Abstract
Epileptic encephalopathies are severe often intractable seizure disorders where epileptiform abnormalities contribute to a progressive disturbance in brain function. Often, epileptic encephalopathies start in childhood and are accompanied by developmental delay and various neurological and non-neurological comorbidities. In recent years, this concept has become virtually synonymous with a group of severe childhood epilepsies including West syndrome, Lennox-Gastaut syndrome, Dravet syndrome, and several other severe childhood epilepsies for which genetic factors are increasingly recognized. In the last 5 years, the field has seen a virtual explosion of gene discovery, raising the number of bona fide genes and possible candidate genes for epileptic encephalopathies to more than 70 genes, explaining 20-25% of all cases with severe early-onset epilepsies that had otherwise no identifiable causes. This review will focus on the phenotypic variability as a characteristic aspect of genetic epilepsies. For many genetic epilepsies, the phenotypic presentation can be broad, even in patients with identical genetic alterations. Furthermore, patients with different genetic etiologies can have seemingly similar clinical presentations, such as in Dravet syndrome. While most patients carry mutations in SCN1A, similar phenotypes can be seen in patients with mutations in PCDH19, CHD2, SCN8A, or in rare cases GABRA1 and STXBP1. In addition to the genotypic and phenotypic heterogeneity, both benign phenotypes and severe encephalopathies have been recognized in an increasing number of genetic epilepsies, raising the question whether these conditions represent a fluid continuum or distinct entities.
Key Words: Epileptic encephalopathy, Genotypic heterogeneity, Next-generation sequencing, Phenotypic heterogeneity, Whole-exome sequencing
Overview of the Epileptic Encephalopathies
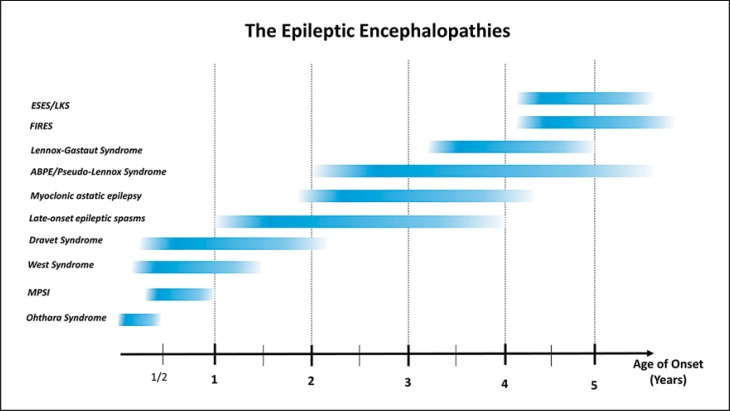
Epilepsies are amongst the most common neurological disorders affecting 5-8/1,000 individuals worldwide [Banerjee et al., 2009]. Up to one-third of epilepsies are refractory to medical treatment, and a significant proportion of childhood intractable epilepsies have significant neurodevelopmental comorbidities, such as developmental delay, plateauing, or regression. These conditions, in which the epileptiform abnormalities are thought to significantly contribute to the overall brain disturbance, are referred to as epileptic encephalopathies. This term is widely accepted by the epilepsy community to encompass a range of clinical syndromes characterized by severe epilepsies of childhood (fig. 1).
Fig. 1.
Age distribution of various epileptic encephalopathies. ABPE = Atypical benign partial epilepsy; ESES = electrical status epilepticus in sleep; FIRES = febrile infection-related epilepsy syndrome; LKS = Landau-Kleffner syndrome; MPSI = migrating partial seizures of infancy.
From Idiopathic West Syndrome to Genetic Epileptic Encephalopathies
Classically, the term epileptic encephalopathy has been used for epilepsy syndromes such as West syndrome with its classical triad of infantile spasms, hypsarrhythmia, and developmental regression. As a fraction of children with infantile spasms have prior normal development, regress during the period of active epilepsy and recover upon receiving antiepileptic medication, this presentation of West syndrome is the paradigmatic example of an epileptic encephalopathy.
However, in most patients with severe epilepsy and in most genetic epilepsies, distinguishing between the contributions of the epileptic abnormality and of the underlying disease is difficult [Guerrini, 2006]. For example, in CDKL5 encephalopathy, one of the most common causes of infantile spasms, most patients also have prior developmental delay [Archer et al., 2006]. However, the contribution of the often intractable epileptic activity to the overall developmental outcome in those patients is not fully clear. In other conditions such as Dravet syndrome due to mutation in SCN1A, developmental plateauing is frequently observed in the second year of life [Harkin et al., 2007]. Again, the developmental trajectory often appears unrelated to the overall seizure burden, and the epileptiform activity in patients with Dravet syndrome is not as prominent and continuous as seen in other epileptic encephalopathies [Nabbout et al., 2013]. In both conditions, the underlying genetic etiology seems to independently drive both the neurodevelopmental and epilepsy phenotypes rather than causing excessive epileptic activity, which can then lead to secondary neurodevelopmental comorbidities. While the precise contribution of the seizure burden to the neurodevelopmental comorbidities is difficult to quantify, both conditions are conceptually referred to as epileptic encephalopathies, given that the epilepsy phenotype is a prominent feature of the disease and is the primary treatment target.
