Abstract
Candida albicans biofilm formation is an important virulence factor in the pathogenesis of disease, a characteristic which has been shown to be heterogeneous in clinical isolates. Using an unbiased computational approach we investigated the central metabolic pathways driving biofilm heterogeneity. Transcripts from high (HBF) and low (LBF) biofilm forming isolates were analysed by RNA sequencing, with 6312 genes identified to be expressed in these two phenotypes. With a dedicated computational approach we identified and validated a significantly differentially expressed subnetwork of genes associated with these biofilm phenotypes. Our analysis revealed amino acid metabolism, such as arginine, proline, aspartate and glutamate metabolism, were predominantly upregulated in the HBF phenotype. On the contrary, purine, starch and sucrose metabolism was generally upregulated in the LBF phenotype. The aspartate aminotransferase gene AAT1 was found to be a common member of these amino acid pathways and significantly upregulated in the HBF phenotype. Pharmacological inhibition of AAT1 enzyme activity significantly reduced biofilm formation in a dose-dependent manner. Collectively, these findings provide evidence that biofilm phenotype is associated with differential regulation of metabolic pathways. Understanding and targeting such pathways, such as amino acid metabolism, is potentially useful for developing diagnostics and new antifungals to treat biofilm-based infections.
Candida bloodstream infections (CBSI) are the third most frequent infection in intensive care units and are associated with unacceptable rates of morbidity and mortality (up to 50%)1,2, as well as high healthcare costs3,4,5. The presence of indwelling devices, such as central venous catheters, profoundly impacts these associated problems. Moreover, poor patient prognosis is impacted by delayed diagnostics and limited antifungal options6. The recalcitrant nature of biofilms reduces the effectiveness of antifungal regimens, thereby complicating the clinical management of CBSI7. Candida albicans is the most prevalent and pathogenic of the Candida species involved in CBSI, which is typified by its capacity to form robust biofilms. Therefore, identifying the molecular mechanisms that contribute to C. albicans biofilm formation and developing new strategies to prevent CBSI is vital for minimising mortality rates and healthcare costs.
C. albicans is an opportunistic pathogen that frequently uses the biofilm lifestyle as a mode of protection against the host immune system and chemotherapeutic intervention. C. albicans capacity for physiological adaptation permits nutrient assimilation and growth in a variety of host environments8. A key component of C. albicans adaptation is filamentous growth, which is stimulated by a variety of environmental and physiological factors, including the carbohydrate source, amino acid starvation, hypoxia, elevated levels of CO2, pH and temperature8,9,10. Metabolic adaptation to such conditions, as well as antifungal agents and stress conditions, promote C. albicans pathogenicity, which includes yeast-hypha morphogenesis, phenotypic switching, adhesins, invasins, and secreted hydrolases8, factors all important to the biofilm phenotype11. In fact, these biofilm related processes are driven by six key transcriptional regulators as part of a complex transcriptional circuitry12. While this level of global control is understood, how these relate to central metabolic processes is limited. Although several anabolic and catabolic pathways, such as those involved in carbon and amino acid metabolisms, have been shown to play a pivotal role in C. albicans pathogenicity, the key pathways associated with biofilm formation remain unclear13,14.
It has recently been shown that biofilm forming ability of candidaemia isolates, and the choice of antifungal drug used for treatment, is significantly associated with patient mortality15,16. Subsequent studies by our group identified that the biofilm phenotype is heterogeneous, and stratification of clinical isolates as either high- or low- biofilm formers (HBF and LBF) has been shown to be a good predictor of clinical outcomes, where the HBF were shown to significantly correlate with mortality, unlike LBF17. While significant and elegant efforts have been made to unravel the mechanisms of biofilm formation12, these have been limited to laboratory strains, which have limited resemblance to the biological heterogeneity exhibited by clinical isolates16,17. Therefore, there is solid rationale to use clinically derived strains for studies that can explain why certain clinical C. albicans biofilms grow to be much more pathogenic (HBF) than others (LBF).
In this study, we evaluated the hypothesis that metabolic adaptation plays an important role in C. albicans biofilm heterogeneity, where differential expression of certain C. albicans genes or pathways is the primary factor that separates the LBF from the HBF. To do so, we performed a comparative RNA sequencing (RNA-Seq) analysis on samples derived from the bloodstream isolates of candidaemia patients, which were previously identified to be HBF or LBF. To put our results from the differential expression analysis into a biologically-interpretable and metabolic context, we first created a global network of interacting C. albicans genes by combining all C. albicans pathway maps in the KEGG database18. In this global network, in order to highlight the region that shows the strongest differential expression, we used a bioinformatics workflow that identifies the subnetwork of the most significantly differentially expressed genes between the LBF and HBF. This network-based approach has been shown to recover relevant biological pathways that were significantly deregulated in single-species as well as multi-microbial communities19,20.
To validate our computational findings experimentally, we selected four genes from the identified subnetwork, and performed real-time quantitative PCR (qPCR) experiments. Using qPCR, the increase or the decrease in the expression of these four genes in the HBF was confirmed using a different set of clinical isolates. Furthermore, we performed an inhibition experiment using the AAT1 gene, a central node in the subnetwork involved in several amino acid pathways, to evaluate the effect of its inhibition on biofilm formation and C. albicans survival. Overall, this study addresses a major gap in our understanding of the metabolic pathways and their role in clinically important C. albicans biofilm entity. Elucidating this essential element of pathogenesis provides invaluable information to identify metabolic biomarkers for early biofilm diagnostics and paves the way for the development of personalised therapeutic strategies.
Results
RNA-Seq analysis
In order to identify the differentially expressed genes between the C. albicans LBF and HBF, we performed a comparative RNA-Seq analysis on pre-characterised clinical isolates17. Sequencing the transcriptome of these six samples (each isolate in triplicate) resulted in nearly 141 million raw RNA-Seq reads (Table 1). Following quality control and filtering reads from non-coding RNAs, a high percentage of raw reads was mapped to the C. albicans genome (87.4% and 95% of LBF and HBF reads, respectively). The percentage of reads that were mapped to the regions of the genome annotated with KEGG genes21, was also high in both LBF (66%) and HBF (74%), indicating that the coding genome regions were mostly covered with the genes in our database. Out of 7281 genes, 6312 were found to be expressed in both conditions, among which 1007 and 783 were significantly upregulated (P < 0.05) with a minimum 2-fold change in the LBF and HBF, respectively (Fig. 1).
