Abstract
Hepatocellular cancer (HCC) remains a significant therapeutic challenge due to poorly understood molecular basis. In the current study, we investigate two independent cohorts of 249 and 194 HCC cases for any combinatorial molecular aberrations. Specifically we assessed for simultaneous HMET expression or hMet activation and CTNNB1 mutations to address any concomitant Met and Wnt signaling. To investigate cooperation in tumorigenesis, we co-expressed hMet and β-catenin point-mutants (S33Y or S45Y) in hepatocytes using sleeping beauty (SB) transposon/transposase and hydrodynamic tail vein injection and characterized tumors for growth, signaling, gene signatures and similarity to human HCC. Missense mutations in exon-3 of CTNNB1 were identified in subsets of HCC patients. Irrespective of amino acid affected, all exon-3 mutations induced similar changes in gene expression. Concomitant HMET overexpression or hMet activation, and CTNNB1 mutations, were evident in 9-12.5% of HCCs. Co-expression of hMet and mutant-β-catenin led to notable HCC in mice. Tumors showed active Wnt and hMet signaling with evidence of glutamine synthetase and cyclin-D1 positivity and MAPK/ERK, AKT/Ras/mTOR activation. Introduction of dominant-negative TCF4 prevented tumorigenesis. The gene expression of mouse tumors in hMet-mutant-β-catenin showed high correlation with subsets of human HCC displaying concomitant hMet activation signature and CTNNB1 mutations. In conclusion, we have identified co-operation of hMet and β-catenin activation in a subset of HCC patients and modeled this human disease in mice with a significant transcriptomic intersection. This model will provide novel insight into the biology of this tumor and allow us to evaluate novel therapies as a step towards precision medicine.
Keywords: β-catenin, mutations, hepatocellular cancer, HGF, Met, sleeping beauty, liver tumor, AKT, Erk, transposon, TCGA, precision medicine
Introduction
Hepatocellular carcinoma (HCC) is the 5th commonest malignancy in men and 9th commonest malignancy in females. It is also 2nd most fatal cancer worldwide. In U.S.A., HCC increased 72 percent between 2003 and 2012 and the rate of deaths due to liver cancer continues to increase faster than any other type of cancer (1). So far, treatment options for HCC are limited (2-4). Surgical resection or liver transplantation is the only curative treatment for HCC, which may be relevant in early stages. Sorafenib, a multikinase inhibitor, is the only chemotherapeutic drug available for unresectable HCC, but has limited efficacy and poor tolerability (5, 6).
Most HCCs are associated with the presence of chronic liver disease like hepatitis B virus (HBV), HCV, alcohol liver disease, non-alcoholic fatty lover disease, diabetes, Aflatoxin and others. Chronic injury incites hepatocyte death, inflammation, oxidative stress and fibrosis, which eventually leads to proliferation of remnant hepatocytes. Hepatocyte proliferation in such adverse milieu leads to DNA damage, survival and expansion of clones of resistant cells and eventually tumorigenesis, which is driven by heterogeneous molecular signaling pathways. Thus, for developing successful therapeutics for HCC, it is paramount to not only identify specific molecular aberrations that drive subsets of HCC, but also generate successful animal models to validate, address biology and eventually test therapies for that tumor.
Met, an oncogene encoding a tyrosine kinase-type growth factor receptor with an affinity for hepatocyte growth factor (HGF) is known to be upregulated in several HCC cases (7). Due to this, its potential as a therapeutic target in HCC has been increasingly discussed. Missense mutations affecting exon-3 of β-catenin gene (CTNNB1) that lead to stabilization and activation of β-catenin are also observed frequently in HCC patients (8). Some studies have addressed the cooperation of the two pathways in liver tumors although a comprehensive study in patients and animals is lacking.
In the current study we investigate if a subset of HCC cases showed simultaneous HMET overexpression or activation along with activating CTNNB1 mutations. Querying The Cancer Genome Atlas (TCGA) and a recently published French cohort (9, 10), we identified a group of patients representing around 9-12.5% of HCC that show simultaneous hMet activation and CTNNB1 mutations. Next, we co-expressed hMet and exon-3 point mutant-β-catenin in hepatocytes using sleeping beauty (SB) transposon/transposase and hydrodynamic tail vein injection. We observed well-differentiated HCC within 6-9 weeks after injection. The tumors demonstrate activation of hMet and Wnt signaling. More importantly, we observed a significant overlap of genes that were upregulated in our mouse model and human HCC displaying concomitant hMet and β-catenin activation. Lastly, an inability of β-catenin-TCF4 to bind its target genes in this model completely prevented tumorigenesis. Thus, our animal model successfully recapitulates a subset of human HCC and will be invaluable to study tumor biology and test novel therapies.
Materials and Methods
Constructs and reagents
Various plasmids used in the current study are described in the online supplement.
Mice and Hydrodynamic tail vein injections
A list of all mice used in the study is provided in Supplementary table 1. Additional details are available in the online supplement.
Immunohistochemical staining
All the primary antibodies used in the present study are listed in Supplementary Table 2. Additional details are available in the online supplement.
Western blot analysis
Antibodies used in this study are listed in Supplementary Table 3. Additional details are available in the online supplement.
Microarray analysis
Liver tissues from control male FVB mice (n=4) and tumor bearing mice were used for RNA extraction and microarray as described in online supplement.
Human sample analysis
The LIHC (Liver hepatocellular carcinoma) RNAseq expression and exome sequencing results were downloaded from The Cancer Genome Atlas (TCGA) synapse data portal (https://www.synapse.org/#%21Synapse:syn2812961) (11). Additional details are provided in the online supplement.
HMET mRNA expression was also quantified in 249 frozen HCC and 5 normal liver tissues previously described by Nault et al (9). The major clinical and pathological features are summarized in Supplementary Table 4. mRNA expression was analyzed as previously described (9). Expression was normalized using 2-DDCT, with R18S and normal liver tissues. Absolute HMET expression in individual HCC sample in this cohort is listed in Supplementary Table 5. Statistical comparisons were made using two-tailed Wilcoxon matched pairs signed rank-test.
Results
Mutations affecting exon-3 of β-catenin in HCC gene display similar expression of most altered genes irrespective of affected amino acid residues
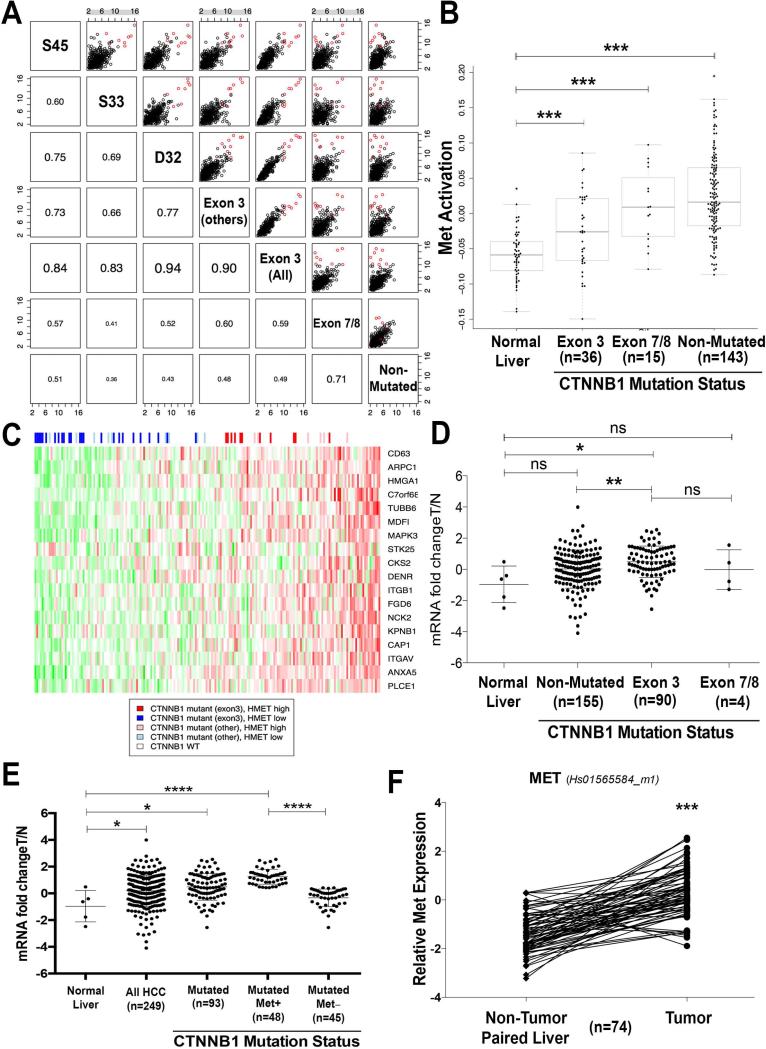
Missense mutations in CTNNB1 are frequent in HCC. To identify mutations in β-catenin gene in the TCGA database, we downloaded and analyzed the LIHC exome sequencing results as described in methods (12). Of the 194 HCCs with available exome sequence, 51 showed mutations in CTNNB1. Of these 51, 36 showed missense mutations in the well-known exon-3 region of CTNNB1. In this group, the most frequent amino acid residue affected was Aspartic acid 32 (D32) (n=13) followed by Serine 33 (S33) (n=8), S45 (n=5) and other residues (n=10). The remaining 15 HCCs showed mutations affecting exon 7/8 that have been recently reported (10). To address signaling outcome of these varying mutations, we next assessed gene expression changes in various mutant groups relative to normal liver. The changes were calculated and the fold-changes of up-regulated genes were compared as a scatter plot and corresponding Pearson correlations are given in the lower part of the grid plot (Figure 1A). Based on most affected genes, mutations affecting S45, S33, D32 or other amino acids in exon-3 all correlate well with each other and to all exon-3 mutations (Pearson coefficient range 0.6-0.94) thus showing that while all HCC samples were broadly similar, CTNNB1 exon-3 mutations show highest similarity to each other irrespective of amino acid residue being affected. Thus, irrespective of amino acid being affected by missense mutations in exon-3 of CTNNB1, the downstream molecular signaling is comparable.
