Abstract
This study was conducted to investigate whether polymorphisms of genes involved in glycolysis are associated with the prognosis of patients with non-small cell lung cancer (NSCLC) after surgical resection. Forty-four single nucleotide polymorphisms (SNPs) of 17 genes in glycolytic pathway were investigated in a total of 782 patients with NSCLC who underwent curative surgical resection. The association of the SNPs with overall survival (OS) and disease free survival (DFS) were analyzed. Among the 44 SNPs investigated, four SNPs (ENO1 rs2274971A > G, PFKM rs11168417C > T, PFKP rs1132173C > T, PDK2 rs3785921G > A) were significantly associated with survival outcomes in multivariate analyses. When stratified by tumor histology, three SNPs (ENO1 rs2274971A > G, PFKM rs11168417C > T, and PDK2 rs3785921G > A) were significantly associated with OS and/or DFS only in squamous cell carcinoma, whereas PFKP rs1132173C > T exhibited a significant association with survival outcomes only in adenocarcinoma. When the four SNPs were combined, OS and DFS decreased as the number of bad genotypes increased (Ptrend = 8 × 10−4 and 3 × 10−5, respectively). Promoter assays showed that ENO1 rs2274971G allele had significantly higher promoter activity compared to the rs2274971A allele. The four SNPs, especially ENO1 rs2274971A > G, may be useful for the prediction of prognosis in patients with surgically resected NSCLC.
Lung cancer is the leading cause of cancer deaths worldwide, with an average 5-year survival rate of 16%1. Although surgery is the best treatment modality for potentially curing early stage non-small cell lung cancer (NSCLC), a large proportion of patients die from disease recurrence. Pathologic stage is the most important predictor of prognosis after surgical resection of NSCLC. However, patients with the same pathologic stage have different risk of recurrence and death2. Therefore, many studies have focused on prognostic biomarkers for more precise prognostication of patients after surgery.
Metabolic reprogramming is one of the hallmarks of cancer3. Dependence on glycolysis under normoxic conditions, or aerobic glycolysis which is often referred to as the Warburg effect, is regarded as one of the characteristic changes in tumor cellular bioenergetics4. Given that glycolysis is much less efficient than oxidative phosphorylation in the production of adenosine triphosphate, cancer cells adapt to this disadvantage by increasing glucose uptake to facilitate increased glucose consumption4. The increased glucose uptake in cancer cells compared to in normal cells is widely exploited in positron emission tomography imaging using 18F-fluorodeoxyglucose (18F-FDG). The major oncogenes such as RAS, MYC, and HIF-1α are key inducers of glycolysis and glucose transporters, whereas tumor suppressor TP53 is known to suppress glycolytic flux5,6. In addition to energy production, the metabolic intermediates of glycolysis play a pivotal role in the production of building blocks such as amino acids, lipids, and nucleic acids in proliferating cancer cells, thus conferring a growth advantage4,5. The accumulation of glycolytic intermediates promotes the pentose phosphate pathway, resulting in the generation of NADPH, a major reducing agent important in redox homeostasis and drug detoxifying reactions in cancer cells7. Glycolytic enzymes and metabolic intermediates of aerobic glycolysis also have important functions beyond glycolysis to facilitate the growth and survival of cancer cells8. For example, the mitochondrial membrane-bound hexokinase 2 antagonizes the pro-apoptotic machinery, providing a survival advantage to cancer cells9. For pyruvate kinase, the M2 isoform has been reported to promote tumorigenesis and provide a selective growth advantage for tumor cells10. In addition, a glycolytic intermediate fructose-1, 6 bisphosphate plays an anti-apoptotic role in cancer cells by maintaining the cytochrome c in a reduced, inactive state11. Taken together, the oncogenic regulation of aerobic glycolysis and various roles of the components of the glycolytic pathway highlight the biological significance of glycolysis in cancer.
In this study, we hypothesized that polymorphisms of genes involved in glycolysis affect energy production, macromolecular biosynthesis and other non-glycolytic functions in cancer cells, thus playing a role in determining the prognosis of patients with lung cancer. To test this hypothesis, we evaluated the association of genetic variants in the glycolytic pathway with the prognosis of lung cancer patients undergoing surgical resection.
Results
Patient characteristics and clinical predictors
The clinical and pathologic characteristics of the patients and association with overall survival (OS) and disease-free survival (DFS) are shown in Table 1. Univariate analysis showed that age (log-rank P [PL-R] = 2 × 10−3), sex (PL-R = 4 × 10−4), smoking status (PL-R = 3 × 10−4), diabetes (PL-R = 0.03), and pathologic stage (PL-R = 1 × 10−11) were significantly associated with OS. Only pathologic stage was significantly associated with DFS (PL-R = 2 × 10−15).
Table 1. Univariate analysis for survival outcomes by clinicopathological features.
| Variables | No. of cases | Overall survival |
Disease free survival |
||||
|---|---|---|---|---|---|---|---|
| No. of deaths (%)a | 5Y-OSR (%)b | Log-rank P | No. of events (%)a | 5Y-DFSR (%)b | Log-rank P | ||
| Overall | 782 | 208 (26.6) | 62 | 340 (43.5) | 45 | ||
| Age (years) | |||||||
| < 65 | 383 | 88 (23.0) | 69 | 2 × 10−3 | 162 (42.3) | 48 | 0.14 |
| ≥ 65 | 399 | 120 (30.1) | 55 | 178 (44.6) | 41 | ||
| Sex | |||||||
| Male | 572 | 173 (30.2) | 59 | 4 × 10−4 | 261 (45.6) | 42 | 0.10 |
| Female | 210 | 35 (16.7) | 71 | 79 (37.6) | 52 | ||
| Smoking status | |||||||
| Never | 232 | 40 (17.2) | 74 | 3 × 10−4 | 90 (38.8) | 50 | 0.15 |
| Ever | 550 | 168 (30.6) | 57 | 250 (45.5) | 43 | ||
| Diabetes | |||||||
| No | 619 | 150 (24.2) | 66 | 0.03 | 259 (41.8) | 47 | 0.19 |
| Yes | 152 | 51 (33.6) | 51 | 73 (48.0) | 36 | ||
| Histological types | |||||||
| SCC | 341 | 103 (30.2) | 60 | 0.17 | 146 (42.8) | 48 | 0.22 |
| AC | 425 | 99 (23.3) | 63 | 184 (43.3) | 42 | ||
| LCC | 16 | 6 (37.5) | 59 | 10 (62.5) | 35 | ||
| Pathologic stage | |||||||
| I | 378 | 59 (15.6) | 76 | 1 × 10−11 | 107 (28.3) | 60 | 2 × 10−15 |
| II | 227 | 81 (35.7) | 52 | 116 (51.1) | 39 | ||
| IIIA | 177 | 68 (38.4) | 47 | 117 (66.1) | 20 | ||
| Adjuvant therapyc | |||||||
| No | 184 | 72 (39.6) | 49 | 0.58 | 102 (56.0) | 37 | 0.36 |
| Yes | 220 | 77 (34.7) | 50 | 131 (59.0) | 25 | ||
| Body mass index | |||||||
| < 25 | 464 | 107 (23.1) | 65 | 200 (43.1) | 41 | ||
| ≥ 25 | 183 | 45 (24.6) | 61 | 0.85 | 75 (41.0) | 46 | 0.40 |
Abbreviations: 5Y-OSR, five year-overall survival rate; 5Y-DFSR, five year-disease free survival rate; SCC, squamous cell carcinoma; AC, adenocarcinoma; LCC, large cell carcinoma.