Table 1. The vast majority of reads in both low (LBF) and high biofilm formers (HBF) passed the quality control.
| Nr. of samples | Nr. of raw reads | % of reads after QC | % reads after t/rRNA removal | % mapped reads to genome | % mapped reads to KEGG genes | |
|---|---|---|---|---|---|---|
| All samples | 6 | 140,631,025 | 99.32 | 97.80 | 91.40 | 70.34 |
| LBF | 3 | 68,484,992 | 98.75 | 96.14 | 87.40 | 66.21 |
| HBF | 3 | 72,146,033 | 99.86 | 99.38 | 95.20 | 74.26 |
A large portion was mapped to the KEGG genes that were used to annotate the C. albicans genome. Percentages are relative to the number of raw reads.
Figure 1. Differential expression of C. albicans genes in the low (LBF) and high biofilm formers (HBF) groups.
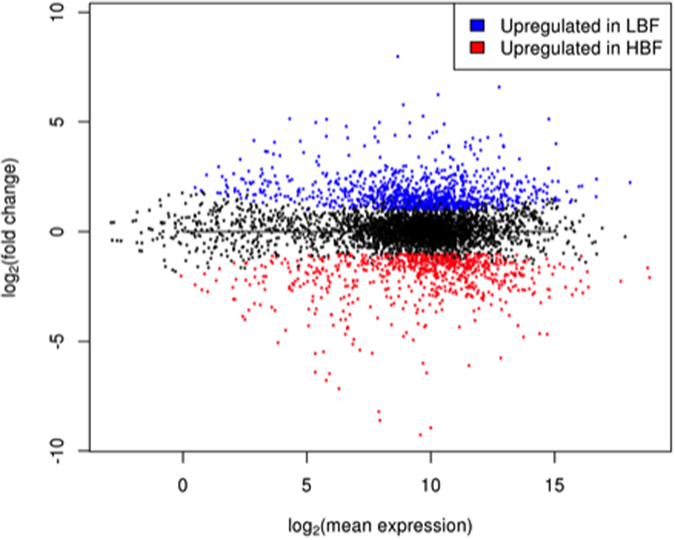
Log fold changes are plotted against the log mean expression of 6312 C. albicans genes. The blue and red dots denote the genes that were upregulated by a minimum 2-fold change in expression (P < 0.05) in the LBF (1007) and HBF (783) respectively.
An overview of differentially expressed functions
To obtain an overview of functional differences, we categorised the genes that showed a minimum 2-fold upregulation (P < 0.05) in their expression in the LBF (1007) and HBF (783) into Gene Ontology processes22 (Supp. Fig. 1). These functional categories included a range of processes including filamentous growth, biofilm formation, cellular transport, response to stress, and pathogenesis.
In both the LBF and HBF, the biofilm formation process represented only 4% of the number of significantly upregulated genes (Supp. Fig. 1). However, when certain genes in this functional category were investigated, those associated with a yeast state were found to be upregulated in the LBF (YWP1, log2 fold change = 8.6; NRG1, log2 fc = 3.7), whereas the HBF had hyphal specific gene expression increased (HWP1, log2 fc = 6; HYR1, log2 fc = 3.8; and ALS3, log2 fc = 2.8). These expression patterns are consistent with the yeast and hyphal phenotypes presented in LBF and HBF, respectively. Similar to the biofilm formation process, there was no difference in the percentage of upregulated genes associated with cell adhesion and hyphal growth in the LBF and HBF, which in total accounted for only 4% of upregulated genes in each group.
The three functional groups that contained the greatest number of significantly upregulated genes were transport, response to chemicals, and response to stress, which accounted for 52% (LBF) and 48% (HBF) of the total number of genes analysed for this section (1790). In the LBF, the greatest change in expression, namely 21%, was found in genes related to stress response (Fig. 1B). Genes associated with cellular transport were the mostly up-regulated ones in the HBF, accounting for 20% of genes (Supp. Fig. 1). A common limitation of differential gene expression analyses is the human bias for investigating the genes related to preconceived hypotheses and those reported in the existing literature. An expected finding was the higher expression of HWP1 in the HBF, as this is a hyphal-specific gene known to be involved in biofilm formation23,24. Reciprocally, the yeast wall protein (YWP1) was significantly upregulated in the LBF isolates. Cells lacking this are more adhesive and can form thicker biofilms25. Moreover, we identified ECE1 was significantly upregulated (log2 6-fold) in the HBF, which codes for the novel candidalysin peptide toxin that has recently been reported to induce epithelial cell damage26. SAP8 was also upregulated in the HBF phentotype, which is a protease previously identified to be highly expressed in mature biofilms of denture stomatitis patients27. Additionally, a glucose transporter gene, HGT9, was also upregulated in the HBF, and from studies we know that glucose has a direct effect on C. albicans adhesion and biofilm formation28. Therefore, the upregulation of HGT9 expression found in this study may be due to the increased utilisation of carbon sources in HFB isolates. Despite these potential genes of interest, we identified a large number of other genes (n = 1790) that were differentially expressed between the LBF and HBF, which is in line with other RNA-Seq studies looking at differentially expressed genes in C. albicans planktonic cells compared to those grown in a biofilm (n = 1599)12. While we can manually try and make sense of the biological data one gene at a time, our ability to decipher what the global impact of all the changes mean in the context of one another is limited.
In this analysis, many genes are grouped into a single functional category and some genes are even associated with multiple categories, making this a very coarse-grained approach. The HWP1 example shows that only looking at the high level categories does not help gain an understanding of the key players involved, and may indeed lead to a biased approach to analysis through selection of perceived biofilm related genes. Hence, this type of analysis is less suitable for pinpointing the most important factors in the metabolic network that contribute to biofilm heterogeneity.
Network analysis identifies the significant changes
In order to obtain a more informative, accurate picture of the mechanisms involved in C. albicans biofilm formation, we performed a computational analysis that takes into account the full network of C. albicans pathways present in the KEGG database. This approach is able to identify the most significantly differentially expressed parts in this full network, and can be summarised in three main steps: i) constructing a global pathway map of interacting genes, ii) assigning a positive or a negative numerical score to each gene in the network, which reflects the significance of the gene’s differential expression, and iii) applying a search algorithm on the score-annotated network for identifying the maximum-scoring subnetwork. This subnetwork corresponds to the region in the global network where the (interacting) genes with the most significant differential expression are located.
To this end, we integrated 104 C. albicans-specific KEGG pathway maps that contain the molecular interaction and reaction gene networks for functional categories, including metabolism, genetic and environmental information processing and cellular processes into a single, global C. albicans gene network (2960 genes). After removing the genes that did not interact with any other genes from the network, the global network consisted of 1721 nodes (genes) and 8197 undirected edges (gene interactions).