In addition, when considering the genetic aspects of childhood epilepsy, the concept of idiopathic West syndrome may be of limited clinical value. While a genetic etiology in severe epilepsies can be securely implicated in up to one-third of patients, this does not apply to patients with idiopathic self-resolving West syndrome. In these patients, no genetic etiology has been identified to date. The lack of gene discovery in patients with idiopathic self-resolving epilepsy can be attributed to at least 2 factors: the lack of inclusion of these patients in published gene discovery studies, and the fact that other genetic inheritance models such as dominant inheritance with incomplete penetrance may potentially be involved, which makes gene discovery more challenging. This suggests that while we borrow the concept of epileptic encephalopathy from patients where the contribution of the abnormal epileptic activity can be clearly delineated, there is little evidence to date that these condition share similar underlying causes and pathophysiology.
In summary, the epilepsy community applies the concept of epileptic encephalopathy to a heterogeneous group of severe disorders, which have epilepsy and EEG findings as prominent features of the disease. However, for most known genetic epileptic encephalopathies, neurodevelopmental comorbidities are a primary feature of the genetic disease rather than secondary to the temporary disturbance of brain function as a result of excessive epileptiform activity. This concept has helped raise awareness for this group of disorders and has allowed for joint cohort studies to probe the underlying genetic etiology [EuroEPINOMICS-RES Consortium et al., 2014].
Given this conceptual overlap with genetic disorders primarily associated with neurodevelopmental conditions such as intellectual disability or autism, these conditions have shared genetic etiologies. This overlap is partially due to the heterogeneous clinical presentations of some of the genetic alterations. In addition, some of the overlap is due to ascertainment bias, depending on which phenotypic feature was initially the driving force behind the diagnostic work-up. For example, it can be assumed that many patients with intellectual disability due to mutations in SCN1A have a variant of Dravet syndrome [Harkin et al., 2007]. Within an increasing understanding of causative genes and their associated phenotypic features, the true differences between primary genetic neurodevelopmental disorders and primary genetic epileptic encephalopathies is slowly emerging [Epi4K Consortium et al., 2013].
History of Gene Discovery in Epileptic Encephalopathies
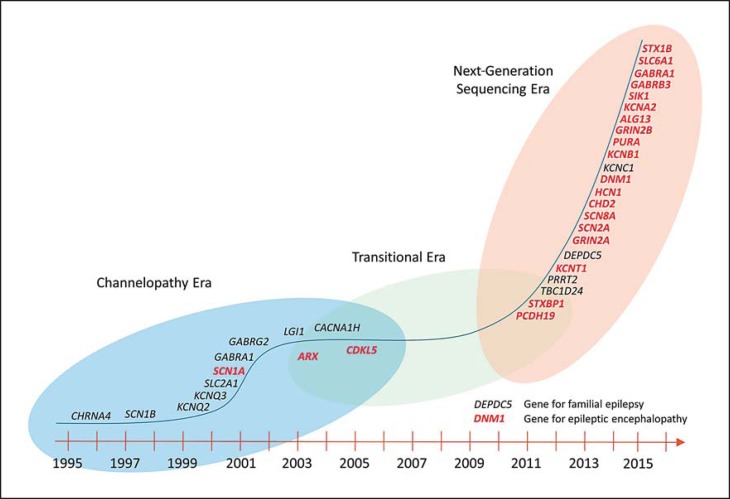
The history of gene discovery in epileptic encephalopathies is intertwined with the overall dynamics of gene discovery in epilepsies in general, which occurred largely in 3 stages: a stage of gene discovery in familial epilepsies, a period of relative stagnation, and the current era of massive parallel sequencing that was ushered in by large-scale studies for copy number variants (fig. 2; table 1).
Fig. 2.
Timeline of gene discovery in human epilepsies.
Table 1.
Examples of genes for epileptic encephalopathies
| Gene | Major epilepsy phenotype | Year of discovery, reference |
|---|---|---|
| SCN1A | Dravet syndrome | Escayg et al., 2000 |
| CDKL5 | atypical Rett syndrome | Kalscheuer et al., 2003 |
| STXBP1 | Ohtahara syndrome | Saitsu et al., 2008 |
| SCN2A | infantile spasms | Heron et al., 2002 |
| SCN8A | infantile spasms | Veeramah et al., 2012 |
| KCNT1 | malignant migrating partial seizures of infancy | Heron et al., 2012 |
| KCNQ2 | neonatal epileptic encephalopathy | Singh et al., 1998 |
| DNM1 | infantile spasms | EuroEPINOMICS-RES Consortium et al., 2014 |
| TBC1D24 | various epileptic encephalopathy phenotypes | Falace et al., 2010 |
| CABRA1 | Dravet syndrome | Cossette et al., 2002 |
| GR1N2A | epilepsy-aphasia syndromes | Reutlinger et al., 2010 |
| SLC6A1 | myoclonic astatic epilepsy | Carvill et al., 2015 |
| CHD2 | photosensitive epileptic encephalopathy | Carvill et al., 2013 |
Lessons from the Era of Family Studies
During the initial era of family studies, many genes underlying relatively mild dominantly inherited familial epilepsies had been discovered including SCN1A, SCN1B, KCNQ2, KCNQ3, and GABRG2 [Steinlein, 2004]. While these gene discoveries initially had no direct link to the more severe epileptic encephalopathies, they laid the general pathophysiological concept, such as the channelopathy concept of the human epilepsies [Steinlein, 2004], which eventually provided the backdrop for the more recent gene discoveries. In addition, the familial epilepsy genes discovered in the early 2000s have frequently been rediscovered in more severe phenotypes [Claes et al., 2003; Patino et al., 2009; Weckhuysen et al., 2012; Carvill et al., 2013, 2014], adding to the impression that many genes implicated in human epilepsies can result in a range of different epilepsy phenotypes, from mild to severe clinical presentations.