aRow percentage.
bFive year-overall survival rate and five year-disease free survival rate, proportion of survival derived from Kaplan-Meier analysis.
cIn pathologic stages II + IIIA: 182 cases received adjuvant chemotherapy alone, 11 cases received adjuvant radiotherapy alone, and 27 cases received both chemotherapy and radiotherapy.
Associations between SNPs and survival outcomes
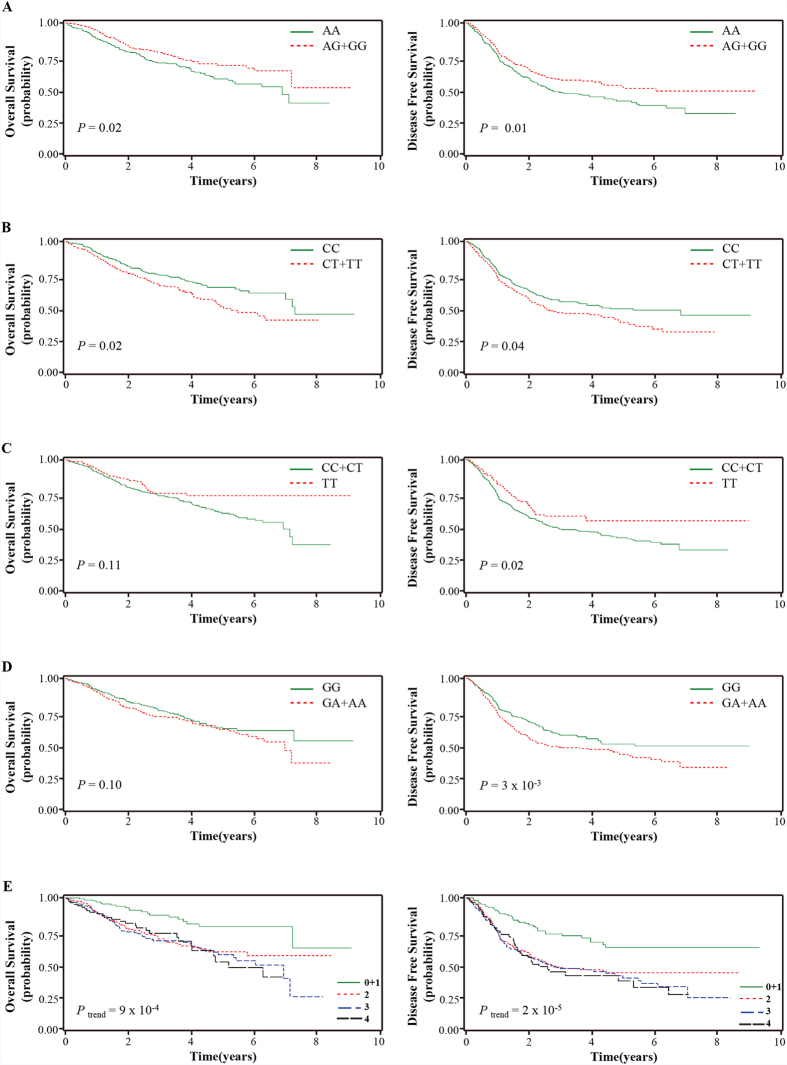
Among the genotyped 54 single nucleotide polymorphisms (SNPs), 44 were analyzed in this study after excluding one SNP with a <95% call rate and nine showing deviation from Hardy-Weinberg equilibrium. The SNP ID, gene information, base change, minor allele frequencies, and P values for OS and DFS of the 44 SNPs are shown in Supplementary Table 1. Among the 44 SNPs analyzed, four SNPs (Enolase 1 [ENO1] rs2274971A > G, phosphofructokinase, muscle [PFKM] rs11168417C > T, phosphofructokinase, platelet [PFKP] rs1132173C > T, and pyruvate dehydrogenase kinase, isoenzyme 2 [PDK2] rs3785921G > A) were significantly associated with survival outcomes. ENO1 rs2274971A > G and PFKM rs11168417C > T were significantly associated with OS (adjusted hazard ratio [aHR] = 0.70, 95% confidence interval [CI] = 0.52–0.95, P = 0.02; and aHR = 1.44, 95% CI = 1.09–1.91, P = 0.01, under the dominant model, respectively) and DFS (aHR = 0.73, 95% CI = 0.58–0.92, P = 7 × 10−3; and aHR = 1.25, 95% CI = 1.00–1.56, P = 0.05, under the dominant model, respectively, Table 2 and Fig. 1A–D). PFKP rs1132173C > T and PDK2 rs3785921G > A were significantly associated with DFS (aHR = 0.73, 95% CI = 0.55–0.98, P = 0.03, under the recessive model; and aHR = 1.40, 95% CI = 1.12–1.75, P = 3 × 10−3, under the dominant model, respectively). We further analyzed the effect of the four SNPs on survival outcomes according to tumor histology. Three SNPs (ENO1 rs2274971A > G, PFKM rs11168417C > T, and PDK2 rs3785921G > A) were significantly associated with OS and/or DFS in squamous cell carcinoma (SCC) but not in adenocarcinoma (AC), whereas PFKP rs1132173C > T exhibited a significant association with survival outcomes in AC but not in SCC (Table 3 and Supplementary Figure 1A–D). The significant difference in survival outcomes according to tumor histology was determined by a test for heterogeneity (P for heterogeneity test < 0.05, for OS and/or DFS in all four SNPs).
