Abstract
Development of the intestinal microbiota during early life serves a key regulatory stage in establishing the host-microbial relationship. This symbiotic relationship contributes to developing host immunity and maintaining health throughout the life span. This study was to develop an approach to colonize conventionally raised mice with a model probiotic bacterium, Lactobacillus rhamnosus GG (LGG), and determine the effects of LGG colonization on intestinal development and prevention of colitis in adulthood. LGG colonization in conventionally raised was established by administering LGG to pregnant mice starting at gestational day 18 and pups at postnatal day 1 to day 5. LGG colonization promoted bodyweight gain and increased diversity and richness of the colonic mucosa-associated microbiota prior to weaning. Intestinal epithelial cell proliferation, differentiation, tight junction formation and mucosal IgA production were all significantly enhanced in LGG colonized mice. Adult mice colonized with LGG showed increased IgA production and decreased susceptibility to intestinal injury and inflammation induced in the dextran sodium sulphate model of colitis. Thus, neonatal colonization of mice with LGG enhances intestinal functional maturation and IgA production and confers life long health consequences on protection from intestinal injury and inflammation. This strategy might be applied for benefiting health in the host.
Keywords: functional maturation of the intestine, IgA, the intestinal microbiota
INTRODUCTION
Colonization of the intestinal mictobiota in humans, occurring in utero,1 is significantly affected by microbial exposures at birth.2 Post-natal colonization is a dynamic and genetically controlled process, leading to establishing a symbiotic relationship between the host and the microbiota.3 This relationship provides an ecosystem that includes a nutrient-rich environment for bacteria, while conferring to the host important contributions, including nutrient metabolism, functional maturation, and the development of immunity.4
Clinical studies have revealed that interruption of the host-microbial relationship leads to several diseases, such as inflammatory bowel disease (IBD), consisting of ulcerative colitis and Crohn’s disease.5–7 Antibiotic treatment has been shown to alter the gut microbiome in humans8 and the antibiotic exposure in childhood is associated with an increased risk of developing IBD.9 Animal studies have demonstrated several defects in development of intestinal morphology, motility, epithelial proliferation and differentiation and the immune system in germ-free animals.10 These defects in germ-free animals can be restored through monoassociation.11 Altering the intestinal microbiota during early development by antibiotic treatment has long-term metabolic consequences even when the microbiota returns to a normal population structure after the cessation of antibiotics.12 These findings emphasize the important contribution of the intestinal microbiota to intestinal homeostasis during development and throughout the life span.
Probiotics have shown clinical benefits on humans of different ages. For example, preliminary evidence suggests that probiotics have efficacy for treatment of infants and young children with infectious diarrhea13, 14 and prevention of antibiotic-associated diarrhea.14 However, a number of meta-analyses vary in their conclusions on the effectiveness of probiotics on disease prevention and treatment.15 Therefore, hypothesis-driven clinical trials using optimal strains, dose, and delivery and treatment protocols are needed.
Several animal studies have suggested that short-term treatment of probiotics, such as LGG,16 L. reuteri,17 and E. coli Nissel 1917,18 enhanced proliferation, migration, differentiation, barrier function formation, and immune defense in intestinal epithelial cells in mice. These findings underscore the importance of identifying the effects of probiotics on intestinal development and disease prevention in the later stage of life.
LGG, a naturally occurring gram-positive bacterium, was originally isolated from the healthy human intestine. LGG has powerful adhesive properties for intestinal cells.19 Perinatal administration of LGG, L acidophilus La-5, and Bifidobacterium animalis subsp. lactis Bb-12 to mothers led to colonization of LGG, but not the other two bacteria, in the children at 10 days and at 3 months of age, which indicates that LGG has high ability to transfer from the mother to the child and colonize in the child.20 LGG has been widely used as a nutritional supplement. Clinical studies have demonstrated that perinatal LGG supplementation to mothers reduced the incidence of eczema at 2, 4 and 7 years of life in at-risk children.21–23 Feeding infants with formulas supplemented with LGG was well tolerated and supported growth.24 LGG has also shown beneficial effects on treating and/or preventing several disorders, including diarrhea and atopic dermatitis.25 To further understand the mechanisms underlying the effects of LGG on disease prevention and treatment, we have demonstrated that LGG and the LGG-derived protein, p40, prevented cytokine-induced apoptosis, preserved barrier function, and up-regulated mucin production in cultured intestinal epithelial cells and in tissue culture models, thereby prevention and treatment of experimental colitis.26–28
Here, we colonized conventionally raised neonatal mice with LGG and provided evidence that LGG colonization promoted growth, epithelial proliferation, differentiation, tight junction formation, IgA production, and maturation of the intestinal microbiota during development and decreased susceptibility to intestinal injury and colitis in adult mice. These results support the efficacy of administration of LGG at early life for enhancing intestinal functional maturation and long-term health consequences in adults.
RESULTS
Generation of an approach for colonization of conventionally raised mice with LGG
The evidence that the microbial exposure at birth shapes the acquisition and structure of the initial microbiota in newborns2 supports the importance of early exposure of microbes for colonization. Furthermore, studies in germ-free mice showed that there was a time window for colonization by caecal contents.29 Therefore, we first generated an approach for colonization of conventionally raised mice with LGG.
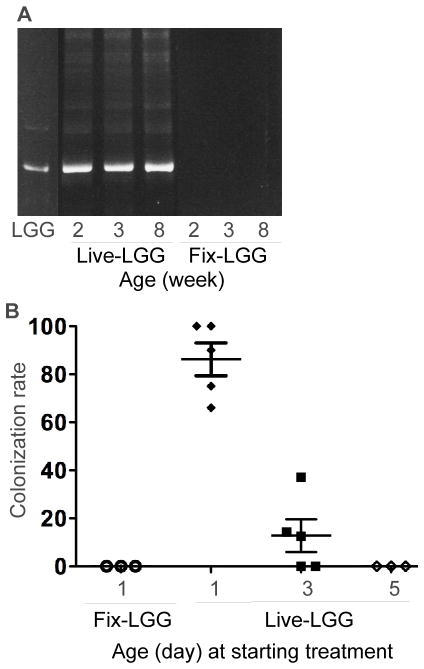
We treated wt pregnant mice with live LGG (Live-LGG) from gestation day 18 to delivery and newborn mice starting at postnatal day 1, 3, and 5 for 5 days. Glutaraldehyde-fixed LGG (Fix-LGG) was used as non-colonization control in this study. Colonization of LGG was defined by recovery of LGG from cultured mouse fecal bacteria using DNA fingerprint analysis 30, 31. PCR analysis was performed to amplify 16S rRNA bacterial genes using specific primers for Lactobacillus rhamnosus 31. DNA from LGG was used as a marker for the DNA fingerprint analysis. The migration profile of PCR amplified products from cultured fecal bacteria of 2, 3 and 8-week old mice treated with Live-LGG from postnatal day 1 to 5 showed the specific band for LGG (Figure 1A). The PCR products were further analyzed by direct sequencing to verify the presence of LGG in LGG-colonized mice. In mice treated with Live-LGG from postnatal day 1 to 5, 100% of mice with LGG detected in feces at 2 and 3-week old had LGG in feces at 8-week old, which indicates that neonatal LGG colonization can persist to adulthood. We did not find the LGG-specific PCR band from mice treated with Fix-LGG for the same period of time (Figure 1A). We also verified the presence of LGG in mouse feces by quantitative PCR (qPCR) analysis using LGG specific primers, as previously reported.32 Results from qPCR analysis showed that the amount of LGG in feces from LGG-colonized mice identified by the DNA fingerprint analysis was significantly higher than that in mice without LGG colonization (Supplemental Figure 1).