One major gene for epileptic encephalopathies that emerged from the era of family studies was the ARX gene [Strømme et al., 2002]; a gene initially identified in families with X-linked mental retardation and epilepsy. ARX, coding for the X chromosome-linked Aristaless-related homeobox gene, was found as the causative gene in a range of X-linked conditions including lissencephaly, agenesis of the corpus callosum with abnormal genitalia, syndromic and nonsyndromic intellectual disability, and infantile spasms without brain malformation. Even though ARX has been a major candidate gene for refractory epileptic encephalopathies, the overall proportions of genetic epilepsy that can be attributed to ARX is probably relatively small. However, as the ARX genetic alteration that is mainly associated with infantile spasms is a polyalanine expansion [Strømme et al., 2002], it is typically not detected by next-generation sequencing. Therefore, the true frequency may be underreported.
The Era of Candidate Studies - Identifying the Classical Genes for Epileptic Encephalopathies
While human epilepsy gene discovery slowly stagnated at the turn of the millennium, candidate gene research and studies detecting genetic alterations based on unusual genetic recombination events were able to identify some of the most common genes for human epilepsies, namely SCN1A in Dravet syndrome and CDKL5 in a variant Rett syndrome [Escayg et al., 2000; Weaving et al., 2004]. Both strategies took advantage of the technology that was available at the time before systematic genome-wide screens for structural rearrangements through genome-wide microarrays or next-generation sequencing technologies became feasible. SCN1A in Dravet syndrome was identified through a candidate gene sequencing approach in severe fever-related epileptic encephalopathy patients [Escayg et al., 2000]. This was based on the observation that milder forms of familial fever-associated epilepsies had previously been implicated in genetic epilepsy with febrile seizures plus (GEFS+) where mutations in SCN1A have been identified [Escayg and Goldin, 2010]. While candidate gene studies in the field of epilepsy were usually underpowered and unable to securely implicate a given gene due to the underlying heterogeneous architecture of the human epilepsies, the discovery of SCN1A in Dravet syndrome was exceptional. There is an unusually strong genotype-phenotype correlation between SCN1A and Dravet syndrome, and the frequency of this subtype of epileptic encephalopathies has been much higher than initially expected. By 2016, it is assumed that Dravet syndrome has a population frequency of 1:22,000 [Bayat et al., 2015], suggesting that this syndrome alone accounts for a significant proportion of epileptic encephalopathies. The CDKL5 gene was identified through sequencing of the breakpoints of a balanced de novo translocation [Kalscheuer et al., 2003] in 2 female patients with severe intellectual disability and infantile spasms. In retrospect, in the era of candidate gene studies, both findings were unusually successful taking advantage of low-hanging fruits, given the very strong genotype-phenotype correlation in SCN1A and a unique recombination event in CDKL5.
The Era of Genome-Wide Studies - from Microarrays to Exome Sequencing
The era of genome-wide screening technologies opened up the door to many of the recent gene findings in epileptic encephalopathies, starting with genes identified through microdeletions and ending with an ongoing stream of novel gene discoveries through exome sequencing technologies (table 1). Genome-wide technologies have offered the possibility to screen large cohorts of patients in parallel allowing the detection of recurrent alterations that are still rare in a particular patient cohort. These technologies also offered the possibility to replace the strategy of candidate gene analysis by hypothesis-free approaches.
The STXBP1 gene is the prime example of genes that were identified in the early stages of genome-wide studies through systematic testing for genome-wide duplications and deletions in patients with severe epilepsies. After a de novo microdeletion spanning STXBP1 was identified in a proband with Ohtahara syndrome, a severe infantile epileptic encephalopathy [Saitsu et al., 2008], additional deletions and mutations were subsequently identified in other patients with early-onset epileptic encephalopathies. To date, STXBP1 is one of the most common genes for epileptic encephalopathies characterized by a wide phenotypic spectrum with infantile spasms and severe intellectual disability as the main consistent features [Stamberger et al., 2016]. Likewise, the GRIN2A gene, currently one of the most common genes for epilepsy-aphasia syndromes, was initially discovered as the gene in a critical region of 3 patients with epileptic encephalopathies and overlapping microdeletions [Reutlinger et al., 2010]. The CHD2 gene was discovered both through follow-up studies of identified solitary microdeletions in patients and through the first systematic exome studies in the field [Suls et al., 2013].
With the advent of next-generation sequencing studies, the focus on gene discovery in epileptic encephalopathies shifted toward systematic family-based exome sequencing to identify de novo mutations on an exome-wide scale. Studies applying these technologies, most notably the Epi4K study [Epi4K Consortium et al., 2013], were able to emphasize the role of known genes and also to provide evidence for additional candidates. Novel genes such as GABRB3 and DNM1 were strongly linked to epilepsy based on the identification of statistically significant enrichment of de novo variants in several individuals with disease (table 2) [EuroEPINOMICS-RES Consortium et al., 2014]. Study paradigms like this have become increasingly relevant in the era of genomic sequencing, given the large-scale data produced by massive parallel sequencing technologies.
Table 2.