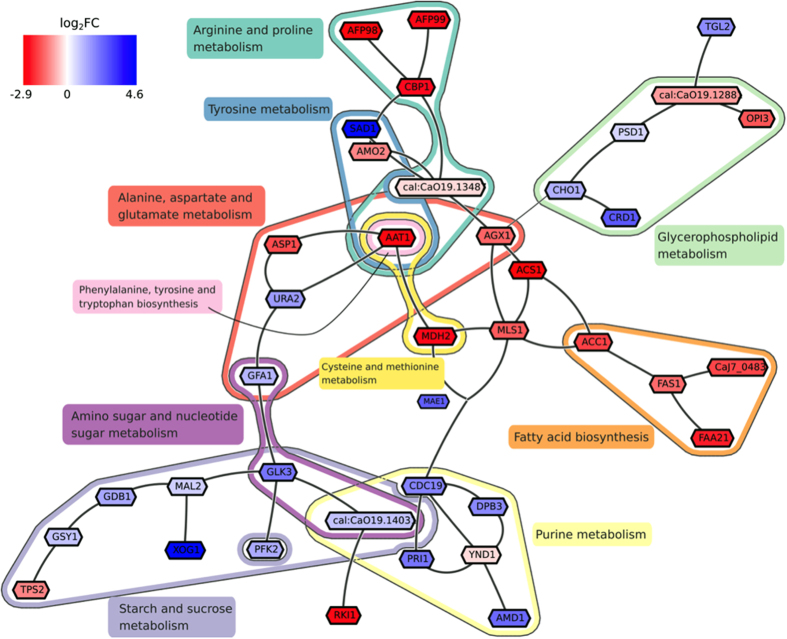
Taking the overlap between the genes that were expressed in both LBF and HBF samples with those in the global C. albicans network resulted in a final network of 859 genes and 1997 gene interactions. Next, we used a strict P value threshold of 10−10: genes in the network with smaller P values were assigned positive scores, while genes with larger P values were assigned negative scores (see Methods). Correspondingly, we assigned 59 of 859 genes in the network positive (node) scores, while the rest of the genes attained negative scores. The Heinz network search algorithm was applied on the network to calculate the maximum-scoring sub-network as described elsewhere20. The identified subnetwork consisted of 39 genes (Supp. Table), of which 20 were down- and 19 were upregulated in the HBF (Fig. 2). The total number of pathways associated with these genes was 41 (Table 2), among which (i) arginine and proline metabolism, (ii) pyruvate metabolism and (iii) fatty acid metabolism were predominantly upregulated in the HBF samples (Fig. 2). In contrast, purine metabolism and starch and sucrose metabolism were mostly up-regulated in the LBF samples (Fig. 2).
Figure 2. The maximum-scoring subnetwork identified in the global C. albicans metabolic network.
The subnetwork corresponds to the region in the global metabolic gene network where the most significantly differentially expressed genes between the LBF and HBF are located. The colours red and blue with gene names denote the fold change (upregulation in HBF and LBF, respectively). Different coloured lines indicate different metabolic pathways (labelled near each pathway). This computationally-identified subnetwork suggests the aspartate aminotransferase (AAT1) to be a key player in biofilm heterogeneity. Moreover it highlights the importance of amino acid metabolic pathways.
Table 2. KEGG pathways that are associated with the genes in the maximum-scoring sub network.
| KEGG Pathway ID | Cumulative pathway score | Pathway name | Gene IDs |
|---|---|---|---|
| cal00330 | 198.9823 | Arginine and proline metabolism | cal:CaO19.13487,AAT1,CBP1,AFP99,AFP98 |
| cal00230 | 132.1469 | Purine metabolism | YND1, AMD1, CDC19, PRI1, DPB3, cal:CaO19.14031, |
| cal00500 | 110.8155 | Starch and sucrose metabolism | GLK3, MAL2, GSY1, TPS2, GDB1, cal:CaO19.14031, XOG1 |
| cal00410 | 104.9783 | beta-Alanine metabolism | cal:CaO19.13487, CBP1, AMO2 |
| cal00250 | 59.83122 | Alanine, aspartate and glutamate metabolism | ASP1, AAT1, AGX1, GFA1, URA2 |
| cal04146 | 56.56852 | Peroxisome | FAA21, CaJ7_0483, AGX1 |
| cal03030 | 54.2462 | DNA replication | PRI1, DPB3 |
| cal00071 | 54.03803 | Fatty acid degradation | FAA21, CaJ7_0483, cal:CaO19.13487, SAD1 |
| cal00061 | 43.38386 | Fatty acid biosynthesis | FAA21, CaJ7_0483, ACC1, FAS1 |
| cal00350 | 42.3465 | Tyrosine metabolism | AAT1, SAD1, AMO2 |
| cal00360 | 39.59607 | Phenylalanine metabolism | AAT1, AMO2 |
| cal00400 | 36.1654 | Phenylalanine, tyrosine and tryptophan biosynthesis | AAT1 |
| cal00270 | 28.13576 | Cysteine and methionine metabolism | AAT1, MDH2 |
| cal00520 | 24.9472 | Amino sugar and nucleotide sugar metabolism | GLK3, GFA1, cal:CaO19.14031 |
| cal00240 | 23.8103 | Pyrimidine metabolism | YND1, PRI1, DPB3, URA2 |
| cal00564 | 21.70483 | Glycerophospholipid metabolism | PSD1, CHO1, OPI3, CRD1, cal:CaO19.12881 |
| cal00030 | 20.34725 | Pentose phosphate pathway | RKI1, PFK2, cal:CaO19.14031 |
| cal00460 | 12.2906 | Cyanoamino acid metabolism | ASP1 |
| cal03420 | 11.9899 | Nucleotide excision repair | DPB3 |
| cal03410 | 11.9899 | Base excision repair | DPB3 |
| cal03018 | 2.67299 | RNA degradation | PFK2 |
| cal00051 | −2.79647 | Fructose and mannose metabolism | GLK3, PFK2 |
| cal00561 | −5.29207 | Glycerolipid metabolism | cal:CaO19.13487, cal:CaO19.12881, TGL2 |
| cal00020 | −8.02964 | Citrate cycle (TCA cycle) | MDH2 |
| cal00565 | −8.28607 | Ether lipid metabolism | cal:CaO19.12881 |
| cal00100 | −8.28607 | Steroid biosynthesis | cal:CaO19.12881 |
| cal00590 | −8.28607 | Arachidonic acid metabolism | cal:CaO19.12881 |
| cal00592 | −8.28607 | alpha-Linolenic acid metabolism | cal:CaO19.12881 |
| cal00260 | −9.61059 | Glycine, serine and threonine metabolism | CHO1, AGX1, AMO2 |
| cal00640 | −15.0766 | Propanoate metabolism | ACS1, ACC1 |
| cal00040 | −15.1134 | Pentose and glucuronate interconversions | cal:CaO19.13487 |
| cal00280 | −15.1134 | Valine, leucine and isoleucine degradation | cal:CaO19.13487 |
| cal00053 | −15.1134 | Ascorbate and aldarate metabolism | cal:CaO19.13487 |
| cal00340 | −15.1134 | Histidine metabolism | cal:CaO19.13487 |
| cal00310 | −15.1134 | Lysine degradation | cal:CaO19.13487 |
| cal00380 | −15.1134 | Tryptophan metabolism | cal:CaO19.13487 |
| cal00680 | −16.4076 | Methane metabolism | ACS1, AGX1, PFK2 |
| cal00052 | −24.6707 | Galactose metabolism | GLK3, MAL2, PFK2, cal:CaO19.14031 |
| cal00630 | −32.8304 | Glyoxylate and dicarboxylate metabolism | MDH2, AGX1, MLS1 |
| cal00010 | −36.1765 | Glycolysis/Gluconeogenesis | GLK3, ACS1, cal:CaO19.13487, CDC19, SAD1, PFK2, cal:CaO19.14031 |
| cal00620 | −59.867 | Pyruvate metabolism | ACS1, cal:CaO19.13487, ACC1, CDC19, MDH2, MAE1, MLS1 |
The cumulative score is the sum of the scores of genes in the pathway that were found in the maximum-scoring sub network.
