–12202 |
Out: outer pair of primers; IN: inner pair of primers: sense; AS: antisense.
Cloning and sequencing
PCR-amplified fragments were either directly sequenced or cloned into pGM-T vector (Tiangen, Beijing) before being sequenced by Sangon Biotech Company (Shanghai, China). Every clone was sequenced twice using T7 primers. Complete genome sequence for SD-15 isolate was obtained by joining the fragment sequences obtained above.
Sequence analysis
Sequence analysis was performed using Lasergene 7 software (Madison, WI USA). Alignment analysis of the nucleotide sequence for SD-15 isolate with known BVDV strains was performed by CLUSTAL W algorithm [38]. Phylogenetic trees were constructed based on either the 5’-UTR sequence or the complete genome sequence using neighbor-joining method [39]. Sequence identity of SD-15 to pestiviruses were analyzed and calculated by DNASTAR software [40]. Different genotypes of pestivirus strains were randomly selected from Table 3 and used for multiple alignment analysis. Analyses of the deduced amino acid sequences for E2 and Erns were performed using CLUSTAL W algorithm [38]. The potential glycosylation sites within the E2 were analyzed by the NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc).
Table 3. The pestivirus strain used for alignment and phylogenetic analysis.
| Stains | Countries | Accession No | Collection date | Subtype | Host |
|---|---|---|---|---|---|
| GS5 | China | KJ541471 | 2013 | 1a | Bovine |
| Singer | Argentina | DQ088995 | 2007 | 1a | - |
| Oregon C24V | England | AF091605 | 1998 | 1a | - |
| SD1 | USA | M96751 | - | 1a | - |
| NADL | USA | M31182 | 1988 | 1a | - |
| Egy/Ismailia/2014 | Egypt | KR029825 | 2014 | 1b | Bovine |
| 12F004 | South Korea | KC963967 | 2012 | 1b | Cattle |
| CP7 | Germany | U63479 | 1987 | 1b | Cattle |
| AU526 | USA | KF835697 | 2013 | 1b | Buffy goat |
| CC13B | China | KF772785 | 2013 | 1b | Cattle |
| Bega-like | Australia | KF896608 | 2012 | 1c | Bovine |
| 10JJ-SKR | South Korea | KC757383 | 2010 | 1d | Cattle |
| Carlito | Switzerland | KP313732 | 2014 | 1e | Cattle |
| Suwancp | Switzerland | KC853440 | 1993 | 1k | Bovine |
| Suwacp | Switzerland | KC853441 | 1993 | 1k | Bovine |
| KS86-1cp | Japan | AB078952 | - | 1J | Bovine |
| KS86-1ncp | Japan | AB078950 | - | 1J | Bovine |
| ZM-95 | China | AF526381 | 1995 | 1m | Swine |
| SD15 | China | KR866116 | 2015 | 1m | Bovine |
| Camel-6 | China | KC695810 | 2013 | 1q | Camelus bactrianus |
| SD0803 | China | JN400273 | 2008 | 1q | Pig |
| SH-28 | China | HQ258810 | 2009 | 2a | Pig |
| HLJ-10 | China | JF714967 | 2011 | 2a | Cattle |
| JZ05-1 | China | GQ888686 | 2005 | 2a | Cattle |
| XJ-04 | China | FJ527854 | 2004 | 2a | Cattle |
| Hokudai-Lab/09 | Japan | AB567658 | 2010 | 2b | Bovine |
| SD1301 | China | KJ000672 | 2012 | 2b | Cattle |
| NRW19-13-8_Dup(+) | Germany | HG426490 | 2013 | 2c | Bos Taurus |
| Potsdam 1600 | Germany | HG426491 | 2000 | 2c | Bos Taurus |
| SH2210-23 | Germany | HG426494 | 2010 | 2c | Bos Taurus |
| VOE 4407 | Germany | HG426495 | 2007 | 2c | Bos Taurus |
| BD31 | USA | U70263 | - | BDV | Lamb |
| X818 | Germany | AF037405 | - | BDV | Sheep |
| Gifhorn | Germany | KF925348 | 2000 | BDV | Pig |
| H2121 | Germany | GU270877 | 2002 | BDV | Rupicapra rupicapra |
| Aveyron | Germany | KF918753 | 1984 | BDV | Sheep |
| Brescia | Switzerland | AF091661 | - | CSFV-1 | - |
| cF114 | China | AF333000 | - | CSFV-1 | - |
| JL1(06) | China | EU497410 | 2006 | CSFV-1 | Swine |
| Shimen | China | AF092448 | - | CSFV-1 | - |
| Heb52010 | China | JQ268754 | 2010 | CSFV-2 | Swine |
| Alfort/Tuebingen | Germany | J04358 | - | CSFV-2 | - |
| HEBZ | China | GU592790 | 2009 | CSFV-2 | Swine |
| JSZL | China | KT119352 | 2014 | CSFV-2 | Pig |
| PC11WB | South Korea | KC149991 | 2011 | CSFV-2 | Wild boar |
| Zj0801 | China | FJ529205 | 2008 | CSFV-2 | Swine |
Results
BVDV is the causative agents for the outbreak
Investigation on the outbreak showed that cattle purchased from Inner Mongolia autonomous region were those to first show watery diarrhea after they were transported from the Inner Mongolia to Shandong province and introduced to the cattle farm. The sick cattle were characterized with pyrexia, oral mucous ulcer and severe diarrhea with mucous and hemorrhage. Several days later, cattle in original herd began to show the clinical signs. By the end of the outbreak, approximately 67% (214/320) of cattle manifest clinical signs and 60% (128/214) of the diseased cattle succumbed to death within 10–15 days. No obvious effect was observed for the majority of sick cattle after they were treated with gentamycin or tobramycin, suggesting the outbreak is likely associated with viral agents.