Previously implicated genes with insufficient evidence for their role in the disease
| Gene | Year of discovery, reference | Comment |
|---|---|---|
| EFHC1 | Suzuki et al., 2004 | initial variants found to be population variants |
| GABRD | Dibbens et al., 2004 | not confirmed in studies after initial discovery, no reported de novo mutations in epilepsy |
| SCN9A | Singh et al., 2009 | variable gene, lack of association |
| CLCN2 | Haug et al., 2003 | not confirmed in studies after initial discovery |
| CACNA1H | Chen et al., 2003 | not confirmed in studies after initial discovery, no reported de novo mutations in epilepsy |
| SRPX2 | Roll et al., 2006 | not confirmed in studies after initial discovery, initial families were found to have other genetic variants |
Paradigm Shifts
Next-generation sequencing technologies have shifted the focus of genomic science from data generation to data interpretation. While genetics has traditionally been a relatively resource-scarce field, the flood of genomic data generated by exome and genome sequencing has overcome the prior bottleneck of limited data generation at the expense of a new interpretation bottleneck - simply, we are generating more data that we can analyze, given the current knowledge. With the arrival of massive amounts of data, novel paradigms have been established in the field largely reflecting a growing sense of caution of not overinterpreting genomic findings in the absence of confirmatory data or statistical association. In the absence of appropriate guidelines for variant interpretation and gene-disease associations in the genetics community, wrong assertions about genes and variants became very abundant in the literature, posing a major interpretation challenge for research and clinical laboratories. For example, in 2016, previously implicated genes including CACNA1H, SCN9A, EFHC1, CLCN2, GABRD, or SRPX2 were no longer considered causative genes after careful assessment of the available evidence linking those genes to disease (table 2) [Pal and Helbig, 2015; Subaran et al., 2015]. These examples suggest that the current wave of gene discovery is paralleled by a wave of gene retirement, constantly refining the list of possible genetic etiologies based on an evolving catalogue of criteria [Richards et al., 2015].
Genotype-Phenotype Correlations
Necessity to Consider Genotype-Phenotype Correlations
Starting with the discovery of familial epilepsy syndromes, the broad range of phenotypes in families has been puzzling. For example, in families with GEFS+ due to mutations in SCN1A, a broad spectrum of phenotypes ranging from unaffected individuals, individuals with simple febrile seizures to patients with severe epileptic encephalopathies can be observed [Miller and Sotero de Menezes, 1993; Goldberg-Stern et al., 2014]. Therefore, while the clinical validity of many epilepsy genes is beyond doubt, the factors influencing the phenotypic expression remain to be determined. Classically, the genetic variation in the gene itself is examined as a first step, asking the question whether particular mutations or genetic alteration preferentially associate with a particular phenotype. Such an observation has been made for a small subset of genetic epilepsies. For example, loss-of-function variants such as nonsense, invariant splice site, or frameshift mutations in SCN1A are primarily seen in patients with Dravet syndrome and rarely in milder familial cases [Guerrini et al., 2010]. Likewise, in the ARX gene, polyalanine expansions have a strong association with infantile spasms with unremarkable MRI imaging in contrast to genetic alterations that result in lissencephaly [Strømme et al., 2002]. Furthermore, in some familial epilepsy syndromes such as benign familial epilepsies due to mutations in SCN2A or SCN8A, the milder familial cases are due to particular recurrent mutations [Berkovic et al., 2004; Gardella et al., 2016]. On a larger scale, however, a broad phenotypic spectrum is a common feature of genetic epilepsies, and key phenotypic differences can even be observed between individuals with the same genetic alteration.
Genes with Benign and Severe Epilepsy Phenotypes
An emerging number of epilepsy genes are found to underlie both benign and severe phenotypes (table 3). Although SCN1A and SCN1B were historically the first 2 genes known to cause GEFS+ with milder self-limited familial epilepsies, more severe phenotypes have subsequently been discovered [Claes et al., 2001; Patino et al., 2009]. SCN1A is the major gene for Dravet syndrome, an infantile-onset fever-associated epileptic encephalopathy. Homozygous variants in SCN1B have also been found in patients with Dravet syndrome [Ogiwara et al., 2012]. Mutations in KCNQ2 which are known to cause benign familial neonatal seizures [Biervert et al., 1998; Singh et al., 1998] have also been found in severe neonatal epileptic encephalopathies, and the latter phenotype is likely to be much more common than the benign familial form [Weckhuysen et al., 2012, 2013]. Mutations in SCN2A were initially identified in families with benign familial neonatal-infantile seizures [Berkovic et al., 2004]. Subsequently, de novo mutations in SCN2A were found to be one of the most common causes of epileptic encephalopathies [Nakamura et al., 2013; Howell et al., 2015]. In addition to these examples where severe phenotypes were recognized after the initial identification of mild familial forms, the opposite sequence of discovery has also been observed for some genes including SCL2A1 and SCN8A.
Table 3.
Genetic etiologies in human epilepsies with both mild and severe phenotypes
| Gene | Mild phenotype | Severe phenotype |
|---|---|---|
| SCN1A | genetic epilepsy with febrile seizures plus | Dravet syndrome |
| SCN2A | benign familial neonatal-infantile seizures | neonatal epileptic encephalopathy |
| SCN8A | benign familial neonatal-infantile seizures | early-onset epileptic encephalopathy |
| KCNQ2 | benign familial neonatal seizures | neonatal epileptic encephalopathy |
| SLC2A1 | familial focal epilepsy | neonatal epileptic encephalopathy (De Vivo syndrome) |
| GABRA1 | juvenile myoclonic epilepsy | early-onset epileptic encephalopathy |
| GABRG2 | genetic epilepsy with febrile seizures plus | early-onset epileptic encephalopathy |
| GRIN2A | epilepsy-aphasia syndromes | early-onset epileptic encephalopathy |
| PRRT2 | benign familial infantile seizures | early-onset epileptic encephalopathy |
SLC2A1, coding for GLUT1 as the main glucose transporter across the blood-brain barrier, was initially identified in a severe neonatal epileptic encephalopathy sometimes referred to as De Vivo syndrome [De Vivo et al., 1991]. However, it has also become clear that mutations in SCL2A1 can be found in patients with early-onset absence epilepsies and in familial epilepsies with milder focal and generalized epilepsies as a presenting feature [Suls et al., 2009; Mullen et al., 2010]. This observation has prompted many clinicians to reconsider the possibility of GLUT1 deficiency in patients with refractory generalized or focal epilepsies, given that patients with GLUT1 deficiency favorably react to the ketogenic diet [De Vivo et al., 1991]. For SCN8A, a further ion channel gene initially implicated in severe early-onset epileptic encephalopathies [Veeramah et al., 2012], a recent study has identified a mild familial epilepsy syndrome with benign infantile convulsions and paroxysmal choreoathetosis due to a recurrent SCN8A mutation, implicating yet another common epileptic encephalopathy gene in a more benign epilepsy syndrome [Gardella et al., 2016].