Validation of RNA-Seq results
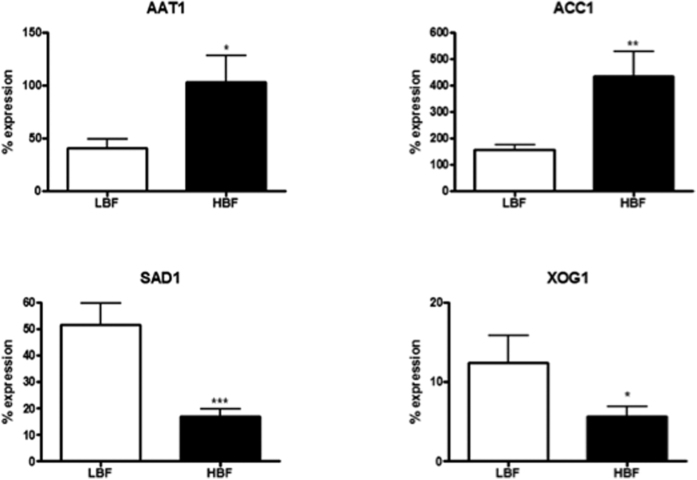
To validate the findings from our RNA-Seq analysis, we performed real-time quantitative PCR (qPCR) on 9 additional HBF and LBF isolates for four genes, AAT1, ACC1, SAD1 and XOG1, which are members of the key KEGG pathways that were found by our network analysis (Fig. 3). Two of the selected genes, SAD1 and XOG1, involved in amino acid and sugar metabolism, respectively, were significantly upregulated (P < 0.05) in the LBF. AAT1 and ACC1, involved in amino acid and fatty acid metabolism, respectively, were upregulated (P < 0.05) in the HBF. For all selected genes, qPCR results were in agreement with the results from the RNA-seq analysis (Fig. 3): the expression levels of SAD1 and XOG1 were found to be 3-fold (P < 0.001) and 2-fold (P < 0.05) upregulated in the LBF. Furthermore, both AAT1 and ACC1 were found to be significantly upregulated by 3-fold in the HBF (P < 0.05 and P < 0.01, respectively).
Figure 3. Validation of RNA-seq data.
Expression of selected genes (AAT1, ACC1, SAD1 and XOG1) was assessed by qPCR analysis of RNA isolated from 24 h biofilms of 9 HBF and 9 LBF clinical isolates. Graphs show percentage expression of each gene compared to a housekeeping gene ACT1. *P < 0.05, **P < 0.01, ***P < 0.001.
AAT1 drives the biofilm phenotype
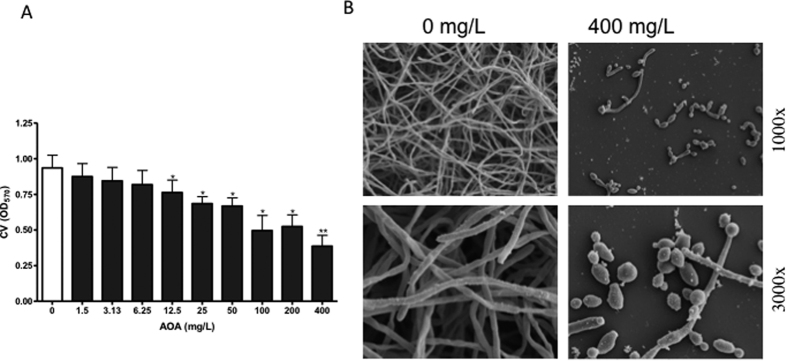
The maximum-scoring subnetwork identified by our bioinformatic workflow highlighted an overall upregulation of genes in different amino acid metabolism pathways in the HBF, therefore, these genes were investigated further. The gene AAT1, coding for aspartate aminotransferase, which plays a central role in different amino acid metabolism pathways, was found to be significantly upregulated in the HBF (Fig. 2). The impact of inhibition of the AAT enzyme activity by the aminoxy acetate (AOA) inhibitor on C. albicans biofilm formation was then examined. At all tested concentrations, AOA had no toxic effect on C. albicans growth. Next, biofilm formation by the HBF (n = 6) in the presence of different concentrations of AOA was assessed. There was significant reduction in biofilm formation in the presence of AOA in a dose dependant manner (Fig. 4A), confirming an apparent role of AAT in biofilm formation. Conversely, no significant reduction in biofilm biomass was found with LBF isolates (n = 5) in the presence of AOA (Supp. Fig. 3). Analysis of the biofilm phenotype by scanning electron microscopy showed striking differences with more yeast and pseudohyphal cells with AOA treatment compared to the filamentous multidimensional architecture in untreated controls (Fig. 4B). This suggests that AAT plays a role in modulating biofilm formation via regulating filamentation and morphogenesis in C. albicans. Furthermore, biochemical assessment of secreted AAT levels with different clinical isolates confirmed that a significantly higher level of AAT was produced by HBF compared to LBF biofilms (Fig. 5).