Table 2. Overall survival and disease free survival according to the ENO1 rs2274971A > G, PFKM rs11168417C > T, PFKP rs1132173C > T, and PDK2 rs3785921G > A genotypes.
| Polymorphism/genotype | No. of Cases (%)a | Overall survival |
Disease free survival |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of deaths (%)b | 5Y-OSR (%)c | HR (95% CI)d | Pd | No. of Event (%)b | 5Y-DFSR (%)c | HR (95% CI)d | Pd | ||
| ENO1 rs2274971e | |||||||||
| AA | 448 (59.1) | 128 (28.6) | 58 | 1.00 | 205 (45.8) | 41 | 1.00 | ||
| AG | 266 (35.1) | 63 (23.7) | 67 | 0.73 (0.54–1.00) | 0.05 | 105 (39.5) | 50 | 0.73 (0.58–0.93) | 0.01 |
| GG | 44 (5.8) | 8 (18.2) | 76 | 0.55 (0.27–1.12) | 0.10 | 15 (34.10 | 59 | 0.69 (0.41–1.17) | 0.17 |
| Dominant | 310 (40.9) | 71 (22.9) | 68 | 0.70 (0.52–0.95) | 0.02 | 120 (38.7) | 51 | 0.73 (0.58–0.92) | 7 × 10−3 |
| Recessive | 714 (94.2) | 191 (26.8) | 61 | 0.61 (0.30–1.24) | 0.17 | 310 (43.4) | 44 | 0.78 (0.46–1.31) | 0.34 |
| Codominant | 0.73 (0.57–0.94) | 0.02 | 0.77 (0.64–0.94) | 0.01 | |||||
| PFKM rs11168417e | |||||||||
| CC | 471 (61.1) | 109 (23.1) | 67 | 1.00 | 186 (39.5) | 49 | 1.00 | ||
| CT | 264 (34.2) | 88 (33.3) | 53 | 1.49 (1.11–1.99) | 8 × 10−3 | 132 (50.0) | 40 | 1.28 (1.02–1.61) | 0.04 |
| TT | 36 (4.7) | 8 (22.2) | 62 | 1.11 (0.54–2.29) | 0.78 | 14 (38.9) | 38 | 1.03 (0.59–1.77) | 0.93 |
| Dominant | 300 ((38.9) | 96 (32.0) | 54 | 1.44 (1.09–1.91) | 0.01 | 146 (48.7) | 39 | 1.25 (1.00–1.56) | 0.05 |
| Recessive | 735 (95.3) | 197 (26.8) | 62 | 0.95 (0.47–1.93) | 0.89 | 318 (43.3) | 46 | 0.94 (0.55–1.60) | 0.81 |
| Codominant | 1.28 (1.02–1.61) | 0.04 | 1.15 (0.96–1.38) | 0.13 | |||||
| PFKP rs1132173e | |||||||||
| CC | 207 (26.9) | 57 (27.5) | 65 | 1.00 | 90 (43.5) | 46 | 1.00 | ||
| CT | 401 (52.1) | 113 (28.2) | 57 | 1.06 (0.76–1.48) | 0.72 | 184 (45.9) | 41 | 1.09 (0.84–1.42) | 0.50 |
| TT | 162 (21.0) | 32 (19.8) | 73 | 0.83 (0.53–1.29) | 0.41 | 57 (35.2) | 55 | 0.77 (0.55–1.08) | 0.14 |
| Dominant | 563 (73.1) | 145 (25.8) | 62 | 1.00 (0.73–1.37) | 0.99 | 241 (42.8) | 45 | 0.99 (0.78–1.27) | 0.96 |
| Recessive | 608 (79.0) | 170 (28.0) | 60 | 0.80 (0.54–1.17) | 0.25 | 274 (45.1) | 43 | 0.73 (0.55–0.98) | 0.03 |
| Codominant | 0.93 (0.76–1.15) | 0.49 | 0.90 (0.77–1.06) | 0.20 | |||||
| PDK2 rs3785921e | |||||||||
| GG | 342 (44.3) | 85 (24.9) | 64 | 1.00 | 133 (38.9) | 49 | 1.00 | ||
| GA | 356 (46.1) | 95 (26.7) | 62 | 1.31 (0.97–1.76) | 0.08 | 163 (45.8) | 42 | 1.39 (1.10–1.76) | 5 × 10−3 |
| AA | 74 (9.6) | 23 (31.1) | 59 | 1.34 (0.82–2.17) | 0.24 | 36 (48.7) | 40 | 1.44 (0.98–2.11) | 0.06 |
| Dominant | 430 (55.7) | 118 (27.4) | 62 | 1.31 (0.99–1.75) | 0.06 | 199 (46.3) | 42 | 1.40 (1.12–1.75) | 3 × 10−3 |
| Recessive | 698 (90.4) | 180 (25.8) | 63 | 1.17 (0.74–1.85) | 0.50 | 296 (42.4) | 46 | 1.22 (0.85–1.74) | 0.29 |
| Codominant | 1.20 (0.98–1.49) | 0.08 | 1.26 (1.07–1.49) | 6 × 10−3 | |||||
Abbreviations: 5Y-OSR, five year-overall survival rate; 5Y-DFSR, five year-disease free survival rate; HR, hazard ratio; CI, confidence interval.
aColumn percentage.
bRow percentage.
cFive year-overall survival rate and five year-disease free survival rate, proportion of survival derived from Kaplan-Meier analysis.
dHRs, 95% CIs, and their corresponding P values were calculated using multivariate Cox proportional hazard models, adjusted for age, gender, smoking status, diabetes, tumor histology, pathologic stage, adjuvant therapy, and body mass index.
eGenotype failures: 24 case for rs2274971, 11 case for rs11168417, 12 case for rs1132173, and 10 case for rs3785921.
Figure 1.
Overall survival and disease free survival according to ENO1 rs2274971A > G (A), PFKM rs11168417C > T (B), PFKP rs1132173C > T (C), and PDK2 rs3785921G > A (D) genotypes, and the number of bad genotypes of the four SNPs (E). P values in the multivariate Cox proportional hazard model.
Table 3. Overall survival and disease free survival according to the ENO1 rs2274971A > G, PFKM rs11168417C > T, PFKP rs1132173C > T, and PDK2 rs3785921G > A genotypes stratified by tumor histology.
