Abstract
A group studying acute lung injury observed an increased percentage of neutrophils in the bronchoalveolar lavage (BAL) fluid of mice. BAL was performed, and lung samples were collected sterilely from 5 C57BL/6 mice that had been bred inhouse. Pure colonies of bacteria, initially identified as Bordetella hinzii were cultured from 2 of the 5 mice which had the highest percentages of neutrophils (21% and 26%) in the BAL fluid. Subsequent sequencing of a portion of the ompA gene from this isolate demonstrated 100% homology with the published B. pseudohinzii sequence. We then selected 10 mice from the investigator's colony to determine the best test to screen for B. pseudohinzii in the facility. BAL was performed, the left lung lobe was collected for culture and PCR analysis, the right lung lobe and nasal passages were collected for histopathology, an oral swab was collected for culture, and an oral swab and fecal pellets were collected for PCR analysis. B. pseudohinzii was cultured from the oral cavity, lung, or both in 8 of the 10 mice analyzed. All 8 of these mice were fecal PCR positive for B. pseudohinzii; 7 had increased neutrophils (5% to 20%) in the BAL fluid, whereas the 8th mouse had a normal neutrophil percentage (2%). Active bronchopneumonia was not observed, but some infected mice had mild to moderate rhinitis. B. pseudohinzii appears to be a microbial agent of importance in mouse colonies that can confound pulmonary research. Commercial vendors and institutions should consider colony screening, routine reporting, and exclusion of B. pseudohinzii.
Abbreviations: BAL, bronchoalveolar lavage; CRISPR, clustered regularly interspaced short palindromic repeats
Bordetella hinzii is a nonfermentative, gram-negative rod that is a commensal organism in the respiratory tract of chickens and other fowl. There are few descriptions of B. hinzii infection in the literature, except for 6 case reports of opportunistic infections in humans1,2-4,8,13 and 2 case reports of pulmonary disease in mice in Japan.6,7 Within the last 2 y, 3 publications have addressed possible new species within the Bordetella genus, including 2 that are closely related to B. hinzii.9,10,12 One of the possible new species has been labeled as ‘genogroup 16’ and has been isolated from both a healthy mouse and from one with cystic fibrosis.10,12 The other proposed novel species has been given the provisional name of B. pseudohinzii.9 B. pseudohinzii has transcriptionally active clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated systems that are not found in any other recognized Bordetella species.9
The published reports from Japan demonstrate that B. hinzii is a potential murine pathogen. The first Japanese report6 described a female C57BL/6 mouse that had clinical signs of sneezing and chattering. Histology of the nasal cavity and lungs demonstrated rhinitis, tracheitis, and peribronchiolar bronchopneumonia, with hyperplasia of bronchus-associated lymphoid tissue. The B. hinzii isolate from the index case was intranasally inoculated and caused clinical signs in Jcl:ICR and NOD/ShiJcl (NOD-SCID) mice. The pattern of disease in the ICR mice was similar to that of the C57BL/6 mouse. NOD-SCID mice had both bronchopneumonia and interstitial pneumonia (that extended to the terminal respiratory bronchioles). Subsequently the same authors performed a survey of mouse facilities7 in Japan and determined the overall prevalence of B. hinzii (using postmortem tracheal swab culture) to be 1.6%. However, of the facilities where B. hinzii was present, the prevalence was approximately 31.8%. Of the 195 mice that were culture-positive for B. hinzii, 7 had gross lung lesions with a histopathologic diagnosis of bronchopneumonia with bronchus-associated lymphoid tissue hyperplasia; however, none of these 7 mice were reported to have antemortem clinical signs. A recent report suggests that the B. hinzii isolate described in the index case may have been misidentified and that it is closely related to Bordetella genogroup 16.10
None of the 4 major commercial vendors of mice report B. hinzii or B. pseudohinzii on their routine health monitoring reports, although the 2014 AALAS–FELASA working group position statement on international rodent health reports lists B. hinzii as an agent of potential importance in mice.11 The current report is the first publication to document mice infected with B. pseudohinzii in the United States and to address the potential negative effect of B. pseudohinzii on pulmonary research. Furthermore, this investigation describes the histologic lesions that appear to be associated with murine B. pseudohinzii infection and the procedures used in collaboration with a commercial diagnostic laboratory to validate an antemortem PCR screening test using pooled fecal pellets.