Figure 4. Inhibition of AAT activity reduces biofilm formation.
C. albicans biofilms (n = 6) were formed in the presence of serially diluted AOA (Aminoxy acetate [AAT inhibitor]), concentration range from 0–400 mg/L. After 24 h incubation, biofilm biomass was assessed by crystal violet (CV) assay. (A) Graph shows CV absorbance. *P < 0.05, **P < 0.01. (B) Biofilms grown on thermonox coverslips in the presence or absence of 400 mg/L of AOA were fixed and processed for scanning electron microscope imaging. Micrographs show the biofilm phenotype at 1000x and 3000x magnification.
Figure 5. Biochemical validation of AAT enzyme activity levels in HBF and LBF isolates.
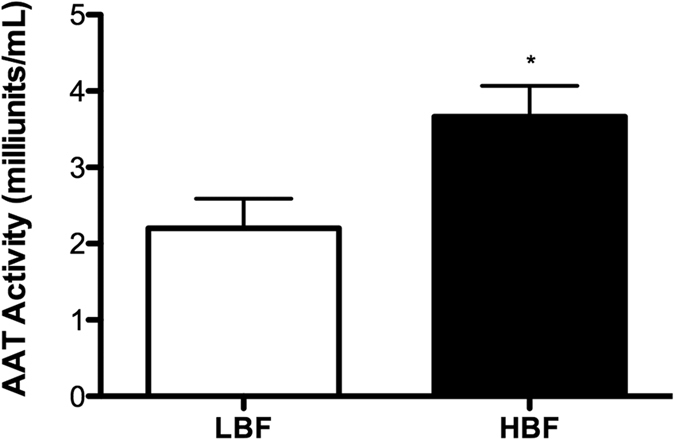
Levels of the AAT enzyme activity in 24 h grown HBF (n = 5) and LBF (n = 5) biofilms were assessed using a colorimetric assay. The bar graph shows AAT activity standardised to biofilm biomass. *P < 0.05.
Discussion
Understanding the molecular mechanisms involved in the phenotypic differences between C. albicans clinical isolates is an important avenue of investigation for developing and improving clinical management strategies, both in terms of antifungal therapy and the development of new diagnostic strategies. We previously reported that C. albicans biofilm heterogeneity is an important clinical entity17, and that extracellular DNA (eDNA) release contributes to biofilm-associated antifungal resistance29. However, this somewhat targeted investigational approach fails to take into account the global factors driving these paradoxical phenotypes that we observe clinically. Here, we identified the differential expression of several metabolic pathways that potentially contribute to C. albicans biofilm heterogeneity. This study is the first to use an integrated KEGG pathway analysis to reveal biologically important pathways in C. albicans, which has highlighted the importance of aspartate amino transferase in C. albicans biofilm development, and identified its potential as a therapeutic target for improving clinical management.
In order to avoid this limitation and focus on novel, promising, targets for improving the management of biofilm-based infections, we conducted an advanced network-based analysis. Using this approach, we found various amino acids, pyruvate and fatty acid metabolism pathways to be central pathways upregulated in the HBF. The increased intracellular level of pyruvate, pentoses and amino acids in early biofilm phase contributes to increased biomass in maturation phase30. Fatty acids have diverse functions in the cells as they are important constituents of lipids, plays a role in energy storage, integrity and dynamics of cell membrane and adhesion to host cells31. Conversely, purine, starch and sucrose metabolic pathways were predominantly expressed in LBF.
Various amino acid pathways, including arginine, proline, purine, alanine, aspartate and glutamate metabolisms were shown to be considerably upregulated in the HBF. The exact regulatory mechanism behind this amino acid metabolism is not yet clear. It is known that amino acid starvation activates the general amino acid control (GCN) response, and that GCN4-regulated amino acid metabolism is required for normal biofilm formation by C. albicans13. In response to amino acid starvation, GCN4 regulates a number of genes, which in turn activates amino acid biosynthesis and morphogenesis in C. albicans and Saccharomyces cerevisiae9. Interestingly, the AAT1 gene that encodes aspartate aminotransferase, was found to be a key member of different amino acid pathways that were upregulated in the HBF. AAT1 was shown to be a soluble protein in hyphae, and knocking out AAT1 gene does not impact C. albicans viability32,33. The mechanism of activation of AAT1 and its role in biofilm formation is not yet clear to us, but we hypothesise that this acts downstream of the GCN4 pathway. Our data suggests that it may have residual morphological effects, as also reported for GCN49, and this may interfere with biofilm stability as a result. However, further mechanistic studies are required to confirm this. Nevertheless, pharmacological inhibition of AAT was shown to significantly reduce the C. albicans biofilm biomass, confirming its apparent role in biofilm formation and its potential as an antifungal target. Our biochemical assay showed a significantly higher AAT level (P < 0.05) among HBF isolates compared to LBF, suggesting that AAT has the potential to be used diagnostically to differentiate the two phenotypes.
Collectively, using an integrated approach to analyse clinically important C. albicans isolates, we were able to link various metabolic pathways to defined biofilm phenotypes. A dedicated computational approach allowed us to dissect high-dimensional RNA-Seq output and to recover the essential pathway biology. The genes that were identified to be the key components were subsequently confirmed (AAT1, ACC1, SAD1 and XOG1) and validated (AAT1) in vitro experiments. AAT1 and ACC1 were found to be upregulated in HBF and SAD1 and XOG1 were upregulated in LBF. Previous studies have investigated the role of some of these genes in biofilm formation. For example, XOG1 gene shown to have no impact on cell wall glucan content of biofilm cells, nor it is necessary for filamentation or biofilm formation34, which is in agreement with our data. ACC1 encodes acetyl-coenzyme-A carboxylases, which have been shown to be upregulated in biofilm cells. Its transcription may indirectly depend on oxygen availability, but also on the availability of inositol in S. cerevisiae35.
Our ultimate aim from these studies and moving forward is to identify a potential target for biofilm diagnostics and antifungal development. Here, we highlighted the importance of amino acid metabolism in the HBF. The key enzyme of this metabolism, AAT, was also shown to be a potential target for the management of biofilm-based infections. Future integrated OMICs approaches will support these investigations.