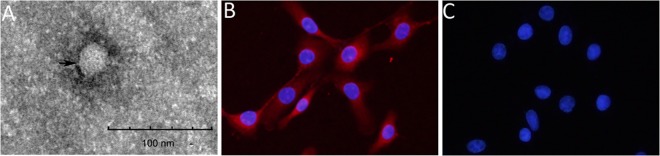
To define the agents for outbreak, feces and spleen samples from four sick or dead cattle were collected and processed for electron microscope (EM) examination or for viral nucleic acid detection. After EM observation, viruses with a size of 30–50 nm were found in all samples (Fig 1A). As shown in Table 4, after PCR amplification, no fragments were obtained using the primers for rinderpest virus, malignant catarrhal fever virus and bluetongue virus; while fragments with expected size of 420 bp were amplified from all four cattle examined using BVDV-specific primers, suggesting that BVDV is likely the agents associated with the outbreak. Sequencing the fragments from all four diseased cattle showed they contained the same nucleotide sequences except cattle 3 has one nucleotide mutation in relation to other three cattle (not shown), further confirming that BVDV likely is responsible for this outbreak.
Fig 1. SD-15 is a noncytopathic BVDV isolate responsible for the outbreak.
Viral particle was observed by electron microscopy (A). BVDV antigen was detected in MDBK cells infected by the inoculum by indirect immunofluorescence assay (B). Normal MDBK cells was detected by IFA (C). The scale bar was indicated as 100 nm.
Table 4. Detection of BVDV viruses from sick cattle with RT-PCR.
| Cattle No | BVDV | RPV | BTV | BMCFV | ||||
|---|---|---|---|---|---|---|---|---|
| Spleen | Feces | Spleen | Feces | Spleen | Feces | Spleen | Feces | |
| Calf-1 | + | + | - | - | - | - | - | - |
| Calf-2 | + | + | - | - | - | - | - | - |
| Calf-3 | + | + | - | - | - | - | - | - |
| Calf-4 | + | + | - | - | - | - | - | - |
BVDV: bovine viral diarrhea virus; RPV: rinderpest virus; BTV: bluetongue virus; BMCFV: bovine malignant catarrhal fever virus; +: positive; -: negative
SD-15 is a noncytopathic biotype BVDV isolate
To isolate the virus, spleen samples were processed and inoculated into MDBK cells. After 5 passages, no obvious cytopathic effects were observed for all four samples. However, the specific immunofluroscent signals were detected in the cytoplasm of infected cells using BVDV E2 antibody generated against the recombinant E2 protein (Fig 1B). No immunofluroscent signal was detected in the mock MDBK cells (Fig 1C). To assure the above results, PCRs were performed and fragments were obtained from infected cells, and revealed they were indeed BVDV-specific sequence (not shown). No fragments were amplified from normal cell controls. Those results further confirm the isolated viruses were BVDVs. The representative BVDV strain was designated as BVDV SD-15 and used for further characterization.
Complete genome sequence of SD-15 and its encoded polyprotein
Since SD-15 was noncytopathic BVDV demonstrated to be associated with an outbreak of such an unusual high morbidity and mortality, it is necessary to unveil its complete genome sequence in an attempt to explore the molecular characters. PCR were performed and the complete genome sequence of SD-15 was obtained by joining the overlapped fragment sequences. After assembling the sequences, the complete genome of SD-15 was revealed to consist of 12285 nucleotides including a 5’-UTR of 300 nt, 3’-UTR of 200 nt, and an open reading frame (ORF) encoding a large precursor polyprotein of 3900 amino acids. Analysis of the deduced amino acid sequence showed that SD-15, like the other BVDV strains, had a conserved domain of NS3 polyprotein peptidase C31 located at 1060–1241 aa, a conserved domain of NS2 peptidase at 882–1016 aa, and the Npro endopeptidase C53 at position of 4–115 aa. Similar to the majority of BVDV-1 strains, SD-15 encoded a polyprotein putatively consisting of 4 structural proteins and 7 nonstructural proteins. No insertion/deletion or recombination was revealed between boundary of NS2 and NS3 nucleotide sequence. The complete genomic sequence of SD-15 was deposited to GenBank with an accession no KR866116.
