Abstract
The purple sea urchin (Strongylocentrotus purpuratus) genome sequence contains a complex repertoire of genes encoding innate immune recognition proteins and homologs of important vertebrate immune regulatory factors. To characterize how this immune system is deployed within an experimentally tractable, intact animal, we investigate the immune capability of the larval stage. Sea urchin embryos and larvae are morphologically simple and transparent, providing an organism-wide model to view immune response at cellular resolution. Here we present evidence for immune function in five mesenchymal cell types based on morphology, behavior and gene expression. Two cell types are phagocytic; the others interact at sites of microbial detection or injury. We characterize immune-associated gene markers for three cell types, including a perforin-like molecule, a scavenger receptor, a complement-like thioester-containing protein and the echinoderm-specific immune response factor 185/333. We elicit larval immune responses by (1) bacterial injection into the blastocoel and (2) seawater exposure to the marine bacterium Vibrio diazotrophicus to perturb immune state in the gut. Exposure at the epithelium induces a strong response in which pigment cells (one type of immune cell) migrate from the ectoderm to interact with the gut epithelium. Bacteria that accumulate in the gut later invade the blastocoel, where they are cleared by phagocytic and granular immune cells. The complexity of this coordinated, dynamic inflammatory program within the simple larval morphology provides a system in which to characterize processes that direct both aspects of the echinoderm-specific immune response as well as those that are shared with other deuterostomes, including vertebrates.
Animal immunity comprises integrated systems of detection, communication and effector mechanisms that are distributed across the organism and mediate a broad set of interactions with the microbial world, including protection from pathogens and maintenance of symbiotic relationships.1 The complexity of these interactions drives rapid evolution within some arms of the immune system,2 whereas other elements are conserved across phyla.3 To study the integration of these evolutionarily labile and more stable systems, some invertebrate organisms offer unique experimental advantages (for example, reduced anatomical complexity, lower diversity of associated microbiota, optical transparency and efficient transgenesis). Because rapid evolutionary divergence and gene loss are common traits of immune gene evolution, phylogenetic position is a critical consideration in choosing a model. Invertebrate deuterostomes provide novel perspectives on animal immunity in general and contribute to understanding the evolutionary origins of vertebrate immunity.
Elie Metchnikoff4, 5 first described phagocytosis based on his observations of cells surrounding foreign bodies in starfish and sea urchin larvae. Since that work, investigations carried out in embryos and larvae of sea urchins and other echinoderms have contributed to many areas of biology, including cell biology, developmental biology and molecular biology,6 and have led to highly detailed gene regulatory network models of development.7, 8 This work is possible because of efficient techniques for transgenesis and gene perturbation in this model, as well as the morphological simplicity and optical transparency of embryonic and larval stages that allow for detailed imaging in living organisms. The sequenced genome of the purple sea urchin (Strongylocentrotus purpuratus) lends additional depth to these contributions.9 This genome encodes a large repertoire of genes with homologs involved in immunity in other species,10, 11 including representatives of transcription factor families that direct vertebrate hematopoiesis12, 13 and lineage-specific expansions of gene families that encode pattern recognition receptors.14 Using the genome sequence as a foundation, we can now return to Metchnikoff's model of cellular immunology with a suite of modern molecular tools to address fundamental questions of animal immunity. This provides the opportunity to investigate both novel immune functions and those that are part of the deuterostome heritage shared with vertebrates.15
To characterize how the complex immune system encoded in the genome functions, we focus here on the larval stage of the purple sea urchin that has a well-characterized biphasic life history.16 In this species, free-living embryos undergo gastrulation by 48 h post fertilization (hpf). During prism stage (72 hpf), the archenteron (primitive gut) elongates to form the mouth and feeding begins by 4–5 days post fertilization (DPF). At the onset of feeding, larvae are ~200 μm in length and consist of ~4000 total cells. Larvae feed for ~10 weeks as planktotrophic suspension feeders,17 and then undergo metamorphosis and settlement as a juvenile.18 Larval morphology includes a skeleton that maintains larval shape, several types of mesenchymal cells (the immune component of which is described in detail here) that populate the blastocoel (body cavity) and a simple gut. The larval gut is composed of an epithelial monolayer in which two sphincters delineate three compartments: foregut, midgut and hindgut.19, 20
These relatively long-lived, free-swimming, feeding larvae rely on a competent immune system to successfully interact with the rich microbiota of their seawater environment.21 In the purple sea urchin, gene expression studies have identified an array of immune transcription factors, receptors, effectors and signal mediators that are expressed at this stage.10, 22, 23 Although the purple sea urchin larval immune response has not been described in detail previously, several larval mesenchymal cell types exhibit morphological and genetic similarities with the well-characterized adult immune cells (coelomocytes).10 Recent studies have investigated cellular immune response in the larvae of other echinoderms. Studies in starfish bipinnaria larvae in which foreign particles were injected into the blastocoel identified mesenchymal cells with immune functions, including migratory behavior and phagocytic selectivity.24, 25 In the green sea urchin Lytechinus variegatus, mesenchyme cells acquire the ability to phagocytose yeast cells shortly after they delaminate at the mid-gastrula stage.26
From this work, it is clear that in echinoderm larvae, some immune functions can be ascribed to a heterogeneous set of mesenchyme cells that derive from a single developmental territory (the non-skeletogenic mesoderm (NSM), also known as secondary mesenchyme).27 This ring-shaped territory is patterned during blastula stage to give rise to several cell lineages,27 including pigment cells28 and a group of nonpigmented cell types that are collectively known as blastocoelar cells.29 The well-characterized regulatory events that direct this specification process30, 31, 32 include functional parallels involving transcription factors with homologs in vertebrate hematopoiesis (for example, Gata1/2/3 and Scl).12
Here we analyze the larval immune response in the purple sea urchin from the perspective of these mesenchymal cells. We characterize five distinct larval cell types and define their role in immune response on the basis of cell behavior, morphology and gene expression. We investigate the response of these cells to intrablastocoelar injection of foreign particles and perturbed bacterial exposure in the gut. We establish an immune response model using the marine bacterium Vibrio diazotrophicus (first isolated from the gut of the congeneric green sea urchin S. droebachiensis33) that elicits immune activity at both the cellular and molecular levels. Although this response involves contributions from several tissues, we specifically focus here on the role of the mesenchymal immune cells. We find that although the larva is morphologically simple, it possesses a heterogeneous set of immune cell types that dynamically interact to resolve immune challenge. These data complement studies of the sea urchin adult immune response and highlight both similarities and differences in the cellular and molecular immunity between these two very different morphological forms that share a common genome.
RESULTS
The larval immune response is mediated by a heterogeneous set of cell types
Previous work in echinoderm larvae indicates that mesenchymal cells (typically blastocoelar cells) are capable of phagocytosing foreign particles.4, 10, 25, 26 In the purple sea urchin, immune function in mesenchymal cells is consistent with developmental expression of the Gata1/2/3 and Scl transcription factors that also play important roles in vertebrate hematopoiesis.12 Although the morphology of some of these cell types has been previously described (primarily from a developmental viewpoint),27, 29, 34 specific immune functions have not been assigned to any of the mesenchymal cells. To characterize these cells from an immune perspective, we observe larvae under several conditions of immune challenge. These include typical laboratory conditions, exposure to specific bacteria in either the sea water or direct blastocoelar injection or culturing larvae in oceanic sea water. Using time-lapse microscopy, we here characterize five morphologically distinct cell types that exhibit immune properties including surveillance-like motility, phagocytic capability and participation in specific immune cell/cell interactions (Figure 1 and Supplementary Table S1). To further delineate these cells, we characterize the expression of cell type-specific immune gene markers (Figure 2). The morphological and transcriptional characteristics of these cell types are outlined below.
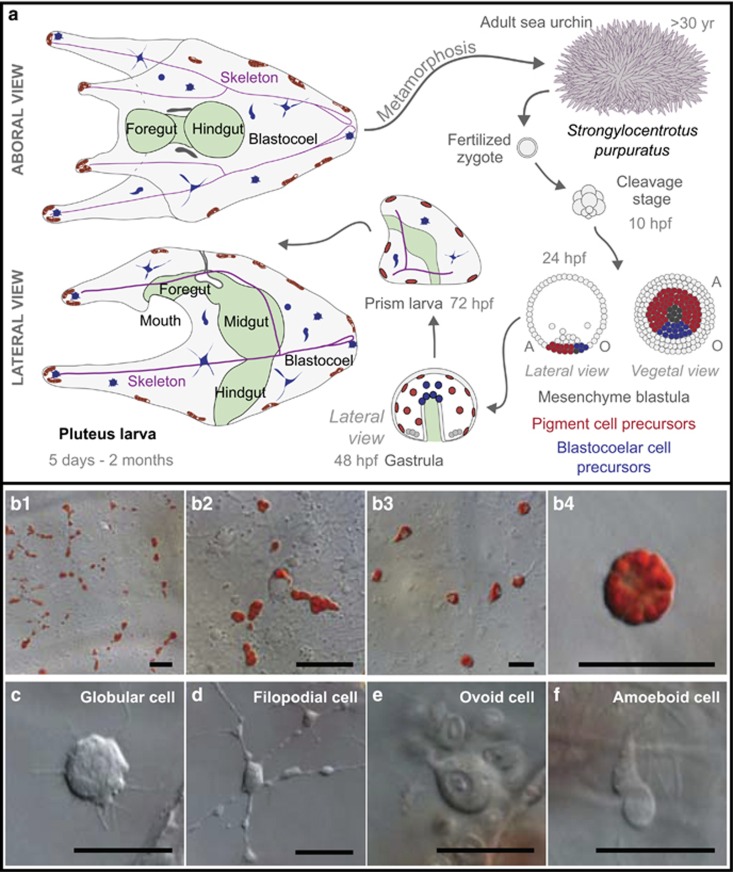
Figure 1.