Case History
A research group studying acute lung injury observed an increased percentage of neutrophils in the BAL fluid from their mice and contacted the veterinary staff. The increase in neutrophils was not strain-specific, and not all mice from the same cage had increased neutrophils. The initial case work-up consisted of performing a gross necropsy and collecting BAL fluid for cytology and the left lung lobe for microbiologic culture from 5 of the investigator's inhouse-bred 8- to 12-wk-old C57BL/6 mice. These mice were selected because most of the investigator's lines were on a C57BL/6 background and because the investigative group uses 8- to 12-wk-old mice for experiments. The animals were anesthetized by using an intraperitoneal injection of ketamine (100 mg/kg) and xylazine (5 mg/kg). Aseptic BAL was performed by tracheostomy with a sterile surgical approach. The hair on the ventral neck was shaved, and the skin was prepared by alternating povidone–iodine and alcohol. Toe pinches were performed to ensure a surgical plane of anesthesia. A 5-mm midline incision was performed to expose the trachea. The trachea was intubated with a sterile 20-gauge, 1-inch intravenous catheter. The thoracic cavity and ribs were opened to visualize the lungs during BAL. Sterile Hanks balanced salt solution (1 mL) was instilled into the lungs; after 10 s, the fluid was aspirated back into the syringe. Cytology was performed to evaluate the neutrophil percentage in the BAL fluid.
All lungs appeared normal on gross examination, but an organism initially identified as B. hinzii was cultured from the lungs of 2 of the 5 animals and was confirmed through PCR testing. Subsequent sequencing of a portion of the ompA gene of this isolate demonstrated that it had 100% homology with the published B. pseudohinzii ompA sequence (see following section). Among the 5 mice sampled, the BAL neutrophil percentages were 2%, 6%, 16%, 21%, and 26%; the investigators considered 4% neutrophils or less to be normal. B. pseudohinzii was cultured from the mice with the 2 highest percentages of neutrophils in the BAL fluid, demonstrating an association between a high neutrophil percentage in the BAL fluid and B. pseudohinzii infection. To establish colony infection prevalence, a study was conducted in collaboration with IDEXX BioResearch (Columbia, MO) to determine the best antemortem test to screen mice for B. pseudohinzii.
Materials and Methods
Facility and husbandry.
The Biologic Resources Laboratory is the centralized animal facility at the University of Illinois at Chicago, an AAALAC-accredited institution. The facility tests for and excludes Helicobacter spp., Mycoplasma pulmonis, pinworms, fur mites, murine norovirus, mouse rotavirus, mouse hepatitis virus, mouse parvoviruses, minute virus of mice, pneumonia virus of mice, reovirus 3, Sendai virus, ectromelia, lymphocytic choriomeningitis virus, murine adenovirus types 1 and 2, polyomavirus, and Theiler murine encephalomyelitis virus. All of the mice were group-housed in autoclaved static microisolator cages with autoclaved corncob bedding, autoclaved nesting enrichment (Cotton square, Ancare, Bellmore, NY), and autoclaved municipal water bottles, with weekly cage changes. Rooms had a 14:10-h light cycle, and mice had unrestricted access to an irradiated diet (Rodent Diet 7912, Envigo Teklad, Indianapolis, IN). All animal work performed was covered by University of Illinois at Chicago IACUC-approved protocols.
Animals.
Ten animals (representing 7 strains of mice) were selected from the investigator's colony for the study. The qualification for selection was that the mice had to be 8 to 12 wk old, to match the age specification for experiments performed by the investigator.
Sample collection.