Figure 1. Relationship of β-catenin gene mutations and HMET expression and/or activation in human HCC.
A. HCC samples were divided into classes based on their CTNNB1 mutation status. Gene expression changes relative to normal liver were calculated and the fold-changes of up-regulated genes were compared as a scatter plot. Corresponding Pearson correlations are given in the lower part of the grid plot. While all HCC samples are broadly similar, CTNNB1 exon-3 mutations show the highest degree of similarity. Genes that were up-regulated to a greater extent in CTNNB1 exon-3 mutated samples are highlighted in red.
B. MET activation signature was compared across normal liver and tumor samples from different CTNNB1 mutation classes. All tumor groups are significantly different from normal liver (***p-value of <0.005).
C. Heatmap of the human TCGA samples depicting CTNNB1 mutation status and inferred MET activation. The genes were taken from “KAPOSI_LIVER_CANCER_MET_UP” with the expression values standardized to mean 0 and standard deviation 1. These genes appear to be highly correlated in this dataset further confirming that they represent a coherent MET downstream response. The samples were ordered by their eigengene value which summarizes the overall expression of this geneset and was used to define the HMEThigh subset. Of the 194 samples, 51 had a CTNNB1 mutation, for 36 the mutations were in exon-3 and of those 11 were considered HMEThigh.
D. For quantification of HMET mRNA expression, qRT-PCR was performed using Taqman assay (HS-01565584_m1) in 5 normal livers, 90 HCC cases that were mutated for CTNNB1 in exon-3, 4 HCCs that were mutated in exon7/8 of CTNNB1 and 155 that had no mutations in CTNNB1 (NS-not significant, *P<0.05, **P<0.01, Mann and Whitney test).
E. mRNA expression of HMET in HCC was quantified using qRT-PCR which was performed by Taqman assay in 5 normal livers (NL) and 249 HCC. Overall HMET expression was significantly higher in HCC versus normal group. HMET expression was significantly higher in the CTNNB1-mutated HCC Among the CTNNB1-mutated group of HCC, a subgroup showed significantly higher expression of HMET than the other (*P<0.05, **P<0.01, ****P<0.0001, Mann and Whitney test).
F. mRNA levels of MET expression in 74 HCC samples with CTNNB1 mutations and non tumor paired liver tissues were assessed by qRT-PCR that was performed using Taqman assay (*** P<0.0001, two-tailed Wilcoxon matched pairs signed rank-test).
Subsets of HCC patients show concomitant HMET overexpression or activation along with β-Catenin gene mutations
Next, LIHC RNAseq expression results from TCGA were downloaded and analyzed as described in methods to identify hMet pathway activation. We used the KAPOSI_LIVER_CANCER_MET_UP (13) gene set from mSigDB (14) and found that this set of genes highly correlated with human TCGA samples (see methods). HMET activation signature was compared across normal liver and HCCs with CTNNB1 exon-3 mutations, exon-7/8 mutations and CTNNB1 non-mutated. All tumor groups were significantly different from the normal liver (p<0.005) (Figure 1B). To identify cases with simultaneous hMet and β-catenin activation, a Heatmap of the human TCGA samples depicting CTNNB1 mutational status and inferred HMET activation was constructed (Figure 1C). Of the 194 HCC samples, 90 were considered HMET-high. Intersecting this set with the CTNNB1 mutation status yielded 17 samples that were both HMET-high and CTNNB1 mutant (exon-3 and exon-7/8) (Figure 1C). Further subgrouping shows 11/194 tumors with simultaneous hMet activation and CTNNB1 exon-3 mutations and 6/194 with simultaneous hMet activation and CTNNB1 exon-7/8 mutations. Thus in TCGA dataset, around 9% of all HCC samples show concomitant CTNNB1 mutations (exon-3 and exon-7/8) and hMet activation.
We next analyzed CTNNB1 mutations and HMET mRNA expression in another independent cohort consisting of 5 normal livers and 249 HCC samples as discussed in methods (Supplementary Figure 4) (9). In this series, missense mutations in CTNNB1 were identified in 93 tumors (Supplementary Figure 5). Of these 93, 90 were located in exon-3 and 4 were in exon-7/8 (one sample had mutations in both exon-3 and −7/8). When assessed for HMET expression, a significant overexpression was observed in CTNNB1 exon-3 mutated HCC group as compared to normal livers (p<0.05) or non-CTNNB1 mutated group (p<0.01) (Figure 1D). No significant differences were observed in HMET expression in exon-7/8 CTNNB1-mutated versus either normal livers or exon-3 CTNNB1-mutated group. (Figure 1D). When combined, exon-3 and exon-7/8 CTNNB1 mutant HCCs continued to be significantly higher in expression of HMET than non-CTNNB1-mutated group (p<0.05) or normal liver controls (p<0.05) (Figure 1E). Further analysis of CTNNB1-mutated group of HCC revealed two categories, one with high and another with low HMET expression. Highly significant differences in HMET expression were evident between these two groups (P<0.0001) (Figure 1E). We also assessed CTNNB1-mutated and non-mutated HCC for fold-change in HMET expression in tumor versus normal livers (Supplementary Table 5). Of the 155 non-CTNNB1-mutated HCC, 12 tumors showed greater than 3-fold and 17 showed between 2-3 fold increase in HMET expression when compared to controls. Thus, 29 of the 155 or 18.6% of the non-CTNNB1-mutated HCC showed greater than 2-fold increase in HMET when compared to controls. Out of the 93 CTNNB1-mutated HCC, 9 tumors showed greater than 3-fold increase while 22 tumors showed between 2-3 fold increase in HMET expression when compared to normal livers. Thus 31 of the 93 or 33.3% of CTNNB1-mutated HCC had greater than 2-fold increase in HMET. Overall, 12.5% of all HCC in this cohort showed simultaneous CTNNB1 mutations and enhanced expression of HMET by at least 2-fold.
In addition to comparing normal livers and HCC samples, we also assessed HMET expression in available 74 tumors and non-tumor paired livers in CTNNB1 mutated HCC. A significantly higher expression of HMET was observed in tumors versus corresponding non-tumor liver tissues (P<0.0001) (Figure 1F).
Thus in 2 independent datasets of 194 and 249 HCC cases, around 9-12.5% of showed simultaneous hMet activation or enhanced HMET expression along with CTNNB1 mutations suggesting cooperation of the two pathways in human HCC.
Concomitant expression of hMet and mutant β-catenin genes leads to liver tumor development in mice
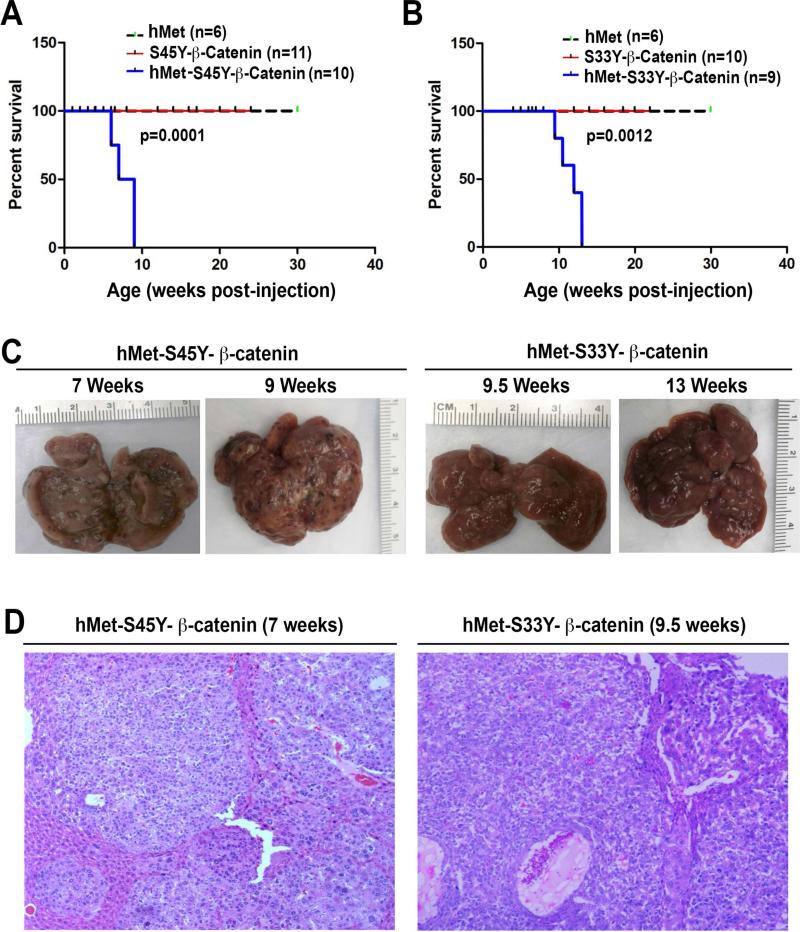
To investigate the significance of these observations in patients, we introduced sleeping beauty (SB) transposase along with SB transposon expressing hMet (with a v5 tag) alone, S45 to tyrosine (Y) mutant form of β-catenin (S45Y-β-catenin) (with a Myc tag) alone, S33Y-β-catenin (with a Myc tag) alone or combination of hMet with either form of mutant β-catenin in the mouse liver using a hydrodynamic tail vein injection as described in methods. Mice injected with hMet or any of the two β-catenin mutants alone, did not show any morbidity even after 22 weeks (Figure 2A and 2B). In striking contrast, co-expression of hMet and S45Y-β-catenin (henceforth referred to as hMet-S45Y-β-catenin) or hMet and S33Y-β-catenin (referred hereon as hMet-S33Y-β-catenin), showed notable morbidity due to increased abdominal girth and decreased physical activity (Figure 2A and 2B). The livers from these mice showed substantive multifocal disease with multiple tumor nodules as shown in representative images (Figure 2C). The tumors in advanced stages showed evidence of intratumoral hemorrhage and bile-filled vesicles. Further analysis of kinetics of tumor development revealed that while hMet-S45Y-β-catenin mice showed notable macroscopic tumors by 6-7 weeks, a comparable tumor burden was evident in hMet-S33Y-β-catenin mice by around 9-9.5 weeks (Figure 2C). At these respective time points, greater than 90% of the hepatic tissue was replaced by tumors that were surrounded by flattened hepatocytes flanking the lesions (Figure 2D). Further, the tumor histology in hMet-S45Y-β-catenin and hMet-S33Y-β-catenin was comparable and consisted predominantly of well-differentiated HCC. The tumors were composed of basophilic hepatocytes often accompanied by cytoplasmic vacuoles containing lipids. Tumor foci showed cytological atypia and trabecular disorganization. In advanced stages (≥9 weeks in hMet-S45Y-β-catenin and ≥13 hMet-S33Y-β-catenin) the HCCs showed areas of necrosis. While this histology was representative of 95% of all tumors, in less than 5%, we also observed cholangiocarcinoma that was composed of primitive ducts and showed stromal reaction (not shown). Multiple tumors developed in every hMet-S45Y-β-catenin and hMet-S33Y-β-catenin mice and HCC was detected in every animal investigated (Supplementary Table 1).