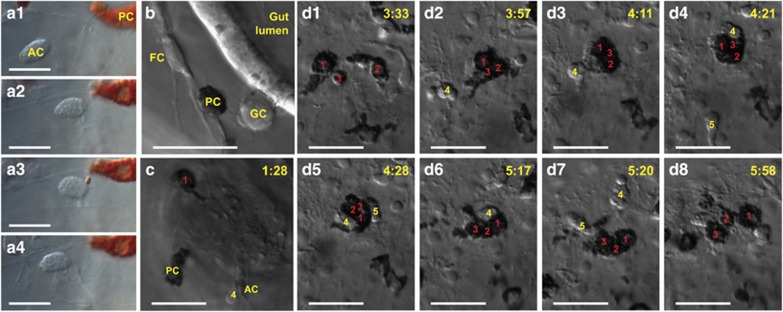
Purple sea urchin larvae are morphologically simple yet have several immune cell types. (a) The purple sea urchin has a biphasic life history. Although many sea urchin species have similar life cycles, the times shown apply to S. purpuratus. Adult animals have life spans of several decades. Fertilized zygotes develop through a rapid cleavage stage to form a mesenchyme blastula of ~800 cells by 24 hpf. In this stage, the NSM is partitioned along the oral (O; blue)/aboral (A; red) axis. In the aboral NSM pigment cell precursors ingress into the blastocoel at ~30 hpf. Oral NSM cells are marked by expression of Gata1/2/3 and Scl homologs and differentiate later into several blastocoelar cell types as they ingress at ~42 hpf (see c–f). Larvae are characterized by a tripartite gut (foregut, midgut and hindgut) and a calcite skeleton. Pigment cells are typically apposed to the ectoderm. The blastocoel is populated with several morphologically distinct types of blastocoelar cells. (b–f) Five types of immune cells are present in sea urchin larva. (b) Pigment cells have two morphologies. A collection of pigment cells near the ectoderm (b1, b3) and a single pigment cell (b2, b4) are shown. In their resting state, pigment cells are stellate (b1, b2). In response to immune stimuli, they become rounded (b3, b4). (c–f) Morphology and behavior define four types of blastocoelar cells. These include (c) globular cells, (d) a subset of filopodial cells, (e) ovoid cells and (f) amoeboid cells. Details of these cells are found in Supplementary Table S1. Scale bar represents 20 μm.
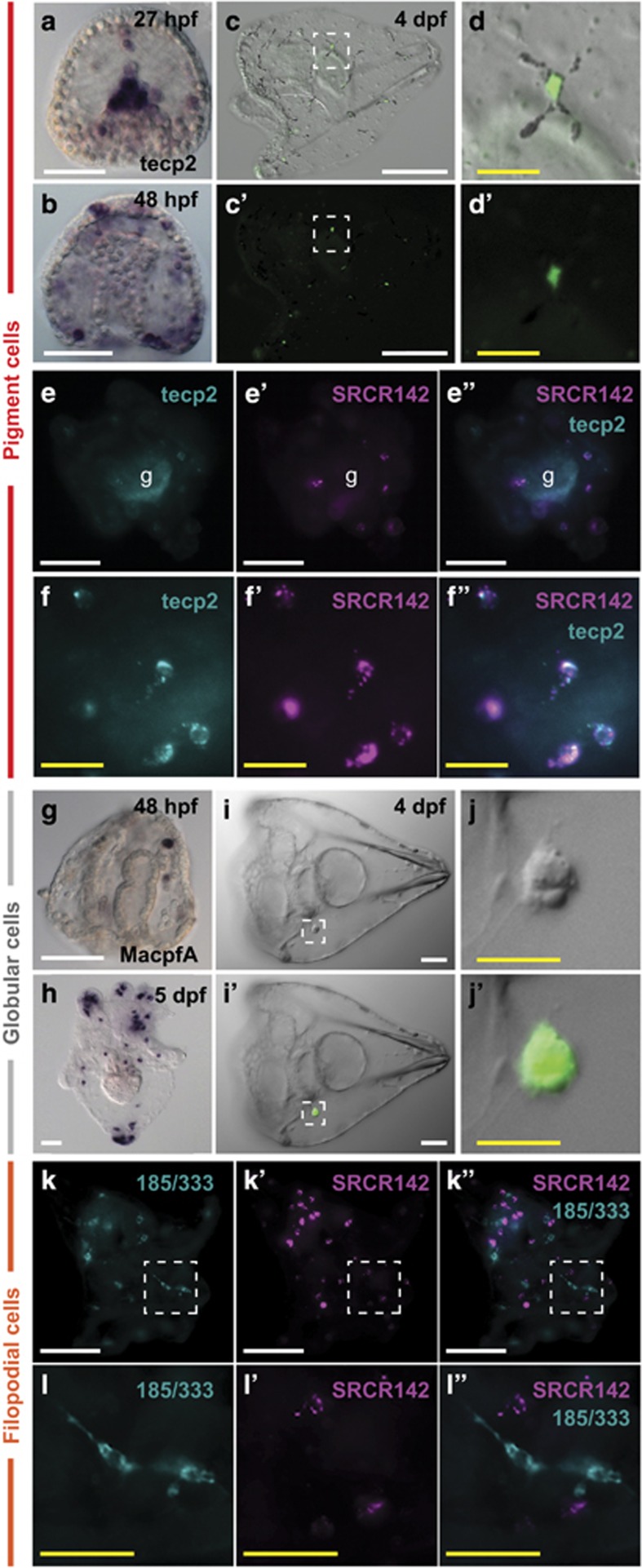
Figure 2.
Mesenchymal immune cells specifically express immune genes during development and in response to bacterial challenge. (a–d) The tecp2 transcript is expressed in pigment cells. (a) WMISH localizes tecp2 expression to the secondary mesenchyme cells ingressing into the blastocoel at 27 hpf and (b) within cells populating the blastocoel and at the ectoderm at 48 hpf. A GFP reporter construct localizes tecp2 expression to pigment cells. (c, d) A larva (4 DPF) expressing GFPtecp2 is shown (c; fluorescence channel c′); the box indicates the inset in (d; fluorescence channel d′). See Supplementary Figure S2 for a detailed time course of WMISH and GFP expression. (e, f) Fluorescent WMISH colocalizes SRCR142 to some tecp2+ pigment cells. tecp2 (green) and srcr142 (pink) (g, indicates diffuse gut autofluorescence; e, e′, f, f′: single fluorescence channels; e′′, f′′ both channels). (g–j) The macpfA.2 transcript localizes to globular cells. WMISH indicates that macpfA2 expression is present in several cells at 48 hpf (g). In larvae (10 DPF), macpfA2+cells are evident throughout the blastocoel as well as the larval arms and apex (h). In embryos transgenic for a macpfA2 GFP reporter, GFP fluorescence is restricted to globular cells (i′, j′). The inset shown in (j) is indicated by the white box in (i) (i, j, DIC; i′, j′, DIC/fluorescence overlay). (k, l) The srcr142 and 185/333 transcripts are expressed in distinct cell populations. Fluorescent WMISH was performed using probes specific for srcr142 (pink) and 185/333 (green) on larvae (10 DPF) exposed to V. diazotrophicus for 24 h. No colocalization was evident between these two transcripts. The location of the 185/333+ cells within the blastocoel as well as their morphology suggests that these cells are filopodial cells. (k, k′, l, l′) Single fluorescence channels; (k′′, l′′) channel overlay. (l) blowup from white box in (k). White scale bars indicate 50 μm; yellow scale bars indicate 20 μm.
Pigment cells
Pigment cells are red, granular, mesenchymal cells that, under typical laboratory conditions, are closely apposed to the aboral ectoderm where they maintain a relatively even distribution with concentrations in the arm tips and apex (Figures 1a and b). These cells move within the plane of the ectoderm using pseudopodial extensions. Larvae (10 DPF) have 50–80 stellate pigment cells, each with ~40 granules distributed around the nucleus and in 2 to 4 pseudopodia (Figure 1b). The granules, which are typically 1–2 μM in diameter, encapsulate echinochrome A, a naphthoquinone35 that can react to form peroxide at extracellular calcium concentrations.36 Pigment cells resemble adult red spherule cells that play a major role in mediating immune response and wound healing after metamorphosis.37
In addition to a suite of previously characterized developmental factors and enzymes implicated in pigment production,38,39 we identified two genes related to those with immune functions in other animals that are specifically expressed in pigment cells using whole-mount in situ hybridization (WMISH) and fluorescent protein reporters (see Figures 2a–f and Supplementary Figures S1a and c and S2). These include a thioester-containing protein (tecp2; SPU_019422) and a scavenger receptor (srcr142; SPU_018429). The tecp2 transcript encodes a protein with five domains associated with the complement system and α-2-macroglobulin (PF07678; Supplementary Figure S1c). Quantitative PCR profiles show that tecp2 expression is initiated by 24 hpf, peaks at 36 hpf and is maintained at low levels in the pluteus larva (84 hpf; Supplementary Figure S2a). WMISH localizes tecp2 expression to aboral NSM (pigment cell precursors) in the mesenchyme blastula (24 hpf; Supplementary Figure S2b) and pigment cells as they ingress into the blastocoel (27–48 hpf; Figures 2a and b and Supplementary Figures S2b–e). To confirm these data in live animals, a tecp2 reporter was constructed in which green fluorescent protein (GFP) expression is driven by the 3.1 kb region upstream of the tecp2 start of transcription (Supplementary Figure S1b). Larvae (4 DPF) transgenic for this construct exhibit GFP expression only within pigment cells (Figures 2c and d).
The purple sea urchin genome includes a complex array of over 200 genes that encode multiple scavenger receptor cysteine-rich (SRCR) domains.10, 14 In vertebrates, scavenger receptors exhibit a broad range of functions, including pathogen recognition and clearance.40 The srcr142 transcript encodes multiple nearly identical SRCR domains. Transcript prevalence measurements show that srcr142 expression initiates at 18 hpf (Supplementary Figure S2f). WMISH colocalizes the expression of srcr142 with tecp2 (Figures 2e and f). Srcr142 is also coexpressed with polyketide synthase (pks; Supplementary Figure S2g) another pigment cell marker.39 Notably, not all pigment cells express both the srcr142 and tecp2 transcripts, indicating some level of heterogeneity. This may reflect either dynamic changes in transcript levels or more stable cell populations (Supplementary Figure S2h).