SD-15 belongs to BVDV-1m subgenotype
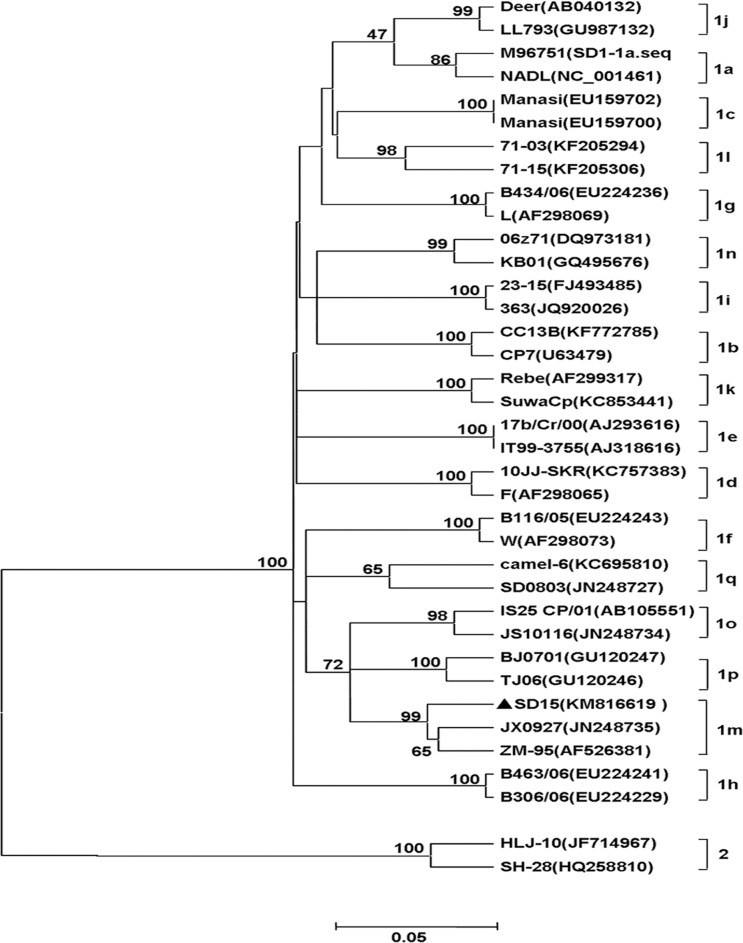
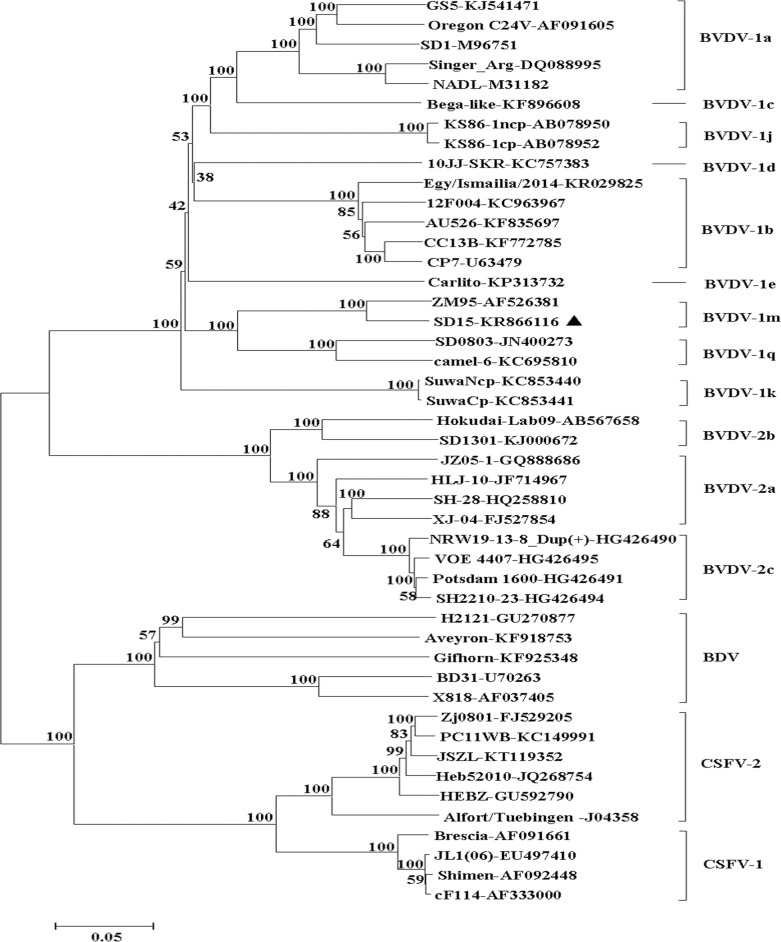
To determine the genotype of SD-15, phylogenetic analysis using neighbor-joining method was performed by analyzing the 5’-UTR sequence of SD-15 with the corresponding sequences of the 36 representative BVDV. As shown in Fig 2, the SD-15, together with ZM-95 and JX0927 was clustered to BVDV-1m subgenotype. Similar result was obtained after analysis of the complete genome sequence of SD-15 with 46 representative pestiviruses listed in Table 3, where four major distinct clusters including BVDV-1, BVDV-2, BDV and CSFV were divided, and SD-15 was grouped to a BVDV-1m subgenotype with ZM-95 within BVDV-1 isolates. Those findings further establish the phylogenetic status of SD-15 as BVDV-1m (Fig 3).
Fig 2. Phylogenetic analysis of SD-15 isolates based on 5’-UTR sequences.
Phylogenic relationship of SD-15 to pestiviruses was generated by analyzing the 5’-UTR sequences from 36 representative BVDV isolates using neighbor-joining method. SD-15, together with BVDV ZM-95 and JX0927 was clustered to BVDV-1m subgenotype and marked as solid triangle.
Fig 3. Phylogenetic analysis of SD-15 with pestiviruses based on the complete genomic sequences.
The complete genome sequence of SD-15 was aligned with representative pestiviruses and phylogenetic tree was generated using the neighboring-joining methods by MEGA 5.2 software. Four major clusters including CSFV, BDV, BVDV-2 and BVDV-1 with different subgenotypes were formed and indicated, respectively. SD-15 together with ZM-95 was grouped in BVDV-1m subgenotype.