Prior to sample collection, all mice were anesthetized by using intraperitoneal ketamine (100 mg/kg) and xylazine (5 mg/kg). Oral swabs were collected for culture (BBL Cultureswab, Becton Dickinson, Franklin Lakes, NJ) and PCR (FLOQSwabs, Copan, Murrieta, CA). Toe pinching was performed to ensure a surgical plane of anesthesia. A BAL cytology sample was collected as described earlier. The left lung lobe was collected aseptically for culture and PCR analysis, and the right lung lobes were insufflated with 10% neutral buffered formalin and placed in 10% neutral buffered formalin for histopathology. The head was placed in 10% neutral buffered formalin for histology of the nasal cavity. Two fecal pellets were collected from the descending colon for PCR analysis.
BAL fluid cytology.
The recovered lavage fluid was centrifuged (500 × g for 20 min), and the cell pellet was resuspended in 200 μL Hanks balanced salt solution. The BAL fluid was processed by using cytocentrifugation (Cytospin 3, Shandon Instruments, Pittsburgh, PA) and stained (Diff-Quik, Dade Behring, Düdingen, Switzerland). BAL cell differential counts were determined by using morphologic criteria under a light microscope under a 40× objective, with evaluation of approximately 500 cells per slide.
Bacterial culture, identification and antibiotic sensitivity.
Microbiologic culture, identification, and antibiotic sensitivity testing were performed at IDEXX BioResearch (Columbia, MO). Lung tissue samples were placed in sterile microcentrifuge tubes with sterile PBS and disrupted by using polypropylene pestles (Pellet pestles, Sigma–Aldrich, St Louis, MO). Oral swabs and disrupted lung tissues were cultured on BBL Trypticase Soy Agar with 5% sheep blood (TSA II plates) and BBL Chocolate II Agar (Becton Dickinson, Franklin Lakes, NJ). Cultures were incubated at 37 °C with 7% CO2.
Bacterial colonies were analyzed by matrix-assisted laser desorption–ionization time-of-flight mass spectrometry, which was performed by using a Microflex mass spectrometer (Bruker Daltronics, Billerica, MA) with flexControl software. A sterile toothpick was used to harvest a bacterial sample from each colony and to directly distribute the bacteria inside a 3-mm diameter etched circle on a stainless steel target (Bruker Daltronics). Target spots were overlaid with 1 μL of a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile–2.5% trifluoroacetic acid matrix (Bruker Daltronics) and allowed to air dry at room temperature. Species-level identification was obtained by automated analysis by using MALDI BioTyper software (Bruker Daltronics) that compared the spectra for each isolate with an integrated reference database.
Antibiotic sensitivity testing was performed by using an automated system (Vitek 2, bioMérieux SA, Marcy l’Étoile, France); AST-GN65 test cards were inoculated according to the manufacturer's instructions. Because B. pseudohinzii and B. hinzii are not included in the Vitek 2 database, isolates were run as both B. avium and B. bronchiseptica, and the resulting antibiograms were identical.
PCR and sequence analysis.
Total nucleic acids were extracted from mouse lungs, fecal pellets, and swabs of the oral cavity (FLOQSwabs, Copan) according to standard protocols for a commercially available platform (One-For-All Vet Kit, Qiagen, Valencia, CA). The B. hinzii/pseudohinzii PCR assay was based on the IDEXX BioResearch proprietary service platform (IDEXX Laboratories, Westbrook, ME). Briefly, the B. hinzii/pseudohinzii real-time PCR assay targets a region of the outer membrane protein A (ompA) gene that is conserved among all B. hinzii/pseudohinzii genomic sequences deposited in GenBank, uses a FAM–TAMRA-labeled hydrolysis probe, and has an analytical sensitivity of between 1 and 10 template copies per PCR reaction. A hydrolysis-probe–based real-time PCR assay targeting a housekeeping gene (18S rRNA or 16S rRNA) was used to determine the amount of genomic DNA present in the test sample, to confirm DNA integrity, and to ensure the absence of PCR inhibitors. Diagnostic real-time PCR analysis and amplification of the product for sequence analysis were performed by using standard primer and probe concentrations with a commercially available master mix (LC480 ProbesMaster, Roche Applied Science, Indianapolis, IN) on a commercially available real-time PCR platform (LightCycler 480, Roche).