Globular cells
These distinct cells are 10–15 μm in diameter and are filled with large, flattened vesicles. Based on the expression of a perforin-like marker (macpfA2; discussed below), these cells are observed in both motile (Figure 1c) and sessile forms (Supplementary Table S1). Using time-lapse microscopy, we find two to five motile globular cells within the blastocoelar cavity. These cells migrate at a relatively constant rate (an average of 2.1 μm min−1) with surveillance-like behavior. Sessile globular cells are localized to the blastocoelar space within the larval arms and apex. Globular cells display dynamic, short-lived filopodial projections that transiently interact with pigment cells, gut and ectodermal epithelial cells and other blastocoelar cell types.
We find that globular cells are marked by expression of the gene encoding a perforin-like factor, macpfA2. The purple sea urchin genome contains six Macpf subfamilies (MacpfA–G; 21 total genes).10 Macpf proteins are characterized by a conserved MACPF domain (Membrane Attack Complex/PerForin; PF01823; Supplementary Figures S1d–f), which is present in vertebrate terminal complement pathway proteins (C6–C9), perforins41 and Mpeg1/Perforin-2.42 MacpfA2 expression initiates in the late gastrula stage (45–48 hpf) and increases until the onset of feeding (Supplementary Figure S3a). WMISH localizes macpfA2 transcripts to 1–2 cells at late gastrula (Supplementary Figure S3b). The number of cells and proportion of larvae that express macpfA2 increases until 10 DPF, when larvae have an average of 10.3 macpfA2+ cells (Figure 2g and Supplementary Figure S3b). To visualize expression of this gene in vivo, a reporter was constructed in which GFP expression was driven by the 5.1 kb region upstream of the macpfA2 translation start site (Supplementary Figure S1e). Fluorescence is observed in both motile globular cells within the blastocoel and also sessile forms in the arms and apex using time-lapse microscopy on embryos transgenic for this reporter (Figures 2i and j). An expressed sequence tag with a similar expression pattern in Hemicentrotus pulcherrimus43 (Genbank: AU274129) corresponds to the 3′ untranslated region of a macpfA type gene homolog.
Phagocytic filopodial cells
A heterogeneous set of cells distributed throughout the blastocoel and across the basal surface of ectoderm and gut epithelia form multiple interconnected, syncytial networks (Figure 1d).29 These cells are characterized by small cell bodies (5–7 μm) and 2 to 5 filopodial projections that extend 10–50 μm. Time-lapse imaging reveals that cells within these networks extend and retract filopodia to break and re-form connections. A subset of these filopodial cells are phagocytic and express the immune effector genes known as 185/333. This gene family encodes a diverse family of echinoid-specific effector proteins that are acutely upregulated in response to immune challenge in a subset of adult coelomoctyes.44 These genes are similarly upregulated following immune challenge in larval immune cells (Figures 2k and l). Expression of 185/333 is never colocalized with srcr142, tecp2 or macpfA2, excluding expression in pigment and globular cells. The number and distribution of 185/333+ cells indicate that these transcripts are expressed in a subset of filopodial cells.
Ovoid cells
This second morphological type of phagocytic cell arises in the blastocoel under some conditions of immune challenge (Figure 1e). These oval-shaped cells are 10–15 μm on their long axis and appear to be independent of the blastocoelar syncytia. When present, only a few of these cells are observed within the larva. These motile, granular cells accumulate at the site of blastocoelar microinjection of microbial particles and may rapidly emerge from a morphological transformation of filopodial cells.
Amoeboid cells
These comma-shaped cells are ∼10 by 5 μm in size (Figure 1f). The most striking feature of amoeboid cells is their rapid migration throughout the larva, consistent with previous descriptions.34 Cell tracking measurements indicate that in uninfected larvae, amoeboid cells have an average velocity of 5.2 μm min−1, 2–3 times faster than the pigment and globular cells (that move at 1.5 and 2.1 μm min−1, respectively). Although amoeboid cells are typically located within the blastocoel, they are sometimes observed near the ectoderm. These cells quickly migrate to sites of bacterial exposure and interact with the gut epithelium and other immune cell types, especially pigment cells. Amoeboid cell morphology and behavior resembles the amoeboid form of the colorless spherule cells, a component of the adult coelomocyte repertoire.37
These specialized immune cells differentially contribute to immune response. This suggests the presence of a multi-armed, distributed immune system that is coordinated across the morphologically simple larva by mechanisms that include interactions among immune cell types, phagocytosis and cell-specific effector gene expression. These genes serve as both markers and indicators of immune activation. Notably, like the previously characterized pigment cell genes, tecp2 and srcr142 expression is initiated in the mid-blastula, whereas the constitutive macpfA2 and inducible 185/333 markers are expressed only after gastrulation as blastocoelar cells differentiate.
Larval immunocytes respond to the introduction of foreign particles in the blastocoel
To characterize the behavior of these cells during immune response, we microinjected bacterial cells and yeast-derived particles (Escherichia coli, V. diazotrophicus and Zymosan A (a glucan preparation derived from yeast cells)) into the blastocoel and monitored larval cell behavior using fluorescent imaging and time-lapse microscopy (Figure 3). Each type of particle elicited distinct responses. Zymosan A particles within the blastocoel trigger rapid phagocytosis by filopodial and ovoid cells (Figure 3a and Supplementary Video S1). Blastocoelar injection of E. coli did not typically induce a strong response, although these cells are sometimes phagocytosed by filopodial cells (Figures 3b and c). In contrast, introduction of V. diazotrophicus into the blastocoel induces a robust response (Figures 3d and e). When injected into the blastocoel, these bacteria are quickly agglutinated. Pigment and in some cases amoeboid cells migrate to concentrations of bacteria near the injection site within minutes. Filopodial cells are the primary mediators of phagocytosis (Supplementary Video S2). Enumeration of the bacterial cells in time-lapse videos of two injected larvae indicate that within 10 min, 11–18% of the bacteria are phagocytosed; by 2 h, the numbers of bacteria are significantly reduced (only 11–23% of original bacteria remain; Figure 3e). Amoeboid cells interact with the phagocytic filopodial cells, but were not observed to phagocytose. Similarly, pigment cells actively interact with several cell types in the vicinity of microbial particles, although phagocytosis is not typically observed. These data illustrate a complex cellular response involving interactions among several cell types within the blastocoel that varies with challenge.
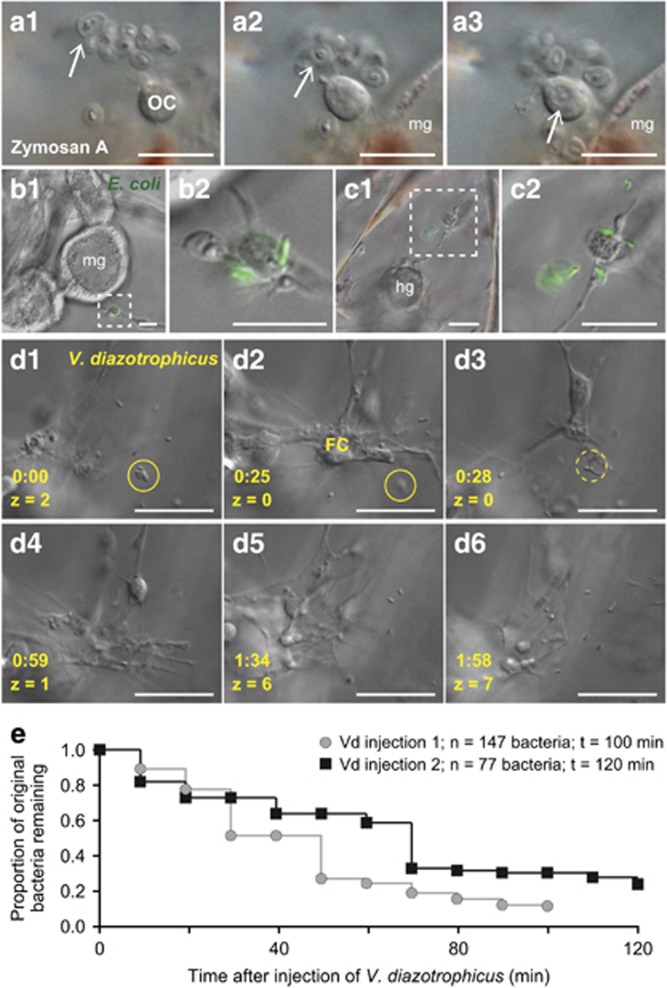
Figure 3.
Larval blastocoelar cells respond to foreign particles in the blastocoel. (a) Zymosan A is phagocytosed by ovoid cells in an injected larva. Three time points extracted from time-lapse images are shown (a1–a3). One of the Zymosan A particles is indicated by a white arrow. (b, c) Injection of two larvae with E. coli DH10B strain expressing GFP. The insets (b2, c2) are enlargements of the dashed boxes in (b1, c1) showing phagocytosis by filopodial cells. (d1–d6) Injection of V. diazotrophicus. A cluster of V. diazotrophicus is indicated with a yellow circle (d1–d3). The dotted lines in (d3) indicate phagocytosis of this cluster. Times are indicated in yellow (h:min). Images in (a) are stills from Supplementary Video S1. (e) Larvae phagocytose the majority of V. diazotrophicus within 2 h of injection. Larvae were injected with V. diazotrophicus and subjected to time-lapse imaging for up to 2 h. Bacteria were enumerated in 10 min intervals and plotted as the proportion of the initial bacterial count. FC, filopodial cell; hg, hindgut; mg, midgut; OC, ovoid cell. All scale bars are 20 μm.
Larval immunocytes respond to bacterial perturbation at the gut epithelium
To assess the role of the gut in mediating larval immunity, we investigated the response to bacteria introduced into the surrounding sea water. Of several bacteria assayed, exposure to V. diazotrophicus elicited a robust and stereotypic cellular response. During this reaction, pigment cells migrate from the basal surface of the ectoderm and accumulate at the gut epithelium (Figures 4a and b). The initiation of cell migration is apparent at 6 h of exposure and by 24 h, nearly all larvae have pigment cells near the gut epithelium. In addition, after 6 h, the gut epithelium thickens and the lumen diameter is reduced to ~25–30% of its unexposed size (corresponding to a volume reduction of up to 98% Figures 4c and d). This is maintained throughout exposure, but gut morphology returns to normal after removing bacteria from the environment. Because this exposure does not result in severe mortality (<5% mortality at 24 h of exposure at up to 107 cells per ml and <20% at 108 cells per ml), we use this inflammatory response as a controllable, reproducible model for further studies.