Figure 1. Detection of LGG in mouse fecal bacteria.
Fecal bacteria collected from mice at indicated ages with Live-LGG and Fix-LGG treatment from postnatal day 1 to 5 were cultured in MRS broth. DNA was isolated from cultured bacteria for PCR analysis to amplify L. rhamnosus specific 16s rRNA gene. PCR products were separated on DGGE to determine DNA migration profiles (A). Feces of Live-LGG and Fix-LGG treatment were prepared from the same mouse at indicated ages. First lane contains the ladder composed of the PCR product from LGG. LGG colonization was defined as detection of LGG in fecal bacteria. The colonization rate of individual litter was determined by the percentage of mice with LGG colonization in the total number of mice from the same litter. The colonization rate of individual litter detected in 3-week old mice treated with Live-LGG and Fix-LGG starting at indicated age (postnatal day) for 5 days is shown (B).
The colonization rate, which was detected in individual litter at 3-week old, was 70%–100%, 0%–40%, and 0, in mice receiving Live-LGG from postnatal day 1 to 5, 3 to 8, and 5 to 10, respectively (Figure 1B). Thus, these results suggest that colonization of conventionally raised mice with LGG is age-dependent. Mice treated with Live-LGG and Fix-LGG from postnatal days 1 to 5 were used as the LGG colonization model and non-colonization control, respectively, in this study.
LGG colonization promotes growth, intestinal epithelial cell proliferation, differentiation and tight junction formation during development in mice
Since it has been reported that feeding infants with formulas supplemented with LGG supported growth24 and affected development of the intestinal microbiota,33 we next examined whether LGG colonization in mice promoted growth and maturation of the intestinal microbiota during development.
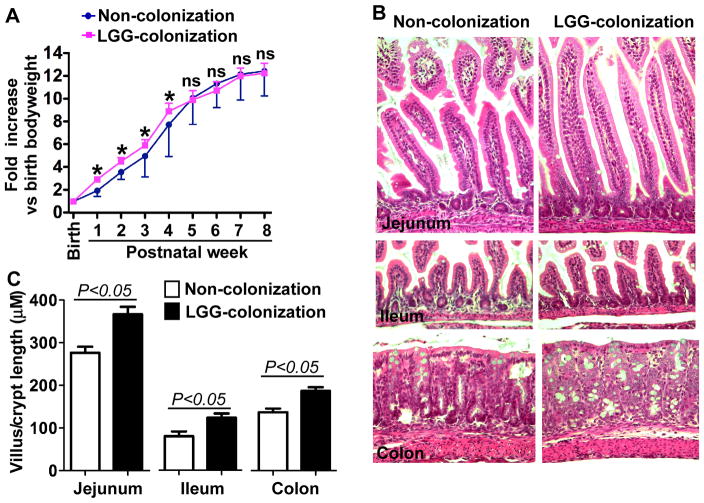
Mice with LGG colonization showed increased bodyweight gain from 1 to 4-weeks of age, but no differences were found in older mice, as compared to non-colonized mice (Figure 2A). Consistent with this result, LGG colonized mice exhibited increased length of villi and depth of crypts at 2-week old (Figure 2B–C). However, we did not find any difference in these parameters between LGG-colonized and non-colonized mice at 8-week old (data not shown). These results indicate a beneficial effect of LGG colonization on intestinal growth during early life.
Figure 2. Colonization of conventionally raised mice with LGG accelerates growth and development of intestinal microbiota before weaning.
Mice were treated with live LGG from postnatal day 1 to day 5 and colonization was confirmed by detecting LGG in fecal bacteria, as described in Figure 1. Fixed LGG was used to treat mice as non-colonization control. Bodyweight was recorded. Bodyweight at birth was set as 1 (A). *p<0.05 compared to the control group at the same age. Indicated intestinal tissues from 2-week old mice were prepared for H&E staining (B) and the length of villus and depth of crypt were measured in at least 50 villi and crypts/mouse (C). In A–C, Non-colonization: n=17, LGG-colonization: n=15.
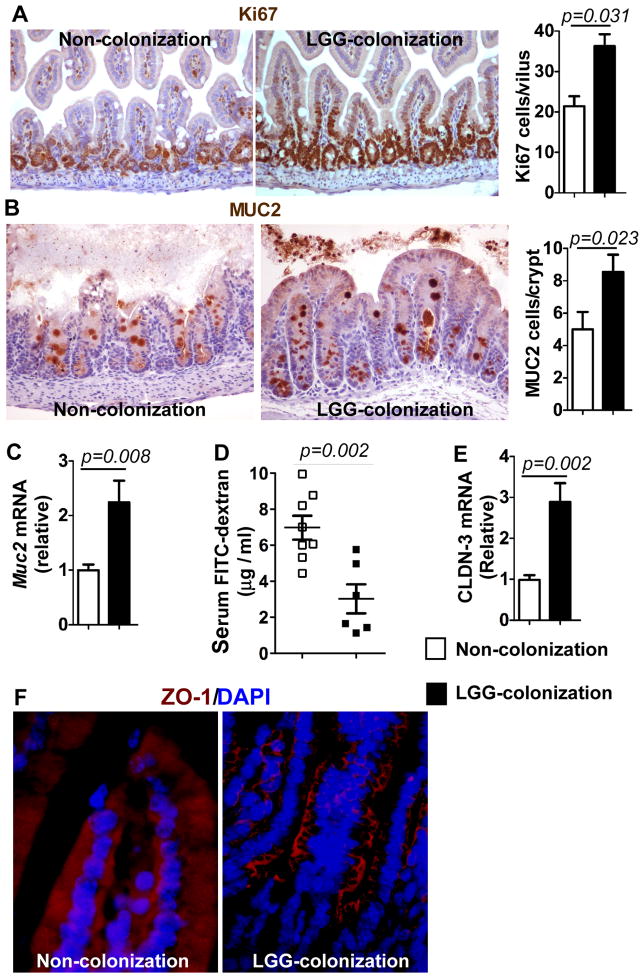
Functional maturation of the gastrointestinal tract completes by the third postnatal week in rodents. Thus, we examined the effects of LGG colonization on regulation of intestinal epithelial cell proliferation, differentiation, and barrier function formation in 2-week old pups. Intestinal tissue sections were immunostained using antibodies against Ki67 and MUC2 to detect proliferation and differentiation, respectively. The numbers of Ki67 (Figure 3A) and MUC2-positive cells (Figure 3B) were increased significantly in pups with LGG colonization. In addition, Muc2 gene expression in colonic epithelial cells isolated from pups with LGG colonization was significantly increased (2.1±0.5 fold, Figure 3C), as compared to that in non-colonized mice.