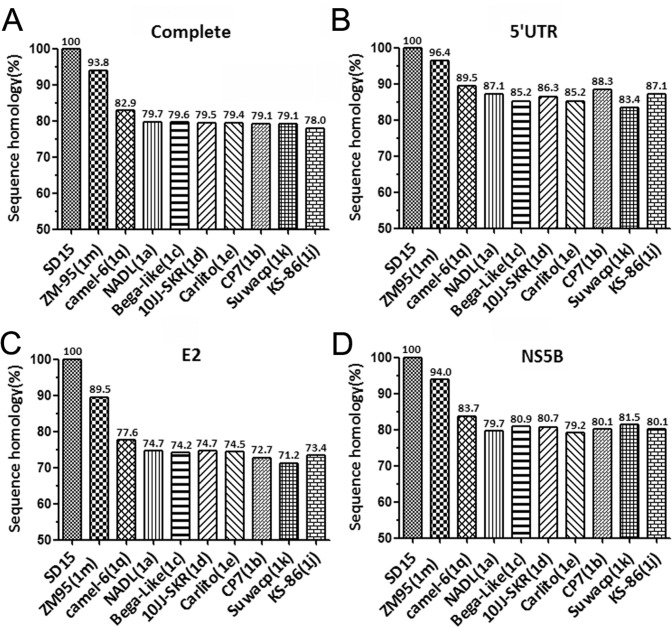
SD-15 had the highest sequence identity with ZM-95 and a diversified sequence homology with other BVDV-1
To determine the sequence homology of SD-15 with other BVDV-1 isolates, the complete sequence, 5’-UTR, E2 and NS5B sequences of SD-15 were aligned with corresponding sequences of BVDV-1 strains, respectively. As shown in Fig 4A, the complete genomic sequence identity of SD-15 was ranging from 78% to 93.8% with other BVDV-1 isolates, where it had the highest homology of 93.8% with ZM-95 isolate, a BVDV-1m strain originally isolated in 1995 from pigs showing clinical signs and lesions similar to classical swine fever in Inner Mongolia region. Similar patterns were also observed when 5’-UTR, E2 and NS5B genes were used for the analysis (Fig 4B–4D). Those results clearly demonstrated a closer relationship of SD-15 to the ZM-95 than other BVDV-1 strains. While the majority of SD-15-encoded genes shared higher sequence identity with ZM-95, the E2 had only 89.5% sequence homology with ZM-95 (Fig 4C), suggesting that the E2 coding region diversified more from the ZM-95 strains than other regions. It was also noted that the 5’-UTR sequence of SD-15 was relatively conserved among BVDV-1 strains in relation to other regions (Fig 4B).
Fig 4. SD-15 is evolutionarily closed to ZM-95 and had diversified sequence homology with other BVDV-1 subgenotypes.
The complete genome (A), 5’-UTR (B), E2 (C) and NS5B (D) nucleotide sequences of SD-15 were aligned with different BVDV-1 subgenotypes by the DNASTAR software. Sequence identities of SD-15 genes with other BVDV-1 subgenotypes were shown beyond the bars, respectively.
Unique glycosylation site revealed in the E2 of SD-15 strain
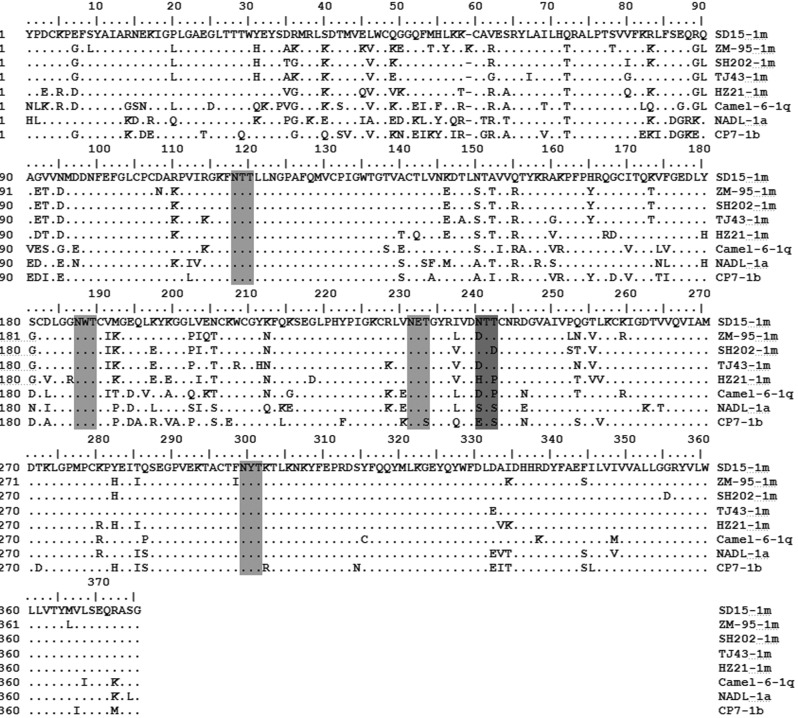
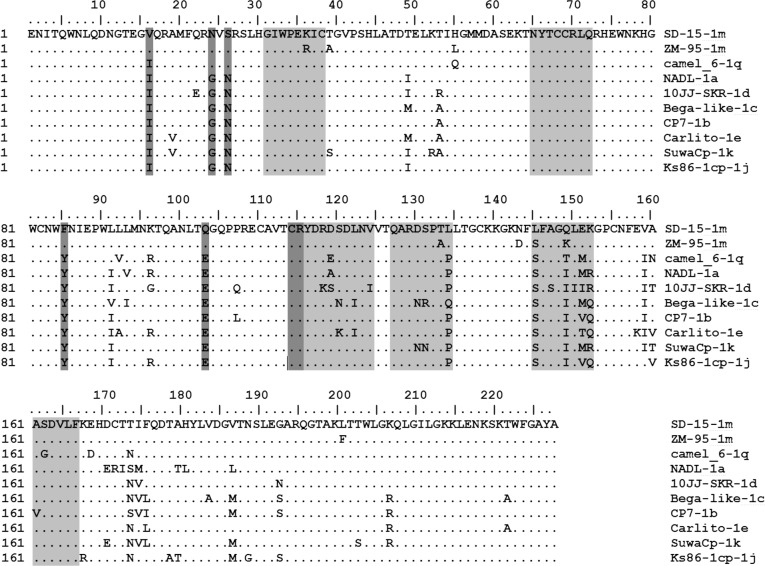
The above results indicate that SD-15-encoded E2 was evolved faster compared with other SD-15-encoded genes. To further determine the diversity of E2 gene, alignment analysis of E2 amino acid sequences of SD-15 with representative BVDV-1 strains (1a, 1b, 1c, 1d, 1e, 1j, 1k, 1m, 1q) was performed. As illustrated in Fig 5, two highly conserved regions within the E2 sequence were revealed to locate at amino acid position of 99–136 and 286–364, respectively, indicating the important role of these conserved regions as reported previously [41]. Moreover, three variable regions were located at positions 1–63, 81–94 and 137–215 (Fig 5), suggesting those regions likely evolved faster than other regions within E2 during BVDV evolution.