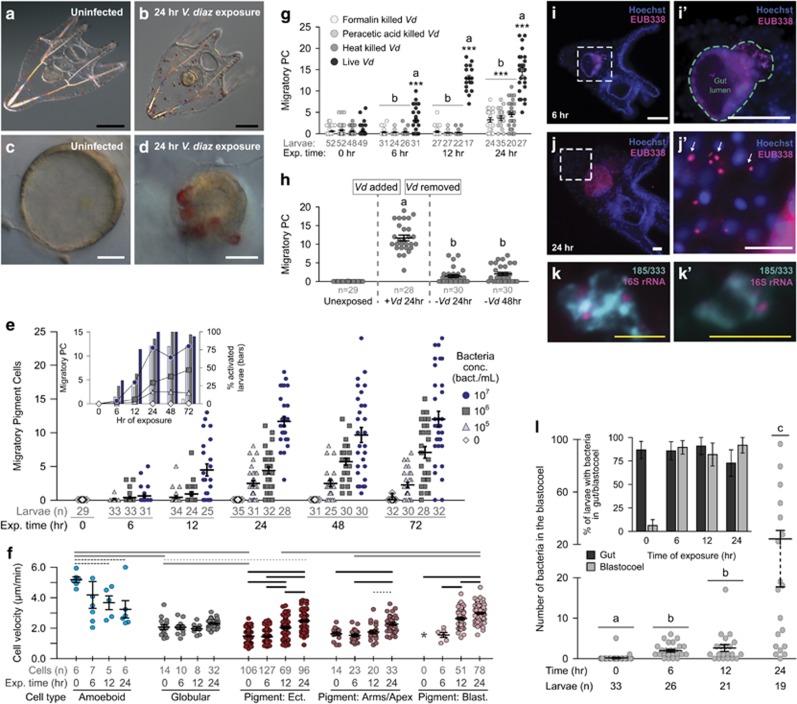
Figure 4.
Seawater exposure to V. diazotrophicus induces a system-wide larval cellular response. (a–d) The larval immune response includes migration of immune cells to the gut epithelium and changes in gut morphology. Before challenge the pigment cells are at the aboral ectoderm and the gut is extended (a, c). After 24 h of exposure to V. diazotrophicus, the larval gut is reduced in size and the gut epithelium thickens (b, d). (e) Pigment cell migration is dependent on bacterial concentration. Larvae exposed to increasing concentrations of V. diazotrophicus were monitored for pigment cell migration over the course of 72 h. Each point represents the number of pigment cells not associated with the ectoderm within an individual larva (including cells within the blastocoel and those associated with the gut). Inset shows the corresponding average cell migration over time (dots) and the proportion of larvae that have migratory pigment cells (bars). (f) Immune cell velocity is affected by exposure to V. diazotrophicus. Larvae were subjected to time-lapse imaging after 0, 6, 12 and 24 h of exposure to V. diazotrophicus. Each point represents the velocity of an individual cell (all cells were tracked in n=3 larvae for each time point). Cells were classified as either amoeboid (blue), globular (gray) or pigment cells located in either the ectoderm (dark red), larval arms or apex (red) or within the blastocoel (pink). Pigment cells were absent from the blastocoels of uninfected larvae (*). Error bars indicate the mean±s.e.m. Black bars indicate comparisons of a single cell type across time points; gray bars indicate comparisons of different cell types (solid lines, P<0.001; dashed lines, P<0.05). (g) Activation of larval cellular response requires live bacteria. Larvae (7 DPF) were exposed to V. diazotrophicus neutralized by three different methods (as well as live bacteria). Pigment cell migration was quantified after 24 h of exposure. ***Significant difference relative to the corresponding unexposed set within exposure types; Letter groupings (a, b) indicate significant differences within time groups (all P<0.0001). (h) Larvae return to pre-exposure state following removal of bacteria. Pigment cell migration was assessed after 24 h of exposure to V. diazotrophicus, and at 24 and 48 h following removal of bacteria. Significantly different groupings are indicated (a, b; P<0.0001). (i–l) Bacteria infiltrate the blastocoel during V. diazotrophicus exposure. Larvae were exposed to V. diazotrophicus and analyzed in fluorescent in situ hybridization (FISH) experiments using a Cy5-conjugated 16S RNA probe (EUB338, pink). Nuclei are counterstained with Hoechst (blue). The fluorescent signal in the gut (i, j) is the result of autofluorescence. Results are representative of six experiments. White squares indicate the regions shown in more detail (h′, i′). The gut is outlined with a dashed line in (i′). White arrows highlight bacteria in the blastocoel. (k, k') Bacteria are evident within 185/333+ cells in the blastocoel. Larvae exposed to V. diazotrophicus were used in WMISH using probes specific for 185/333 (green) and 16S rRNA from V. diazotrophicus (pink). Enlargements from two representative larvae are shown. (l) The number of bacteria within the larval blastocoel increase over time in response to exposure to V. diazotrophicus. Each data point represents the number of bacteria counted within the blastocoel of an individual larva. Results are pooled from two sets of larval samples in six experiments. Statistical groupings are indicated (a, b, c; P<0.0001). Inset shows the frequency of larvae with any bacteria in their gut (dark gray) or blastocoel (light gray). Sample size (n) is indicated under the abscissa in (e–h). All error bars indicate s.e.m. Black scale bars indicate 100 μm; white scale bars indicate 20 μm; yellow bars indicate 10 μm. Significance in all cases was determined using an unpaired, two-tailed Student's t-test.
The pigment cell migration response is proportional to the concentration of bacteria in the sea water and is reversible (Figures 4e and h). Bacterial concentration affects both the proportion of responding animals and also the number of migratory pigment cells within each larva. At 24 h of exposure with 107 bacteria per ml, 100% of larvae have pigment cells within the blastocoel or interacting with the gut epithelium (larvae have an average of 11.7 migratory pigment cells). At 105 bacteria per ml, 80% of larvae exhibit a more muted inflammatory response (on average, 3.1 migratory pigment cells per larva; Figure 4e). To characterize response resolution, larvae exposed to V. diazotrophicus for 24 h were observed following removal of bacteria. After 24 h of exposure, larvae exhibited the expected response, but after a subsequent 24 h in filtered sea water, most pigment cells returned to the ectoderm (Figure 4h). The levels do not reach baseline, possibly because of low levels of residual bacteria.
To investigate the contribution of living bacteria, larvae were exposed to V. diazotrophicus killed using one of three methods (heat, formalin or peracetic acid; 108 cells per ml) and pigment cell migration was quantified. At 24 h of exposure, some pigment cell migration is evident in response to killed bacteria, although significantly less than that induced by live bacteria (Figure 4g). Minimal migration in response to killed bacteria may result from high concentrations of bacterial-derived molecules in the sea water. These data indicate that live bacteria are required to induce a robust cell migration response.
Exposure to V. diazotrophicus also affects the motility of amoeboid and pigment cells (Figure 4f). Amoeboid cells, which typically migrate rapidly through the blastocoel, slow down significantly (from 5.2 μm min−1 in uninfected larvae to 1.9 μm min−1 at 24 h of exposure). This is likely the result of increased interactions with other mesenchymal cell types (discussed below). In contrast, pigment cells, which are typically relatively static in the aboral ectoderm (1.5 μm min−1), begin to move more rapidly (3.0 μm min−1 at 24 h of exposure). Notably, this effect is apparent not only within the pigment cells that have migrated to the blastocoel, but also in those that remain near the ectoderm.
The fate of the bacteria within the larva was assessed using fluorescent in situ hybridization (Figures 4i–l) with a Cy5-labeled oligonucleotide probe that hybridizes to part of the 16S rRNA sequence that is conserved across most bacterial species (EUB338).45 Bacteria are always evident the gut lumen. However, after 6 h of exposure to V. diazotrophicus, bacteria are also evident within the blastocoel, with increasing levels up to 24 h (Figure 4l). Nuclear counterstaining indicates that some bacteria are very close to cell nuclei (consistent with phagocytosis), whereas other bacterial cells are isolated in the blastocoel. WMISH colocalizes cells expressing the immune activation marker 185/333 with the V. diazotrophicus cells, indicating that 185/333+ cells phagocytose bacteria within the blastocoel (Figure 4k).
Immune cell interactions throughout the larva
Several types of canonical interactions occur among larval immune cells. In immune quiescent animals, pigment cells are evenly distributed near the ectoderm;28 although pseudopods make transient contact, cell bodies are rarely in close contact. Under nonchallenge conditions, pigment and amoeboid cells occasionally exhibit specific but brief interactions (Figure 5a and Supplementary Video S3). In response to immune challenge (either intrablastocoelar injection (Figure 3) or seawater exposure to bacteria (Figure 4)), these interactions occur more frequently. Amoeboid cells, globular cells and pigment cells interact directly with the gut epithelium (Figures 5b and c). In some cases, amoeboid cells and pigment cells form tight, dynamic associations near the ectoderm that are maintained for several hours (Figures 5c and d and Supplementary Videos S4 and S5). Globular cells are generally independent from other immune cells (they occasionally make contact with pigment cells) but often interact with both ectodermal and gut epithelia (Supplementary Video S6).
Figure 5.