Figure 3. LGG colonization enhances proliferation, differentiation, and tight junction formation in 2-week old mice.
Mice were treated as described in Figure 2. Proliferation (Ki67, A) in the small intestine and differentiation (MUC2, B) in the colon were assessed using immunostaining. The numbers of positively stained cells were shown. Real-time PCR analysis of Muc2 expression in colonic epithelial cells was shown (C). Barrier formation was detected using the in vivo FITC-dextran assay. The FITC-dextran level in serum is shown (D). Real-time PCR analysis was performed to detect CLND-3 expression in the small intestinal epithelial cells (E). Paraffin-embedded small intestinal tissues were used to determine ZO-1 distribution by immunohistochemistry using an anti–ZO-1 antibody and FITC-labeled secondary antibody and visualized using fluorescence microcopy (red staining). Nuclei were stained with DAPI (blue staining) (F). In C and E, the average of mRNA expression levels in the non-colonized mice was set as 100%, and the mRNA expression level of each mouse was compared to this average. In A–C and E, n-7 in each group. Images in F represent 5 mice in each group.
While development of barrier function occurs before birth, there is ongoing maturation postnatally. We have reported that LGG-derived p40 prevents cytokine and hydrogen peroxide disruption of tight junctions in vivo and in vitro.27, 34, 35 Thus, we next assessed the tight junction formation in LGG-colonized mice using three approaches. The in vivo permeability assay was performed to examine barrier function by measuring serum FITC-dextran level after injection of FITC-dextran into the small intestine. The serum FITC-dextran in LGG-colonized mice was significantly lower than that in non-colonized mice (Figure 3D), which indicates maturation of barrier function by LGG colonization. LGG treatment has been found to increase mRNA expression of a tight junction protein, claudin 3 (CLDN3) in neonatal mice 16. We found that mRNA expression of CLDN3 was increased in neonatal mice with LGG colonization (Figure 3E). Immunostaining of ZO-1 showed that LGG colonization increased ZO-1 membrane localization in the ilium (Figure 3F). These data suggest that LGG colonization promotes tight junction formation in neonatal mice.
In total, these results indicate that LGG colonization has the potential effects on promoting intestinal functional maturation through promoting proliferation, differentiation and tight junction formation during development in mice.
LGG colonization enhances IgA production during development and in adulthood
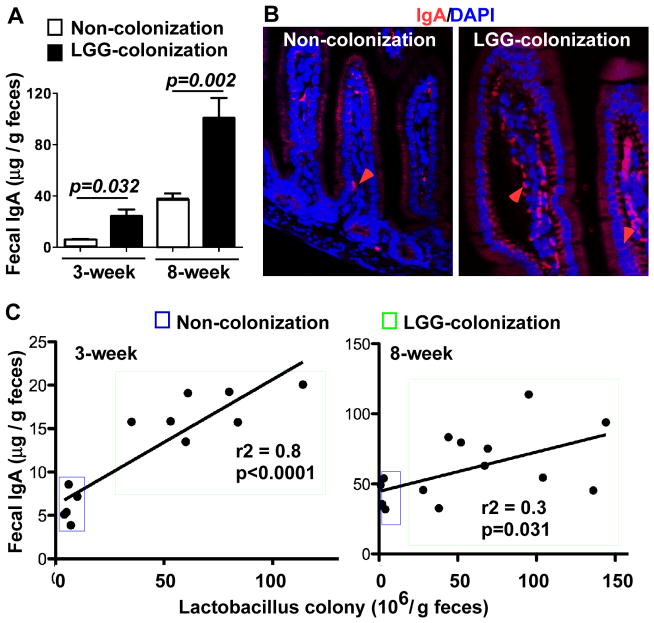
IgA in the gastrointestinal tract plays an important role in maintenance of the homeostatic community of commensal bacteria and protects the host against pathogenic infections.36 Probiotics have been reported to strengthen the immune response by increasing production of IgA.37, 38 Thus, we examined the effects of LGG colonization on IgA production. The IgA level in feces was measured using ELISA. Fecal IgA levels in 3- and 8-week old mice with LGG colonization were significantly higher than those of age-matched non-colonized mice (Figure 4A). Immunostaining showed that LGG colonization increased the number of IgA expressing cells in lamina propria of the small intestine (Figure 4B). The number of fecal Lactobacillus colonies was examined by culturing fecal bacteria on MRS agar. The fecal sIgA level directly correlated with the number of Lactobacillus in feces in 3 and 8 week-old mice with LGG colonization (Figure 4C), which further suggests that LGG colonization plays a role in regulation of IgA production in the intestine.
Figure 4. LGG colonization increases production of IgA in the intestine.
Mice were treated as described in Figure 2 and feces were collected at the indicated ages. Fecal sIgA levels were determined using ELISA assay. N=10 in non-colonization group, N=14 in LGG-colonized group (A). The small intestinal tissues from 3-week old pups were prepared for IgA immunostaining (B). Red arrows indicate IgA positively stained lamina propria cells. Images in B represent 5 mice in each group. Feces were collected from mice with indicated ages and cultured on MRS agar to determine the number of Lactobacillus colonies. The number of fecal Lactobacillus colonies was plotted against the fecal sIgA level for each mouse. The lines illustrate the best-fit linear regressions with the correlation coefficient and significance as indicated (C).
LGG colonization benefits maturation of the intestinal microbiota during development
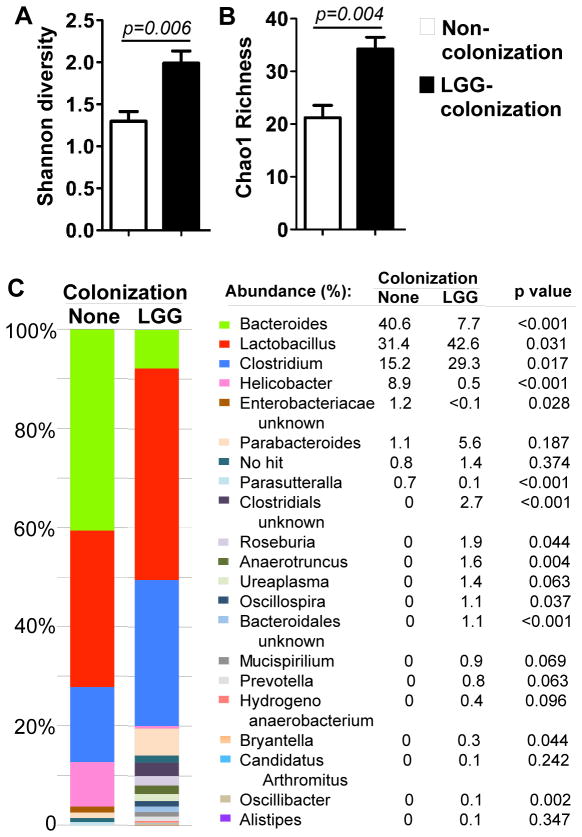
We next studied the impact of evolution of LGG colonization on development of the intestinal microbiota. Community profiling of the colonic mucosally-adherent bacteria isolated from 3 and caecal bacteria from 8-week old mice was assessed by pyrosequencing of amplified bacterial 16S rDNA. LGG colonization increased Shannon diversity and Chao1 richness in 3-week old mice (Figure 5A–B). There were no differences of richness and diversity of microbiota in between LGG-colonized and non-colonized mice in 8-weeks old mice (Supplemental Figure 2A–B). It should be noted that the diversity and richness of the intestinal microbiota in 3-week old LGG-colonized mice showed similarity to that of 8-week old mice with and without LGG colonization.