Materials and Methods
Culture conditions and standardisation
This study utilised Candida albicans strain SC5314 and a series of routine patient anonymised clinical bloodstream isolates (n = 30) collected under the approval of the NHS Scotland Caldicott Gaurdians, as part of candidaemia epidemiology surveillance study17,36. C. albicans clinical bloodstream isolates previously identified to have high biofilm formation (HBF [n = 15]) and low biofilm formation (LBF [n = 15]) were used throughout this study17,36. Isolates were stored in Microbank® vials (Pro-Lab Diagnostics, Cheshire, UK) at −80 °C until sub-cultured onto Sabouraud’s dextrose agar (SAB [Sigma-Aldrich, Dorset, UK]). Isolates were propagated in yeast peptone dextrose (YPD) medium (Sigma-Aldrich, Dorset, UK), washed by centrifugation and re-suspended in RPMI-1640 (Sigma-Aldrich, Dorset, UK) to 1 × 106 cells/mL, as described previously37. Biofilms were grown in polystyrene plates or 75 cm2 tissue culture flasks in RPMI for 24 h at 37 °C.
RNA extraction and sequencing
Following incubation, biofilms were washed with PBS before a cell scraper was used to dislodge the biomass, which was homogenised using a bead beater, and RNA extracted using the TRIzol™ (Life Technologies, Paisley, UK) method as described previously38. Total RNA was DNase (Qiagen, Crawley, UK) treated and purified using an RNeasy MiniElute clean up kit (Qiagen, Crawley, UK), as per manufacturer’s instructions. RNA was quantified and quality assessed using a NanoDrop spectrophotometer (ND-1000, ThermoScientific, Loughborough, UK). The integrity of the RNA was assessed using an agarose gel to visualise the two distinct rRNA bands. Each isolate was grown in triplicate and a minimum of 10 μg of total RNA was submitted for each sample and sent for sequencing to The GenePool (Edinburgh, UK). RNA integrity was assessed using a Bioanalyzer where an RIN value > 7.0 was deemed acceptable for RNA-Seq using Illumina 50 base pair sequencing.
Pre-processing of RNA-Seq data
The reads that had an overlap of at least 8 bp with the adapter sequence (Illumina TruSeq v3 i7) were filtered out using cutadapt v1.7.1 with default parameters39. Quality-trimming on the remaining reads was done using sickle v1.31 (available at https://github.com/najoshi/sickle) with an average quality score of 15 over a 5-bp sliding window. Reads that were trimmed by sickle to a length shorter than 30 bp were discarded. Subsequently, reads originating from non-coding RNAs were filtered out using sortMeRNA v1.99-beta40 with default parameters and databases.
Candida albicans genome and its annotation
The diploid genome sequence of Candida albicans SC5314 (assembly 22) was downloaded (February 2015) from the Candida Genome Database45, http://www.candidagenome.org/and was converted to a haploid genome by discarding one of the chromosomes in each homologous chromosome pair. This haploid reference was annotated using the C. albicans genes in the KEGG database21 as follows: (i) The nucleotide sequences of 14,661 C. albicans SC5314 genes in the KEGG database were clustered at 85% sequence identity using the USEARCH41 algorithm (-cluster_fast option). The resulting 7,281 non-homologous genes were mapped to the haploid C. albicans genome using the UBLAST algorithm (E-value 10−6, query coverage 0.95) to annotate the RNA-coding regions in the genome in the form of a GTF file using in-house scripts. Subsequently, STAR RNA-Seq aligner42 was used to generate index files for the haploid genome using the GTF annotations. The pre-processed reads were then aligned to the indexed genome using the STAR RNA-Seq aligner with default parameters.
Differential expression analysis
Following mapping, for each sample, the number of reads that were unambiguously mapped to a gene was estimated by parsing the STAR RNA-Seq alignment output by HTSeq43 v.0.6.1 (default parameters). Next, an unpaired differential expression analysis (LBF vs HBF) using DESeq2 v1.6.144 was performed with default settings as described in the package vignette. Briefly, the package normalizes read counts to account for different library sizes and uses negative binomial generalized linear models to determine a conservative set of genes that are differentially expressed across experimental conditions.
Functional annotation
We determined the Gene Ontology22 functional processes of significantly upregulated genes that showed a minimum 2-fold increase in their expression in the HBF and LBF. To do so, the list of genes were used as input for the GO Slim Mapper tool of the Candida Genome Database45 with the preselection of the following processes: biofilm formation, cell adhesion, cell budding, cell wall organization, cellular homeostasis, filamentous growth, hyphal growth, interspecies interaction between organisms, pathogenesis, pseudohyphal growth, response to chemical, response to drug, response to stress, signal transduction and transport.
C. albicans gene network
The KEGG pathway maps of C. albicans SC5314 were used to create a global functional interaction network of C. albicans genes. To this end, gene relations defined in the individual pathways, such as acting on the same substrate, were used to derive the interactions between genes, which were then merged into a single network. The global gene network for the HBF-LBF comparison was then obtained by taking the intersection of genes in the global C. albicans network and those genes that were tested for differential expression, followed by the removal of non-interacting genes.
Identifying the key metabolic subnetwork
Following the works of Dittrich, et al.46 and May, et al.47, the computational analysis presented here aims at identifying the set of connected (i.e. interacting) C. albicans genes with the most significant deregulation across the LBF and HBF groups. In other words, the result is a subnetwork within the global C. albicans network, which contains the connected genes of most significant fold changes. This requires (i) the annotation of genes (nodes) in the global network with positive and negative numerical scores that reflect the significance of the difference in expression across conditions, followed by (ii) the application of a network search algorithm that can identify the maximum-scoring region in the network. In order to perform these steps, first, we used the BioNet R package48 to fit a statistical model to the P value distribution of fold changes (see Supp. Fig. 2., for details, see Pounds and Morris49). Next, a false-discovery rate of 10−9 was used to determine a P value threshold (10−10), according to which, the statistical model was used to calculate positive and negative weights for genes with P values smaller and larger than the threshold, respectively. Once the genes (nodes) in the network were annotated with such numerical scores, the Heinz algorithm46 was used to calculate the maximum-scoring subnetwork that delineated the most significantly deregulated region in the larger network. The subnetwork was visualised using the Cytoscape50 plugin eXamine51.