Immune cells interact with the gut epithelium and other cell types. (a) Pigment cells and amoeboid cells interact. The amoeboid cell (labeled AC) approaches the pigment cell (PC; a1, a2) that briefly extends a single granule-filled pseudopod to interact with the amoeboid cell (a3, duration of contact is ∼1 min), and then retreats (a4, see Supplementary Video S3). (b) Several immune cell types interact after microbial disturbance. A pigment cell (PC) interacts with a filopodial cell (FC) in the blastocoel, whereas a globular cell (GC) interacts with the gut epithelium in a larva exposed to V. diazotrophicus. (c) Pigment cells and an amoeboid cell interact with the gut epithelium. Two pigment cells (PC, and 1) and an amoeboid cell (AC4). The pigment cells extend and retract pseudopodia to interact with the gut epithelium in an immune-challenged larva (24 h of V. diazotrophicus exposure). Images shown are captured from a 6 h time-lapse microscopy video of a larva infected with V. diazotrophicus after 24 h exposure (Supplementary Video S4). (d1–d8) Pigment cells and amoeboid cells form a long-lasting tight cluster near the ectoderm. Three pigment cells (1–3) interact near the ectoderm. Cells 1 and 4 are the same cells shown in (c) that have traveled from the gut back to the ectoderm. These cells form a close association, and are shortly joined by two amoeboid cells (cells 4 and 5; d2 and d4). These five cells interact dynamically for ~2 h before dispersing (d8). The times (shown in yellow) indicate the time (h:mm) from the start of the imaging. Scale bars indicate 20 μm.
Larval immune cells respond transcriptionally to bacterial exposure
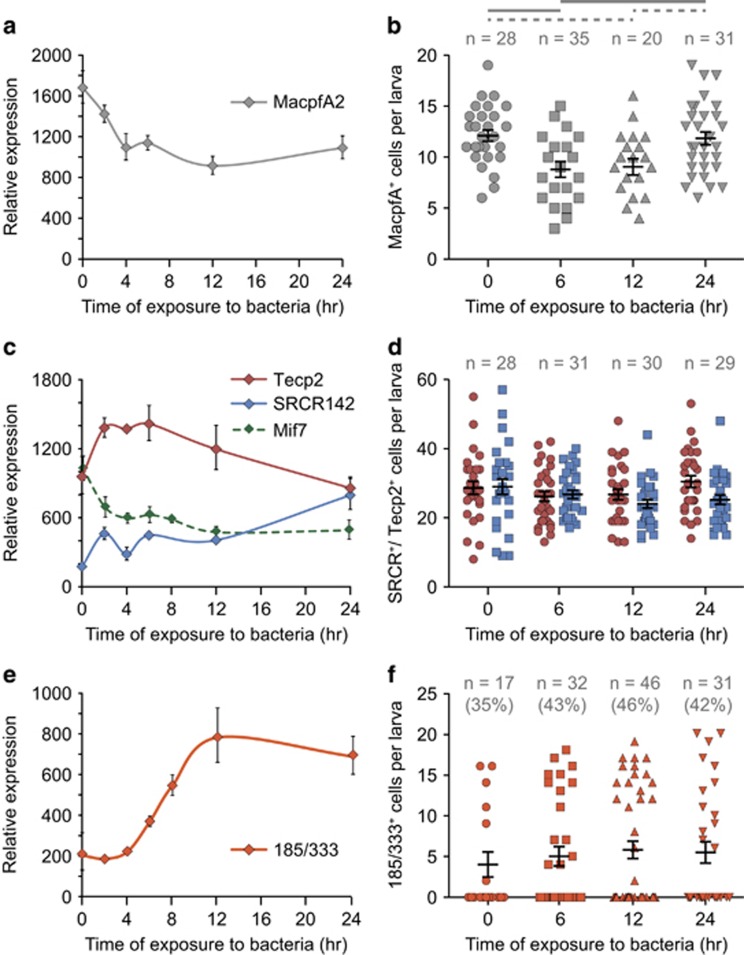
Coincident with this cellular response, V. diazotrophicus induces system-wide changes in gene activity, including genes expressed in subsets of larval immune cells (Figure 6). In response to bacterial exposure, transcript levels for the globular cell gene macpfA2 decrease moderately to 66% of the level in uninfected animals by 4 h (Figure 6a). This coincides with a slight but consistent decrease in the number of cells expressing this gene (an average of 12.1 macpfA2+ cells in uninfected larvae vs 8.8 after 6 h; Figure 6b). This reduction in globular cell number explains the decrease in macpfA2 transcripts. By 24 h of exposure, the number of macpfA2+ cells returns to the uninfected state and transcript levels begin to increase.
Figure 6.
Larvae respond to bacteria by altering gene expression levels in immune cells. Larvae were infected with V. diazotrophicus and used in quantitative PCR (qPCR) analyses to assess transcript prevalence (a, c, e) and WMISH to count number of cells (b, d, f) expressing the globular cell marker macpfa2 (a, b), pigment cell markers srcr142, tecp2 and mif7 (c, d) and filopodial cell marker 185/333 (e, f). Proportions of larvae that have 185/333+ cells are indicated in parentheses in (f). Error bars are assessed based on four qPCR replicates. Student's t-test indicates that the numbers of cells that express macpfA2 are statistically different at the time points of exposure (thick line, P<0.005; dashed line, P<0.05). Sample size of larvae counted (n) is indicated.
The pigment cell transcripts srcr142 and tecp2 are each upregulated after exposure, although with different kinetics (tecp2 is induced within 2 h of exposure to bacteria and returns to baseline at 24 h; srcr142 expression is not elevated until after 12 h; Figure 6c). WMISH shows that the numbers of cells expressing tecp2 and srcr142 remain constant throughout exposure (Figure 6d), indicating that these genes are transcriptionally upregulated. In contrast, another pigment cell marker, macrophage inhibitory factor 7 (mif7),38 is downregulated approximately twofold by 2 h of exposure (Figure 6c). Downregulation of this chemokine may be involved in the later pigment cell migration. These differential expression profiles may reflect unique roles for these genes within the larval immune response.
Finally, the 185/333 genes are strongly upregulated later in response to bacterial exposure (Figures 6e and f). In uninfected larvae, 185/333 expression is typically low and varies among animals (using highly sensitive fluorescent in situ hybridization techniques, we see that 35.3% of uninfected larvae have an average of 11.3 cells that express 185/333 at low levels; Figure 6f). During exposure, the number of cells expressing 185/333 does not increase, but transcript prevalence rises significantly. The similar numbers of 185/333Lo cells in quiescent state and 185/333Hi cells after immune challenge suggests that this subset of filopodial cells is specialized and differentiated from other cells with similar morphology even in the absence of immune challenge.
Larvae exhibit a similar response after exposure to other microbes
To examine interactions between sea urchin larvae and the broad spectrum of bacterial species in sea water, larvae were grown in filtered (0.22 μm) artificial sea water until the onset of feeding and then cultured in freshly collected natural sea water that was filtered using a 25 μm Nitex mesh (Wildco, Yulee, FL, USA) to remove predators but retain bacteria, viruses and food sources. By 96 h of exposure, 78.6% of larvae exhibit at least one pigment cell that is not associated with the ectoderm. These observations suggest that under more natural conditions, low levels of immune activation are more common than in the low microbial load laboratory settings, and that this activation is similar in character but present at a reduced extent relative to our observations after V. diazotrophicus exposure.
To investigate the response to additional bacterial species, we isolated bacteria from larvae exposed to freshly collected, 25 μm filtered sea water. Larvae (7 DPF) were washed to eliminate unassociated bacteria, homogenized and plated on marine broth agar. Bacterial colonies were grown at 15 °C and characterized by 16S rRNA sequencing. In parallel, 16S rRNA sequences were amplified directly from homogenized larvae to identify unculturable species. Over the course of several surveys, we have identified 89 bacterial types (Supplementary Table S3), of which 37 were culturable. Of these, the majority (78 isolates) were γ-proteobacteria, consistent with other surveys of seawater microbial content.46
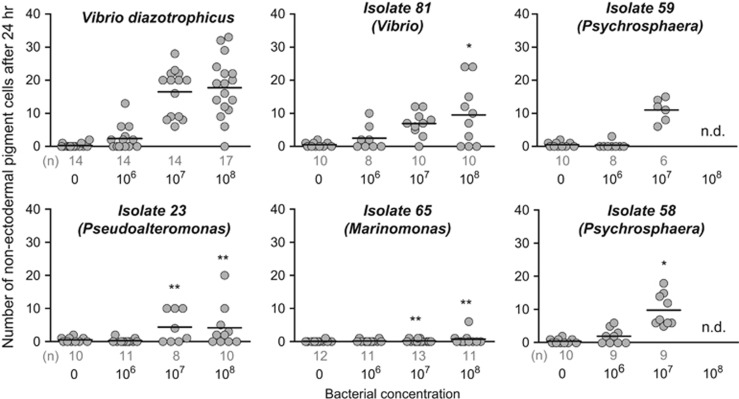
Six strains were used to assess the specificity of the larval cellular immune response (Figure 7). Pigment cell migration was quantified in larvae exposed to varying bacterial concentrations (104–108 bacteria per ml). Two isolates elicited similar responses, although less robust, relative to V. diazotrophicus (a Vibrio (isolate 81) and a Pseudoalteromonas (isolate 23); Figure 7). One Marinomonas-like species (isolate 65) did not induce cell migration, mirroring previous experiments using E. coli (after 24 h of exposure to 108 E. coli per ml, larvae exhibited an average of 0.43 pigment cells in the blastocoel). Two Psychrosphaera-like strains (isolates 58 and 59) caused lower levels of cell migration than V. diazotrophicus (106–107 per ml; Figure 7). Finally, a V. splendidus-like strain (isolate 85) was lethal to larvae even at very low concentrations (100% larval mortality was observed within 24 h at 104 per ml).
Figure 7.
Larvae differentially respond to immune challenge with several bacterial isolates. Larvae were exposed to varying concentrations of five of the isolated bacterial strains (see Supplementary Table S2 for details). The numbers of pigment cells within the blastocoel (including those associated with the gut) were counted after 24 h of exposure. Experiments at 108 bacteria per ml were not determined (n.d.) for isolates 58 and 59 because of bacterial clumping. Asterisks indicate significant differences in pigment cell migration compared with the corresponding concentration of V. diazotrophicus according to the two-tailed unpaired Student's t-test (*P⩽0.05; **P⩽0.005). Sample size (n) is indicated under the abscissa.