A bacterial isolate from an affected mouse underwent sequence analysis. A portion of the ompA gene was amplified by using sequencing primers that flank the region targeted by the B. hinzii/pseudohinzii real-time PCR assay. The amplicon generated by the sequencing primers was purified and sequenced in both directions by using Sanger DNA sequencing (GENEWIZ, South Plainfield, NJ). Contig assembly was performed by using Sequencher software (Gene Codes, Ann Arbor, MI), and the consensus sequence was compared with sequences deposited in the GenBank and Whole-Genome-Shotgun contig databases by using BLAST software.
Histology.
Fixed nasal passages were decalcified with a solution of 10% sodium citrate and 25% formic acid. Lung and decalcified nasal passages were processed for paraffin embedment, blocked in wax, and cut into 5-μm sections. For all mice, 3 sections of the nasal cavity were examined (sectioned in accordance to standards used by the National Toxicology Program, sections I, II, and III). Tissue sections were stained with hematoxylin and eosin, and stained slides were permanently cover-slipped. Stained sections were examined for abnormalities.
Sentinel testing.
Female Crl:CD1(ICR) sentinel mice (age, 15 wk) had been in the facility for 3 mo when fecal samples were collected. Mice were housed in sterile microisolator cages that received dirty bedding from colony cages during weekly cage changes. On average, there was one sentinel cage (3 mice per cage) for every 60 colony cages. Eight sentinel cages from the investigator's colony (housed in 2 different rooms) were tested. All mice in each sentinel cage were gently restrained, and 1 or 2 fecal pellets per mouse were collected and pooled together. All samples were processed for PCR analysis as described earlier.
Statistics.
A 2×2 table was generated (Table 1) to determine the sensitivity, specificity, negative predictive value, and positive predictive value of using fecal PCR compared with the ‘gold standard’ of culture. The following formulas were used (top left box, a; top right box, b; bottom left box, c; and bottom right box, d): sensitivity, a / (a + c); specificity, d / (b + d); positive predictive value, a / (a + b); and negative predictive value, d / (c + d).
Table 1.
Fecal PCR compared with culture of lung or oral cavity (n = 10)
| Culture results |
|||
| Positive | Negative | ||
| PCR results | |||
| Positive | 8 | 0 | |
| Negative | 0 | 2 | |
Results
PCR and sequence analysis.
A bacterial isolate from an affected mouse was identified as B. hinzii by matrix-assisted laser desorption–ionization time-of-flight mass spectrometry and real-time PCR analysis and underwent nucleic acid sequencing. The 362-bp fragment displayed 100% identity to the only available B. pseudohinzii ompA sequence (GenBank Whole-Genome-Shotgun sequence JHEP02000002.1) and 98% identity to available B. hinzii ompA sequences (GenBank accession numbers CP012076.1, CP012077.1, AM748263.1, AM748264.1, and AM748265.1).
No mice were PCR-positive for B. pseudohinzii by oral swab. One mouse was PCR-positive for B. pseudohinzii from the left lung lobe. Eight mice were PCR-positive for B. pseudohinzii according to fecal PCR analysis, including the mouse that was PCR-positive from the lung lobe sample (Table 2). All mice positive by fecal PCR analysis were culture-positive according to samples from the lung or oral cavity (or both). The 2 mice from which B. pseudohinzii was not cultured were PCR-negative for B. pseudohinzii from all collection sites. The B. pseudohinzii-positive fecal samples yielded an estimated 50 to 100 copies per PCR test.
Table 2.