Figure 5. LGG colonization modulates LGG colonization benefits maturation of the intestinal microbiota in 3-week old mice.
Mice were treated as described in Figure 2. Genomic DNA was extracted from colonic tissues of 3-week old pups to assess the community composition of the mucosally-adherent microbiota using 454-based 16S rRNA amplicon library sequencing. Shannon diversity (A) and Chao1 richness (B), and detailed relative abundance of bacterial taxa at the genus level (C) are shown. n=5 in each group.
The functional maturation of the intestinal microbiome in humans includes compositional shifts in the abundances of major microbial taxa, with the intestinal microbiota of the young resembling that of the adult by approximately 3 years of age.39 To gain greater insight into the effects of LGG colonization on maturation of mouse intestinal microbiome, we analyzed the abundance of specific taxa of the microbiota for comparing the microbial composition in 3-week old mice to that in 8-week old mice with and without LGG colonization. Genus-level analysis showed that major taxonomic groups with abundance from high to low were Bacteroides, Lactobacillus, Clostridium, and Helicobacter in 3-week old non-colonized mice (Figure 5C), which were shifted to Lactobacillus, Clostridium, and Bacteroides in 8-week old non-colonized mice (Supplemental Figure 2C). Thus, estabolisment of the intestinal microbiota in adult mice is a dynamic process, with significant change of the microbial compositions, such as decrease of Bacteroides in adult mice.
LGG colonization stimulated a significant expansion of Lactobacillus and Clostridium and contraction of Bacteroides and Helicobacter in 3-week old mice (Figure 5C). Although LGG colonization in 8-week old mice caused a significant expansion of Lactobacillus, Bacteroides, and Blautia, and contraction of Parasutteralla and Aldercreutzia (Supplemental Figure 2C), the pattern of abundance of the major bacteria was not change. It should be noted that LGG colonization resulted in changing the bacterial composition in 3-week old mice to a similar abundance pattern in 8-week old non-colonized and colonized mice.
Thus, the results that LGG colonization in mice elicited a shift of the diversity, richness, and the composition of the intestinal microbiota at early stage of life to an adult-like microbiota suggest that LGG colonization facilitates maturation of the intestinal microbiota, which may exert beneficial effects on intestinal development.
LGG colonization prevents DSS-induced intestinal injury and inflammation in adult mice
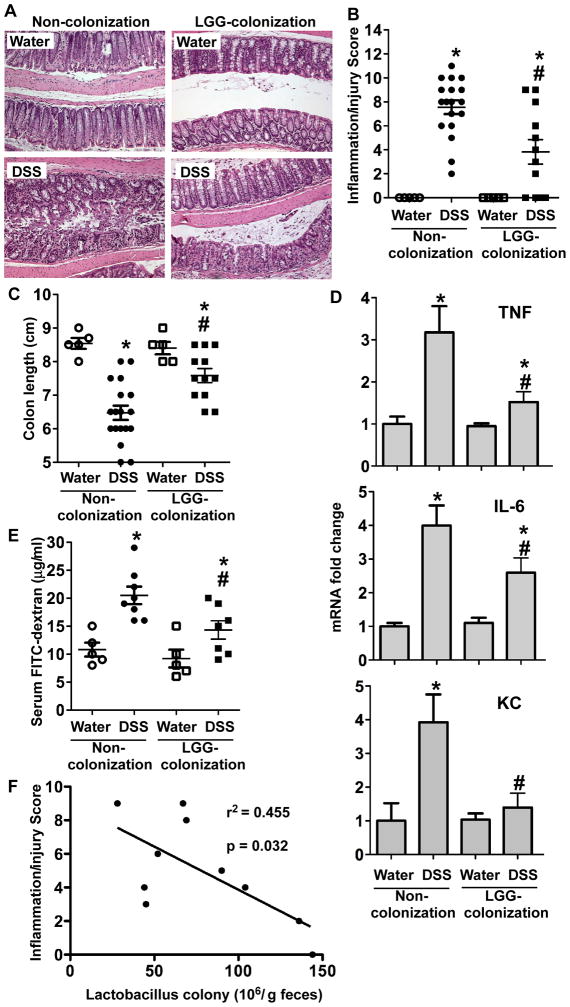
It has been reported that immune effects of microbial exposure at early-life are persist into later stage of life and associated with prevention of IBD in humans40 and animals.41 Therefore, we asked if neonatal LGG colonization prevented intestinal inflammation in adult mice. 8-week old mice were treated with 3% dextran sodium sulphate (DSS) in drinking water for 4 days. DSS treatment induced colonic injury and acute colitis with massive ulceration, crypt damage, and severe inflammation in non-colonized mice (score: 7.68±1.5). These abnormalities were significantly reduced in mice with LGG colonization (score: 3.81±1.1, p<0.05) (Figure 6A–B). DSS-induced shortening of the colon, a feature of intestinal inflammation, in non-colonized mice was prevented in LGG-colonized mice (Figure 6C).
Figure 6. Colonization with LGG prevents colitis in adult conventionally raised mice.
Mice were treated as described in Figure 2. 8-week old mice were treated with 3% DSS in drinking water for 4 days. Paraffin-embedded colon sections were stained with H&E for light microscopic assessment of epithelial damage and inflammation (A). The inflammation/injury scores are shown (B). The length of colon was measured (C). mRNA was isolated from the colonic tissues for real-time PCR analysis of indicated cytokine mRNA expression levels. The average of cytokine mRNA expression levels in the water group of non-colonized mice was set as 100%, and the mRNA expression level of each mouse was compared to this average (D). Intestinal permeability was determined using an in vivo permeability assay on the fourth day of DSS treatment. FITC-dextran level in sera is shown (E). *p<0.01 compared to the water-treated mice in the non-colonization group. # p<0.05 compared to the non-colonized mice with DSS treatment. Feces were collected from LGG-colonized mice before DSS treatment and cultured on MRS agar to determine the number of Lactobacillus colonies. The number of fecal Lactobacillus colony was plotted against the inflammation and injury score for each mouse. The lines illustrate the best-fit linear regressions with the correlation coefficient and significance as indicated (E).