Fig 5. A unique glycosylation site revealed within SD-15-encoded E2 amino acid sequence.
Potential glycosylation sites within E2 amino acid sequence of SD-15 were predicted using NetNGlyc 1.0 Server software. An extra and unique glycosylation site was revealed at 240NTT only in SD-15 strain in addition to four conserved glycosylation sites shared with all BVDV-1 strains examined. The glycosylation sites shared by all BVDV-1 examined were less lightly shadowed than the unique glycosylation site in SD-15.
To further characterize E2 protein sequence, the potential glycosylation sites were analyzed by the NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc). As shown in Fig 5, four potential glycosylation sites Asn-X-Ser/Thr were found in the E2 protein for the majority of BVDV-1 genotypes (1a, 1b, 1c, 1d, 1e, 1j, 1k, 1m, 1q), where three glycosylation sites (117 NTT,186 NWT and 298 NYT) were highly conserved and one site (230 NET) was different only in CP7 BVDV-1 strain. It is surprising to note that an additional glycosylation site (240 NTT) was found in C-terminus of E2 only in SD-15 strain. Whether this extra glycosylation site in SD-15 affects pathogenicity or antigenicity of SD-15 strain is unknown and the subject for future investigation.
SD-15-encoded Erns had a unique mutation and linear epitope
Seven linear epitopes of Erns protein were well-characterized previously [42]. They include the epitope 31GIWPEKIC38, 65NYTCCKLQ72, 127QARNRPTT134, 145SFAGTVIE152, 161VEDILY166, 114CRYDKNTDVNV124 and 116YDKNTDVNV124. Since Erns is structural glycoprotein that possess the intrinsic ribonuclease activity involved in virus attachment and entry into target cells, analysis of the sequence variation will enhance the understanding of virus evolution. Therefore, diversity of Erns amino acid sequence of BVDV-1 genotypes (1a, 1b, 1c, 1d, 1e, 1j, 1k, 1m, 1q) was analyzed by comparison of SD-15 with those of other BVDV strains. As shown in Fig 6, the amino acid sequence of Erns of SD-15 was much conserved than E2 protein within different BVDV-1 genotypes. Seven linear epitopes were also observed in SD-15 strain, in which the linear epitopes 145SFAGTVIE152 was highly variable and the remains were conserved. The residues W33, L71, Q127, N130, S145, and G148 were conserved in all BVDV-1 strains examined except SD-15 strain, where the S145L was observed only in SD-15 isolate, suggesting the conserved function of those amino acid residues. In addition, several unique mutations including residues I16V, Y85F, E103Q in the Erns of BVDV-1m, residues G24N and N26S in BVDV-1m and BVDV-1q were found.
Fig 6. Unique amino acid and linear epitope mutations revealed in the SD-15-encoded Erns protein.
Analysis of the deduced amino acid sequence of SD-15-encoded Erns was performed to explore the variability of linear epitopes in relation to known BVDV-1 strains. Out of seven linear epitopes of Erns mapped, six were conserved among BVDV-1 genotypes and one was variable. Unique amino acid mutations and linear epitope mutations were also observed only in BVDV-1m subgenotype and highlighted.
Discussion
BVD is one of the most significant diseases impacting the cattle industry worldwide. While reports on BVDV outbreak increased significantly in China in the last 15 years, they mainly focused on the BVDV genotyping using the 5’-UTR sequence regions [33, 43], resulting in the molecular aspects and genome sequences of BVDV related to those outbreak neglected. In this study, we characterized a novel BVDV isolate SD-15 from cattle population manifesting severe diarrhea and mucosal ulcers. Epidemiological investigation traced the cattle back to Zemeng county of Inner Mongolia autonomous region, where the first BVDV-1m strain ZM-95 was isolated. ZM-95 is a cytopathogenic BVDV strain isolated by our laboratory from pigs showing clinical signs and pathological lesions similar to classical swine fever twenty year ago in China [7]. SD-15 is a noncytopathic BVDV strain isolated in this study from cattle newly introduced to Shandong from Zemeng County of the Inner Mongolia autonomous regions. The complete genome sequence analysis and molecular characterization of SD-15 demonstrated that SD-15, together with ZM-95, belongs to BVDV-1m subgenotype that is currently reported only in China. The biotype difference between ZM-95 (CP) and SD-15 (NCP) was likely due to long-term interactive adaption of virus to host. Since BVDV-1m was exclusively reported in China, we assume that BVDV-1m had been existing in cattle population for long time and evolved to infect other species such as pigs, sheep and deer. This is evidenced by our previous etiological investigation that both cattle and sheep in the Inner Mongolia region had a very high BVDV infection [44]. The finding that the majority of BVDV-1 strains from other regions in China were BVDV-1m [33, 45], indicating BVDV-1m strains were likely to spread widely in China. Although we had no direct evidence bridging ZM-95 and SD-15 BVDV strain currently, the results that ZM-95 and SD-15 were isolated from pigs and cattle, respectively, in the same region of the Inner Mongolia indicate that BVDV-1m strain is likely the main subgenotype circulating in the Inner Mongolia. Since Inner Mongolia region was the hub of cattle trade and cattle industry in China, the BVDV-1m strains detected in other regions in China are probably originated from Inner Mongolia.