For the KCNT1 gene, a different and unusual phenotypic spectrum is emerging. This gene is implicated in a severe early-onset epileptic encephalopathy referred to as malignant migrating partial seizures of infancy [Barcia et al., 2012]. However, mutations in this gene have also been found in families with severe frontal lobe epilepsy [Heron et al., 2012]. Surprisingly, both conditions may exist in the same family [Møller et al., 2015]. While the clinical spectrum of KCNT1 does not range from mild to severe in the same way as the other epilepsy genes do, this novel phenotypic spectrum deserves mentioning as it may provide a unique insight into the role and function of the causative gene.
In summary, even though the clinical presentations of benign epilepsies and severe epileptic encephalopathies are vastly different, several genes are known to cause both conditions. This indicates that both severe epileptic encephalopathies and familial epilepsies do not exist in isolation, but that the careful examination of both ends of the spectrum can be mutually informative.
Fluid Overlap versus Distinct Entities
Are the various entities associated with major epilepsy genes distinct entities or do they occur on a fluid spectrum ranging from benign to severe phenotypes? The genetic approaches to human epilepsies have always had a strong ascertainment bias. While the main focus of early genetic studies was family studies, the current explosion of genetic findings is heavily biased towards severe treatment-resistant epilepsies. Therefore, both the benign end and severe end of the putative spectrum for each gene are heavily overrepresented and currently provide the impression that the phenotypes associated with genes such as SCN1A, SCN2A, SCN8A, KCNQ2, or SCL2A1 may represent distinct entities at both ends of the spectrum rather than a continuum. To address the question of the full phenotypic spectrum, the example of SCN1A may be instructive. Given the overall frequency of SCN1A mutations in fever-associated epilepsies in general, this gene has frequently been tested and identified in familial cases, providing a comprehensive overview of the associated phenotypes [Miller and Sotero de Menezes, 1993; Helbig, 2015]. The SCN1A example shows a continuum of phenotypes ranging from simple febrile seizures to a more severe epilepsy syndrome, arguing for a continuum rather than mutation-specific entities that do not overlap. This gradient is somewhat reflected in the functional alterations of the various SCN1A variants, ranging from mild alterations to complete loss of function [Meng et al., 2015]. Also, common variants in SCN1A have been found to be associated with various epilepsy phenotypes as common risk factors [Kasperaviciute et al., 2013; Feenstra et al., 2014; International League Against Epilepsy Consortium on Complex Epilepsies, 2014]. With increasing genetic studies in common epilepsies, the middle ground between the extremes of mild familial epilepsies and severe epilepsy encephalopathies will be increasingly explored. It will be interesting to see to what extent the SCN1A example also holds true for other genetic etiologies or whether they represent distinct entities.
Future Directions
Genotypes, Phenotypes and Modifiers
Epileptic encephalopathies are characterized by genetic and phenotypic heterogeneity. A similar epilepsy phenotype such as West syndrome, Lennox-Gastaut syndrome, or Dravet syndrome can be caused by different genes, and mutations in a given gene can result in various phenotypes. While some genes and variants allow for a precise genotype-phenotype correlation, the spectrum of phenotypes associated with a given gene is often unexplained and not necessarily related to a particular variant in that gene. Therefore, the majority of phenotypic heterogeneity in genetic epilepsies is unexplained, both with regards to the epilepsy phenotype and other neurodevelopmental comorbidities. For example, truncating variants in SCN1A can result in the full clinical picture of Dravet syndrome including developmental plateauing resulting in severe intellectual disability. However, some patients with truncating SCN1A mutations and similarly severe epilepsies have a normal or near-normal intellect, even though this constellation is assumed to be very rare. The reason for this heterogeneity is not understood. Genetic modifiers are typically implicated, but the study paradigms to successfully address these questions still need to be developed.
Patient Registries, Natural History Studies, and Personalized Medicine
Despite a strong emphasis on epilepsy phenotypes, many clinical aspects of genetic epilepsies remain unknown. The increasing interest to embark on systematic treatment studies for personalized medicine approaches in genetic epilepsies have highlighted the need to better elucidate the constant and variable features of genetic epilepsies and to identify ideal points for intervention. This has rekindled interest in the field to understand the natural history of epileptic encephalopathies and in creating patient registries and databases in collaboration with an emerging active patient community for many of the known epilepsy genes.
In summary, over the last decade, the epileptic encephalopathies have made transition from disorders with a largely unknown etiology to a broadening group of distinct genetic entities that can be identified in up to one-third of all patients. Phenotypic variability is an emerging theme for genetic epilepsies that will have a profound impact on personalized medicine approaches for these conditions.
Disclosure Statement
The authors have no conflicts of interest to declare.