| Polymorphism/genotype | Overall survival |
PHb | Disease free survival |
PHb | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SCC |
AC |
SCC |
AC |
|||||||
| HR (95% CI)a | Pa | HR (95% CI)a | Pa | HR (95% CI)a | Pa | HR (95% CI)a | Pa | |||
| ENO1 rs2274971 | ||||||||||
| Dominant | 0.43 (0.26–0.70) | 6 × 10−4 | 1.10 (0.73–1.65) | 0.65 | 4 × 10−3 | 0.53 (0.36–0.78) | 1 × 10−3 | 0.95 (0.70–1.28) | 0.73 | 0.02 |
| Recessive | 0.58 (0.21–1.60) | 0.29 | 0.65 (0.24–1.79) | 0.41 | 0.88 | 0.84 (0.39–1.81) | 0.65 | 0.77 (0.38–1.57) | 0.47 | 0.87 |
| Codominant | 0.52 (0.34–0.78) | 2 × 10−3 | 1.01 (0.72–1.40) | 0.97 | 0.01 | 0.62 (0.45–0.87) | 5 × 10−3 | 0.93 (0.72–1.20) | 0.57 | 0.06 |
| PFKM rs11168417 | ||||||||||
| Dominant | 1.82 (1.22–2.72) | 4 × 10−3 | 1.15 (0.76–1.74) | 0.50 | 0.12 | 1.90 (1.35–2.67) | 2 × 10−4 | 0.96 (0.71–1.30) | 0.78 | 3 × 10−3 |
| Recessive | 1.14 (0.46–2.83) | 0.77 | 0.54 (0.13–2.21) | 0.39 | 0.38 | 0.89 (0.39–2.02) | 0.77 | 0.90 (0.42–1.95) | 0.80 | 0.98 |
| Codominant | 1.50 (1.10–2.06) | 0.01 | 1.05 (0.74–1.51) | 0.78 | 0.15 | 1.48 (1.14–1.91) | 3 × 10−3 | 0.96 (0.74–1.24) | 0.75 | 0.02 |
| PFKP rs1132173 | ||||||||||
| Dominant | 1.00 (0.63–1.57) | 0.98 | 0.97 (0.61–1.54) | 0.90 | 0.93 | 1.01 (0.69–1.48) | 0.98 | 0.93 (0.66–1.30) | 0.67 | 0.75 |
| Recessive | 1.25 (0.74–2.10) | 0.41 | 0.48 (0.26–0.89) | 0.02 | 0.02 | 0.96 (0.62–1.50) | 0.87 | 0.57 (0.38–0.85) | 6 × 10−3 | 0.09 |
| Codominant | 1.07 (0.79–1.46) | 0.65 | 0.80 (0.59–1.07) | 0.14 | 0.18 | 0.99 (0.77–1.27) | 0.94 | 0.81 (0.66–1.00) | 0.05 | 0.23 |
| PDK2 rs3785921 | ||||||||||
| Dominant | 1.32 (0.88–2.00) | 0.18 | 1.23 (0.81–1.86) | 0.34 | 0.81 | 1.59 (1.12–2.26) | 9 × 10−3 | 1.19 (0.87–1.61) | 0.27 | 0.22 |
| Recessive | 1.58 (0.88–2.86) | 0.13 | 0.88 (0.42–1.83) | 0.72 | 0.22 | 1.89 (1.15–3.10) | 0.01 | 0.78 (0.45–1.36) | 0.38 | 0.02 |
| Codominant | 1.30 (0.96–1.74) | 0.09 | 1.09 (0.80–1.49) | 0.57 | 0.42 | 1.49 (1.16–1.91) | 2 × 10−3 | 1.05 (0.84–1.32) | 0.67 | 0.04 |
Abbreviations: SCC, squamous cell carcinoma; AC, adenocarcinoma; HR, hazard ratio; CI, confidence interval.
aHRs, 95% CIs, and their corresponding P values were calculated using multivariate Cox proportional hazard models, adjusted for age, gender, smoking status, diabetes, pathologic stage, adjuvant therapy, and body mass index.
bPH, P for heterogeneity test.
Combined effects of the four SNPs on survival outcomes
We performed an exploratory analysis to investigate the combined effects of ENO1 rs2274971A > G, PFKM rs11168417C > T, PFKP rs1132173C > T, and PDK2 rs3785921G > A, which showed a significant association with survival outcomes in the individual SNP analysis. Because ENO1 rs2274971 AA, PFKM rs11168417 CT + TT, PFKP rs1132173 CC + CT, and PDK2 rs3785921 GA + AA were associated with worse survival outcomes, we considered these as bad genotypes and then evaluated their combined effects by grouping the patients based on the number of bad genotypes in each patient. Because there was no death in the zero bad genotype group, we combined the zero and one bad genotype groups. Compared with the reference group that had 0–1 bad genotypes, OS and DFS decreased as the number of bad genotypes increased (Ptrend = 8 × 10−4 for OS; and Ptrend = 3 × 10−5 for DFS; Table 4 and Fig. 1E). In SCC, the combined analysis of ENO1 rs2274971 AA, PFKM rs11168417 CT + TT, and PDK2 rs3785921 GA + AA, which were associated with worse survival outcomes in SCC, also showed a significant decrease in OS and DFS as the number of bad genotypes increased (Ptrend = 2 × 10−5 for OS; and Ptrend = 5 × 10−7 for DFS; Table 4 and Supplementary Figure 1E).
Table 4. Combined effects of the SNPs on overall survival and disease free survival.
| No. of bad genotypesa | No. of cases (%)b | Overall survival |
Disease free survival |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of deaths (%)c | 5Y-OSR (%)d | P (L-R) | HR (95% CI)e | Pe | No. of events (%)c | 5Y-DFSR (%)d | P (L-R) | HR (95% CI)e | Pe | ||
| All casesf | |||||||||||
| 0 or 1g | 145 (19.5)f | 20 (13.8) | 79 | 5 × 10−4 | 1.00 | 38 (26.2) | 63 | 6 × 10−7 | 1.00 | ||
| 2 | 279 (37.6) | 78 (28.0) | 60 | 2.26 (1.36–3.74) | 2 × 10−3 | 126 (45.2) | 44 | 2.02 (1.40–2.93) | 2 × 10−4 | ||
| 3 | 231 (31.1) | 68 (29.4) | 58 | 2.59 (1.55–4.32) | 3 × 10−4 | 107 (46.3) | 40 | 2.35 (1.62–3.43) | 8 × 10−6 | ||
| 4 | 88 (11.8) | 27 (30.7) | 53 | 2.62 (1.46–4.70) | 1 × 10−3 | 45 (51.1) | 38 | 2.37 (1.54–3.67) | 1 × 10−4 | ||
| Ptrend | 8 × 10−4 | 3 × 10−5 | |||||||||
| 0 + 1g | 145 (19.5)f | 20 (13.8) | 79 | 3 × 10−5 | 1.00 | 38 (26.2) | 63 | 9 × 10−7 | 1.00 | ||
| 2 + 3 + 4 | 598 (80.5) | 173 (29.0) | 58 | 2.44 (1.51–3.92) | 3 × 10−4 | 278 (46.5) | 41 | 2.20 (1.56–3.10) | 8 × 10−6 | ||
| SCCh | |||||||||||
| 0 | 41 (12.4) | 4 (9.8) | 86 | 2 × 10−3 | 1.00 | 7 (17.1) | 74 | 3 × 10−6 | 1.00 | ||
| 1 | 120 (36.3) | 31 (25.8) | 66 | 3.28 (0.99–10.87) | 0.05 | 45 (37.5) | 54 | 3.04 (1.28–7.21) | 0.01 | ||
| 2 | 116 (35.1) | 40 (34.5) | 52 | 5.30 (1.62–17.37) | 6 × 10−3 | 56 (48.3) | 38 | 4.56 (1.94–10.73) | 5 × 10−4 | ||
| 3 | 54 (16.3) | 24 (44.4) | 46 | 7.67 (2.28–25.80) | 1 × 10−3 | 32 (59.3) | 37 | 6.76 (2.77–16.50) | 3 × 10−5 | ||
| Ptrend | 2 × 10−5 | 5 × 10−7 | |||||||||
Abbreviations: 5Y-OSR, five year-overall survival rate; 5Y-DFSR, five year-disease free survival rate; HR, hazard ratio; CI, confidence interval; PL-R, Log-rank P; SCC, squamous cell carcinoma; AC, adenocarcinoma.