Validation of the RNA-Seq analysis
C. albicans clinical isolates exhibiting LBF (n = 9) and HBF (n = 9) were selected for the analysis of genes related to biofilm formation. Biofilms were grown and RNA was extracted as described elsewhere36. Next, cDNA was synthesised using High Capacity RNA to cDNA kit (Life Technologies, Paisley, UK) in a MyCycler PCR machine (Bio-Rad Laboratories, Hertfordshire, UK), following manufacturer’s instructions. The primers for the four selected genes from the KEGG pathway analysis are listed in Table 3. Cycle conditions consisted of 2 min at 50 °C, 10 min at 95 °C and forty cycles of 15 s at 95 °C and 60 s at 60 °C. All qPCR assays were performed in duplicate using MxProP Quantitative PCR machine and MxProP 3000 software (Stratagene, Amsterdam, Netherlands) and controls consisted of reactions in which reverse transcriptase template were absent. Gene expression was calculated using the ΔCt method where the genes of interest were normalised to the housekeeping gene ACT1.
Table 3. Real-time PCR primers used in this study.
| Gene | Direction | Primer sequence (5′→3′) |
|---|---|---|
| AAT1 | Forward | CATTGGCTCCACCAGACAAG |
| Reverse | TCTCTATAAGCACCAACCCCC | |
| SAD1 | Forward | AGGTCTAGGTGCAACTTCGC |
| Reverse | CAGGGTACCCCAGAATGAGC | |
| XOG1 | Forward | CCAAGTGTTTTCCGGTGGTG |
| Reverse | TCCCAACCCCAGTTACAAGC | |
| ACC1 | Forward | TGGAGATTAAGAGTTACTGGTGC |
| Reverse | GATAGCACGCAATGGGAACG |
Validation of aminotransferase activity
Aspartate amino transferase enzyme activity was measured using a commercial assay kit (Sigma, Dorset, UK), by following the manufacturer’s instructions. To assess the impact of AAT inhibition on C. albicans biofilm formation, we used an inhibitor called aminoxyacetate (AOA [Sigma, Dorset, UK]). C. albicans clinical isolates (n = 12) were grown for 24 h in the presence of serially diluted concentration of AOA (ranging from 0 to 400 mg/L) in RPMI at 37 °C, with an untreated control included on each plate for comparison. After incubation, the biofilms were washed in PBS and their biomasses were quantified using a crystal violet technique previously described by our group17. Growth curve analysis was also performed for isolates grown in the presence of AOA (100 and 400 mg/L) by measuring absorbance at a wavelength of 530 nm for 24 h in YPD and RPMI media. No significant change in endpoint growth levels were found in the presence of AOA at any tested concentrations, though a slight delay in growth rate in RPMI (data not shown).
Scanning Electron microscopy
C. albicans type strain SC5314 was grown directly onto Thermanox™ coverslips (Nunc, Roskilde, Denmark) for 24 h in the presence and absence of AOA (400 mg/L) After incubation period, biofilms were carefully washed with PBS and processed for SEM as previously described52. Briefly, samples were fixed in 2% paraformaldehyde, 2% glutaraldehyde, and 0.15% [wt/vol] alcian blue in 0.15 M sodium cacodylate (pH 7.4). The biofilms were sputter coated with gold and viewed under a JEOL JSM-6400 scanning electron microscope.
Additional Information
How to cite this article: Rajendran, R. et al. Integrating Candida albicans metabolism with biofilm heterogeneity by transcriptome mapping. Sci. Rep. 6, 35436; doi: 10.1038/srep35436 (2016).
Supplementary Material
Acknowledgments
This work was supported by the Wellcome Trust Strategic Award for Medical Mycology and Fungal Immunology 097377/Z/11/Z. We are grateful to microbiology colleagues throughout Scotland for clinical isolates collection.
Footnotes
Author Contributions R.R., L.S., R.K. participated in isolate collection, study design, and carried out the experimental studies and were responsible for the manuscript preparation. A.M., B.W.B., S.A. and J.H. devised and performed computational pipelines, and undertook statistical analysis, and were responsible for the manuscript preparation. C.W., B.L.J., K.V.B. and C.A.M. participated in study design, data interpretation and helped draft the manuscript. G.R. conceived the study, participated in study design, data analysis and was responsible for writing and submission of the final manuscript. All authors read and approved the manuscript.
References
- Wisplinghoff H. et al. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39, 309–317, doi: 10.1086/421946 (2004). [DOI] [PubMed] [Google Scholar]
- Pfaller M. A. & Diekema D. J. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20, 133–163, doi: 10.1128/CMR.00029-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler M. J., Pellegrini D. C. & Safdar N. Attributable mortality of central line associated bloodstream infection: systematic review and meta-analysis. Infection 43, 29–36, doi: 10.1007/s15010-014-0689-y (2015). [DOI] [PubMed] [Google Scholar]
- Yousif A., Jamal M. A. & Raad I. Biofilm-based central line-associated bloodstream infections. Adv Exp Med Biol 830, 157–179, doi: 10.1007/978-3-319-11038-7_10 (2015). [DOI] [PubMed] [Google Scholar]
- Eggimann P., Que Y. A., Revelly J. P. & Pagani J. L. Preventing invasive Candida infections. Where could we do better? J Hosp Infect 89, 302–308, doi: 10.1016/j.jhin.2014.11.006 (2015). [DOI] [PubMed] [Google Scholar]
- Kollef M., Micek S., Hampton N., Doherty J. A. & Kumar A. Septic shock attributed to Candida infection: importance of empiric therapy and source control. Clin Infect Dis 54, 1739–1746, doi: 10.1093/cid/cis305 (2012). [DOI] [PubMed] [Google Scholar]
- Ramage G., Rajendran R., Sherry L. & Williams C. Fungal biofilm resistance. International journal of microbiology 2012, 528521, doi: 10.1155/2012/528521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. J., Brown G. D., Netea M. G. & Gow N. A. Metabolism impacts upon Candida immunogenicity and pathogenicity at multiple levels. Trends Microbiol 22, 614–622, doi: 10.1016/j.tim.2014.07.001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi G. et al. Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J 21, 5448–5456 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P. E. Growth of Candida albicans hyphae. Nat Rev Microbiol 9, 737–748, doi: 10.1038/nrmicro2636 (2011). [DOI] [PubMed] [Google Scholar]
- Nobile C. J. & Johnson A. D. Candida albicans Biofilms and Human Disease. Annual review of microbiology 69, 71–92, doi: 10.1146/annurev-micro-091014-104330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C. J. et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148, 126–138, doi: 10.1016/j.cell.2011.10.048 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sanchez S. et al. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryotic cell 3, 536–545 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay A. K. et al. Analysis of Candida albicans mutants defective in the Cdk8 module of mediator reveal links between metabolism and biofilm formation. PLoS genetics 10, e1004567, doi: 10.1371/journal.pgen.1004567 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbarello M. et al. Biofilm production by Candida species and inadequate antifungal therapy as predictors of mortality for patients with candidemia. Journal of clinical microbiology 45, 1843–1850, doi: 10.1128/JCM.00131-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbarello M. et al. Risk factors and outcomes of candidemia caused by biofilm-forming isolates in a tertiary care hospital. PLoS One 7, e33705, doi: 10.1371/journal.pone.0033705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran R. et al. Biofilm formation is a risk factor for mortality in patients with Candida albicans bloodstream infection - Scotland, 2012-2013. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases, doi: 10.1016/j.cmi.2015.09.018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. & Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- May A. et al. metaModules identifies key functional subnetworks in microbiome-related disease. Bioinformatics 32, 1678–1685, doi: 10.1093/bioinformatics/btv526 (2016). [DOI] [PubMed] [Google Scholar]