Acknowledgment
I.H. is supported by intramural funds of the University of Kiel, by a grant from the German Research Foundation (HE5415/3-1) within the EuroEPINOMICS framework of the European Science Foundation and additional grants of the German Research Foundation (DFG, HE5415/5-1, HE 5415/6-1), German Ministry for Education and Research (01DH12033, MAR 10/012), and grants by the German chapter of the International League Against Epilepsy (DGfE) and the Genetics Commission of the International League Against Epilepsy.
References
- 1.Archer HL, Evans J, Edwards S, Colley J, Newbury-Ecob R, et al. CDKL5 mutations cause infantile spasms, early onset seizures, and severe mental retardation in female patients. J Med Genet. 2006;43:729–734. doi: 10.1136/jmg.2006.041467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee PN1, Filippi D, Allen Hauser W. The descriptive epidemiology of epilepsy - a review. Epilepsy Res. 2009;85:31–45. doi: 10.1016/j.eplepsyres.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barcia G, Fleming MR, Deligniere A, Gazula VR, Brown MR, et al. De novo gain-of-function KCNT1 channel mutations cause malignant migrating partial seizures of infancy. Nat Genet. 2012;44:1255–1259. doi: 10.1038/ng.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayat A, Hjalgrim H, Møller RS. The incidence of SCN1A-related Dravet syndrome in Denmark is 1:22,000: a population-based study from 2004 to 2009. Epilepsia. 2015;56:e36–39. doi: 10.1111/epi.12927. [DOI] [PubMed] [Google Scholar]
- 5.Berkovic SF, Heron SE, Giordano L, Marini C, Guerrini R, et al. Benign familial neonatal-infantile seizures: characterization of a new sodium channelopathy. Ann Neurol. 2004;55:550–557. doi: 10.1002/ana.20029. [DOI] [PubMed] [Google Scholar]
- 6.Biervert C, Schroeder BC, Kubisch C, Berkovic SF, Propping P, et al. A potassium channel mutation in neonatal human epilepsy. Science. 1998;279:403–406. doi: 10.1126/science.279.5349.403. [DOI] [PubMed] [Google Scholar]
- 7.Carvill GL, Heavin SB, Yendle SC, McMahon JM, OʼRoak BJ, et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet. 2013;45:825–830. doi: 10.1038/ng.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvill GL, Weckhuysen S, McMahon JM, Hartmann C, Møller RS, et al. GABRA1 and STXBP1: novel genetic causes of Dravet syndrome. Neurology. 2014;82:1245–1253. doi: 10.1212/WNL.0000000000000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carvill GL, McMahon JM, Schneider A, Zemel M, Myers CT, et al. Mutations in the GABA transporter SLC6A1 cause epilepsy with myoclonic-atonic seizures. Am J Hum Genet. 2015;96:808–815. doi: 10.1016/j.ajhg.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Lu J, Pan H, Zhang Y, Wu H, et al. Association between genetic variation of CACNA1H and childhood absence epilepsy. Ann Neurol. 2003;54:239–243. doi: 10.1002/ana.10607. [DOI] [PubMed] [Google Scholar]
- 11.Claes L, Del-Favero J, Ceulemans B, Lagae L, Van Broeckhoven C, de Jonghe P. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet. 2001;68:1327–1332. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claes L, Ceulemans B, Audenaert D, Smets K, Löfgren A, et al. De novo SCN1A mutations are a major cause of severe myoclonic epilepsy of infancy. Hum Mutat. 2003;21:615–621. doi: 10.1002/humu.10217. [DOI] [PubMed] [Google Scholar]
- 13.Cossette P, Liu L, Brisebois K, Dong H, Lortie A, et al. Mutation of GABRA1 in an autosomal dominant form of juvenile myoclonic epilepsy. Nat Genet. 2002;31:184–189. doi: 10.1038/ng885. [DOI] [PubMed] [Google Scholar]
- 14.De Vivo DC, Trifiletti RR, Jacobson RI, Ronen GM, Behmand RA, Harik SI. Defective glucose transport across the blood-brain barrier as a cause of persistent hypoglycorrhachia, seizures, and developmental delay. N Engl J Med. 1991;325:703–709. doi: 10.1056/NEJM199109053251006. [DOI] [PubMed] [Google Scholar]
- 15.Dibbens LM, Feng HJ, Richards MC, Harkin LA, Hodgson BL, et al. GABRD encoding a protein for extra- or peri-synaptic GABAA receptors is a susceptibility locus for generalized epilepsies. Hum Mol Genet. 2004;13:1315–1319. doi: 10.1093/hmg/ddh146. [DOI] [PubMed] [Google Scholar]
- 16.Epi4K Consortium, Epilepsy Phenome/Genome Project, Allen AS, Berkovic SF, Cossette P, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escayg A, Goldin AL. Sodium channel SCN1A and epilepsy: mutations and mechanisms. Epilepsia. 2010;51:1650–1658. doi: 10.1111/j.1528-1167.2010.02640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escayg A, MacDonald BT, Meisler MH, Baulac S, Huberfeld G, et al. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat Genet. 2000;24:343–345. doi: 10.1038/74159. [DOI] [PubMed] [Google Scholar]