aENO1 rs2274971 AA, PFKM rs11168417 CT + TT, PFKP rs1132173 CC + CT, and PDK2 rs3785921 GA + AA.
bColumn percentage.
cRow percentage.
dFive year-overall survival rate and five year-disease free survival rate, proportion of survival derived from Kaplan-Meier analysis.
eHRs, 95% CIs, and their corresponding P values were calculated using multivariate Cox proportional hazard models, adjusted for age, gender, smoking status, diabetes, tumor histology, pathologic stage, adjuvant therapy, and body mass index.
fCombined genotypes of rs11168417, rs2274971, rs3785921, rs1132173. Thirty nine patients with missing genotype data at any of the four SNPs.
g21 cases for zero bad genotype and 124 cases for one bad genotype. Because zero bad genotype had no death, zero and one bad genotypes were combined.
hCombined genotypes of rs11168417, rs2274971, rs3785921. Ten patients with missing genotype data at any of the three SNPs.
Effect of rs2274971A > G on promoter activity of ENO1
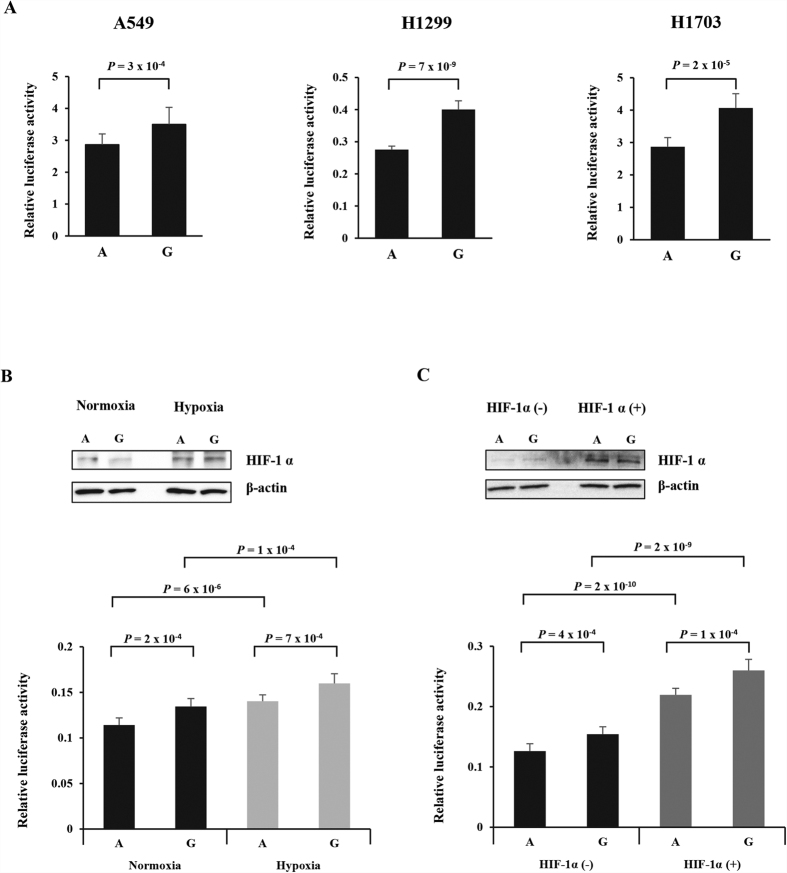
The effect of the rs2274971A > G polymorphisms on the promoter activity of ENO1 was investigated in a luciferase assay. For this analysis, we generated pGL3-Basic-ENO1 constructs containing rs2274971A > G and transfected the constructs into NSCLC cells (A549, H1299, and H1703). The rs2274971G allele exhibited significantly higher promoter activity than the rs2274971A allele (P = 3 × 10−4, P = 7 × 10−9, and P = 2 × 10−5, respectively, Fig. 2A). Because ENO1 expression is known to be induced by hypoxia and the SNP site in the fragment cloned into pGL3-basic vector is predicted to lie in a HIF-1α binding site according to SNPinfo website (https://snpinfo.niehs.nih.gov/cgi-bin/snpinfo/tfbs.cgi? 2_rs2274971), we further performed promoter assay under hypoxia and HIF-1α overexpression. For HIF-1α overexpression, we cotransfected pGL3-Basic-ENO1 constructs into A549 cells with pEGFP-HIF-1α. The luciferase activity was significantly increased by hypoxia and HIF-1α overexpression (Fig. 2B,C). Under hypoxia and HIF-1α overexpression, promoter activity was consistently higher in rs2274971G allele than the rs2274971A allele (P = 7 × 10−4, Fig. 2B, and P = 1 × 10−4, Fig. 2C). These results suggest that the rs2274971G allele is associated with higher expression of ENO1 compared with rs2274971A allele and that hypoxia and HIF-1α upregulate ENO1 expression.
Figure 2. Functional analysis of the ENO1 rs2274971A > G by dual luciferase reporter assay.
NSCLC cells (A549, H1299, and H1703) were transfected with pRL-SV40 and pGL3-basic-ENO1 rs2274971A or G allele-containing constructs (A). At 24 h posttransfection, A549 cells were exposed to hypoxia for 24 h (B). For HIF-1α overexpression, we cotransfected pRL-SV40 and pGL3-basic-ENO1 constructs into A549 cells with pEGFP-HIF-1α (C). Luciferase activity was measured using Dual-Luciferase® Reporter Assay System. Firefly luciferase activity was normalized by pRL-SV40 Renilla luciferase activity. Each bar represents mean ± S.E.M. from three independent experiments carried out in quadruplicate. P value, Student’s t-test.