To further understand the effects of LGG colonization on colitis, markers of colitis in this model were examined. Increased proinflammatory cytokine production, such as TNF, KC, and IL-6, is a hallmark of DSS-induced colitis.42, 43 Therefore, we tested the effects of LGG colonization on inflammatory cytokine production in the DSS-induced colitis model. We used Real-Time PCR analysis of RNA isolated from the colon to detect proinflammatory cytokine mRNA levels. TNF, KC, and IL-6 mRNA levels were significantly increased in non-colonized mice, which were reduced in LGG colonized mice (Figure 6D). Disruption of intestinal integrity is a character of DSS-induced intestinal injury and colitis. DSS induced increased permeability, as determined by the level of FITC-dextran in the serum, in non-LGG colonized mice, which was attenuated in mice with LGG colonization (Figure 6E).
More importantly, the number of Lactobacillus in feces was negatively correlated with the inflammation/injury score in mice with LGG colonization (Figure 6F). These results indicate that the effects of LGG colonization on intestinal development, maturation of the microbiota, and IgA production are associated with long-term health outcomes for protection against intestinal inflammation.
DISCUSSION
One important factor that affects the actions of probiotics in vivo is the bioavailability. In most animal studies mice are given LGG via daily gavage. However, the viability of probiotics in the intestinal tract delivered through the oral route is a concern for assessing probiotic clinical efficacy. Therefore, approaches that assure survival of probiotics in the gastrointestinal tract could increase the clinical efficacy of probiotics. Thus, our study was focused on developing a model of LGG colonization in conventional mice and investigating its effects on intestinal development and the potential life long health consequences.
The microbial environment at birth, which is shaped by the mode of delivery, significantly affects colonization.2 This evidence suggests that there is a narrow window of opportunity shortly after birth for colonization. In fact, studies have shown that LGG persisted in the stools only for 7 to 14 days after discontinuing feeding infants with formula supplemented with LGG for 7 days.44 In our study, by introduction of LGG to mice from postnatal day 1 to 5, we developed a mouse model of LGG colonization in conventionally raised mice, which could persist in 8-week old mice. These findings suggest that postnatal day 1 to 5 is a proper time frame for bacterial colonization in conventionally raised mice. Currently, most reported studies were focused on colonization of germ-free animals with bacteria. However, human and animals are naturally colonized by commensal bacteria after birth and it is questionable whether studies using germ-free animals can be extrapolated to the physiological condition. Thus, successful colonization of conventional mice with LGG provides a useful strategy to study the role of commensal bacteria and probiotics in regulation of intestinal development and protective immunity under physiological conditions.
Currently, studies regarding host factors that contribute to bacterial colonization are limited. Polysaccharide A (PSA) from Bacteroides fragilis has been reported to activate the TLR pathway in CD4+ T cells to promote immunologic tolerance for B. fragilis colonization in germ-free mice.45 Since we have found that LGG-derived p40 activates EGF receptor activation in intestinal epithelial cells,27, 46 we conducted experiments to colonize LGG in Egfrwa5 mice with defective EGFR kinase. However, none of Egfrwa5 mice (n=7) could be colonized with LGG. This evidence suggests that EGF receptor signaling may play a role in LGG colonization. We have reported that activation of EGF receptor by p40 stimulated mucin production26 and increased Muc2 expressing cells in LGG-colonized mice were found in this study. Mucin has been shown to mediate bacterial growth. Both commensal and pathogenic bacteria benefit from their ability to regulate mucin synthesis and/or secretion in host goblet cells.47, 48. Thus, LGG-stimulated mucin production may contribute to LGG colonization.
The process for developing initial microbiota community is driven by complex interactions, including the mother’s microbiota, mode of delivery, type of feeding, the environment, and use of antibiotics and disease.49 In addition to oligosaccharides in breast milk, which have been shown to support the establishment of a healthy microbiota in breast-fed infants,50, 51 mother-derived secretory IgA in breast milk has been reported to shape the composition of the intestinal microbiota in neonatal mice at the age of weaning, as compared to that in neonates that did not receive maternal secretory IgA. These differences were magnified when the mice reached adulthood.52 Furthermore, a study using different genetic background mice showed that BALB/c mice had significant difference in abundance of bacterial species and higher diversity as compared to C57BL/6 mice, which was correlated with higher abundance and diversity of IgAs in BALB/c mice than C57BL/6 mice.53 This study also demonstrated polyreactive IgAs-mediated entrance of non-invasive bacteria to Peyer’s patches contributed to the induction of bacteria-specific IgA and the establishment of a positive feedback loop of IgA production.53 Thus, up-regulation of IgA production by LGG colonization may play roles in increasing diversity and richness of the microbiota in neonatal mice. However, our results showed that LGG colonization elicited changes in bacterial composition, but did not affect the diversity and richness of the microbiota in adult mice. Similar results were reported in a human study that perinatal supplementation of LGG to mothers had no major impacts on later microbiota development in children.54 Development of the intestinal microbiota is a dynamic process, the microbiota diversity in adults is genetically and environmentally driven.55 LGG colonization-induced effects may not serve as a major factor determining microbiota diversity in adult mice, although increased IgA production may mediate promoting maturation of the intestinal microbiota in neonatal mice, as shown in our study. In addition to IgA production, LGG colonization may exert other functions that contribute to regulation of the intestinal microbiota at the early stage and in adulthood, which is worthy of further investigation.
Results from this study suggest that LGG colonization led to several outcomes that may contribute to maintaining health and disease prevention. LGG colonization increased the abundance of Lactobacillus and Clostridium in 3-week old mice. Studies have demonstrated that Faecalibacterium prausnitzii, an intestinal bacterium belonging to the Clostridium cluster, was reduced in abundance in Crohn’s disease,56 and Clostridium butyricum, as a probiotic, induces interleukin-10 production in macrophages in inflamed mucosa and prevents acute experimental colitis.57 This evidence supports the contribution of LGG colonization to host health through regulation of the intestinal microbiota.
Our results demonstrate that the effects of LGG colonization on increased IgA production can persist in adult mice. Mechanisms underlying IgA in breast milk promotes lifelong intestinal homeostasis have been demonstrated in mice. Early exposure to maternal secretory IgA prevented the translocation of aerobic bacteria from the neonatal gut into draining lymph nodes and resulted in ameliorating DSS-induced colonic damage in adult mice, which was associated with up-regulation of expression of gens involved in cell metabolism and repair intestinal epithelial cells.52 Thus, this evidence supports the possibility that increased IgA production by LGG colonization may contribute to decreasing susceptibility to colitis in adults.
It is important to study the mechanisms by which LGG colonization increases IgA production. A proliferation-inducing ligand (APRIL), a tumor necrosis factor family member, plays a role in directing B cell class switching towards IgA secreting plasma cells.58, 59 Our preliminary studies have shown that p40, a LGG derived protein,26–28 up-regulated APRIL expression in intestinal epithelial cells, which contributed to promoting IgA production by B cells (un-published data). This mechanism may be involved in LGG colonization-enhanced IgA production because APRIL expression in small intestinal epithelial cells was up-regulated in LGG-colonized mice (Supplemental Figure 3).