- 19.EuroEPINOMICS-RES Consortium; Epilepsy Phenome/Genome Project; Epi4K Consortium De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am J Hum Genet. 2014;95:360–370. doi: 10.1016/j.ajhg.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falace A, Filipello F, La Padula V, Vanni N, Madia F, et al. TBC1D24, an ARF6-interacting protein, is mutated in familial infantile myoclonic epilepsy. Am J Hum Genet. 2010;87:365–370. doi: 10.1016/j.ajhg.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feenstra B, Pasternak B, Geller F, Carstensen L, Wang T, et al. Common variants associated with general and MMR vaccine-related febrile seizures. Nat Genet. 2014;46:1274–1282. doi: 10.1038/ng.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardella E, Becker F, Møller RS, Schubert J, Lemke JR, et al. Benign infantile seizures and paroxysmal dyskinesia caused by an SCN8A mutation. Ann Neurol. 2016;79:428–436. doi: 10.1002/ana.24580. [DOI] [PubMed] [Google Scholar]
- 23.Goldberg-Stern H, Aharoni S, Afawi Z, Bennett O, Appenzeller S, et al. Broad phenotypic heterogeneity due to a novel SCN1A mutation in a family with genetic epilepsy with febrile seizures plus. J Child Neurol. 2014;29:221–226. doi: 10.1177/0883073813509016. [DOI] [PubMed] [Google Scholar]
- 24.Guerrini R. Epilepsy in children. Lancet. 2006;367:499–524. doi: 10.1016/S0140-6736(06)68182-8. [DOI] [PubMed] [Google Scholar]
- 25.Guerrini R, Cellini E, Mei D, Metitieri T, Petrelli C, et al. Variable epilepsy phenotypes associated with a familial intragenic deletion of the SCN1A gene. Epilepsia. 2010;51:2474–2477. doi: 10.1111/j.1528-1167.2010.02790.x. [DOI] [PubMed] [Google Scholar]
- 26.Harkin LA, McMahon JM, Iona X, Dibbens L, Pelekanos JT, et al. The spectrum of SCN1A-related infantile epileptic encephalopathies. Brain. 2007;130:843–852. doi: 10.1093/brain/awm002. [DOI] [PubMed] [Google Scholar]
- 27.Haug K, Warnstedt M, Alekov AK, Sander T, Ramírez A, et al. Mutations in CLCN2 encoding a voltage-gated chloride channel are associated with idiopathic generalized epilepsies. Nat Genet. 2003;33:527–532. doi: 10.1038/ng1121. [DOI] [PubMed] [Google Scholar]
- 28.Helbig I. Genetic causes of generalized epilepsies. Semin Neurol. 2015;35:288–292. doi: 10.1055/s-0035-1552922. [DOI] [PubMed] [Google Scholar]
- 29.Heron SE, Crossland KM, Andermann E, Phillips HA, Hall AJ, et al. Sodium-channel defects in benign familial neonatal-infantile seizures. Lancet. 2002;360:851–852. doi: 10.1016/S0140-6736(02)09968-3. [DOI] [PubMed] [Google Scholar]
- 30.Heron SE, Smith KR, Bahlo M, Nobili L, Kahana E, et al. Missense mutations in the sodium-gated potassium channel gene KCNT1 cause severe autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet. 2012;44:1188–1190. doi: 10.1038/ng.2440. [DOI] [PubMed] [Google Scholar]
- 31.Howell KB, McMahon JM, Carvill GL, Tambunan D, Mackay MT, et al. SCN2A encephalopathy: a major cause of epilepsy of infancy with migrating focal seizures. Neurology. 2015;85:958–966. doi: 10.1212/WNL.0000000000001926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.International League Against Epilepsy Consortium on Complex Epilepsies Genetic determinants of common epilepsies: a meta-analysis of genome-wide association studies. Lancet Neurol. 2014;13:893–903. doi: 10.1016/S1474-4422(14)70171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalscheuer VM, Tao J, Donnelly A, Hollway G, Schwinger E, et al. Disruption of the serine/threonine kinase 9 gene causes severe X-linked infantile spasms and mental retardation. Am J Hum Genet. 2003;72:1401–1411. doi: 10.1086/375538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasperaviciute D, Catarino CB, Matarin M, Leu C, Novy J, et al. Epilepsy, hippocampal sclerosis and febrile seizures linked by common genetic variation around SCN1A. Brain. 2013;136:3140–3150. doi: 10.1093/brain/awt233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng H, Xu HQ, Yu L, Lin GW, He N, et al. The SCN1A mutation database: updating information and analysis of the relationships among genotype, functional alteration, and phenotype. Hum Mutat. 2015;36:573–580. doi: 10.1002/humu.22782. [DOI] [PubMed] [Google Scholar]
- 36.Miller IO, Sotero de Menezes MA. SCN1A-related seizure disorders. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, et al., editors. GeneReviews(R) Seattle: University of Washington; 1993. [Google Scholar]
- 37.Møller RS, Heron SE, Larsen LH, Lim CX, Ricos MG, et al. Mutations in KCNT1 cause a spectrum of focal epilepsies. Epilepsia. 2015;56:e114–120. doi: 10.1111/epi.13071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mullen SA, Suls A, De Jonghe P, Berkovic SF, Scheffer IE. Absence epilepsies with widely variable onset are a key feature of familial GLUT1 deficiency. Neurology. 2010;75:432–440. doi: 10.1212/WNL.0b013e3181eb58b4. [DOI] [PubMed] [Google Scholar]