Figure 2. Survival and tumorigenesis in hMet-β-catenin mice.
A. Kaplan Meier curve comparing survival of FVB mice injected with hMet, S45Y-β-catenin or hMet-S45Y-β-catenin. hMet-S45Y-β-catenin-injected mice show significantly reduced survival as compared to the other groups (p=0.0001).
B. Kaplan Meier curve comparing survival of FVB mice injected with hMet, S33Y-β-catenin or hMet-S33Y-β-catenin. hMet-S33Y-β-catenin-injected mice show significantly reduced survival as compared to the other groups (p=0.0012)
C. Gross images of livers from hMet-S45Y-β-catenin and hMet-S33Y-β-catenin at different times after injection shows notable macroscopic disease in both groups.
D. H&E staining of representative liver sections from hMet-S45Y-β-catenin group at 7 weeks and hMet-S33Y-β-catenin group at 9.5 weeks shows presence of well-differentiated HCC throughout the liver. The tumors are composed of basophilic hepatocytes often accompanied by cytoplasmic lipid accumulation and only mild cytological atypia. The tumors showed a solid or macrotrabecular growth pattern. (Magnification: 100X)
Microscopic tumor nodules in hMet-S45Y-β-catenin and hMet-S33Y-β-catenin mice
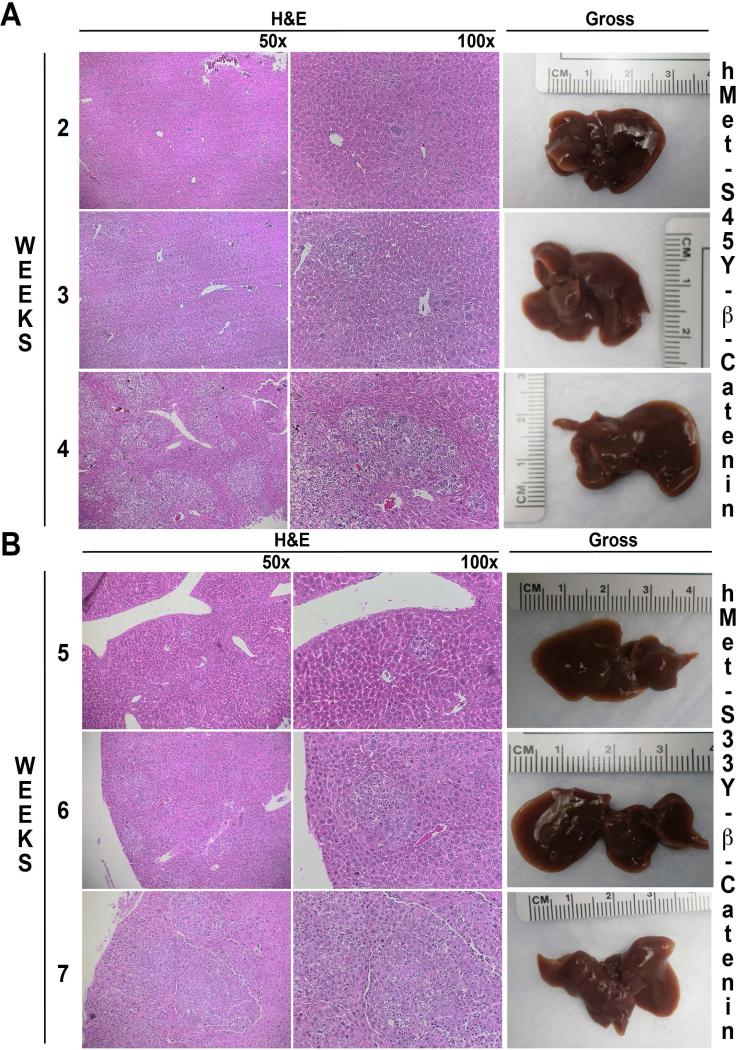
Next we assessed the initiation and progression of tumorigenesis in hMet-S45Y-β-catenin and hMet-S33Y-β-catenin mice. Since macroscopic disease was visible earlier in the hMet-S45Y-β-catenin mice, we assessed liver sections from this group at 2-4 weeks after injection. As seen in figure 3A, while no macroscopic tumors were observed at these time points, there was evidence of microscopic disease as early as 2 weeks. At this stage the lesions consisted of only a few basophilic hepatocytes somewhat larger than the neighboring cells. At 3 weeks, such cells expanded to form small foci of basophilic hepatocytes containing lipid and mild cellular atypia. At 4 weeks, these clusters containing the same types of cells grew further. Similar microscopic foci were evident in the livers of hMet-S33Y-β-catenin mice cells albeit at later time points (5-7 weeks). Specifically, occasional abnormal cells were evident at 5 weeks, while small clusters formed at 6 weeks and progressed further at 7 weeks (Figure 3B).
Figure 3. Early stages of hepatic tumor development in hMet-S45Y-β-catenin and hMet-S33Y-β-catenin mice.
A. In hMet-S45Y-β-catenin livers, isolated hepatocytes that are large and basophilic, are observed at 2 weeks after injection. These cells gradually expand into microscopic foci at 3 and 4 weeks which are composed of basophilic cells with lipid accumulation and occasional mitotic figures while no gross disease is evident.
B. In hMet-S33Y-β-catenin livers, isolated, large and basophilic hepatocytes are observed at 2-4 weeks after injection but do not begin to expand into microscopic foci until 5-7 weeks. The foci are composed of basophilic hepatocytes with lipid accumulation but without any evidence of macroscopic disease.
Tumors in hMet-S45Y-β-catenin and hMet-S33Y-β-catenin mice are composed of dually transfected hepatocytes throughout the disease evolution
Next, we wanted to address that at all stages of tumor development in both hMet-S45Y-β-catenin and hMet-S33Y-β-catenin mice, the nodules are composed of cells that have integrated both hMet and respective β-catenin mutants. Since the β-catenin plasmids injected were tagged with Myc and hMet was V5-tagged, we performed immunohistochemistry for these epitopes at different times after injection. While isolated cells in pericentral region were positive for Myc and V5 at 1 week for hMet-S45Y-β-catenin and 4 weeks for hMet-S33Y-β-catenin, these single cells expanded to small Myc-positive and V5-positive clusters for hMet-S45Y-β-catenin by 3 weeks and hMet-S33Y-β-catenin by 7 weeks (Supplementary Figure 1A and 1B). These foci evolved into tumor nodules and continued to be positive concomitantly for both epitopes as they expanded in size at 6 weeks in hMet-S45Y-β-catenin and at 9 weeks in hMet-S33Y-β-catenin livers (Supplementary Figure 1A and 1B). Most of the hepatic tissue was replaces by Myc and V5 positive nodules in hMet-S45Y-β-catenin livers by 9 weeks and by 13 weeks in hMet-S33Y-β-livers (Supplementary Figure 1A and 1B). Thus the tumors emanate from and continue to be composed of dually transfected cells throughout the tumorigenesis process in the current model.