Results from this study demonstrated that some effects of LGG colonization are limited to the early postnatal period, including accelerating bodyweight gain, and intestinal epithelial cell proliferation and differentiation. In fact, a clinical study demonstrated that perinatal LGG supplementation to mothers did not affect the weight gain in children remaining normal bodyweight over the 10-year follow-up. However, in children who became overweight by the age of 10 years, there was a tendency that the body mass index (BMI)s of children whose mothers received LGG were constantly lower until 7 years of age as compared with children whose mothers received placebo.60 Therefore, LGG colonization is not likely to result in overweight and may have beneficial effects on prevention of obesity in the later life.
In summary, these findings elucidate the impact of neonatal colonization of mice with LGG on intestinal development and long-term consequence in the adult as protection against colitis. The approach of LGG colonization applied in this study should be useful for delivery of other probiotics to increase their bioavailability and beneficial effects on the host.
METHODS
Mice and LGG colonization
LGG (American Type Culture Collection 53103) was cultured in Lactobacillus MRS broth. Fixation of LGG was performed using 0.25% glutaraldehyde in PBS for 15 minutes at room temperature followed by two washings with 0.1 M glycine to inactive free aldehyde and three washings with PBS.61
Paired pregnant C57BL/6 female mice from the same litter were housed in the same cage until gestational day 18. Then pregnant mice were separated into different cages and gavaged with 108 cfu live LGG (Live-LGG) or glutaraldehyde fixed-LGG (Fix-LGG) until delivery. Pups were fed with 107cfu Live-LGG or Fix-LGG for 5 days starting at postnatal day 1, 3, and 5. All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee at Vanderbilt University.
Bodyweight was recorded at birth and every week. Percentage of change compared to the bodyweight at birth was used to evaluate the growth rate. Intestinal tissues from 2- and 3- week old mice were isolated for preparation of paraffin-embedded tissue sections for immunostaining and hematoxylin and eosin (H&E) staining. Villus length and crypt depth were measured using H&E stained sections.
To detect fecal lactobacillus, mouse feces were solubilized in MRS broth and supernatants were cultured on MRS agar. The number of fecal Lactobacillus colony was counted.
Determining LGG colonization by DNA fingerprint analysis
To detect LGG in fecal bacteria, feces were cultured in MRS broth. Genomic DNA was isolated from cultured bacteria using the Wizard Genomic DNA Purification Kit (Promega Corporation). DNA was used as a template for polymerase chain reaction (PCR) amplification of 16s rRNA gene using L. rhamnosus specific primers (CAGACTGAAAGTCTGACGG and GCGATGCGAATTTCTATTATT).30, 31 PCR products were separated using denaturing gradient gel electrophoresis (DGGE) to determine their migration profiles. Genomic DNA from LGG was used as control. Nucleotide sequences of PCR products were further confirmed by direct sequencing using primers for PCR amplification.
LGG colonization was defined as detection of LGG in fecal bacteria using DNA fingerprint analysis. The colonization rate of individual litter was determined by the percentage of mice with LGG colonization in the total number of mice from the same litter.
Fecal IgA detection by ELISA
Fecal homogenates were prepared by solubilizing feces in PBS (10% wt:vol) and saved at −80°C. 96-well plates were coated with an anti-mouse IgA antibody (Sigma-Aldrich) in PBS at 4°C overnight. Nonspecific protein binding was blocked by incubating plates with 1% of bovine serum albumin (BSA) in PBS for 1 hr at room temperature. Plates were then incubated with supernatants of the fecal homogenates for 1 hr, followed by a FITC-conjugated anti-mouse IgA antibody (Sigma-Aldrich) for 1 hr at room temperature. Fluorescent intensity was measured using a fluorescent plate reader. Purified mouse IgA (Sigma-Aldrich) was used for generating a standard calibration curve.
Intestinal permeability assay
To detect permeability in 2-week old mice, the abdomen was opened and a ligation was performed in the ileum at 2 mm above the cecum. FITC-conjugated dextran dissolved in water (4000 mol wt, Sigma-Aldrich) was injected into the ileum at 4 mg/10 g bodyweight. Then the abdomen was closed. In adult mice with DSS treatment, colitis occurred in the colon. A ligation was performed in the colon at 2 mm below the cecum, then FITC-dextran was injected into the colon. Whole blood was collected using heparinized micro-hematocrit capillary tubes via eye bleed 2 h after FITC-dextran administration. Fluorescence intensity in sera was analyzed using a plate reader. The concentration of FITC-dextran in sera was determined by comparing to the FITC-dextran standard curve.
Microbiota analysis
Frozen colonic tissues from 3-week old pups and cecum feces from 8-week old mice were sent to the Research and Testing Laboratory, LLC (Lubbock, Texas) for metagenomics data collection. The 16sRNA samples were extracted on site and subjected to tag-encoded FLX amplicon pyrosequencing at a 3 K average coverage. Raw metagenomics data was analyzed using Qiime62 (http://www.qiime.org) including 454-based 16S rRNA amplicon library sequencing. Diversity was examined from two perspectives, overall richness, which was expressed as the number of operational taxonomic units (OTUs) and quantified using the Chao1 richness estimator, and overall diversity, which was expressed as Shannon Diversity. Measures of diversity were screened for group differences using an analysis of variance (ANOVA). Individual OTUs were examined for significant changes between non-colonized and colonized groups. OTU count data were analyzed using a generalized linear model with a negative binomial distribution. Multivariate differences among groups were evaluated with “Permutational Multivariate Analysis of Variance Using Distance Matrices,” function adonis. For ADONIS, distances among samples first were calculated using unweighted or weighted UniFrac, and then an ANOVA-like simulation was conducted to test for group differences.
Intestinal epithelial cell isolation
Small intestinal and colonic epithelial cells were isolated as described before.27 Detailed experimental procedures are provided in the Supplementary Material.
Real-time PCR assay
Total RNA was isolated from small intestinal, colonic epithelial cells, and colonic tissues for Real-time PCR analysis of MUC2, CLDN3, TNF, IL-6, and KC gene expression. Detailed experimental procedures are provided in the Supplementary Material.
Immunhistochemistry
Paraffin-embedded tissue sections were used for immunohistochemistry to detect cells expression of Ki67, MUC2, and IgA, and ZO-1 localization. Detailed experimental procedures are provided in the Supplementary Material.
Analysis of intestinal injury and colitis
8-week old mice were administered 3% DSS (molecular weight 36–50 kDa, MP Biomedicals) in drinking water for 4 days. Mice were fed with drinking water as control for DSS treatment. Paraffin-embedded colon tissue sections were stained with hematoxylin and eosin for light microscopic examination to assess colon injury and inflammation. Samples from the entire colon were examined by a pathologist blinded to treatment conditions. A modified combined scoring system 63 including degree of inflammation (scale of 0–3) and crypt damage (0–4), percentage of area involved by inflammation (0–4) and crypt damage (0–4), and depth of inflammation (0–3) was applied for assessing intestinal injury and colitis.
Statistical analysis
Statistical significance was determined by one-way ANOVA followed by Newman-Keuls analysis using Prism 5.0 (GraphPad Software, Inc. San Diego, CA) for multiple comparisons and T-test for paired samples. A p value < 0.05 was defined as statistically significant. All data are presented as mean ± S.E.M.