The first BVD was reported in Jilin China in 1980 [27]. Since then, BVD outbreak has not been increasingly reported until the later 1990s’ and early 2000s’. Recently, BVDs were reported in many regions across China and the predominant genotypes/subgenotypes were BVDV-1b and BVDV-1m [15, 34, 35]. Since the phylogenetic analysis clustered the first BVDV strain CC-184, which was isolated in China by our laboratory from the cows imported from Europe, to BVDV-1b [27–28], it is likely that the current BVDV-1b strains reported in China were originally from European countries [28, 46]. Majority of BVDV strains isolated recently from Jilin and other regions in China including BVDV-CC13B, JL-1 isolates were BVDV-1b subgenotype that shared the highest sequence identity with the CP7 strain originally isolated in Germany [21, 47]. Those results further support our hypothesis that predominant BVDV-1b subgenotype currently spread in China is originated from Europe.
Sequencing SD-15 genome revealed a 12,285 nucleotides that encoded a polyprotein consisting of 4 structural proteins and 7 nonstructural proteins. Like the majority of BVDV-1 strains, SD-15 contains no cellular sequence or viral sequence insertion within the boundary NS2/NS3 nonstructural proteins. Alignment analysis using the complete sequence, 5’-UTR, E2 and NS5B showed SD-15 had a sequence identity of 93.8% with ZM95, which is much higher than those of BVDV-1 strains such as BVDV-1a NADL (79.7%) and BVDV-1b CP7 (79.1%), indicating SD-15 had lower sequence homology to the BVDV-1a and BVDV-1b subgenotypes than the rest of the BVDV-1 subgenotype. It is interesting to note that SD-15-encoded E2 had the highest sequence diversity within BVDV-1 viruses, especially at the regions of amino acid residues 1–63, 81–94 and 137–215, indicating that SD-15-encoded E2 evolved faster compared with other SD-15-encoded genes during BVDV evolution.
It has been demonstrated that all three glycoproteins encoded by the pestiviruses are involved in the virus attachments and entry into the target cells [25]. The glycoprotein E2 is essential for virus entry and infectivity via the formation of E1-E2 heterodimer. Erns has intrinsic ribonuclease activity that functions to inhibit the production of type I interferons and help in the development of persistent infection [25, 26]. Studies has demonstrated that variation in the amino acid and antigenic structure, disulfide bond formation, glycosylation, and RNase activity affect the virulence of pestiviruses to the animals. Also, the antigenic difference in glycosylation influence the efficacy of vaccine. As one of the most common forms of protein modifications, glycosylation play an important role in virus infection and alteration of glycosylation sites had significant impact on virus survival, pathogenicity, antigenicity, and transmissibility [25, 48–50]. The discoveries of an extra and unique glycosylation site in E2 of SD-15 strain in addition to four glycosylation sites shared by all BVDV-1 subgenotype suggest that this unique glycosylation site may have an effect on the virulence and pathogenicity of SD-15. The findings of unique amino acid mutations and linear epitope alteration in Erns of SD-15 suggest that these mutations likely also affect the antigenicity and pathogenicity of the SD-15, which is the subject for future exploration.
Acknowledgments
The authors thank Professor Shenghua Yang at the Jilin University for his assistance for EM observations.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the Ministry of Science and Technology of China with a grant no. 2016YFD0500904 to XPW; and by the People's Government of Jilin Province for the Key Scientific and Technology projects with grant no. 20140204064NY to XPW.
References
- 1.Baker J. Bovine viral diarrhea virus: a review. J Am Vet Med Assoc. 1987;190(11): 1449–1458. [PubMed] [Google Scholar]
- 2.Duffell SJ, Harkness JW. Bovine virus diarrhoea-mucosal disease infection in cattle. Vet Rec. 1985;117(10): 240–245. 10.1136/vr.117.10.240 [DOI] [PubMed] [Google Scholar]
- 3.Houe H. Epidemiological features and economical importance of bovine virus diarrhoea virus (BVDV) infections. Vet Microbiol. 1999;64(2–3): 89–107. 10.1016/S0378-1135(98)00262-4 [DOI] [PubMed] [Google Scholar]
- 4.Carman S, van Dreumel T, Ridpath J, Hazlett M, Alves D, Dubovi E, et al. Severe acute bovine viral diarrhea in Ontario, 1993–1995. J Vet Diagn Invest. 1998; 10(1): 27–35. 10.1177/104063879801000106 [DOI] [PubMed] [Google Scholar]
- 5.Kim SG, Anderson RR, Yu JZ, Zylich NC, Kinde H, Carman S, et al. Genotyping and phylogenetic analysis of bovine viral diarrhea virus isolates from BVDV infected alpacas in North America. Vet Microbiol. 2009;136(3–4): 209–216. 10.1016/j.vetmic.2008.10.029 [DOI] [PubMed] [Google Scholar]
- 6.Løken T. Ruminant pestivirus infections in animals other than cattle and sheep. Vet Clin North Am Food Anim Pract. 1995;11(3): 597–614. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Tu C, Li H, Jin K, Xuan H, Chang G, et al. Detection and isolation of bovine viral diarrhea virus from classical swine fever suspected pigs. Chin J Vet Sci. 1996;16(4): 341–345. [Google Scholar]