Evidence of activation of β-catenin signaling and Met downstream AKT/mTOR and Ras/ERK signaling in the tumors in hMet-S45Y-β-catenin and hMet-S33Y-β-catenin mice
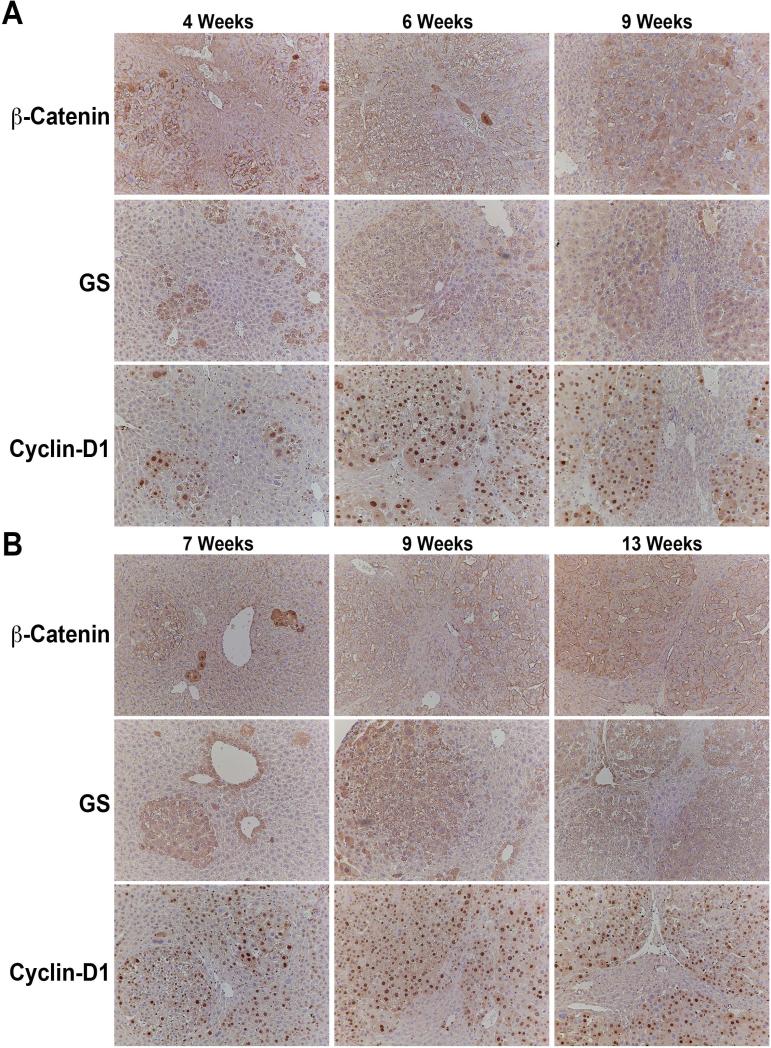
We next investigated the status of Wnt signaling in microscopic and advanced tumors in hMet-S45Y-β-catenin and hMet-S33Y-β-catenin mice. Nuclear and cytoplasmic localization of β-catenin is a good indicator of its activity, although it is not highly sensitive. We observe a subset of tumor cells which show characteristics of β-catenin activation at all stages (Figure 4A and 4B). Intriguingly, we see a very small subset of tumor cells that are very strongly positive for nuclear β-catenin especially at early stages (4-6 weeks in hMet-S45Y-β-catenin and at 7 weeks in hMet-S33Y-β-catenin mice) (Figure 45A and 4B). Glutamine synthetase (GS) is a downstream target of β-catenin and has been shown to be a reliable biomarker of stabilizing β-catenin mutations (15, 16). Indeed small and large tumor foci were consistently and homogeneously positive for this stain at all stages indicating activation of β-catenin in the tumors (Figure 4A and 4B). Similarly, cyclin-D1 is regulated by β-catenin signaling during liver regeneration and in hepatic tumors (17, 18). We noted a clear positivity of small and large tumor foci to be strongly positive for cyclin-D1 in both hMet-S45Y-β-catenin and hMet-S33Y-β-catenin livers at all stages (Figure 4A and 4B).
Figure 4. Evidence of β-catenin activation in hepatic tumors in both hMet-S45Y-β-catenin and hMetS33Y-β-catenin mice.
A. Small and large tumor foci in hMet-S45Y-β-catenin livers at 4, 6 and 9 weeks show a subset of tumor cells with clear cytoplasmic and nuclear β-catenin although the staining was heterogeneous. However, these foci were uniformly and abundantly positive for β-catenin targets Glutamine synthetase (GS) and Cyclin-D1. (Magnification: 100x)
B. Small and large tumor foci in hMet-S33Y-β-catenin livers at 7, 9 and 13 weeks show a subset of tumor cells with clear cytoplasmic and nuclear β-catenin although the staining was heterogeneous. However, these foci were uniformly and abundantly positive for β-catenin targets Glutamine synthetase (GS) and Cyclin-D1. (Magnification: 100x)
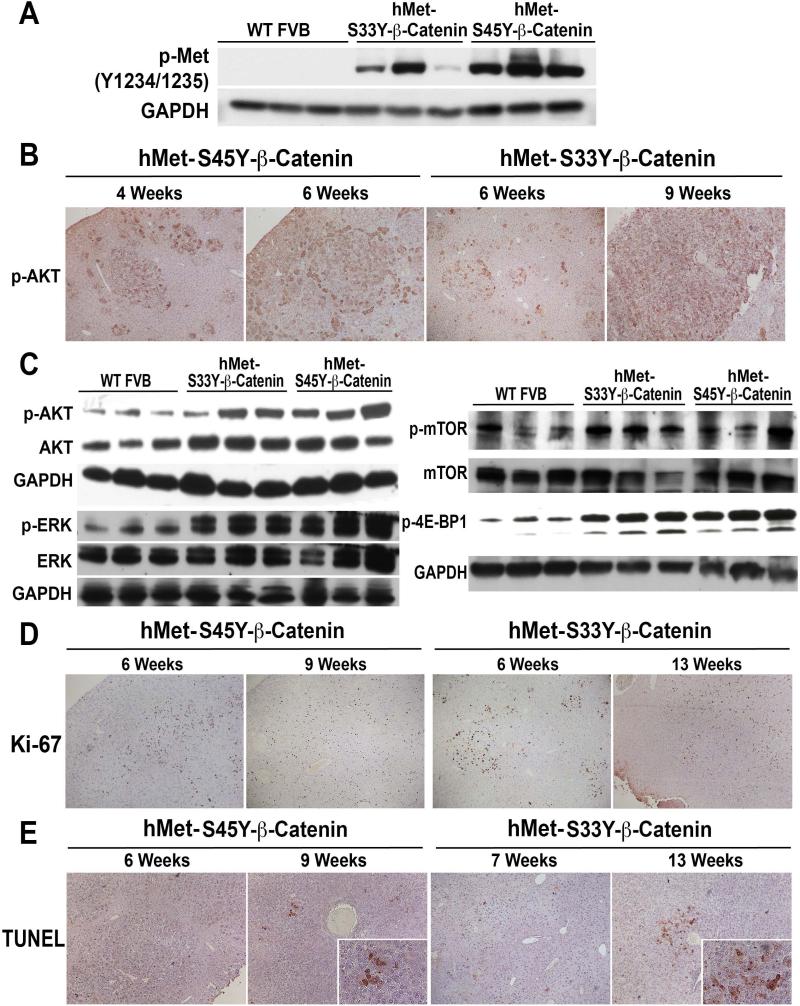
To address Met activation in tumors, we first examined the phosphorylation status of phospho-Met (p-Met) (Y1234/1235). Western blots using whole cell lysates from tumor bearing livers from 6-9 weeks for hMet-S45Y-β-catenin and from 9-13 weeks for hMet-S33Y-β-catenin mice showed a dramatic increase in p-Met levels (Figure 5A). To investigate downstream signaling following Met activation, we assessed the tumors for p-AKT by immunohistochemistry. The tumor foci in hMet-S45Y-β-catenin and hMet-S33Y-β-catenin livers showed strong immunoreactivity to p-AKT (Figure 5B). This increase was also observed in the western blots using whole cell lysates from tumor bearing livers from 6-9 weeks for hMet-S45Y-β-catenin and from 9-13 weeks for hMet-S33Y-β-catenin mice (Figure 5C). Since hMet can also induce ERK signaling downstream of MAPK/PI3K, we next examined the lysates for total and p-ERK. While no change in total ERK was appreciable, a notable increase in p-ERK was observed in lysates from tumor bearing livers from 6-9-week hMet-S45Y-β-catenin and 9-13-week hMet-S33Y-β-catenin mice (Figure 5C). Since PI3K/AKT signaling can lead to mTOR activation, we also assessed tumors for p-mTOR (Ser2448) and downstream p-4E-BP1 (Thr37/46). A notable increase in both p-mTOR and p-4E-BP1 was evident in liver lysates from tumor bearing hMet-S45Y-β-catenin and hMet-S33Y-β-catenin mice (Figure 5C). Thus tumors occurring in hMet-S45Y-β-catenin and hMet-S33Y-β-catenin mice show activation of Wnt and Met signaling.
Figure 5. Evidence of Met/AKT/mTOR and Met/ERK activation, cell proliferation and enhanced cell survival in hepatic tumors in both hMet-S45Y-β-catenin and hMet-S33Y-β-catenin mice.
A. Whole cell lysates from tumor-bearing livers of hMet-S45Y-β-catenin (4-9 weeks) and hMet-S33Y-β-catenin (7-13 weeks) mice showed increased levels of p-Met (Tyr1234/1235) as compared to wild-type (WT) FVB livers in representative Western blot analysis. Equal loading was verified by GAPDH.