Supplementary Material
Acknowledgments
Grant Support
This work was supported by National Institutes of Health (NIH) grants R01DK081134 (F.Y.), R01DK56008 and R01DK54993 (D.B.P.), National Nature Science Foundation of China grants 81470796 (F.Y.) and 81300272 (H.C.), Tianjin Research Program of Application Foundation and Advanced Technology of China grant 15JCZDJC36600 (F.Y.), and core services performed through Vanderbilt University Medical Center’s Digestive Disease Research Center supported by NIH grant P30DK058404.
Footnotes
Disclosures
The authors disclose no conflicts of interest.
References
- 1.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237ra265. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 4.Walker WA. Initial intestinal colonization in the human infant and immune homeostasis. Ann Nutr Metab. 2013;63:8–15. doi: 10.1159/000354907. [DOI] [PubMed] [Google Scholar]
- 5.Haberman Y, Tickle TL, Dexheimer PJ, Kim MO, Tang D, Karns R, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124:3617–3633. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen R, Russell RK, Reiff C, Louis P, McIntosh F, Berry SH, et al. Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn’s but not in ulcerative colitis. Am J Gastroenterol. 2012;107:1913–1922. doi: 10.1038/ajg.2012.335. [DOI] [PubMed] [Google Scholar]
- 7.Michail S, Durbin M, Turner D, Griffiths AM, Mack DR, Hyams J, et al. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm Bowel Dis. 2012;18:1799–1808. doi: 10.1002/ibd.22860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raymond F, Ouameur AA, Deraspe M, Iqbal N, Gingras H, Dridi B, et al. The initial state of the human gut microbiome determines its reshaping by antibiotics. ISME J. 2016;10:707–720. doi: 10.1038/ismej.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kronman MP, Zaoutis TE, Haynes K, Feng R, Coffin SE. Antibiotic exposure and IBD development among children: a population-based cohort study. Pediatrics. 2012;130:e794–803. doi: 10.1542/peds.2011-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg RD. The indigenous gastrointestinal microflora. Trends Microbio. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 11.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 12.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Preidis GA, Hill C, Guerrant RL, Ramakrishna BS, Tannock GW, Versalovic J. Probiotics, enteric and diarrheal diseases, and global health. Gastroenterology. 2011;140(1):8–14. doi: 10.1053/j.gastro.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guandalini S. Probiotics for prevention and treatment of diarrhea. J Clin Gastroenterol. 2011;45(Suppl):S149–153. doi: 10.1097/MCG.0b013e3182257e98. [DOI] [PubMed] [Google Scholar]
- 15.Sanders ME, Guarner F, Guerrant R, Holt PR, Quigley EM, Sartor RB, et al. An update on the use and investigation of probiotics in health and disease. Gut. 2013;62:787–796. doi: 10.1136/gutjnl-2012-302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel RM, Myers LS, Kurundkar AR, Maheshwari A, Nusrat A, Lin PW. Probiotic bacteria induce maturation of intestinal claudin 3 expression and barrier function. Am J Pathol. 2012;180:626–635. doi: 10.1016/j.ajpath.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preidis GA, Saulnier DM, Blutt SE, Mistretta TA, Riehle KP, Major AM, et al. Probiotics stimulate enterocyte migration and microbial diversity in the neonatal mouse intestine. FASEB J. 2012;26:1960–1969. doi: 10.1096/fj.10-177980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker S, Oelschlaeger TA, Wullaert A, Vlantis K, Pasparakis M, Wehkamp J, et al. Bacteria regulate intestinal epithelial cell differentiation factors both in vitro and in vivo. PloS one. 2013;8:e55620. doi: 10.1371/journal.pone.0055620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forestier C, De Champs C, Vatoux C, Joly B. Probiotic activities of Lactobacillus casei rhamnosus: in vitro adherence to intestinal cells and antimicrobial properties. Res Microbiol. 2001;152:167–173. doi: 10.1016/s0923-2508(01)01188-3. [DOI] [PubMed] [Google Scholar]
- 20.Dotterud CK, Avershina E, Sekelja M, Simpson MR, Rudi K, Storro O, et al. Does Maternal Perinatal Probiotic Supplementation Alter the Intestinal Microbiota of Mother and Child? J Pediatr Gastroenterol Nutr. 2015;61:200–207. doi: 10.1097/MPG.0000000000000781. [DOI] [PubMed] [Google Scholar]
- 21.Kalliomaki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet. 2001;357:1076–1079. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 22.Kalliomaki M, Salminen S, Poussa T, Arvilommi H, Isolauri E. Probiotics and prevention of atopic disease: 4-year follow-up of a randomised placebo-controlled trial. Lancet. 2003;361:1869–1871. doi: 10.1016/S0140-6736(03)13490-3. [DOI] [PubMed] [Google Scholar]
- 23.Kalliomaki M, Salminen S, Poussa T, Isolauri E. Probiotics during the first 7 years of life: a cumulative risk reduction of eczema in a randomized, placebo-controlled trial. J Allergy Clin Immunol. 2007;119:1019–1021. doi: 10.1016/j.jaci.2006.12.608. [DOI] [PubMed] [Google Scholar]
- 24.Scalabrin DM, Johnston WH, Hoffman DR, P’Pool VL, Harris CL, Mitmesser SH. Growth and tolerance of healthy term infants receiving hydrolyzed infant formulas supplemented with Lactobacillus rhamnosus GG: randomized, double-blind, controlled trial. Clin Pediatr. 2009;48:734–744. doi: 10.1177/0009922809332682. [DOI] [PubMed] [Google Scholar]
- 25.Doron S, Snydman DR, Gorbach SL. Lactobacillus GG: bacteriology and clinical applications. Gastroenterol Clin North Am. 2005;34:483–498. doi: 10.1016/j.gtc.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Cao H, Liu L, Wang B, Walker WA, Acra SA, et al. Activation of Epidermal Growth Factor Receptor Mediates Mucin Production Stimulated by p40, a Lactobacillus rhamnosus GG-derived Protein. J Biol Chem. 2014;289:20234–20244. doi: 10.1074/jbc.M114.553800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan F, Cao H, Cover TL, Washington MK, Shi Y, Liu L, et al. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J Clin Invest. 2011;121:2242–2253. doi: 10.1172/JCI44031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007;132:562–575. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen CH, Nielsen DS, Kverka M, Zakostelska Z, Klimesova K, Hudcovic T, et al. Patterns of early gut colonization shape future immune responses of the host. PloS one. 2012;7:e34043. doi: 10.1371/journal.pone.0034043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan LL, Hagen KE, Tannock GW, Korver DR, Fasenko GM, Allison GE. Detection and identification of Lactobacillus species in crops of broilers of different ages by using PCR-denaturing gradient gel electrophoresis and amplified ribosomal DNA restriction analysis. Appl Environ Microbiol. 2003;69:6750–6757. doi: 10.1128/AEM.69.11.6750-6757.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walter J, Heng NC, Hammes WP, Loach DM, Tannock GW, Hertel C. Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Appl Environ Microbiol. 2003;69:2044–2051. doi: 10.1128/AEM.69.4.2044-2051.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahlroos T, Tynkkynen S. Quantitative strain-specific detection of Lactobacillus rhamnosus GG in human faecal samples by real-time PCR. J Appl Microbio. 2009;106:506–514. doi: 10.1111/j.1365-2672.2008.04018.x. [DOI] [PubMed] [Google Scholar]