Discussion
This study was conducted to investigate whether genetic polymorphisms in the glycolytic pathway affect the prognosis of patients with NSCLC after surgical resection. Among the 44 SNPs evaluated, the ENO1 rs2274971A > G, PFKM rs11168417C > T, PFKP rs1132173C > T, and PDK2 rs3785921G > A polymorphisms were significantly associated with survival outcomes. More importantly, combined analysis of the four SNPs more precisely predicted prognosis. In addition, stratified analysis by tumor histology suggested that genetic polymorphisms in the glycolytic pathway exert a different prognostic effect according to tumor histology of NSCLC. These results suggest that the glycolytic pathway plays an important role in determining the prognosis of early stage NSCLC. SNP analysis of glycolytic genes may be useful for predicting prognosis after surgical resection in NSCLC, thereby help to refine therapeutic decisions.
In the present study, ENO1 rs2274971A > G, PFKM rs11168417C > T, PFKP rs1132173C > T, and PDK2 rs3785921G > A polymorphisms in glycolytic pathway were associated with survival outcomes after surgery, suggesting that the SNPs in glycolytic pathway may be prognostic markers in early stage NSCLC. However, because glycolysis involves sequential reactions mediated by a series of enzymes interacting in the glycolytic process, it is unlikely that the prognosis of lung cancer patients can be determined by a single genetic polymorphism in the glycolytic pathway. Instead, a pathway-based multigenic approach may amplify the effects of individual polymorphisms and have better resolution in predicting prognosis12,13. Although four SNPs were associated with survival outcomes in individual SNP analysis, considering the borderline CI and multiple comparisons issue, the impact of any individual SNP on survival outcomes may be marginal. When the adverse genotypes of four SNPs were combined, these genotypes additively contributed to decreased survival, enabling more precise prediction of prognosis. Our results suggest that combined analysis of multiple SNPs in relevant pathways has a higher potential value for identifying SNPs with prognostic significance.
Enolase (2-phospho-D-glycerate hydrolase) is one of the key glycolytic enzymes that catalyzes the reversible conversion of 2-phosphoglycerate to phophoenolpyruvate. The non-neuronal enolase (α-enolase, ENO1) plays an important role in several physiological processes depending on the cellular localization14. ENO1 is mainly localized in the cytoplasm to participate in glycolysis. In addition to its glycolytic enzyme activity, it is expressed on the cell surface and acts as a plasminogen receptor to bind and activate plasminogen and promote degradation of the extracellular matrix for cell migration and cancer metastasis15. In contrast, ENO1 can be alternatively translated into a c-Myc promoter binding protein, which is localized to the nucleus to bind to the c-Myc promoter and suppress the transcription of c-Myc14,16, suggesting a tumor suppressor function. Overexpression of ENO1 has been correlated with tumor progression and poor prognosis in many tumor tissues17,18, whereas its down-regulation has also been reported and associated with poor prognosis in several cancers16,19. In the present study, the ENO1 rs2274971G allele, which exhibited significantly higher promoter activity compared to the rs2274971A allele, was associated with better survival outcomes, suggesting that ENO1 has a potential tumor suppressor function. However, ENO1 rs2274971A > G was not significantly associated with clinicopathological features such as age, sex, smoking status, diabetes, tumor histology, pathologic stage, and histologic grade (data not shown).
A key step in glycolysis, the phosphorylation of fructose 6-phosphate to fructose 1, 6 bisphosphate, is catalyzed by phosphofructokinase-1 (PFK-1) and commits glucose to the glycolytic pathway. PFK-1 activity is rate-limiting, critical in determining glycolytic flux, and tightly regulated20,21. PFK-1 is a tetramer composed of three types of subunits, M (muscle), L (liver), and P (platelet), which are encoded by three different genes, PFKM, PFKL, and PFKP, respectively. PFK-1 tetrameric isoenzymes have various subunit compositions, and thus different kinetic properties and activities, according to the tissue type22. PFK-1 activity is increased in response to proliferation signals and is correlated with elevated glycolysis in proliferating cells21. Elevated PFK-1 activity is induced by oncogenic signals or HIF-1α activation, which is characteristic of cancer cells21,23. PDK phosphorylates and inhibits pyruvate dehydrogenase, preventing pyruvate from entering mitochondrial matrix to form acetyl-CoA for the Krebs cycle. Increased expression or activity of PDK may play a role in the metabolic shift away from mitochondrial oxidation to facilitate the glycolytic process in cancer24,25. PDK is frequently upregulated in tumor tissues by MYC, HIF activation, or TP53 loss25,26,27. PDK2 is the most widely distributed and best characterized isoform among the four PDK isoforms28. Particularly, PDK2 is the most sensitive to dichloroacetic acid, which activates pyruvate dehydrogenase by inhibiting PDK and shifts the metabolism of cancer cells from glycolysis to oxidative phosphorylation. Dichloroacetic acid has been shown to have anti-cancer effect in preclinical studies29,30, and has been tested in early clinical trials in several cancers31,32. PFKP rs1132173 (Phe309Phe) is a synonymous SNP, and PFKM rs11168417 and PDK2 rs3785921 are intronic variants. Although synonymous SNPs and intronic SNPs have been considered non-functional, studies have reported that synonymous variations may affect protein translation efficiency, structure, and function, as well as splicing events, mRNA stability, and microRNA binding33,34. Recent results of the Encyclopedia of DNA Elements (ENCODE) project provided evidence that genetic variations in non-coding DNA, such as intronic SNP, play an important role in the regulation of gene expression35. An alternative explanation is that the association between the SNPs and survival outcomes may be due to linkage disequilibrium (LD) with other functional variants. Future studies are needed to understand the biologic mechanism of the observed associations between the SNPs and survival outcomes.
Interestingly, stratified analysis by tumor histology suggested that polymorphisms in glycolytic pathway genes may play a different prognostic role according to the tumor histology of NSCLC. This finding is comparable to those of previous studies36,37 showing that glucose metabolism differs between SCC and AC of the lung: SCC was associated with higher 18F-FDG uptake and higher levels of glycolysis-related markers compared to AC. Further studies are required to understand the role of glycolysis in the pathogenesis and prognosis of NSCLC, particularly the differential role in SCC and AC. There were several limitations to this study. First, this study did not provide direct evidence that the four genes are involved in the development and progression of lung cancer, limiting the applicability to the biological mechanism of the observed associations between the SNPs and survival outcomes. Second, all patients were treated at the same hospital, which may have led to a selection bias. Third, this study included only a Korean patient population, and thus the results may not be generalizable for other ethnic groups.
In conclusion, the analysis of glycolytic pathway gene polymorphisms, particularly combined analysis, may be useful for predicting patient prognosis after surgery, thereby helping to refine therapeutic decisions in NSCLC. Future studies are warranted to understand the biological mechanism of our findings and validate our results in a larger number of patients, including diverse ethnic groups.