B. Tumors in hMet-S45Y-β-catenin livers at 4 and 6 weeks and in hMet-S33Y-β-catenin livers at 6 and 9 weeks show tumors to be strongly positive for phospho-AKT (Ser473). (Magnification: 100X)
C. Whole cell lysates from tumor-bearing livers of hMet-S45Y-β-catenin (4-9 weeks) and hMet-S33Y-β-catenin (7-13 weeks) mice showed increased levels of p-AKT (Ser473) and p-ERK (Thr202/Tyr204) as compared to normal wild-type (WT) FVB livers in representative Western blot analysis. Total levels of AKT and ERK were comparable and equal loading was verified by GAPDH.
D. Whole cell lysates from tumor-bearing livers of hMet-S45Y-β-catenin (4-9 weeks) and hMet-S33Y-β-catenin (7-13 weeks) mice showed increased levels of p-mTOR (Ser2448) and p-4E-BP1 (Thr37/46) as compared to normal wild-type (WT) FVB livers in representative western blot analysis. Total levels of mTOR were comparable and equal loading was verified by GAPDH.
E. Tumors in hMet-S45Y-β-catenin livers at 6 and 9 weeks and in hMet-S33Y-β-catenin livers at 6 and 13 weeks showed strong immunoreactivity to Ki-67 suggesting several tumor cells to be in S-phase of cell cycle. (Magnification: 50X)
F. Tumors in hMet-S45Y-β-catenin livers at 4 weeks and in hMet-S33Y-β-catenin livers at 7 weeks showed no TUNEL-positive nuclei suggesting no cell death. However at advanced stages of tumorigenesis as at 9 weeks in hMet-S45Y-β-catenin livers and at 13 weeks in hMet-S33Y-β-catenin livers, there were several cells in the tumors that showed TUNEL positivity supporting ongoing cell death. (Magnification: 50X; Inset-200X)
Next, we determined the biological response to the active β-catenin and hMet signaling. We examined the number of tumor cells in S-phase by immunohistochemistry for Ki-67. The tumor nodules were strongly positive for Ki-67 in both hMet-S45Y-β-catenin and hMet-S33Y-β-catenin livers especially at early stages of tumor development (Figure 5D). Fewer cells were Ki-67-positive especially at advanced stages of hepatic tumor development such as 9 weeks in hMet-S45Y-β-catenin and 13 weeks in hMet-S33Y-β-catenin mice (Figure 5D). We also assessed cell death by immunohistochemistry for TUNEL. Very few to no TUNEL-positive cells were evident in early and middle stages of hepatic tumor development in either hMet-S45Y-β-catenin or hMet-S33Y-β-catenin mice (Figure 5E). However, at advanced stage, several TUNEL-positive tumor cells were evident within the tumor nodules in both hMet-S45Y-β-catenin and hMet-S33Y-β-catenin livers (Figure 5E). These coincided with the presence of necrotic foci in advanced stages in both groups of animals and may be a result of combination of excessive tumor burden, lack of space for growth, compression and lack of sufficient vasculature within the large tumors.
Intact β-catenin-TCF4 binding is essential for tumorigenesis in hMet-S45Y-β-catenin and hMet-S33Y-β-catenin mice
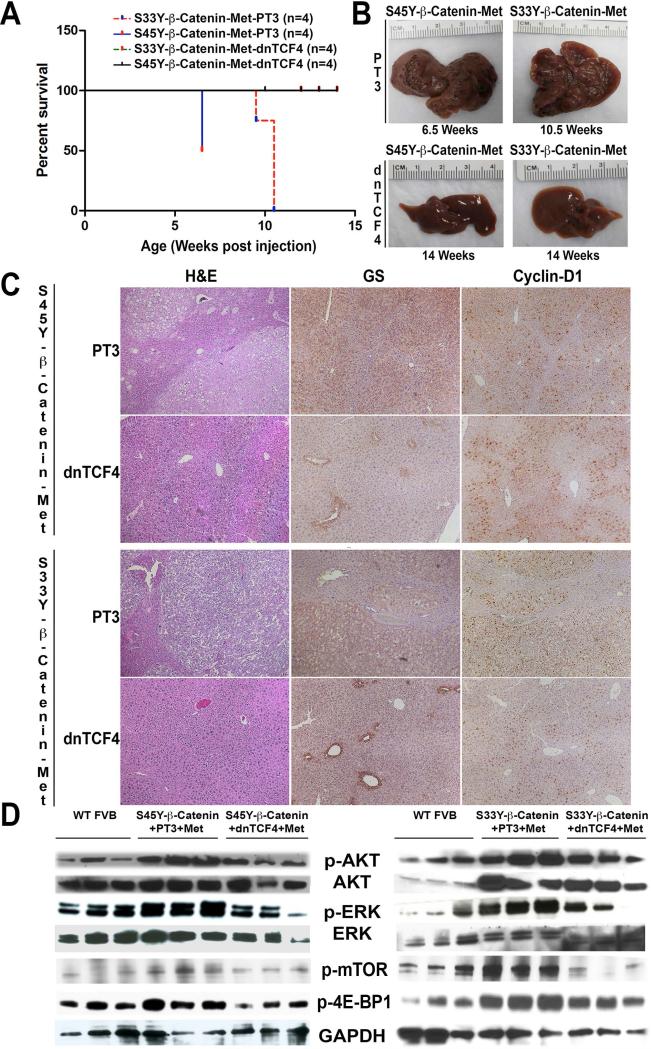
As a transcriptional co-activator, β-catenin functions via interaction with transcription factors to promote gene expression. TCF4 is one such prototypical transcription factor. To determine whether TCF4 is required for hMet-β-catenin induced tumors and also address of β-catenin therapeutic targeting may be of essence in the current model, we co-delivered dnTCF4 or control plasmid (pT3) at the time of the hydrodynamic tail vein injections of hMet-S45Y-β-catenin and hMet-S33Y-β-catenin plasmids. Introduction of dnTCF4 but not pT3 led to a dramatic difference in survival of the injected mice (Figure 6A). While S45Y-β-catenin-pT3-hMet and S33Y-β-catenin-pT3-hMet mice already had a notable macroscopic disease at 6.5 weeks and 10.5 weeks respectively, there was a complete abrogation of tumor development in S45Y-β-catenin-dnTCF4-hMet and S33Y-β-catenin-dnTCF4-hMet even at 14 weeks after injection (Figure 6B).
Figure 6. Combined injection of hMet-mutant-β-catenin and dnTCF4 abolishes tumor development in mice by preventing β-catenin and Met activation.
A. Kaplan Meier curve comparing survival of FVB mice injected with hMet-S33Y-β-catenin-PT3, hMet-S45Y-β-catenin-PT3, hMet-S45Y-β-catenin-dnTCF4 and hMet-S33Y-β-catenin-dnTCF4 shows significant improvement in survival in the both dnTCF4 groups (p=0.0001).
B. Gross images of livers from hMet-S45Y-β-catenin-PT3, hMet-S33Y-β-catenin-PT3, hMet-S45Y-β-catenin-dnTCF4 and hMet-S33Y-β-catenin-dnTCF4 at the indicated times showing lack of macroscopic disease in both dnTCF4 groups.
C. Hematoxylin and Eosin (H&E) stain and immunohistochemistry for Glutamine synthetase (GS) and Cyclin-D1 in livers sections from 6.5 weeks in hMet-S45Y-β-catenin-PT3 mice and10.5 weeks in hMet-S33Y-β-catenin-PT3 mice show notable tumors composed of lipid containing, basophilic hepatocytes and these foci to be homogeneously positive for both GS and Cyclin-D1. However, no tumors were evident in livers of mice injected with hMet-S45Y-β-catenin-dnTCF4 or hMet-S33Y-β-catenin-dnTCF4 even after 14 weeks by H&E, GS staining which is only pericentral and Cyclin-D1, which is only midzonal, as observed in normal livers. (Magnification: 50X)
D. Western blot analysis using whole cell liver lysates show decreased levels of p-AKT (Ser473), p-ERK (Thr202/Tyr204), p-mTOR (Ser2448) and p-4E-BP1 (Thr37/46) to almost that of wild-type (WT) FVB livers, in the hMet-S45Y-β-catenin-dnTCF4 (14 weeks) livers when compared to the hMet-S45Y-β-catenin-PT3 (6.5 weeks) indicating a notable abrogation of Met signaling (left panel). Similar analysis in the right panel shows a notable abrogation of Met signaling in hMet-S33Y-β-catenin-dnTCF4 (14 weeks) livers when compared to the hMet-S33Y-β-catenin-PT3 (9.5-10.5 weeks). Total levels of AKT and ERK were comparable and equal loading was verified by GAPDH.
E. Whole cell lysates from tumor-bearing livers of hMet-S45Y-β-catenin (4-9 weeks) and hMet-S33Y-β-catenin (7-13 weeks) mice showed increased levels of pmTOR (Ser2448) and p-4E-BP1 (Thr37/46) as compared to normal wild-type (WT) FVB livers in representative western blot analysis. Equal loading was verified by GAPDH.