- 8.Rodríguez-Prieto V, Kukielka D, Rivera-Arroyo B, Martínez-López B, de las Heras AI, Sánchez-Vizcaíno JM, et al. Evidence of shared bovine viral diarrhea infections between red deer and extensively raised cattle in south-central Spain. BMC Vet Res. 2016;12(1): 1 10.1186/s12917-015-0630-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagai M, Sato M, Nagano H, Pang H, Kong X, Murakami T, et al. Nucleotide sequence homology to bovine viral diarrhea virus 2 (BVDV 2) in the 5' untranslated region of BVDVs from cattle with mucosal disease or persistent infection in Japan. Vet Microbiol. 1998;60(2–4): 271–276. 10.1016/S0378-1135(98)00158-8 [DOI] [PubMed] [Google Scholar]
- 10.Becher P, Orlich M, Kosmidou A, Konig M, Baroth M, Thiel HJ. Genetic diversity of pestiviruses: identification of novel groups and implications for classification. Virology. 1999;262(1): 64–71. 10.1006/viro.1999.9872 [DOI] [PubMed] [Google Scholar]
- 11.Rebhun WC, French TW, Perdrizet JA, Dubovi EJ, Dill SG, Karcher LF. Thrombocytopenia associated with acute bovine virus diarrhea infection in cattle. J Vet Intern Med. 1989;3(1): 42–46. 10.1111/j.1939-1676.1989.tb00327 [DOI] [PubMed] [Google Scholar]
- 12.Pellerin C, van den Hurk J, Lecomte J, Tijssen P. Identification of a new group of bovine viral diarrhea virus strains associated with severe outbreaks and high mortalities. Virology. 1994;203(2): 260–268. 10.1006/viro.1994.1483 [DOI] [PubMed] [Google Scholar]
- 13.Luzzago C, Lauzi S, Ebranati E, Giammarioli M, Moreno A, Cannella V, et al. Extended genetic diversity of bovine viral diarrhea virus and frequency of genotypes and subtypes in cattle in Italy between 1995 and 2013. Biomed Res Int. 2014;2014: 147145 10.1155/2014/147145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vilcek S, Durkovic B, Kolesarova M, Paton DJ. Genetic diversity of BVDV: consequences for classification and molecular epidemiology. Prev Vet Med. 2005;72(1–2): 31–35; discussion 215–219. 10.1016/j.prevetmed.2005.08.004 [DOI] [PubMed] [Google Scholar]
- 15.Xue F, Zhu YM, Li JA, Zhu LC, Ren XG, Feng JK, et al. Genotyping of bovine viral diarrhea viruses from cattle in China between 2005 and 2008. Vet Microbiol. 2010;143(2–4): 379–383. 10.1016/j.vetmic.2009.11.010 [DOI] [PubMed] [Google Scholar]
- 16.Giangaspero M, Harasawa R, Weber L, Belloli A. Genoepidemiological evaluation of Bovine viral diarrhea virus 2 species based on secondary structures in the 5 ' untranslated region. J Vet Med Sci. 2008;70(6): 571–580. 10.1292/jvms.70.571 [DOI] [PubMed] [Google Scholar]
- 17.Flores EF, Ridpath JF, Weiblen R, Vogel FSF, Gil LHVG. Phylogenetic analysis of Brazilian bovine viral diarrhea virus type 2 (BVDV-2) isolates: evidence for a subgenotype within BVDV-2. Virus Res. 2002;87(1): 51–60. 10.1016/S0168-1702(02)00080-1 [DOI] [PubMed] [Google Scholar]
- 18.Wang W, Shi X, Tong Q, Wu Y, Xia MQ, Ji Y, et al. A bovine viral diarrhea virus type 1a strain in China: isolation, identification, and experimental infection in calves. Virol J. 2014;11: 8 10.1186/1743-422X-11-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng R, Brock KV. Molecular cloning and nucleotide sequence of a pestivirus genome, noncytopathic bovine viral diarrhea virus strain SD-1. Virology. 1992;191(2): 867–869. [DOI] [PubMed] [Google Scholar]
- 20.Colett MS, Larson R, Gold C, Strick D, Anderson DK, Purchio AF. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology. 1988;165(1): 191–199. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S, Tan B, Ding Y, Wang F, Guo L, Wen Y, et al. Complete genome sequence and pathogenesis of bovine viral diarrhea virus JL-1 isolate from cattle in China. Virol J. 2014;11: 67 10.1186/1743-422X-11-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tautz N, Elbers K, Stoll D, Meyers G, Thiel HJ. Serine protease of pestiviruses: determination of cleavage sites. J Virol. 1997;71(7): 5415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rumenapf T, Unger G, Strauss JH, Thiel HJ. Processing of the envelope glycoproteins of pestiviruses. J Virol. 1993;67(6): 3288–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. [DOI] [PubMed] [Google Scholar]
- 25.Shirato K, Miyoshi H, Goto A, Ako Y, Ueki T, Kariwa H, et al. Viral envelope protein glycosylation is a molecular determinant of the neuroinvasiveness of the New York strain of West Nile virus. J Gen Virol. 2004;85: 3637–3645. 10.1099/vir.0.80247-0 [DOI] [PubMed] [Google Scholar]
- 26.Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, Chen J, et al. The flavivirus precursor membrane-envelope protein complex: Structure and maturation. Science. 2008;319(5871): 1830–1834. 10.1126/science.1153263 [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Liu Z, Wu Y. Isolation and identification of bovine viral diarrhea virus-mucosal disease virus strain Changchun 184. Chin J Vet Sci. 1983;3(2): 546–553. [Google Scholar]
- 28.Wang X, Tu C, Li H, Xuan H, Zhu W, Fei E, et al. Comparison of the main region within P125 gene of bovine viral diarrhea virus. Chin J Vet Sci. 1996;16(6): 546–553. [Google Scholar]