Materials and Methods
Study populations
A total of 782 patients with pathologic stages I, II, or IIIA (micro-invasive N2) NSCLC who underwent curative surgical resection at Kyungpook National University Hospital (KNUH, n = 354) and Seoul National University Bundang Hospital (SNUBH, n = 428) were enrolled in this study. All patients included in this study were ethnic Koreans. None of the patients received chemotherapy or radiotherapy prior to surgery. The pathologic staging of tumors was determined according to the International System for Staging Lung Cancer2. Written informed consent was obtained from all patients prior to surgery at each of the participating institutions. This study was approved by the institutional review boards of KNUH and SNUBH, and was performed in accordance with the research protocol which was approved by the institutional review boards of KNUH and SNUBH.
Selection of SNPs and genotyping
To collect potentially functional polymorphisms in glycolytic pathway genes, we first searched the public SNP database (http://www.ncbi.nlm.nih.gov/SNP) for all SNPs in major glycolytic genes. Next, using the FuncPred utility for functional SNP prediction and TagSNP utility for LD tag SNP selection in the SNPinfo web server (https://snpinfo.niehs.nih.gov/), a total of 54 potentially functional SNPs in 19 glycolytic pathway genes with minor allele frequency ≥ 0.05 in the HapMap JPT data were collected after excluding those in LD (r2 ≥ 0.8). Genomic DNA was extracted from peripheral blood lymphocytes using blood QuickGene DNA whole blood kit S (Fujifilm, Tokyo, Japan). Genotyping was performed using the MassARRAY® iPLEX assay (SEQUENOM Inc., San Diego, CA, USA). Duplicate samples and negative controls were included to ensure the accuracy of genotyping. Approximately 5% of the samples were randomly selected to be genotyped again with a restriction fragment length polymorphism assay by a different investigator and the results were 100% concordant.
Promoter-luciferase constructs and luciferase assay
We investigated whether rs2274971A > G ( + 487 from transcription start site) of ENO1 coding alpha-enolase modulates the promoter activity of the gene by conducting a luciferase assay. A 1151-bp fragment (from −387 to + 764) including rs2274971A > G was synthesized by polymerase chain reaction using genomic DNA. The forward primer with a KpnI restriction site (5′-GGGGTACCAGTGGTGCTTCAACTGGTATC-3′) and reverse primer with a XhoI restriction site (5′-CCGCTCGAGTCAATCAGTTACCTGAGTGC-3′) were used. The polymerase chain reaction products were cloned into the KpnI/XhoI sites of the pGL3-basic vector (Promega, Madison, WI, USA), resulting in pGL3-Basic-ENO1 constructs containing rs2274971A > G. The correct sequence of all clones was verified by DNA sequencing. Human NSCLC cells (A549, H1299, and H1703) were purchased from Korean Cell Line Bank (KCLB), Seoul, Korea, and authenticated by KCLB using short tandem repeat DNA fingerprinting. The cells were transfected with pRL-SV40 vector (Promega) and pGL3-basic vector using Effectene® transfection reagent (Qiagen, Hilden, Germany). The cells were harvested 24 h after transfection and lysates were prepared using a Dual-Luciferase® Reporter Assay System (Promega). Luciferase activity was measured using a SynergyTM HTX Multi-Mode Microplate Reader (BioTek Instruments, Winooski, VT, USA). The results were normalized to those of pRL-SV40 Renilla luciferase activity. All experiments were performed three times in quadruplicate.
Hypoxia condition and HIF-1α overexpression
At 24 h posttransfection with pRL-SV40 and pGL3-basic-ENO1 containing rs2274971A or G allele, A549 cells were exposed to hypoxia for 24 h. For hypoxia, the cells were incubated at 5% CO2, 1% O2, 94% N2 in a hypoxic chamber (Invivo2 400, Ruskinn Technologies, UK) at 37 °C. For HIF-1α overexpression, A549 cells were transfected with pRL-SV40, pGL3-basic-ENO1 containing rs2274971A or G allele, and pEGFP-HIF-1α.
Western blot analysis
Cells were harvested using ProNATM CETi Lysis Buffer containing protease inhibitors and phosphatase inhibitors (Translab, Daejeon, Korea), and protein concentration was determined using the PierceTM BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). Protein samples were resolved by SDS-PAGE and transferred to a pre-wetted polyvinylidene difluoride membranes (Millipore Corporation, Bedford, MA, USA). The membrane was incubated in blocking buffer. Anti-HIF-1α (Novus Biologicals, Littleton, CO, USA) primary antibody was used. The primary β-actin (Santa Cruz Biotech, Santa Cruz, CA, USA) was used as an internal control. Immunoreactive proteins were visualized by SuperSignal™ West Femto Maximum Sensitivity Substrate kit (Thermo Fisher Scientific, Waltham, MA, USA).
Statistical analysis
Differences in the distribution of genotypes according to the clinicopathologic factors of patients were compared using χ2 tests. OS was measured from the day of surgery until the date of death or to the last follow-up date. DFS was estimated from the day of surgery until recurrence or death. The survival estimates were calculated by the Kaplan-Meier method. The differences in OS and DFS across different genotypes were compared by the log-rank test. HRs and 95% CIs were estimated using multivariate Cox proportional hazards models, with adjustment for age, gender, smoking status, tumor histology, pathologic stage, and adjuvant therapy. All analyses were performed with Statistical Analysis System for Windows, version 9.2 (SAS Institute, Cary, NC, USA).
Additional Information
How to cite this article: Lee, S. Y. et al. Genetic Polymorphisms in glycolytic pathway are associated with the prognosis of patients with early stage non-small cell lung cancer. Sci. Rep. 6, 35603; doi: 10.1038/srep35603 (2016).
Supplementary Material
Acknowledgments
This study was supported by the R&D program of MKE/KEIT (10040393, Development and commercialization of molecular diagnostic technologies for lung cancer through clinical validation).
Footnotes
Author Contributions Conceived and designed the experiments: S.Y.L., C.C.J. and J.Y.P. Performed the experiments: C.C.J., J.E.C., M.J.H., D.K.J., S.K.D., S.A.B., H.J.K. and H.-G.K. Analyzed the data and statistical analyses: S.Y.L., C.C.J., K.M.S., J.Y.J., S.H.C., W.K.L. and J.Y.P. Contributed reagents/material/analysis tools: S.Y.L., E.B.L., S.S.Y., Y.S., J.L., S.I.C., C.H.K., S.C., Y.M.L., I.-K.L, S.J. and J.Y.P. Wrote the main manuscript text: S.Y.L., C.C.J. and J.Y.P. All authors reviewed the manuscript.