We next assessed the status of β-catenin and hMet signaling in pT3 and dnTCF4 groups to address if the two signaling pathways were indeed synergistic and hence cooperated in hepatic oncogenesis. As expected, we observed the tumors in S45Y-β-catenin-pT3-hMet (6.5 weeks) and S33Y-β-catenin-pT3-hMet (10.5 weeks) groups to be positive for both GS and cyclin-D1 (Figure 6C). However, there were no microscopic tumors visible in the livers from S45Y-β-catenin-dnTCF4-hMet and S33Y-β-catenindnTCF4-hMet mice even at 14 weeks (Figure 6C). Further, as in a normal adult murine liver, GS in S45Y-β-catenin-dnTCF4-hMet and S33Y-β-catenin-dnTCF4-hMet livers was localized to the pericentral and cyclin-D1 to midzonal hepatocytes only. One isolated small microscopic tumor was observed in one S45Y-β-catenin-dnTCF4-hMet at 14 weeks, which was negative for both GS and cyclin-D1 (not shown), and was the only tumor noted in any of the dnTCF4 group of animals. We also used total cell lysates from whole livers from various groups of animals to investigate hMet downstream signaling. We identified a notable decrease in p-AKT and p-ERK in the S45Y-β-catenin-dnTCF4-hMet and S33Y-β-catenin-dnTCF4-hMet livers as compared to pT3 control group (Figure 6D). Thus, our findings support the relevance of β-catenin-TCF4 signaling in HCC in this model and suggest the significance of its inhibition for therapeutic purposes. Also, these data suggest an important functional cooperation between the β-catenin and hMet signaling as a requirement for tumorigenesis in the current model.
Gene signature of hMet-β-catenin tumors
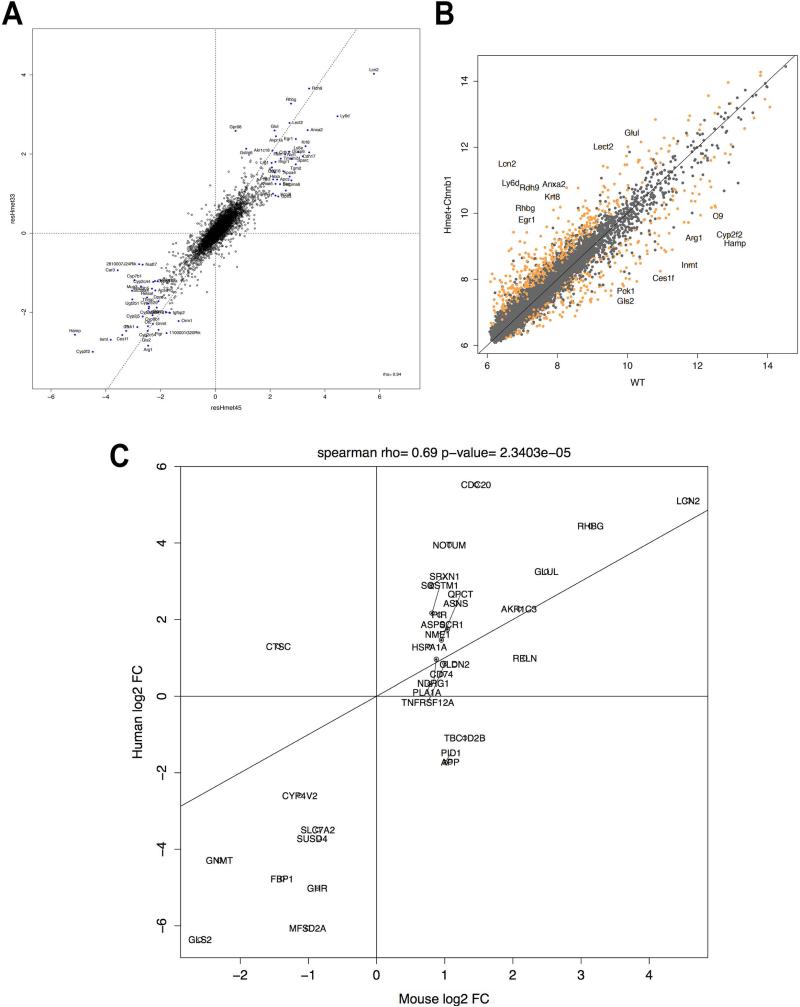
To perform an unbiased molecular profiling, the tumor-bearing hMet-β-catenin livers were subjected to transcriptomic analysis using Affymetrix genearray as described in the methods. Differential expression between normal and tumor samples revealed several known targets of Wnt signaling and growth factor signaling to be upregulated at least 2-fold in the tumors (Table 1). An XY plot of absolute expression values comparing normal and tumor samples also highlights the genes that are significantly differentially expressed at a q-value FDR threshold of 0.001 (Figure 7A). This analysis validates the activation of Wnt and Met signaling as various known targets (Glul, Lect2, Egr1, Lcn2) had a minimum fold change value of 5.62 (2.49 in log2 space). Interestingly, several genes were down regulated in the tumors, some of which are known negative targets of the Wnt signaling in liver (Table 1).
Table 1.
Genes displaying most altered expression in hMET-mutant-β-catenin tumors:
| Symbol | Gene Name | log2 fold change | t-stat | p-value | FDR |
|---|---|---|---|---|---|
| Cdh17 | cadherin 17 | 2.017267382 | 4.040284888 | 0.000560868 | 0.01023877 |
| Akr1c18 | aldo-keto reductase family 1, member C18 | 2.073069573 | 16.56170585 | 8.84801E-14 | 2.53687E-10 |
| Cyp2c39 | cytochrome P450, family 2, subfamily c, polypeptide 39 | 2.119291806 | 3.558053644 | 0.00179297 | 0.020728756 |
| Tmem71 | transmembrane protein 71 | 2.132958811 | 5.691065008 | 1.0667E-05 | 0.00082889 |
| Reln | reelin | 2.185781554 | 7.540923059 | 1.71758E-07 | 3.93966E-05 |
| Slc1a2 | solute carrier family 1 (glial high affinity glutamate transporter), member 2 | 2.298041896 | 5.619734195 | 1.26099E-05 | 0.000871196 |
| Dusp6 | dual specificity phosphatase 6 | 2.37535242 | 5.651331957 | 1.17083E-05 | 0.000849864 |
| Gstm3 | glutathione S-transferase, mu 3 | 2.411624254 | 5.771039914 | 8.84786E-06 | 0.000748988 |
| Egr1 | early growth response 1 | 2.424156044 | 4.615050467 | 0.000139211 | 0.004362179 |
| Krt8 | keratin 8 | 2.429658568 | 4.13071531 | 0.000450478 | 0.008907544 |
| Oat | ornithine aminotransferase | 2.51515967 | 5.936839506 | 6.01771E-06 | 0.000575124 |
| Anxa2 | annexin A2 | 2.561350709 | 7.374367457 | 2.4464E-07 | 5.19573E-05 |
| Avpr1a | arginine vasopressin receptor 1A | 2.685633449 | 9.844745783 | 1.87491E-09 | 1.18488E-06 |
| Gpr98 | G-protein coupled receptor 98 | 2.787243895 | 3.997742288 | 0.000621737 | 0.010771112 |
| Ly6d | lymphocyte antigen 6 complex, locus D | 2.810631756 | 4.41129016 | 0.000228086 | 0.005637571 |
| Gstm6 | glutathione S-transferase, mu 6 | 2.83311938 | 5.608156004 | 1.29577E-05 | 0.000884568 |
| Glul | glutamate-ammonia ligase (glutamine synthetase) | 2.9078866 | 9.791284784 | 2.06629E-09 | 1.18488E-06 |
| Lect2 | leukocyte cell-derived chemotaxin 2 | 2.924657921 | 19.87643846 | 2.15157E-15 | 1.23378E-11 |
| Rhbg | Rhesus blood group-associated B glycoprotein | 3.352856801 | 11.51750787 | 1.05754E-10 | 1.77187E-07 |
| Lcn2 | lipocalin 2 | 3.687152241 | 4.794529001 | 9.02172E-05 | 0.003359314 |
| Rdh9 | retinol dehydrogenase 9 | 3.701351298 | 8.461871276 | 2.598E-08 | 1.14598E-05 |
| Cyp2f2 | cytochrome P450, family 2, subfamily f, polypeptide 2 | −3.290940397 | −4.081753642 | 0.000507249 | 0.009565309 |
| Pck1 | phosphoenolpyruvate carboxykinase 1, cytosolic | −3.083022647 | −4.570893912 | 0.000154918 | 0.00470025 |
| Arg1 | arginase, liver | −2.871591928 | −6.300951716 | 2.60935E-06 | 0.00028947 |
| C9 | complement component 9 | −2.827675119 | −4.528730136 | 0.000171576 | 0.004870659 |
| Dio1 | deiodinase, iodothyronine, type I | −2.60449765 | −6.469442381 | 1.78211E-06 | 0.0002139 |
| Gls2 | glutaminase 2 (liver, mitochondrial) | −2.567159896 | −7.754493321 | 1.09725E-07 | 2.99619E-05 |
| Pigr | polymeric immunoglobulin receptor | −2.420370594 | −8.037974959 | 6.11072E-08 | 2.19005E-05 |
| Otc | ornithine transcarbamylase | −2.389487162 | −7.914902987 | 7.86836E-08 | 2.37472E-05 |
| Gnmt | glycine N-methyltransferase | −2.379866986 | −10.27362174 | 8.70367E-10 | 6.2387E-07 |
| Ces1f | carboxylesterase 1F | −2.368726952 | −4.784839682 | 9.23514E-05 | 0.003394697 |
| Cyp8b1 | cytochrome P450, family 8, subfamily b, polypeptide 1 | −2.367210203 | −7.06132982 | 4.80383E-07 | 8.34748E-05 |
| Inmt | indolethylamine N-methyltransferase | −2.327314044 | −3.770308452 | 0.001077046 | 0.01510056 |
| Cyp2j5 | cytochrome P450, family 2, subfamily j, polypeptide 5 | −2.139532927 | −5.383926046 | 2.20023E-05 | 0.001274424 |
| G6pc | glucose-6-phosphatase, catalytic | −2.107266628 | −3.8578801 | 0.000871951 | 0.013333453 |
Figure 7. A unique gene signature of hMet-mutant-β-catenin tumors correlates with a subset of human HCC that display hMet-activation and mutant-CTNNB1 simultaneously.