- 29.Xu X, Zhang Q, Yu X, Liang L, Xiao C, Xiang H, et al. Sequencing and comparative analysis of a pig bovine viral diarrhea virus genome. Virus Res. 2006;122(1–2): 164–170. 10.1016/j.virusres.2006.05.005 [DOI] [PubMed] [Google Scholar]
- 30.Weng X, Song Q, Wu Q, Liu M, Wang M, Wang J. Genetic characterization of bovine viral diarrhea virus strains in Beijing, China and innate immune responses of peripheral blood mononuclear cells in persistently infected dairy cattle. J Vet Sci. 2015;16(4): 491–500. 10.4142/jvs.2015.16.4.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Y, Wang S, Du R, Wang Q, Sun C, Wang N, et al. Isolation and identification of a bovine viral diarrhea virus from sika deer in china. Virol J. 2011;8: 83 10.1186/1743-422X-8-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong X, Cao X, Zheng F, Chen Q, Zhou J, Yin H, et al. Identification and characterization of a novel subgenotype of bovine viral diarrhea virus isolated from dairy cattle in Northwestern China. Virus Genes. 2013;46(2): 375–376. 10.1007/s11262-012-0861-3 [DOI] [PubMed] [Google Scholar]
- 33.Zhang S, Tan B, Guo L, Wang F, Zhu H, Wen Y, et al. Genetic diversity of bovine viral diarrhea viruses in commercial bovine serum batches of Chinese origin. Infect Genet Evol. 2014;27: 230–233. 10.1016/j.meegid.2014.07.021 [DOI] [PubMed] [Google Scholar]
- 34.Zhong F, Li N, Huang X, Guo Y, Chen H, Wang X, et al. Genetic typing and epidemiologic observation of bovine viral diarrhea virus in Western China. Virus Genes. 2011;42(2): 204–207. 10.1007/s11262-010-0558-4 [DOI] [PubMed] [Google Scholar]
- 35.Xue F, Zhu Y, Li J, Zhu L, Ren X, Feng J, et al. Genotyping of bovine viral diarrhea viruses from cattle in China between 2005 and 2008. Vet Microbiol. 2010;143(2–4): 379–383. 10.1016/j.vetmic.2009.11.010 [DOI] [PubMed] [Google Scholar]
- 36.Wang W, Shi X, Chen C, Wu H. Genetic characterization of a noncytopathic bovine viral diarrhea virus 2b isolated from cattle in China. Virus Genes. 2014;49(2): 339–341. 10.1007/s11262-014-1067-7 [DOI] [PubMed] [Google Scholar]
- 37.Tao J, Wang Y, Wang J, Wang JY, Zhu G. Identification and genetic characterization of new bovine viral diarrhea virus genotype 2 strains in pigs isolated in China. Virus Genes. 2013;46(1): 81–87. 10.1007/s11262-012-0837-3 [DOI] [PubMed] [Google Scholar]
- 38.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24): 4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4): 406–425. [DOI] [PubMed] [Google Scholar]
- 40.Burland TG. DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol. 1999:71–91. [DOI] [PubMed] [Google Scholar]
- 41.El Omari K, Iourin O, Harlos K, Grimes JM, Stuart1 DI. Structure of a Pestivirus Envelope Glycoprotein E2 Clarifies Its Role in Cell Entry. Cell Rep. 2013; 3(1):30–35. 10.1016/j.celrep.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang FI, Deng MC, Huang YL, Chang CY. Structures and Functions of Pestivirus Glycoproteins: Not Simply Surface Matters. Viruses. 2015; 7(7): 3506–3529. 10.3390/v7072783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng M, Ji S, Fei W, Raza S, He C, Chen Y, et al. Prevalence study and genetic typing of bovine viral diarrhea virus (BVDV) in four bovine species in China. PloS one. 2015;10(4): e0121718 10.1371/journal.pone.0121718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, Zhu W, Ren W, Chang G, Hu X, Liu H, et al. Investigation of bovine viral diarrhea-mucosal disease infection in cattle and sheep in Inner Mongolia regions. Chin J Prev Vet Med. 1993;4: 41–42. [Google Scholar]
- 45.Lang Y, Gao S, Du J, Shao J, Cong G, Lin T, et al. Polymorphic genetic characterization of E2 gene of bovine viral diarrhea virus in China. Vet Rec. 2014;174(3–4): 554–559. 10.1016/j.vetmic.2014.10.018 [DOI] [PubMed] [Google Scholar]
- 46.Shi S,Zhang X. bovine viral diarrhea virus-mucosal disease virus was isolated from frozen semen imported from New Zealand. Chin J Vet Dru. 1987;01: 26–27. [Google Scholar]
- 47.Zhu L, Xing Z, Jia C, Wang T, Gai X, Song L, et al. Complete genomic sequence of bovine viral diarrhea virus strain CC13B. Chin J Vet Sci. 2014;34(7): 1065–1071. [Google Scholar]
- 48.Sainz IF, Holinka LG, Lu Z, Risatti GR, Borca MV. Removal of a N-linked glycosylation site of classical swine fever virus strain Brescia Erns glycoprotein affects virulence in swine. Virology. 2008, 370(1):122–129. 10.1016/j.virol.2007.08.028 [DOI] [PubMed] [Google Scholar]
- 49.Fernandez-Sainz I, Holinka LG, Gavrilov BK, Prarat MV, Gladue D, Lu Z, et al. Alteration of the N-linked glycosylation condition in E1 glycoprotein of classical swine fever virus strain Brescia alters virulence in swine. Virology. 2009, 386(1): 210–216. 10.1016/j.virol.2008.12.042 [DOI] [PubMed] [Google Scholar]
- 50.Risatti GR, Holinka LG, Sainz IF, Carrillo C, Lu Z, Borca M. N-linked glycosylation status of classical swine fever virus strain Brescia E2 glycoprotein influences virulence in swine. J Virol. 2007, 81(2): 924–933. 10.1128/JVI.01824-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.