References
- Siegel R., Naishadham D. & Jemal A. Cancer statistics, 2013. CA Cancer J Clin 63, 11–30 (2013). [DOI] [PubMed] [Google Scholar]
- Detterbeck F. C., Boffa D. J. & Tanoue L. T. The new lung cancer staging system. Chest 136, 260–271 (2009). [DOI] [PubMed] [Google Scholar]
- Hanahan D. & Weinberg R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- Hsu P. P. & Sabatini D. M. Cancer cell metabolism: Warburg and beyond. Cell 134, 703–707 (2008). [DOI] [PubMed] [Google Scholar]
- DeBerardinis R. J., Lum J. J., Hatzivassiliou G. & Thompson C. B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7, 11–20 (2008). [DOI] [PubMed] [Google Scholar]
- Yeung S. J., Pan J. & Lee M. H. Roles of p53, MYC and HIF-1 in regulating glycolysis - the seventh hallmark of cancer. Cell Mol Life Sci 65, 3981–3999 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riganti C., Gazzano E., Polimeni M., Aldieri E. & Ghigo D. The pentose phosphate pathway: an antioxidant defense and a crossroad in tumor cell fate. Free Radic Biol Med 53, 421–436 (2012). [DOI] [PubMed] [Google Scholar]
- Ganapathy-Kanniappan S. & Geschwind J. F. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer 12, 152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D. J. et al. Hexokinase II integrates energy metabolism and cellular protection: Akting on mitochondria and TORCing to autophagy. Cell Death Differ 22, 248–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk H. R. et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 (2008). [DOI] [PubMed] [Google Scholar]
- Diaz-Ruiz R. et al. Mitochondrial oxidative phosphorylation is regulated by fructose 1,6-bisphosphate. A possible role in Crabtree effect induction? J Biol Chem 283, 26948–26955 (2008). [DOI] [PubMed] [Google Scholar]
- Lee S. Y. et al. Polymorphisms in DNA repair and apoptosis-related genes and clinical outcomes of patients with non-small cell lung cancer treated with first-line paclitaxel-cisplatin chemotherapy. Lung Cancer 82, 330–339 (2013). [DOI] [PubMed] [Google Scholar]
- Wang W. et al. Genetic variants in cell cycle control pathway confer susceptibility to lung cancer. Clin Cancer Res 13, 5974–5981 (2007). [DOI] [PubMed] [Google Scholar]
- Perconti G. et al. The kelch protein NS1-BP interacts with alpha-enolase/MBP-1 and is involved in c-Myc gene transcriptional control. Biochim Biophys Acta 1773, 1774–1785 (2007). [DOI] [PubMed] [Google Scholar]
- Principe M. et al. Targeting of surface alpha-enolase inhibits the invasiveness of pancreatic cancer cells. Oncotarget 6, 11098–11113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. S., Wu W., Walsh G., Hong W. K. & Mao L. Enolase-alpha is frequently down-regulated in non-small cell lung cancer and predicts aggressive biological behavior. Clin Cancer Res 9, 3641–3644 (2003). [PubMed] [Google Scholar]
- Tsai S. T. et al. ENO1, a potential prognostic head and neck cancer marker, promotes transformation partly via chemokine CCL20 induction. Eur J Cancer 46, 1712–1723 (2010). [DOI] [PubMed] [Google Scholar]
- Zhao M. et al. Enolase-1 is a therapeutic target in endometrial carcinoma. Oncotarget 6, 15610–15627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejeskar K. et al. Introduction of in vitro transcribed ENO1 mRNA into neuroblastoma cells induces cell death. BMC cancer 5, 161 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins C. M., Yang J., Sims H. F. & Gross R. W. Reversible high affinity inhibition of phosphofructokinase-1 by acyl-CoA: a mechanism integrating glycolytic flux with lipid metabolism. J Biol chem 286, 11937–11950 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin A. et al. Regulation of glucose metabolism by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatases in cancer. Exp Mol Pathol 86, 174–179 (2009). [DOI] [PubMed] [Google Scholar]
- Dunaway G. A., Kasten T. P., Sebo T. & Trapp R. Analysis of the phosphofructokinase subunits and isoenzymes in human tissues. Biochem J 251, 677–683 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Sánchez R., Rodríguez-Enríquez S., Marín-Hernández A. & Saavedra E. Energy metabolism in tumor cells. FEBS J 274, 1393–1418 (2007). [DOI] [PubMed] [Google Scholar]
- Sutendra G. & Michelakis E. D. Pyruvate dehydrogenase kinase as a novel therapeutic target in oncology. Frontiers in oncol 3, 38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z. Y., Xiao L., Bode A. M., Dong Z. & Cao Y. Glycolytic genes in cancer cells are more than glucose metabolic regulators. J Mol Med 92, 837–845 (2014). [DOI] [PubMed] [Google Scholar]
- Kim J. W., Tchernyshyov I., Semenza G. L. & Dang C. V. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3, 177–185 (2006). [DOI] [PubMed] [Google Scholar]
- Contractor T. & Harris C. R. p53 negatively regulates transcription of the pyruvate dehydrogenase kinase Pdk2. Cancer Res 72, 560–567 (2012). [DOI] [PubMed] [Google Scholar]
- Roche T. E. & Hiromasa Y. Pyruvate dehydrogenase kinase regulatory mechanisms and inhibition in treating diabetes, heart ischemia, and cancer. Cell Mol Life Sci 64, 830–849 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet S. et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer cell 11, 37–51 (2007). [DOI] [PubMed] [Google Scholar]
- Sutendra G. et al. Mitochondrial activation by inhibition of PDKII suppresses HIF1α signaling and angiogenesis in cancer. Oncogene 32, 1638–1650 (2013). [DOI] [PubMed] [Google Scholar]
- Michelakis E. D. et al. Metabolic modulation of glioblastoma with dichloroacetate. Sci Transl Med 2, 31ra34 (2010). [DOI] [PubMed] [Google Scholar]
- Garon E. B. et al. Dichloroacetate should be considered with platinum-based chemotherapy in hypoxic tumors rather than as a single agent in advanced non-small cell lung cancer. J Cancer Res Clin Oncol 140, 443–452 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman Y. Y., Tuller T., Keinan A. & Ruppin E. Selection for translation efficiency on synonymous polymorphisms in recent human evolution. Genome Biol Evol 3, 749–761 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin J. B. & Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet 12, 32–42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer T. W. et al. Differences in metabolism between adeno- and squamous cell non-small cell lung carcinomas: spatial distribution and prognostic value of GLUT1 and MCT4. Lung Cancer 76, 316–323 (2012). [DOI] [PubMed] [Google Scholar]
- Schuurbiers O. C. et al. Glucose metabolism in NSCLC is histology-specific and diverges the prognostic potential of 18FDG-PET for adenocarcinoma and squamous cell carcinoma. J Thorac Oncol 9, 1485–1493 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.