Results from candidate antemortem screening tests
| Mouse | BAL neutrophils (%) | Oral culture | Lung culture | Oral PCR | Lung PCR | Fecal PCR |
| 1 | 20 | + | + | — | — | + |
| 2 | 5 | — | + | — | + | + |
| 3 | 2 | — | + | — | — | + |
| 4 | 20 | + | — | — | — | + |
| 5 | 2 | — | — | — | — | — |
| 6 | 5 | + | — | — | — | + |
| 7 | 10 | — | + | — | — | + |
| 8 | 20 | — | + | — | — | + |
| 9 | 5 | + | — | — | — | + |
| 10 | 2 | — | — | — | — | — |
BAL.
Of the 10 mice tested, 3 had normal percentages of neutrophils in the BAL fluid (2%), and the remaining 7 mice had elevated percentages of neutrophils in the BAL fluid, ranging from 5% to 20% (Table 2).
Culture.
B. pseudohinzii was cultured from the oral-cavity swabs of 4 mice and from the lungs of 5 mice (including 1 of the 4 that yielded B. pseudohinzii from the oral cavity). B. pseudohinzii was not cultured from 2 mice (Table 2).
Susceptibility.
B. pseudohinzii was susceptible to imipenem, amikacin, gentamicin, tobramycin, marbofloxacin, tetracycline, and trimethoprim–sulfamethoxazole. B. pseudohinzii showed intermediate sensitivity to enrofloxacin and chloramphenicol and was resistant to ampicillin, amoxicillin–clavulanic acid, piperacillin, cefpodoxime, ceftiofur, and nitrofurantoin.
Histology.
Very mild perivascular and peribronchiolar infiltrates (predominantly lymphoid cells with few neutrophils and macrophages) were observed in the lungs of all mice and were considered to be background findings. All mice had mild suppurative inflammation around the vomeronasal organ—a finding that was considered to be another background lesion. Mouse numbers 5 (Figure 1) and 10 were considered to be normal. Mouse numbers 1 and 4 had mild to moderate intraepithelial suppurative inflammation in the nasal cavity. Mouse numbers 2, 6, and 8 had nasal foreign bodies with associated neutrophils and mucus. Mouse numbers 2, 3 (Figure 1), and 9 had very mild rhinitis. Mouse numbers 6, 7, and 8 had mild to moderate suppurative rhinitis. Mouse numbers 2 (Figure 1) and 4 had mild peribronchiolar and mild perivascular infiltrates. Mouse 6 had marked lymphoid hyperplasia around a bronchus and large bronchiole, and mouse 8 had mild to moderate acidophilic granulomatous pneumonia. Histologic descriptions for all mice are provided in Table 3.
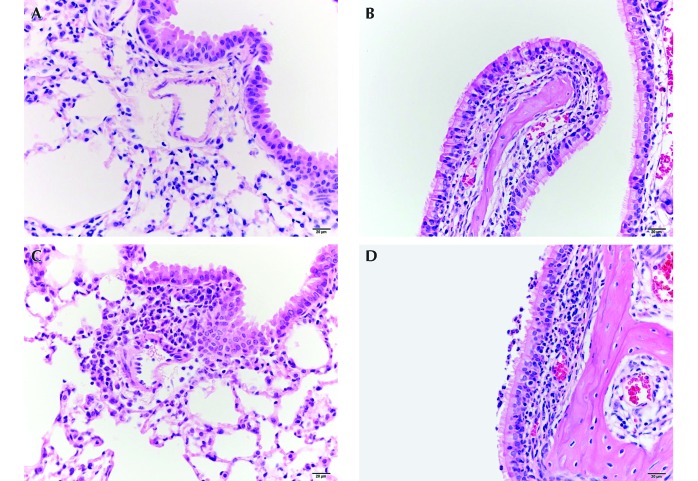
Figure 1.