- Dittrich M. T., Klau G. W., Rosenwald A., Dandekar T. & Muller T. Identifying functional modules in protein-protein interaction networks: an integrated exact approach. Bioinformatics 24, i223–i231, doi: 10.1093/bioinformatics/btn161 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. & Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic acids research 28, 27–30, doi: 10.1093/Nar/28.1.27 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M. A. et al. The Gene Ontology (GO) database and informatics resource. Nucleic acids research 32, D258–D261, doi: 10.1093/nar/gkh036 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staab J. F., Datta K. & Rhee P. Niche-specific requirement for hyphal wall protein 1 in virulence of Candida albicans. PLoS One 8, e80842, doi: 10.1371/journal.pone.0080842 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C. J., Nett J. E., Andes D. R. & Mitchell A. P. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryotic cell 5, 1604–1610, doi: 10.1128/EC.00194-06 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger B. L. Insight into the antiadhesive effect of yeast wall protein 1 of Candida albicans. Eukaryotic cell 11, 795–805, doi: 10.1128/EC.00026-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyes D. L. et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 532, 64–68, doi: 10.1038/nature17625 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G., Coco B., Sherry L., Bagg J. & Lappin D. F. In vitro Candida albicans biofilm induced proteinase activity and SAP8 expression correlates with in vivo denture stomatitis severity. Mycopathologia 174, 11–19, doi: 10.1007/s11046-012-9522-2 (2012). [DOI] [PubMed] [Google Scholar]
- Santana I. L. et al. Dietary carbohydrates modulate Candida albicans biofilm development on the denture surface. PLoS One 8, e64645, doi: 10.1371/journal.pone.0064645 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran R. et al. Extracellular DNA release confers heterogeneity in Candida albicans biofilm formation. BMC microbiology 14, 303, doi: 10.1186/s12866-014-0303-6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeater K. M. et al. Temporal analysis of Candida albicans gene expression during biofilm development. Microbiology 153, 2373–2385, doi: 10.1099/mic.0.2007/006163-0 (2007). [DOI] [PubMed] [Google Scholar]
- Schweizer E. & Hofmann J. Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol Mol Biol Rev 68, 501–517, table of contents, doi: 10.1128/MMBR.68.3.501-517.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez R., Nombela C., Diez-Orejas R. & Gil C. Two-dimensional reference map of Candida albicans hyphal forms. Proteomics 4, 374–382, doi: 10.1002/pmic.200300608 (2004). [DOI] [PubMed] [Google Scholar]
- Xu D. et al. Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog 3, e92, doi: 10.1371/journal.ppat.0030092 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taff H. T. et al. A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathog 8, e1002848, doi: 10.1371/journal.ppat.1002848 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zara G. et al. FLO11 expression and lipid biosynthesis are required for air-liquid biofilm formation in a Saccharomyces cerevisiae flor strain. FEMS Yeast Res 12, 864–866, doi: 10.1111/j.1567-1364.2012.00831.x (2012). [DOI] [PubMed] [Google Scholar]
- Sherry L. et al. Biofilms formed by Candida albicans bloodstream isolates display phenotypic and transcriptional heterogeneity that are associated with resistance and pathogenicity. BMC microbiology 14, 182, doi: 10.1186/1471-2180-14-182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G., Vande Walle K., Wickes B. L. & Lopez-Ribot J. L. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob Agents Chemother 45, 2475–2479 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G., Bachmann S., Patterson T. F., Wickes B. L. & Lopez-Ribot J. L. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J Antimicrob Chemother 49, 973–980 (2002). [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011 17, doi: 10.14806/ej.17.1.200pp.10-12 (2011). [DOI] [Google Scholar]
- Kopylova E., Noe L. & Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217, doi: 10.1093/bioinformatics/bts611 (2012). [DOI] [PubMed] [Google Scholar]
- Edgar R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461, doi: 10.1093/bioinformatics/btq461 (2010). [DOI] [PubMed] [Google Scholar]
- Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T. & Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169, doi: 10.1093/bioinformatics/btu638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis D. O. et al. The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic acids research 40, D667–D674, doi: 10.1093/nar/gkr945 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich M. T., Klau G. W., Rosenwald A., Dandekar T. & Muller T. Identifying functional modules in protein-protein interaction networks: an integrated exact approach. Bioinformatics 24, I223–I231, doi: 10.1093/bioinformatics/btn161 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- May A. et al. metaModules identifies key functional subnetworks in microbiome-related disease. Bioinformatics, doi: 10.1093/bioinformatics/btv526 (2015). [DOI] [PubMed] [Google Scholar]
- Beisser D., Klau G. W., Dandekar T., Muller T. & Dittrich M. T. BioNet: an R-Package for the functional analysis of biological networks. Bioinformatics 26, 1129–1130, doi: 10.1093/bioinformatics/btq089 (2010). [DOI] [PubMed] [Google Scholar]
- Pounds S. & Morris S. W. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics 19, 1236–1242, doi: 10.1093/bioinformatics/btg148 (2003). [DOI] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504, doi: 10.1101/Gr.1239303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkla K. et al. eXamine: exploring annotated modules in networks. BMC bioinformatics 15, 201, doi: 10.1186/1471-2105-15-201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlandsen S. L., Kristich C. J., Dunny G. M. & Wells C. L. High-resolution visualization of the microbial glycocalyx with low-voltage scanning electron microscopy: dependence on cationic dyes. J Histochem Cytochem 52, 1427–1435, doi: 10.1369/jhc.4A6428.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.