DISCUSSION
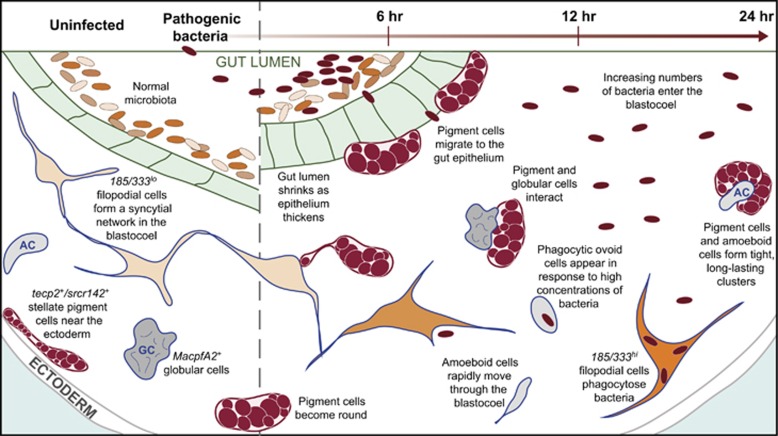
Here we define the immune cell complement of the feeding sea urchin larva and describe the cellular interactions that occur during immune challenge with particular emphasis on gut-associated disturbance. We characterize the morphology and behavior of five types of mesenchymal cells that cooperatively mediate the systemic larval immune response and characterize several cell type-specific gene markers with immune implications (including potential recognition, communication and effector molecules). Furthermore, we describe a gut-associated inflammatory response that includes pigment cell migration and interactions among cell types (summarized in Figure 8).
Figure 8.
The larval response to gut-associated bacterial perturbation occurs in cells and tissues throughout the organism. A graphic timeline of the larval response to pathogenic bacteria in the sea water is shown including changes in the gut morphology, cell behavior and alterations in gene expression levels. Under uninfected conditions, stellate pigment cells are apposed to the ectoderm, low levels of 185/333 expression are evident in a subset of filopodial cells and some cell/cell interactions are rare. Exposure to V. diazotrophicus induces a rapid thickening of the gut epithelium, pigment cell and amoeboid migration to the gut, and upregulation of 185/333. As bacterial cells penetrate the gut epithelium and enter the blastocoel, they are phagocytosed by 185/333+ filopodial cells. AC, amoeboid cell; GC, globular cell.
The larval immune response is mediated by a coordinated battery of distinct cell types
In response to the introduction of bacteria to the blastocoel, several cell types quickly converge at the injection site. Similarly, water-borne bacterial perturbation elicits effects across all immune cell types. The programmed involvement of different cell types suggests a division of labor that relies on specialized cell functions. The migratory, granular pigment cells are central in larval defense. These cells are rapidly recruited to sites of exposure (injections sites or the gut epithelium) and may directly function in immune response by producing antimicrobial compounds and carrying out sterilizing immunity with reactive oxygen species.36 These cells interact closely with bacteria in injection experiments, although phagocytosis is primarily carried out by a subset of filopodial cells and sometimes by ovoid cells. The 185/333Hi filopodial cells colocalize with bacteria that have infiltrated the blastocoel, consistent with phagocytic activity of these cells. Globular cells are implicated in immunity by their surveillance-like behavior and through production of the perforin-like macpfA2. Finally, the amoeboid cells contribute to immune response through long-lasting interactions with pigment cells and phagocytic filopodial cells. Although their exact role in larval immune response remains to be defined, their rapid migration throughout the blastocoel and recruitment to regions where bacteria are present indicate a regulatory role in defense.
The cellular response may function in parallel with systems of secreted soluble factors. Several genes involved in complement pathways in other animals are expressed in the larva, including four C3/4/5 homologs, three Bf/C2 homologs, eight perforin-like factors, and four C-type lectins that are characterized by a single C-type lectin domain and a signal peptide10 (KM Buckley and JP Rast, unpublished data). In adult sea urchins, a C3 homolog acts as an opsonin and is expressed by a subset of coelomocytes.47, 48 We find that when bacteria emerge from the injection needle, they are initially dispersed. Quickly thereafter (typically within ~5 min), the bacteria are in clumps, suggesting rapid agglutination. Together, the specialized functions within these cellular and secreted systems are tightly integrated to mount a coordinated, system-wide larval immune response.
Communication among cell types and tissues in the larval immune response
Although mesenchymal cells are important mediators of the gut-associated larval immune response, initial pathogen recognition most likely occurs in the gut epithelium in our V. diazotrophicus immune response model. Bacterial accumulation within the lumen initiates system-wide transcriptional and cellular changes. For example, we show that downregulation of mif7 in pigment cells occurs within 2 h of exposure to bacteria. This occurs well before we observe bacterial penetration of the gut epithelium. Furthermore, in immune quiescent larvae, limited contact occurs between the gut and immune cells. The spatial disconnect between the bacteria in the gut and the peripheral immune cells suggests information transfer, either by diffusible signals or a cell/cell relay system.
Cytokines and chemokines likely regulate the larval immune response over longer distances. These factors are typically small, quickly evolving molecules and it is difficult to identify homologs outside of the vertebrates.49 The sea urchin genome encodes homologs of several signal systems, including Mif-like factors, interleukin-1 receptor-like proteins, tumor necrosis factor superfamily members and a family of interleukin-17 ligands and their receptors.10 Parallel RNA-sequencing surveys of immune-challenged larvae reveal that many of these molecules are expressed and tightly regulated (KM Buckley and JP Rast, unpublished). Other factors are also likely involved in this immune signaling that are either divergent homologs of those in vertebrates or are unique to the echinoderm lineage.
Larval immune response also appears to be coordinated by cell/cell interactions. In immune quiescent larvae, pigment cells are uniformly spread throughout the aboral ectoderm where they dynamically interact through pseudopodial projections, although cell bodies maintain a fairly even distribution. In contrast, during immune response, motility is affected in several cell types and specific, programmed interactions occur. The clearest example of this is the interactions between pigment cells and the gut epithelium. Others involve intimate and long-lasting associations between pigment and amoeboid cells, the duration of which implies the possibility of complex modes of immune communication. The mechanisms that mediate this communication and its downstream consequences remain unknown, but candidates emerge from surveys of gene expression during immune response.
Deploying two immune systems from one genome
Sea urchin larvae and adults have drastically different anatomy and lifestyles. The larval stage lasts for ∼2 months as compared with many decades as adults. Larvae possess a few hundred immune cells, whereas adults may have tens of millions of circulating coelomocytes (and many more embedded in tissues). Larvae are planktotrophic; adults feed primarily on kelp and other benthic algae. Adult coelomocytes are classified into at least six types,37 including a heterogeneous set of phagocytic and secretory cells, the granular colorless and red spherule cells and vibratile cells.37, 50 Coelomocytes express homologs of genes involved in immune response in other animals, including complement factors,48 a complex SRCR repertoire51 and multigene families of diverse Toll-like receptors.23
In many ways, the larval immune cell repertoire resembles that of the adult. The clearest example is between larval pigment cells and adult red spherule cells. These cell types are nearly identical morphologically, although larval pigment cells are usually smaller and contain fewer granules. The two populations have similar transcription profiles (for example, gcm, a transcription factor and pks, which is involved in pigment production).12 Larval amoeboid cells exhibit similar morphology and behavior to immune activated adult colorless spherule cells; functions for both highly motile cell types remain undefined. Finally, similarities are evident in gene expression between subsets of larval and adult phagocytic cells. The most notable of these are the 185/333 genes that are strongly expressed following immune challenge in subtypes of adult phagocytes44 and larval phagocytic filopodial cells. These cells also share the expression of several regulatory transcription factors (for example, Gata1/2/3 and Scl).12 These transcriptional similarities may reflect regulatory subcircuits that are deployed in immune responses of both stages.
In addition to these similarities, there are several notable differences. For example, there are no clear morphological counterparts for either the adult vibratile coelomocytes in the larva or the larval globular cells in the adult. These cells may be specialized for their respective stages or may be present in altered morphological forms. Differential expression is apparent among the subfamilies of immune receptors expressed by the adult and larval stages.23 This may, in part, reflect the reduced larval cell number and a corresponding reduced capacity to manage recognition complexity. We also observe different but overlapping, suites of effector molecules expressed in the two stages (for example, subfamilies of the Macpf and 185/333 genes). The differential usage of gene family paralogs in each life stage likely reflects specific regulatory requirements. Ultimately, comparisons of the larval and adult cellular immune systems will reveal how the sea urchin deploys different but overlapping immune systems from a common genome in its maximally indirect life history.
Emergence of immune competence in the sea urchin larva
The detailed gene regulatory network model describing embryonic development in the purple sea urchin provides a framework in which to investigate the origin, specification and maintenance of larval immune cells. These cells emerge in two waves: pigment cells, which differentiate early in development, and the suite of blastocoelar cell types, which differentiate slightly later, beginning in the late-gastrula stage.27 Expression of gene markers in each of these cell types is consistent with this developmental pattern. Transcription of pigment cell markers is initiated early (~18 hpf), whereas expression of genes associated with populations of blastocoelar cells is not evident until the Gata1/2/3 and Scl homologs are downregulated (~40 hpf).12 Downregulation of these transcription factors is associated with immune cell differentiation in both the sea urchin embryo and vertebrate hematopoietic stem cells.52 The terminal differentiation of pigment cells and their positioning within the ectoderm coincides with the delamination of blastocoelar cells with phagocytic capability and the beginning of immune competency.4, 26 Ultimately, much of this may be timed so that the immune system is competent at the initiation of feeding.
A muted form of the gut-associated immune response can be activated as early as late gastrula before mouth formation. A more robust inflammatory response is evident later after the onset of feeding. A gene regulatory network approach also provides a strategy to characterize the developmental mechanisms that function across the organism to regulate the maturation and ordered emergence of immune competency. It is unlikely that each compartment of this highly integrated immune system matures autonomously. Rather, it is probable that there is a system of distributed circuitry linked by cell signaling that coordinates the development of the competent immune system as it is actuated across the larval tissues.