(A) The lung of mouse 5, which was negative for B. pseudohinzii, shows minimal inflammatory cell infiltrates around pulmonary vessels and bronchioles. (B) The nasal cavity of the same mouse has minimal neutrophilic infiltrates within the respiratory mucosa of the maxilloturbinate. (C) The lung of mouse 2, which was positive for B. pseudohinzii, shows mild inflammatory cell infiltrates (predominantly lymphocytes and plasma cells) around the pulmonary vessels and bronchioles. (D) The nasal cavity of another mouse positive for B. pseudohinzii (mouse 3) has mild neutrophilic infiltration within the respiratory epithelium and lamina propria and minimal neutrophils in the lumen of the lateral meatus.
Table 3.
Histologic lesion score compared with percentage of neutrophils in the BAL and fecal PCR results
| Lung |
Nasal cavity |
|||||||
| Mouse | Peribronchiolar infiltrates | Perivascular infiltrates | Cellular exudates | Turbinate mucosal infiltrates | Luminal exudates | Foreign bodies | BAL neutrophils (%) | Fecal PCR |
| 1 | — | + | + | ++ | — | — | 20 | + |
| 2 | ++ | ++ | — | ++ | + | + | 5 | + |
| 3 | — | + | — | ++ | + | — | 2 | + |
| 4 | ++ | ++ | — | + | — | — | 20 | + |
| 5 | + | + | — | — | — | — | 2 | — |
| 6 | ++++ | + | — | ++ | ++ to +++ | + | 5 | + |
| 7 | +++ | + | — | ++ to +++ | ++ to +++ | — | 10 | + |
| 8 | ++ | — | +++ | ++ | ++ to +++ | + | 20 | + |
| 9 | — | + | — | ++ | ++ | — | 5 | + |
| 10 | + | + | — | ++ | — | — | 2 | — |
Lesion scores: +, minimal, very mild, or rare; ++, mild; +++, moderate; and ++++, marked.
Sentinel testing.
Of the 8 cages of sentinel mice tested, 7 were fecal PCR-positive for B. pseudohinzii. We did not have PCR data to demonstrate that these sentinel mice were negative for B. pseudohinzii on arrival, but subsequent shipments of sentinel mice from the same vendor were all negative.
Statistics.
The fecal PCR results were compared with the culture results from the lung and oral cavity, with a sample size of 10 mice (Table 1). The sensitivity and specificity of the fecal PCR assay for B. pseudohinzii were 100%; the positive and negative predictive values were 100%.
Discussion
This report is the first description of mice infected with B. pseudohinzii in the United States. What initially began as a case work-up for an investigator has led to new information about a microbial agent that, until now, has not been documented in the field of laboratory animal medicine. Our findings suggest that infection with B. pseudohinzii may confound pulmonary research, and institutions should consider testing for, and excluding, this organism from mouse colonies used to support pulmonary research. Moreover, our investigation determined that fecal PCR analysis is the best antemortem test for B. pseudohinzii and that dirty-bedding sentinels can be used to screen for B. pseudohinzii.
Some mice from the case reports in Japan that were infected with B. hinzii had bronchopneumonia. We did not observe bronchopneumonia in any of our mice that were infected with B. pseudohinzii, but the acidophilic granulomatous pneumonia that we noted in mouse number 8 might represent resolving pneumonia. In this report, 6 of the 8 infected mice had rhinitis, and 3 of the 6 mice with rhinitis also had nasal foreign bodies, which confounded the interpretation of the causality of rhinitis. The 2 mice that were negative for B. pseudohinzii did not have evidence of rhinitis and demonstrated only a few neutrophils around the vomeronasal organ, which were a background lesion observed in all 10 mice.
The high sequence similarity between the Japanese B. hinzii strain 3224 and the genogroup 16 BH370 isolate suggests that the Japanese strain was misidentified.9 Comparison of the partial gyrB gene sequence of strain 32246 (accession no. ab444711.1) with the GenBank Whole-Genome-Shotgun B. pseudohinzii sequence (no. JHEP02000002.1) demonstrates 99% homology, compared with its 95% homology with other B. hinzii sequences. Further sequence comparisons are warranted, but the Japanese isolate may actually be B. pseudohinzii, and therefore B. pseudohinzii might cause a spectrum of disease, ranging from asymptomatic rhinitis to clinical bronchopneumonia.