A. Comparison of log2 fold-changes in gene expression between hMet-S45Y-β-catenin and hMet-S33Y-β-catenin livers shows that the expression programs induced by the two mutants of β-catenin are highly similar and with a Pearson correlation of 0.94 (p-value <1e-16). This allowed us to pool the data together for further analysis.
B. XY plot of absolute expression values comparing WT and hMet-mutant-β-catenin samples. Genes highlighted in orange are significantly differentially expressed at a q-value FDR threshold of 0.001. Two genes that are considered canonical β-catenin targets (Glul encoding for Glutamine Synthetase, and Lect2 encoding for leukocyte cell-derived chemotaxin-2) had a minimum fold change value of 5.62 (2.49 in log2 space) and all genes whose change in expression change exceeded that threshold are labeled.
C. We computed the differential expression of the CTNNB1+HMEThigh group with all other samples and assessed to which extent these gene changes were recapitulated in the mouse model. Taking a set of genes at a permissive cutoff of q-value FDR of 0.1 and minimum log2 fold change of 2 in both mouse and human datasets, we found that the set of changes highly correlated with a Spearman correlation of 0.69 and a p-value of 2.3403e–05.
Next, we compared the gene expression of the tumors occurring in the hMet-S45Y-β-catenin to those in the hMet-S33Y-β-catenin livers. For this we compared log2 fold-changes of the two different Ctnnb1 mutants to WT livers. We observed that the expression programs induced by hMet-S33Y-β-catenin and hMet-S45Y-β-catenin were highly similar with a Pearson correlation of 0.94 (p-value <1e-16) (Figure 7B).
Significant correlation of gene expression of hMet-β-catenin tumors in mice to patients in TCGA database with unique hMet-mutant β-catenin signatures
Lastly, we aimed to determine if the tumors occurring in hMet-β-catenin mice shared molecular signatures with a subset of HCC patients. We used the limma differential expression pipeline to contrast the expression of the 11 HMET-high and CTNNB1 exon-3 mutant human samples relative to the other 177 LIHC samples. To assess the degree of similarity between the human differential expression analysis of this subset of HCC cases and hMet-β-catenin mouse model, we mapped mouse genes to their human orthologs according to the MGI mouse-human orthology database (19) and assessed the fold-change correlation for genes that were significant at and q-value FDR of 0.2 and had a minimum log2 fold-change of 1.7 in both datasets. This analysis revealed a significantly high correlation between the sets of changes in the two groups with a Pearson correlation of 0.69 and a p-value of 2.3403e–05 (Figure 7C). This analysis demonstrates recapitulation of a subset of human HCC in a mouse model at a molecular level and in a time and cost efficient manner.
Discussion
HCC occurs mostly in the backdrop of chronic liver disease where cycles of hepatocyte injury, inflammation and wound healing in the form of hepatic fibrosis and cirrhosis coerce hepatic regeneration to maintain function. Unfortunately, oxidative stress and other less characterized cellular mechanisms that are inherent to the adverse milieu, can often lead to DNA damage and mismatch repair in dividing and proliferating hepatocytes. As a result, the cells accumulate epigenetic and genetic aberrations, which promote clonal growth due to advantages in survival and proliferation, to eventually yield tumors. Identification of such key aberrations that are initiating and driving hepatic tumorigenesis will thus be of essence for the much-needed targeted therapeutics in HCC.
Activation of Wnt signaling has been reported in a subset of HCC cases due to diverse mechanisms ranging from mutations in CTNNB1, AXIN1/2 and others (8). Somatic missense mutations affecting phosphorylation sites or nearby amino acid residues in exon-3 have been reported in 8-40% of all HCC cases. A recent study revealed CTNNB1 mutations in 37% of HCC cases while activation of Wnt/β-catenin signaling was observed in 54% of all HCC cases (10). In our current study, analysis of TCGA database that is composed of patients mostly from the United States, revealed mutations in CTNNB1 in around 26% of all HCC cases (n=194). However, the mutations confined to exon-3 of CTNNB1 were evident in around 19% of cases while mutations in exon-7/8 were evident in around 8% of HCC cases. Further analysis also revealed that based on the expression of most altered genes, all exon-3 CTNNB1 mutations correlated well with each other than with exon-7/8 or non-CTNNB1-mutated HCCs. This indicates that irrespective of which amino acid is altered due to missense mutations in exon-3, the eventual molecular outcome (based on target gene expression) is quite similar. In the second cohort comprising mostly of patients from Europe, revealed mutations in CTNNB1 in around 38% of all HCC cases (n=249). In this group, the mutations confined to exon-3 were 36% versus exon-7/8 that were less than 2%. This analysis reveals some interesting differences in not only the frequency of β-catenin gene mutations but also the coding region affected. While the exact reason for these differences will require additional investigation, it may be a function of varying etiology and geography. Alcohol intake is the most frequent etiology for HCC in the European study, whereas Hepatitis C and B were more frequent associated with HCC in the United States (10, 20).
hMet activation is another aberration frequently reported in a significant subset of HCC cases. The mechanisms of Met-activation appear to be diverse ranging from amplification to overexpression (Reviewed in (7)). In the current study we also identified increased HMET expression and hMet activation in two independent cohorts of patients. While in a group of 249 HCC cases we observed 60 patients with 2-folder or greater expression of HMET, in another cohort we identified a previously published hMet activation signature in 90 of the 194 HCC cases (13). Thus, overall HCC 24% cases showed greater than 2-fold HMET expression whereas 46% showed hMet activation expression signature. Indeed HMET overexpression has been reported in HCC cases ranging from 20-44% and its activation based on target gene expression signature is reported in around 42% of all HCC cases (7, 13).
Intriguingly our analysis also revealed 8-12.5% of all HCC cases in both databases showing concomitant HMET overexpression and CTNNB1 mutations. Previous studies have shown an intriguing relationship between the two pathways. Our group has previously identified a β-catenin-Met complex in hepatocyte and HGF can induce nuclear translocation of β-catenin (21, 22). A transgenic mouse, which overexpresses hMet in hepatocytes, showed appearance of dysplastic foci that evolved into HCC (23). Interestingly, the tumors in this model harbored Ctnnb1 mutations due to unexplained reasons. When SB expression system was used to co-express hMet and amino-terminal (exon-3) truncated mutant-β-catenin (TM-β-catenin), it led to enhanced HCC between 4 weeks with mortality around 12 weeks in around 74% of animals. While TM-β-catenin does represent active form of β-catenin, such truncation has not been reported in human HCC cases (9, 10, 16). In fact, exon-3 truncations in CTNNB1 are only frequent in hepatoblastomas (24). Indeed, when co-expressed, TM-β-catenin and Yes associated protein-1 (YAP1) led to hepatoblastoma in mice (25). Further, TM-β-catenin is distinct in its target gene expression from full-length β-catenin (26). Hence, to truly recapitulate human HCC in mice, we chose to co-express representative point mutants affecting exon-3 of β-catenin gene along with hMet. In TCGA, the number of HCC cases with mutations in CTNNB1 affecting S45 was 5, and those affecting S33 was 8. In the French cohort, there were 20 HCC cases with S45 mutations and 8 cases with S33 mutations. Since the molecular signaling downstream of all exon-3 point mutations was similar, we chose these two residues as representative mutants to co-express with hMet gene.
Indeed co-expression of hMet and either of the two point mutants of β-catenin led to notable HCC in our animal model. The process was quite robust in both mutants although the process of tumorigenesis was accelerated in S45Y-β-catenin. In fact, the isolated hepatocytes or small clusters of such cells co-expressing hMet and mutant β-catenin persisted in S33Y longer than S45Y and eventually these clusters progressed to enlarge and generate larger macroscopic tumors more rapidly in the S45Y mutant of β-catenin. At this time don't know the basis of this disparity. Interestingly, advanced tumors in both mutant models when analyzed by genearray analysis, show highly concordant molecular signatures. Thus, in cooperating with hMet, initiation of tumorigenesis by S33Y- versus S45Y- mutant-β-catenin may be different, but molecular and biological outcome is remarkably similar.
To determine the downstream signaling of mutant β-catenin and hMet, we assessed the tumors at various stages in both models for the canonical targets of the two pathways. Classical β-catenin targets relevant in HCC including GS and cyclin-D1 were clearly upregulated in tumors at all stages (16, 17, 27). The role of GS in glutamine addiction of β-catenin-mutant liver tumors has been shown (28). Also, role of cyclin-D1 in regulating tumor cell proliferation in Met-β-catenin model has been shown (17). hMet signaling is more complex and divergent and can lead to activation of MEK/ERK, PI3K/AKT/mTOR and others (7). hMet activation along with ERK and AKT activation, were visible through the detection of their respective phosphorylated forms. Recently, an important role of focal adhesion kinase (FAK) in mediating Met-TM-β-catenin driven HCC through ERK and AKT activation has been shown (29). hMet activation also led to mTOR activation which was determined by the presence of p-AKT, p-mTOR and p-4E-BP1. Indeed mTOR activation has been observed in HCC cases and its inhibition discussed for therapy (30, 31). What was most intriguing was the extent of overlap of gene expression between human HCC displaying hMet activation and mutant β-catenin signature, and our mouse model. This recapitulation of around 10% of human HCCs in mice makes our model highly relevant for investigation of mechanism and therapies for precision medicine. While tumor xenografts, chemical carcinogenesis and genetic mouse models have traditionally been the gold standards, these have notable limitations like occurrence of non-relevant and random mutations, length of time to tumorigenesis and lack of hepatic environment. Using reductionist approach and combination of SB and hydrodynamic tail vein injection, which allows for successful genomic integration of transgenes into 1-5% of hepatocytes in mice, we are able to induce clinically relevant tumors in a time-effective manner (Reviewed in (32)).