Conclusions
The sea urchin larva is capable of mounting an intricately regulated molecular immune response that involves the coordinated action of several immune cell types and other tissues. This response is not simply an axis of bacterial recognition and phagocytosis coupled with antimicrobial protein production that might be expected in a morphologically simple invertebrate, but instead involves several highly integrated, system-wide pathways that have evolved in parallel within the host over hundreds millions of years and are shared with more complex vertebrates. The larval immune system exhibits distinct similarities to that of the adult, but also differs in terms of cell types and gene expression. Notably, homologs of several vertebrate immune regulatory and signaling systems are also expressed in the sea urchin larval immune response. Future studies will continue to investigate these similarities and to characterize the gene regulatory network systems that coordinate immune response on the scale of the organism. It is likely that much of the causal circuitry that regulates this response is ancient and conserved among phyla and that these findings can be extended to studies in other animal lineages including vertebrates.
METHODS
Animals and larval culture
Adult S. purpuratus animals were obtained from the Point Loma Marine Invertebrate Lab (Lakeside, CA, USA) and maintained as in Solek et al.12 At 4 DPF, larvae were diluted (1 larva per ml) and stirred at 30 r.p.m. Larvae were fed Rhodomonas lens (3000 cells per ml) every other day beginning at 5 DPF. All animal protocols were approved by the Sunnybrook Research Institute Animal Care Committee.
Isolation and identification of larval-associated bacterial species
Larvae (5 DPF) were transferred to sea water (25 μm filtered) collected at the Mount Desert Island Biological Laboratory (MDIBL; Frenchman Bay, Bar Harbor, Maine, USA). Sea water was changed daily. At 10 DPF, larvae were washed through three washes of 0.2 μm filtered artificial sea water, homogenized in 100 μl of filtered sea water (FSW), plated on Marine Broth (MB; Difco, BD Sciences, Mississauga, ON, USA) agar plates and incubated at 15 °C for up to 2 days. Individual colonies were identified by PCR amplification of the 16S rRNA (Supplementary Table S1). PCR products were purified with the QIAQuick PCR Purification Kit (Qiagen, Toronto, ON, Canada) and sequenced. The Ribosomal Database Project53 and SILVA54 were used to classify isolates to the genera level.
Larval bacterial exposure
Rifampicin (Rif)-resistant mutants were generated for Vibrio diazotrophicus (ATCC 33466) through positive selection on LB agar (Sigma Aldrich, Oakville, ON, USA) containing Rif (100 μg ml−1).55 Bacterial conjugation was used to generate an isolate of V. diazotrophicus that expressed GFP.56
Rif-resistant V. diazotrophicus were cultured in LBRif (100 μg ml−1) at 15 °C with shaking at 250 r.p.m. E. coli were cultured at 37 °C in LB. Bacterial strains isolated from Atlantic sea water were grown on MB agar plates at 15 °C and pelleted or scraped off agar plates. Bacteria were counted in a Petroff-Hausser counting chamber and washed three times with FSW before exposure experiments. To assess the reversibility of immune response, larvae were exposed to V. diazotrophicus for 24 h, then transferred through three sequential washes of 0.2 μm filtered artificial sea water to remove bacteria.
For experiments using killed bacteria, bacteria were grown as described above and washed three times in FSW before one of the following treatments. (1) Bacteria were heat-killed in a 30-min incubation at 60 °C.57 (2) Bacteria were formalin-killed with 0.8% formalin for 24 h at room temperature and 24 h at 4 °C58 and washed twice with phosphate-buffered saline. (3) Bacteria were acid-killed with 0.1% peracetic acid for 30 min at room temperature and washed twice with phosphate-buffered saline. Killing efficacy was assessed by plating the neutralized bacteria on LBRif agar plates. In each case, fewer than 10 colonies were recovered per 106 treated bacteria plated.
Blastocoelar injection, microscopy and time-lapse imaging
For injection, bacteria were grown for 16 h (37 °C for E. coli (DH10B); 15 °C for V. diazotrophicus), washed three times in FSW and resuspended at 4 × 106 cells per μl for injection in FSW. Fluorescent tagged Zymosan A (Thermo Fisher Scientific, Waltham, MA, USA) was washed in FSW three times and resuspended at two particles per pl. Larvae were immobilized for injection in one of two ways: (1) placed in Petri dishes and pressed against the edge of a coverslip with the injection needle or (2) injected in a protamine sulfate-treated Petri dishes.
Imaging was performed using a Zeiss Axioplan 2 or Observer Z.1 with either Zeiss HrM or MrC5 cameras. A chilled platform (20/20 Technology, Wilmington, NC, USA) maintained larvae at 15 °C. Larvae were imaged in 20 mm glass-bottomed petri dishes (MatTek, Ashland, MA, USA) and immobilized under a coverslip anchored with double-sided tape (0.1 mm thick, Scotch 665, 3M, Maplewood, MN, USA). Images were obtained using both optical sectioning and temporal acquisition settings in the Axiovision software (Zeiss, Oberkochen, Germany). For time lapse, Z-stacks (1 to 2 μm steps) were taken at intervals of 1 min. Post-acquisition image processing and analysis was done using FIJI.59 Cell tracking was performed with the MTrackJ package.60
Reverse transcription quantitative PCR and RNA sequencing
Larvae were collected using 100 μm Nitex. RNA was extracted with TRIZOL (Thermo Fisher Scientific) following the manufacturer's protocol and purified using the PureLink RNA Mini Kit (Thermo Fisher Scientific). Contaminating DNA was removed using DNAfree (Thermo Fisher Scientific). Complementary DNA was synthesized with Superscript III (Thermo Fisher Scientific) using random hexamers. Quantitative PCR was performed as in Fugmann et al.61 Oligonucleotide sequences are shown in Supplementary Table S1.
In situ hybridization
Larvae were fixed with 4% paraformaldehyde overnight at 4 °C, dehydrated in an ethanol series and stored at −20 °C. The protocol for WMISH was adapted from previous studies.62, 63 Fluorescent WMISH was performed according to Croce and McClay.64 Primers used for generating probes are in Supplementary Table S1.
Bacteria were detected using the 16S rRNA probe EUB338 (5′-Cy5-GCTGCCTCCCGTAGGAGT-3′)45 with a protocol modified from Manz et al.65 As a negative control, the EUB338 sequence was randomly permuted and Cy5-conjugated (5′-Cy5-GTCTTAGGTGCGCCAGCC-3′). Rehydrated larvae were prehybridized in hybridization buffer (HB; 900 mM NaCl, 20 mM Tris HCl, pH 7.5, 0.01% SDS) for 30 min at 46 °C. Hybridization was performed at 46 °C for 18 h using 1 ng μl−1 probe in HB. Larvae were washed twice in HB for 30 min at 48 °C. Larvae were counterstained with Hoechst 33342 (2 nM) for 15–30 min at room temperature. Control experiments indicate that the EUB338 probe binds to several species of marine bacteria, whereas the scrambled negative control probe does not (Supplementary Figure S4).
Reporter constructs
Fluorescent protein reporter constructs were generated for macpfA2 and tecp2 (Supplementary Figure S1). Genomic regions were amplified (Phusion Taq; New England Biolabs, Ipswich, MA, USA) using primers shown in Supplementary Table S1. Products were cloned into pBluescript containing the GFP coding sequence and an SV40 3′ untranslated region. Constructs were linearized with AscI or NotI (New England Biolabs) before zygotic microinjection.
Statistics and sample selection
Sample size was chosen based on pilot experiments to reach statistical significance. Each individual experiment was performed with sibling larvae from a single mating. Multiple matings were analyzed in most cases. Two-tailed t-tests were used to compare treatments. Methods for statistical analysis of quantitative PCR data are as described previously.12 Only larva showing normal developmental morphologies were included in the analyses. Specimens for analysis were randomly chosen by aliquoting from cultures of thousands of embryos or larvae. Subsamples were counted in their entirety.
Acknowledgments
We thank Michele K Anderson for critically reading the manuscript. The research at the Mount Desert Island Biological Laboratories was supported by a New Investigator award. We also thank James Coffman and his laboratory at MDIBL for their support. This research was supported by funding from the Canadian Institutes of Health Research (MOP74667) and the Natural Sciences and Engineering Council (NSERC 312221) to JPR.
The authors declare no conflict of interest.
Supplementary Material
References
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009; 9: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pespeni MH, Garfield DA, Manier MK, Palumbi SR. Genome-wide polymorphisms show unexpected targets of natural selection. Proc R Soc B Biol Sci 2012; 279: 1412–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch TCG, Augustin R, Anton-Erxleben F, Fraune S, Hemmrich G, Zill H. Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol 2009; 33: 559–569. [DOI] [PubMed] [Google Scholar]
- Metchnikoff E. Lectures on the Comparative Pathology of Inflammation: Delivered at the Pasteur Institute in 1891. Starling FA and Starling EH (transl). 1st edn. Kegan Paul, Trench, Trubner and Company: London, UK, 1893.