The lesions that occur due to B. pseudohinzii infection are consistent with the general pathogenesis of Bordetella spp. These organisms initially colonize the nasal cavity, leading to attenuation of the epithelium, damage to ciliated cells, and infiltration with inflammatory cells.5 In addition to colonizing the nasopharynx, the fimbriae of Bordetella spp. allow for the attachment to ciliated cells in the trachea and colonization.5 The extracellular release of tracheal cytotoxin leads to ciliostasis and elicits an inflammatory response.5 The trachea was not examined in the current study but will be collected during future studies. Bordetella organisms are often resistant to a wide range of antibiotics because of the ability to form a biofilm and because Bordetella species can hide from the immune system within phagocytic cells.5 The organisms’ ability to colonize the nasopharynx, form a biofilm, and evade the immune system explains the chronic nature of many Bordetella infections.
The B. pseudohinzii infection in our colony of mice did not cause clinical signs of disease but did appear to have a negative effect on pulmonary research. Of the 15 animals studied, 12 were infected with B. pseudohinzii, and 10 of these mice had increased percentages of neutrophils in the BAL fluid. Our data suggest that B. pseudohinzii infection should be a differential diagnosis for an increased percentage of neutrophils in BAL fluid. However, 2 mice (one from the case history and one from the screening test evaluation) had normal percentages of neutrophils in the BAL fluid but were either culture- or PCR-positive for B. pseudohinzii; therefore, a normal neutrophil count did not rule out B. pseudohinzii infection.
An ideal screening test is noninvasive, inexpensive, reliable, and valid. We evaluated oral swab culture, oral swab PCR analysis, and fecal PCR analysis as potential antemortem screening tests for B. pseudohinzii in 10 mice. The PCR assay used in this study was designed to amplify regions of the ompA gene that were conserved among all B. hinzii sequences deposited in GenBank in 2014. Because B. pseudohinzii and associated ompA sequence data were not reported until 2015,9 it was fortunate that the PCR assay we used in the current study supported the identification of B. pseudohinzii. All mice that were positive according to cultures of the oral cavity or lung were also positive for B. pseudohinzii by fecal PCR analysis. In the 10 mice tested, fecal PCR was 100% specific and sensitive and had positive and negative predictive values of 100%. PCR testing of fecal samples suggests that B. pseudohinzii can be detected in sentinel animals exposed to dirty bedding, consistent with the finding that after 12 wk in the facility, 7 of 8 cages of sentinel mice yielded B. pseudohinzii in their feces. Additional work needs to be done to fully understand the transfer of B. pseudohinzii to dirty bedding sentinels. Given that the B. pseudohinzii-positive fecal samples yielded an estimated 50 to 100 copies per PCR reaction suggests that fecal pellets from 5 to 10 mice can be pooled together for screening purposes. The ability to test for B. pseudohinzii by using pooled fecal-pellet PCR assays allows for a simple, noninvasive method for antemortem testing.
Currently, none of the 4 major commercial vendors provide test results for B. hinzii or B. pseudohinzii on their routine health reports. Personal communication with vendors has revealed that one vendor is performing annual PCR testing for B. hinzii, but these results are not part of routine health reporting and are not accessible. Our study and recent literature suggest that B. pseudohinzii, B. hinzii,6,7 and another similar organism (genotype 1610,12) are potential emerging pathogens. All 3 of these reported agents may actually be B. pseudohinzii, and the potential to confound pulmonary research should be considered for all of them. Until commercial vendors recognize B. pseudohinzii, B. hinzii, and related organisms as microbial agents of importance and include them on routine health-monitoring reports, it will be difficult for research institutions to exclude them from their colonies.