Targeted therapies against key molecular aberrations will be key to precision medicine in Oncology such as use of Crizotinib in non-small cell lung adenocarcinoma with EML4-ALK fusion (33). In the current study, use of dnTCF4 prevented development of tumors despite the presence of mutant β-catenin and hMet. As expected, no downstream β-catenin signaling was evident. Additionally, despite it's presence, there was lack of hMet activation thus demonstrating a true functional synergy between the two pathways in hepatic tumorigenesis. These observations support an important role of suppressing β-catenin signaling as a chemoprophylaxis measure in the relevant subsets of HCC cases. Current studies are ongoing to address the role of β-catenin suppression with or without Met inhibition after the establishment of disease. It should be emphasized that we have already demonstrated the in vivo efficacy of β-catenin suppression in a preclinical HCC model (34). At the same time, specific Met inhibitors such as Tivatinib and other more non-specific receptor tyrosine kinase inhibitors such as Foretinib and Cabozantinib are in various phases of clinical trials (reviewed in (7)).
Supplementary Material
Acknowledgements
This work was funded by National Institutes of Health grants 1R01DK62277 and 1R01DK095498, and Endowed Chair for Experimental Pathology to SPSM.
Footnotes
Conflict of interest: Dr. Monga is a consultant for Dicerna Pharmaceuticals and Abbvie Pharmaceuticals. He has Corporate Research Agreement with Dicerna Pharmaceuticals and Abbvie Pharmaceuticals. None of the content of the current study is a result of any collaboration with the above companies.
References
- 1.Ryerson AB, Eheman CR, Altekruse SF, Ward JW, Jemal A, Sherman RL, Henley SJ, et al. Annual Report to the Nation on the Status of Cancer, 1975-2012, featuring the increasing incidence of liver cancer. Cancer. 2016 doi: 10.1002/cncr.29936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruix J, Boix L, Sala M, Llovet JM. Focus on hepatocellular carcinoma. Cancer Cell. 2004;5:215–219. doi: 10.1016/s1535-6108(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 3.Spangenberg HC, Thimme R, Blum HE. Targeted therapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2009;6:423–432. doi: 10.1038/nrgastro.2009.86. [DOI] [PubMed] [Google Scholar]
- 4.Llovet JM, Bruix J. Novel advancements in the management of hepatocellular carcinoma in 2008. J Hepatol. 2008;48(Suppl 1):S20–37. doi: 10.1016/j.jhep.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 5.Kane RC, Farrell AT, Madabushi R, Booth B, Chattopadhyay S, Sridhara R, Justice R, et al. Sorafenib for the treatment of unresectable hepatocellular carcinoma. Oncologist. 2009;14:95–100. doi: 10.1634/theoncologist.2008-0185. [DOI] [PubMed] [Google Scholar]
- 6.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 7.Giordano S, Columbano A. Met as a therapeutic target in HCC: facts and hopes. J Hepatol. 2014;60:442–452. doi: 10.1016/j.jhep.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Monga SP. beta-Catenin Signaling and Roles in Liver Homeostasis, Injury, and Tumorigenesis. Gastroenterology. 2015;148:1294–1310. doi: 10.1053/j.gastro.2015.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nault JC, De Reynies A, Villanueva A, Calderaro J, Rebouissou S, Couchy G, Decaens T, et al. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145:176–187. doi: 10.1053/j.gastro.2013.03.051. [DOI] [PubMed] [Google Scholar]
- 10.Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.TCGA Synapse Portal [Google Scholar]
- 12.The Cancer Genome Atlas [Google Scholar]
- 13.Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582–1595. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cadoret A, Ovejero C, Terris B, Souil E, Levy L, Lamers WH, Kitajewski J, et al. New targets of beta-catenin signaling in the liver are involved in the glutamine metabolism. Oncogene. 2002;21:8293–8301. doi: 10.1038/sj.onc.1206118. [DOI] [PubMed] [Google Scholar]
- 16.Zucman-Rossi J, Benhamouche S, Godard C, Boyault S, Grimber G, Balabaud C, Cunha AS, et al. Differential effects of inactivated Axin1 and activated beta-catenin mutations in human hepatocellular carcinomas. Oncogene. 2007;26:774–780. doi: 10.1038/sj.onc.1209824. [DOI] [PubMed] [Google Scholar]
- 17.Patil MA, Lee SA, Macias E, Lam ET, Xu C, Jones KD, Ho C, et al. Role of cyclin D1 as a mediator of c-Met- and beta-catenin-induced hepatocarcinogenesis. Cancer Res. 2009;69:253–261. doi: 10.1158/0008-5472.CAN-08-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan X, Behari J, Cieply B, Michalopoulos GK, Monga SP. Conditional deletion of beta-catenin reveals its role in liver growth and regeneration. Gastroenterology. 2006;131:1561–1572. doi: 10.1053/j.gastro.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 19.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE, Mouse Genome Database G. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 2015;43:D726–736. doi: 10.1093/nar/gku967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lafaro KJ, Demirjian AN, Pawlik TM. Epidemiology of hepatocellular carcinoma. Surg Oncol Clin N Am. 2015;24:1–17. doi: 10.1016/j.soc.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Monga SP, Mars WM, Pediaditakis P, Bell A, Mule K, Bowen WC, Wang X, et al. Hepatocyte growth factor induces Wnt-independent nuclear translocation of beta-catenin after Met-beta-catenin dissociation in hepatocytes. Cancer Res. 2002;62:2064–2071. [PubMed] [Google Scholar]
- 22.Zeng G, Apte U, Micsenyi A, Bell A, Monga SP. Tyrosine residues 654 and 670 in beta-catenin are crucial in regulation of Met-beta-catenin interactions. Exp Cell Res. 2006;312:3620–3630. doi: 10.1016/j.yexcr.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tward AD, Jones KD, Yant S, Cheung ST, Fan ST, Chen X, Kay MA, et al. Distinct pathways of genomic progression to benign and malignant tumors of the liver. Proc Natl Acad Sci U S A. 2007;104:14771–14776. doi: 10.1073/pnas.0706578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armengol C, Cairo S, Fabre M, Buendia MA. Wnt signaling and hepatocarcinogenesis: the hepatoblastoma model. Int J Biochem Cell Biol. 2011;43:265–270. doi: 10.1016/j.biocel.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Tao J, Calvisi DF, Ranganathan S, Cigliano A, Zhou L, Singh S, Jiang L, et al. Activation of beta-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology. 2014;147:690–701. doi: 10.1053/j.gastro.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lade A, Ranganathan S, Luo J, Monga SP. Calpain induces N-terminal truncation of beta-catenin in normal murine liver development: diagnostic implications in hepatoblastomas. J Biol Chem. 2012;287:22789–22798. doi: 10.1074/jbc.M112.378224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cieply B, Zeng G, Proverbs-Singh T, Geller DA, Monga SP. Unique phenotype of hepatocellular cancers with exon-3 mutations in beta-catenin gene. Hepatology. 2009;49:821–831. doi: 10.1002/hep.22695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tardito S, Chiu M, Uggeri J, Zerbini A, Da Ros F, Dall'Asta V, Missale G, et al. L Asparaginase and inhibitors of glutamine synthetase disclose glutamine addiction of beta-catenin-mutated human hepatocellular carcinoma cells. Curr Cancer Drug Targets. 2011;11:929–943. doi: 10.2174/156800911797264725. [DOI] [PubMed] [Google Scholar]
- 29.Shang N, Arteaga M, Zaidi A, Stauffer J, Cotler SJ, Zeleznik-Le NJ, Zhang J, et al. FAK is required for c-Met/beta-catenin-driven hepatocarcinogenesis. Hepatology. 2015;61:214–226. doi: 10.1002/hep.27402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen BW, Chen W, Liang H, Liu H, Liang C, Zhi X, Hu LQ, et al. Inhibition of mTORC2 induces cell cycle arrest and enhances the cytotoxicity of doxorubicin by suppressing MDR1 expression in HCC cells. Mol Cancer Ther. 2015 doi: 10.1158/1535-7163.MCT-15-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duvoux C, Toso C. mTOR inhibitor therapy: Does it prevent HCC recurrence after liver transplantation? Transplant Rev (Orlando) 2015;29:168–174. doi: 10.1016/j.trre.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Chen X, Calvisi DF. Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am J Pathol. 2014;184:912–923. doi: 10.1016/j.ajpath.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delgado E, Okabe H, Preziosi M, Russell JO, Alvarado TF, Oertel M, Nejak-Bowen KN, et al. Complete response of Ctnnb1-mutated tumours to beta-catenin suppression by locked nucleic acid antisense in a mouse hepatocarcinogenesis model. J Hepatol. 2015;62:380–387. doi: 10.1016/j.jhep.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.