- Tauber A. Metchnikoff and the phagocytosis theory. Nat Rev Mol Cell Biol 2003; 4: 897–901. [DOI] [PubMed] [Google Scholar]
- Ernst SG. Offerings from an urchin. Dev Biol 2011; 358: 285–294. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci USA 2008; 105: 5955–5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH. A genomic regulatory network for development. Science 2002; 295: 1669–1678. [DOI] [PubMed] [Google Scholar]
- Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC. The genome of the sea urchin Strongylocentrotus purpuratus. Science 2006; 314: 941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino T, Loza-Coll M, Messier C, Majeske AJ, Cohen AH, Terwilliger DP. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol 2006; 300: 349–365. [DOI] [PubMed] [Google Scholar]
- Rast JP, Smith LC, Loza-Coll M, Hibino T, Litman GW. Genomic insights into the immune system of the sea urchin. Science 2006; 314: 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solek CM, Oliveri P, Loza-Coll M, Schrankel CS, Ho ECH, Wang G. An ancient role for Gata-1/2/3 and Scl transcription factor homologs in the development of immunocytes. Dev Biol 2013; 382: 280–292. [DOI] [PubMed] [Google Scholar]
- Schrankel CS, Solek CM, Buckley KM, Anderson MK, Rast JP. A conserved alternative form of the purple sea urchin HEB/E2-2/E2A transcription factor mediates a switch in E-protein regulatory state in differentiating immune cells. Dev Biol (e-pub ahead of print 2 June 2016; doi:10.1016/j.ydbio.2016.05.034). [DOI] [PubMed]
- Buckley KM, Rast JP. Diversity of animal immune receptors and the origins of recognition complexity in the deuterostomes. Dev Comp Immunol 2015; 49: 179–189. [DOI] [PubMed] [Google Scholar]
- Dishaw LJ, Leigh B, Cannon JP, Liberti A, Mueller MG, Skapura DP. Gut immunity in a protochordate involves a secreted immunoglobulin-type mediator binding host chitin and bacteria. Nat Commun 2016; 7: 10617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClay DR. Evolutionary crossroads in developmental biology: sea urchins. Development 2011; 138: 2639–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathmann RR. Larval feeding in echinoderms. Am Zool 1975; 15: 717–730. [Google Scholar]
- Heyland A, Hodin J. A detailed staging scheme for late larval development in Strongylocentrotus purpuratus focused on readily-visible juvenile structures within the rudiment. BMC Dev Biol 2014; 14: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MM, Cruz Smith L, Cameron RA, Urry LA. The larval stages of the sea urchin, Strongylocentrotus purpuratus. J Morphol 2008; 269: 713–733. [DOI] [PubMed] [Google Scholar]
- Burke RD, Chia FS. Morphogenesis of the digestive tract of the pluteus larva of Strongylocentrotus purpuratus: sphincter formation. Int J Invertebr Reprod 1980; 2: 1–12. [Google Scholar]
- Jannasch HW, Jones GE. Bacterial populations in sea water as determined by different methods of enumeration. Limnol Oceanogr 1959; 4: 128–139. [Google Scholar]
- Tu Q, Cameron RA, Worley KC, Gibbs RA, Davidson EH. Gene structure in the sea urchin Strongylocentrotus purpuratus based on transcriptome analysis. Genome Res 2012; 22: 2079–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley KM, Rast JP. Dynamic evolution of toll-like receptor multigene families in echinoderms. Front Immunol 2012; 3: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa R, Matsumoto M, Kaneko H. Characterization of a scavenger receptor cysteine-rich-domain-containing protein of the starfish, Asterina pectinifera: ApSRCR1 acts as an opsonin in the larval and adult innate immune systems. Dev Comp Immunol 2012; 36: 51–61. [DOI] [PubMed] [Google Scholar]
- Furukawa R, Takahashi Y, Nakajima Y, Dan-Sohkawa M, Kaneko H. Defense system by mesenchyme cells in bipinnaria larvae of the starfish, Asterina pectinifera. Dev Comp Immunol 2009; 33: 205–215. [DOI] [PubMed] [Google Scholar]
- Silva JR. The onset of phagocytosis and identity in the embryo of Lytechinus variegatus. Dev Comp Immunol 2000; 24: 733–739. [DOI] [PubMed] [Google Scholar]
- Ruffins SW, Ettensohn CA. A clonal analysis of secondary mesenchyme cell fates in the sea urchin embryo. Dev Biol 1993; 160: 285–288. [DOI] [PubMed] [Google Scholar]
- Gibson AW, Burke RD. The origin of pigment cells in embryos of the sea urchin Strongylocentrotus purpuratus. Dev Biol 1985; 107: 414–419. [DOI] [PubMed] [Google Scholar]
- Tamboline CR, Burke RD. Secondary mesenchyme of the sea urchin embryo: ontogeny of blastocoelar cells. J Exp Zool 1992; 262: 51–60. [DOI] [PubMed] [Google Scholar]
- Ransick A, Davidson EH. Cis-regulatory logic driving glial cells missing: self-sustaining circuitry in later embryogenesis. Dev Biol 2012; 364: 259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna SC, Ransick A, Li E, Davidson EH. Diversification of oral and aboral mesodermal regulatory states in pregastrular sea urchin embryos. Dev Biol 2013; 375: 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc V, Rottinger E, Besnardeau L, Lepage T. Nodal and BMP2/4 signaling organizes the oral-aboral axis of the sea urchin embryo. Dev Cell 2004; 6: 397–410. [DOI] [PubMed] [Google Scholar]
- Guerinot ML, West PA, Lee J V, Colwell RR. Vibrio diazotrophicus sp. nov., a marine nitrogen-fixing bacterium. Int J Syst Bacteriol 1982; 32: 350–357. [Google Scholar]
- Gibson AW, Burke RD. Migratory and invasive behavior of pigment cells in normal and animalized sea urchin embryos. Exp Cell Res 1987; 173: 546–557. [DOI] [PubMed] [Google Scholar]
- McClendon JF. Echinochrome, a red substance in sea urchins. J Biol Chem 1912; 11: 435–441. [Google Scholar]
- Perry G, Epel D. Ca2+-stimulated production of H2O2 from naphthoquinone oxidation in Arbacia eggs. Exp Cell Res 1981; 134: 65–72. [DOI] [PubMed] [Google Scholar]
- Smith LC, Ghosh J, Buckley KM, Clow LA, Dheilly NM, Haug T. Echinoderm immunity. Adv Exp Med Biol 2011; 708: 260–301. [DOI] [PubMed] [Google Scholar]
- Rast JP, Cameron RA, Poustka AJ, Davidson EH. brachyury Target genes in the early sea urchin embryo isolated by differential macroarray screening. Dev Biol 2002; 246: 191–208. [DOI] [PubMed] [Google Scholar]
- Canton J, Neculai D, Grinstein S. Scavenger receptors in homeostasis and immunity. Nat Rev Immunol 2013; 13: 621–634. [DOI] [PubMed] [Google Scholar]
- Calestani C, Rast JP, Davidson EH. Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening. Development 2003; 130: 4587–4596. [DOI] [PubMed] [Google Scholar]
- Tschopp J, Masson D, Stanley KK. Structural/functional similarity between proteins involved in complement- and cytotoxic T-lymphocyte-mediated cytolysis. Nature 1986; 322: 831–834. [DOI] [PubMed] [Google Scholar]
- McCormack R, Podack ER. Perforin-2/Mpeg1 and other pore-forming proteins throughout evolution. J Leukoc Biol 2015; 98: 761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoguchi E, Tokuoka M, Kominami T. In situ screening for genes expressed preferentially in secondary mesenchyme cells of sea urchin embryos. Dev Genes Evol 2002; 212: 407–418. [DOI] [PubMed] [Google Scholar]
- Ghosh J, Buckley KM, Nair SV, Raftos DA, Miller CA, Majeske AJ. Sp185/333: a novel family of genes and proteins involved in the purple sea urchin immune response. Dev Comp Immunol 2010; 34: 235–245. [DOI] [PubMed] [Google Scholar]
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 1990; 56: 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim JA, Koo H, Dennis LN, Kumar R, Ptacek T, Morrow CD. An abundance of Epsilonproteobacteria revealed in the gut microbiome of the laboratory cultured sea urchin, Lytechinus variegatus. Front Microbiol 2015; 6: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clow LA, Raftos DA, Gross PS, Smith LC. The sea urchin complement homologue, SpC3, functions as an opsonin. J Exp Biol 2004; 207: 2147–2155. [DOI] [PubMed] [Google Scholar]
- Gross PS, Clow LA, Smith LC. SpC3, the complement homologue from the purple sea urchin, Strongylocentrotus purpuratus, is expressed in two subpopulations of the phagocytic coelomocytes. Immunogenetics 2000; 51: 1034–1044. [DOI] [PubMed] [Google Scholar]
- Secombes CJ, Wang T, Bird S. The interleukins of fish. Dev Comp Immunol 2011; 35: 1336–1345. [DOI] [PubMed] [Google Scholar]
- Johnson PT. The coelomic elements of sea urchins (Strongylocentrotus. I. The normal coelomocytes; their morphology and dynamics in hanging drops. J Invertebr Pathol 1969; 13: 25–41. [DOI] [PubMed] [Google Scholar]
- Pancer Z. Individual-specific repertoires of immune cells SRCR receptors in the purple sea urchin (S. Purpuratus. Adv Exp Med Biol 2001; 484: 31–40. [DOI] [PubMed] [Google Scholar]
- Pimanda JE, Ottersback K, Knezevic K, Kinston S, Chan WYI, Wilson NK. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc Natl Acad Sci USA 2007; 104: 17692–17697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007; 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 2007; 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupin A, Gualtieri M, Roquet-Banères F, Morichaud Z, Brodolin K, Leonetti J-P. Resistance to rifampicin: at the crossroads between ecological, genomic and medical concerns. Int J Antimicrob Agents 2010; 35: 519–523. [DOI] [PubMed] [Google Scholar]
- Neiman J, Guo Y, Rowe-Magnus DA. Chitin-induced carbotype conversion in Vibrio vulnificus. Infect Immun 2011; 79: 3195–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M, Brown KM, Smith LC. The gene encoding the sea urchin complement protein, SpC3, is expressed in embryos and can be upregulated by bacteria. Dev Comp Immunol 2003; 27: 529–538. [DOI] [PubMed] [Google Scholar]
- Nakhamchik A, Wilde C, Rowe-Magnus DA. Identification of a Wzy polymerase required for group IV capsular polysaccharide and lipopolysaccharide biosynthesis in Vibrio vulnificus. Infect Immun 2007; 75: 5550–5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Methods Enzymol 2012; 504: 183–200. [DOI] [PubMed] [Google Scholar]
- Fugmann SD, Messier C, Novack LA, Cameron RA, Rast JP. An ancient evolutionary origin of the Rag1/2 gene locus. Proc Natl Acad Sci USA 2006; 103: 3728–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minokawa T, Rast JP, Arenas-Mena C, Franco CB, Davidson EH. Expression patterns of four different regulatory genes that function during sea urchin development. Gene Expr Patterns 2004; 4: 3449–456. [DOI] [PubMed] [Google Scholar]
- Ransick A, Rast JP, Minokawa T, Calestani C, Davidson EH. New early zygotic regulators expressed in endomesoderm of sea urchin embryos discovered by differential array hybridization. Dev Biol 2002; 246: 132–147. [DOI] [PubMed] [Google Scholar]
- Croce JC, McClay DR. Dynamics of Delta/Notch signaling on endomesoderm segregation in the sea urchin embryo. Development 2010; 137: 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol 1992; 15: 593–600. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.