B. pseudohinzii should be considered as a microbial agent of importance in murine colonies that might confound pulmonary research and as a potential pathogen. Institutions with pulmonary research programs should consider testing for and excluding this and other related organisms. To raise awareness and to comply with the AALAS–FELASA suggested international health report,11 we urge commercial vendors to routinely test for B. pseudohinzii, B. hinzii, and related organisms; include the test results on health reports, and exclude these organisms from production colonies.
References
- 1.Arvand M, Feldhues R, Mieth M, Kraus T, Vandamme P. 2004. Chronic cholangitis caused by Bordetella hinzii in a liver transplant recipient. J Clin Microbiol 42:2335–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cookson BT, Vandamme P, Carlson LC, Larson AM, Sheffield JVL, Kersters K, Spach DH. 1994. Bacteremia caused by a novel Bordetella species, “B. hinzii. J Clin Microbiol 32:2569–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabre A, Dupin C, Bénézit F, Goret J, Piau C, Jouneau S, Guillot S, Mégraud F, Kayal S, Desrues B, Le Coustumier A, Guiso N. 2015. Opportunistic pulmonary Bordetella hinzii infection after avian exposure. Emerg Infect Dis 21:2122–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fry NK, Duncan J, Edwards MT, Tilley RE, Chitnavis D, Harman R, Hammerton H, Dainton L. 2007. A UK clinical isolate of Bordetella hinzii from a patient with myelodysplastic syndrome. J Med Microbiol 56:1700–1703. [DOI] [PubMed] [Google Scholar]
- 5.Gyles CL, editor. 2010. Pathogenesis of bacterial infections in animals, 4th ed. Ames (IA): Blackwell Publishing.
- 6.Hayashimoto N, Yasuda M, Goto K, Takakura A, Itoh T. 2008. Study of a Bordetella hinzii isolate from a laboratory mouse. Comp Med 58:440–446. [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashimoto N, Morita H, Yasuda M, Ishida T, Kameda S, Takakura A, Itoh T. 2012. Prevalence of Bordetella hinzii in mice in experimental facilities in Japan. Res Vet Sci 93:624–626. [DOI] [PubMed] [Google Scholar]
- 8.Hristov AC, Auwaeter PG, Romagnoli M, Carroll KC. 2008. Bordetella hinzii septicemia in associated with Epstein-Barr virus viremia and an Epstein-Barr viruse associated diffuse large B-cell lymphoma. Diagn Microbiol Infect Dis 61:484–486. [DOI] [PubMed] [Google Scholar]
- 9.Ivanov YV, Shariat N, Register KB, Linz B, Rivera I, Hu K, Dudley EG, Harvill ET. 2015. A newly discovered Bordetella species carries a transcriptionally active CRISPR-Cas with a small Cas9 endonuclease. BMC Genomics 16:863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loong SK, Mahfodz NH, Wali HA, Talib SA, Nasrah SN, Wong PF, Abubakar S. 2016. Molecular and antimicrobial analyses of nonclassical Bordetella isolated from a laboratory mouse. J Vet Med Sci 78:715–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pritchett-Corning KR, Prins JB, Feinstein R, Goodwin J, Nicklas W, Riley L, Federation of Laboratory Animal Science Associations;American Association for Laboratory Animal Science 2014. AALAS/FELASA working group on health monitoring of rodents for animal transfer. J Am Assoc Lab Anim Sci 53:633–640. [PMC free article] [PubMed] [Google Scholar]
- 12.Spilker T, Leber AL, Marcon MJ, Newton DW, Darrah R, Vandamme P, Lipuma JJ. 2013. A simplified sequence-based identification scheme for Bordetella reveals several putative novel species. J Clin Microbiol 52:674–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vandamme P, Hommez J, Vancanneyt M, Monsieurs M, Hoste B, Cookson B, Wirsing von Konig CH, Kersters K, Blackall PJ. 1995. Bordetta hinzii sp. nov., isolated from poultry and humans. Int J Syst Bacteriol 45:37–45. [DOI] [PubMed] [Google Scholar]