- 33.Agarwal R, Sharma N, Chaudhry R, Deorari A, Paul VK, Gewolb IH, et al. Effects of oral Lactobacillus GG on enteric microflora in low-birth-weight neonates. J Pediatr Gastroenterol Nutr. 2003;36:397–402. doi: 10.1097/00005176-200303000-00019. [DOI] [PubMed] [Google Scholar]
- 34.Seth A, Yan F, Polk DB, Rao RK. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP kinase-dependent mechanism. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1060–1069. doi: 10.1152/ajpgi.00202.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoda K, Miyazawa K, Hosoda M, Hiramatsu M, Yan F, He F. Lactobacillus GG-fermented milk prevents DSS-induced colitis and regulates intestinal epithelial homeostasis through activation of epidermal growth factor receptor. Eur J Nutr. 2014;53:105–115. doi: 10.1007/s00394-013-0506-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. 2012;12:821–832. doi: 10.1038/nri3322. [DOI] [PubMed] [Google Scholar]
- 37.Chattha KS, Vlasova AN, Kandasamy S, Esseili MA, Siegismund C, Rajashekara G, et al. Probiotics and colostrum/milk differentially affect neonatal humoral immune responses to oral rotavirus vaccine. Vaccine. 2013;31:1916–1923. doi: 10.1016/j.vaccine.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Vrese M, Rautenberg P, Laue C, Koopmans M, Herremans T, Schrezenmeir J. Probiotic bacteria stimulate virus-specific neutralizing antibodies following a booster polio vaccination. Eur J Nutr. 2005;44:406–413. doi: 10.1007/s00394-004-0541-8. [DOI] [PubMed] [Google Scholar]
- 39.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez-Serrano P, Perez-Calle JL, Perez-Fernandez MT, Fernandez-Font JM, Boixeda de Miguel D, Fernandez-Rodriguez CM. Environmental risk factors in inflammatory bowel diseases. Investigating the hygiene hypothesis: a Spanish case-control study. Scand J Gastroenterol. 2010;45:1464–1471. doi: 10.3109/00365521.2010.510575. [DOI] [PubMed] [Google Scholar]
- 41.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egger B, Bajaj-Elliott M, MacDonald TT, Inglin R, Eysselein VE, Buchler MW. Characterisation of acute murine dextran sodium sulphate colitis: cytokine profile and dose dependency. Digestion. 2000;62:240–248. doi: 10.1159/000007822. [DOI] [PubMed] [Google Scholar]
- 43.Garside P. Cytokines in experimental colitis. Clin Exp Immunol. 1999;118:337–339. doi: 10.1046/j.1365-2249.1999.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petschow BW, Figueroa R, Harris CL, Beck LB, Ziegler E, Goldin B. Effects of feeding an infant formula containing Lactobacillus GG on the colonization of the intestine: a dose-response study in healthy infants. J Clin Gastroenterol. 2005;39:786–790. doi: 10.1097/01.mcg.0000177245.53753.86. [DOI] [PubMed] [Google Scholar]
- 45.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan F, Liu L, Dempsey PJ, Tsai YH, Raines EW, Wilson CL, et al. A Lactobacillus rhamnosus GG-derived soluble protein, p40, stimulates ligand release from intestinal epithelial cells to transactivate epidermal growth factor receptor. J Biol Chem. 2013 doi: 10.1074/jbc.M113.492397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dharmani P, Srivastava V, Kissoon-Singh V, Chadee K. Role of intestinal mucins in innate host defense mechanisms against pathogens. J Innate Immun. 2009;1:123–135. doi: 10.1159/000163037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martens EC, Roth R, Heuser JE, Gordon JI. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J Biol Chem. 2009;284:18445–18457. doi: 10.1074/jbc.M109.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Martin R, Heilig HG, Zoetendal EG, Jimenez E, Fernandez L, Smidt H, et al. Cultivation-independent assessment of the bacterial diversity of breast milk among healthy women. Res Microbiol. 2007;158:31–37. doi: 10.1016/j.resmic.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 51.Martin R, Jimenez E, Heilig H, Fernandez L, Marin ML, Zoetendal EG, et al. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl Environ Microbiol. 2009;75:965–969. doi: 10.1128/AEM.02063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rogier EW, Frantz AL, Bruno ME, Wedlund L, Cohen DA, Stromberg AJ, et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc Natl Acad Sci USA. 2014;111:3074–3079. doi: 10.1073/pnas.1315792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fransen F, Zagato E, Mazzini E, Fosso B, Manzari C, El Aidy S, et al. BALB/c and C57BL/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen-specific IgA and microbiota diversity. Immunity. 2015;43:527–540. doi: 10.1016/j.immuni.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 54.Endo A, Prtty A, Kalliomki M, Isolauri E, Salminen S. Long-term monitoring of the human intestinal microbiota from the 2nd week to 13 years of age. Anaerobe. 2014;28:149–156. doi: 10.1016/j.anaerobe.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 55.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 56.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanai T, Mikami Y, Hayashi A. A breakthrough in probiotics: Clostridium butyricum regulates gut homeostasis and anti-inflammatory response in inflammatory bowel disease. J Gastroenterol 2015. 2015;50:928–935. doi: 10.1007/s00535-015-1084-x. [DOI] [PubMed] [Google Scholar]
- 58.He B, Xu W, Santini PA, Polydorides AD, Chiu A, Estrella J, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26(6):812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 59.Xu W, He B, Chiu A, Chadburn A, Shan M, Buldys M, et al. Epithelial cells trigger frontline immunoglobulin class switching through a pathway regulated by the inhibitor SLPI. Nat Immunol. 2007;8:294–303. doi: 10.1038/ni1434. [DOI] [PubMed] [Google Scholar]
- 60.Luoto R, Kalliomaki M, Laitinen K, Isolauri E. The impact of perinatal probiotic intervention on the development of overweight and obesity: follow-up study from birth to 10 years. Int J Obes. 2010;34:1531–1537. doi: 10.1038/ijo.2010.50. [DOI] [PubMed] [Google Scholar]
- 61.Yan F, Polk DB. Probiotic bacterium prevents cytokine-induced apoptosis in intestinal epithelial cells. The Journal of biological chemistry. 2002;277(52):50959–50965. doi: 10.1074/jbc.M207050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kennedy RJ, Hoper M, Deodhar K, Erwin PJ, Kirk SJ, Gardiner KR. Interleukin 10-deficient colitis: new similarities to human inflammatory bowel disease. Br J Surg. 2000;87(10):1346–1351. doi: 10.1046/j.1365-2168.2000.01615.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.