- 39.Nabbout R, Chemaly N, Chipaux M, Barcia G, Bouis C, et al. Encephalopathy in children with Dravet syndrome is not a pure consequence of epilepsy. Orphanet J Rare Dis. 2013;8:176. doi: 10.1186/1750-1172-8-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura K, Kato M, Osaka H, Yamashita S, Nakagawa E, et al. Clinical spectrum of SCN2A mutations expanding to Ohtahara syndrome. Neurology. 2013;81:992–998. doi: 10.1212/WNL.0b013e3182a43e57. [DOI] [PubMed] [Google Scholar]
- 41.Ogiwara I, Nakayama T, Yamagata T, Ohtani H, Mazaki E, et al. A homozygous mutation of voltage-gated sodium channel β1 gene SCN1B in a patient with Dravet syndrome. Epilepsia. 2012;53:e200–203. doi: 10.1111/epi.12040. [DOI] [PubMed] [Google Scholar]
- 42.Pal D, Helbig I. Commentary: pathogenic EFHC1 mutations are tolerated in healthy individuals dependent on reported ancestry. Epilepsia. 2015;56:195–196. doi: 10.1111/epi.12906. [DOI] [PubMed] [Google Scholar]
- 43.Patino GA, Claes LR, Lopez-Santiago LF, Slat EA, Dondeti RS, et al. A functional null mutation of SCN1B in a patient with Dravet syndrome. J Neurosci. 2009;29:10764–10778. doi: 10.1523/JNEUROSCI.2475-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reutlinger C, Helbig I, Gawelczyk B, Subero JI, Tönnies H, et al. Deletions in 16p13 including GRIN2A in patients with intellectual disability, various dysmorphic features, and seizure disorders of the rolandic region. Epilepsia. 2010;51:1870–1873. doi: 10.1111/j.1528-1167.2010.02555.x. [DOI] [PubMed] [Google Scholar]
- 45.Richards S, Aziz N, Bale S, Bick D, Das S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roll P, Rudolf G, Pereira S, Royer B, Scheffer IE, et al. SRPX2 mutations in disorders of language cortex and cognition. Hum Mol Genet. 2006;15:1195–1207. doi: 10.1093/hmg/ddl035. [DOI] [PubMed] [Google Scholar]
- 47.Saitsu H, Kato M, Mizuguchi T, Hamada K, Osaka H, et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat Genet. 2008;40:782–788. doi: 10.1038/ng.150. [DOI] [PubMed] [Google Scholar]
- 48.Singh NA, Charlier C, Stauffer D, DuPont BR, Leach RJ, et al. A novel potassium channel gene, KCNQ2, is mutated in an inherited epilepsy of newborns. Nat Genet. 1998;18:25–29. doi: 10.1038/ng0198-25. [DOI] [PubMed] [Google Scholar]
- 49.Singh NA, Pappas C, Dahle EJ, Claes LR, Pruess TH, et al. A role of SCN9A in human epilepsies, as a cause of febrile seizures and as a potential modifier of Dravet syndrome. PLoS Genet. 2009;5:e1000649. doi: 10.1371/journal.pgen.1000649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamberger H, Nikanorova M, Willemsen MH, Accorsi P, Angriman M, et al. STXBP1 encephalopathy: a neurodevelopmental disorder including epilepsy. Neurology. 2016;86:954–962. doi: 10.1212/WNL.0000000000002457. [DOI] [PubMed] [Google Scholar]
- 51.Steinlein OK. Genetic mechanisms that underlie epilepsy. Nat Rev Neurosci. 2004;5:400–408. doi: 10.1038/nrn1388. [DOI] [PubMed] [Google Scholar]
- 52.Strømme P, Mangelsdorf ME, Shaw MA, Lower KM, Lewis SM, et al. Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat Genet. 2002;30:441–445. doi: 10.1038/ng862. [DOI] [PubMed] [Google Scholar]
- 53.Subaran RL, Conte JM, Stewart WC, Greenberg DA. Pathogenic EFHC1 mutations are tolerated in healthy individuals dependent on reported ancestry. Epilepsia. 2015;56:188–194. doi: 10.1111/epi.12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suls A, Mullen SA, Weber YG, Verhaert K, Ceulemans B, et al. Early-onset absence epilepsy caused by mutations in the glucose transporter GLUT1. Ann Neurol. 2009;66:415–419. doi: 10.1002/ana.21724. [DOI] [PubMed] [Google Scholar]
- 55.Suls A, Jaehn JA, Kecskés A, Weber Y, Weckhuysen S, et al. De novo loss-of-function mutations in CHD2 cause a fever-sensitive myoclonic epileptic encephalopathy sharing features with Dravet syndrome. Am J Hum Genet. 2013;93:967–975. doi: 10.1016/j.ajhg.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzuki T, Delgado-Escueta AV, Aguan K, Alonso ME, Shi J, et al. Mutations in EFHC1 cause juvenile myoclonic epilepsy. Nat Genet. 2004;36:842–849. doi: 10.1038/ng1393. [DOI] [PubMed] [Google Scholar]
- 57.Veeramah KR, OʼBrien JE, Meisler MH, Cheng X, Dib-Hajj SD, et al. De novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am J Hum Genet. 2012;90:502–510. doi: 10.1016/j.ajhg.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weaving LS, Christodoulou J, Williamson SL, Friend KL, McKenzie OL, et al. Mutations of CDKL5 cause a severe neurodevelopmental disorder with infantile spasms and mental retardation. Am J Hum Genet. 2004;75:1079–1093. doi: 10.1086/426462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weckhuysen S, Mandelstam S, Suls A, Audenaert D, Deconinck T, et al. KCNQ2 encephalopathy: emerging phenotype of a neonatal epileptic encephalopathy. Ann Neurol. 2012;71:15–25. doi: 10.1002/ana.22644. [DOI] [PubMed] [Google Scholar]
- 60.Weckhuysen S, Ivanovic V, Hendrickx R, Van Coster R, Hjalgrim H, et al. Extending the KCNQ2 encephalopathy spectrum: clinical and neuroimaging findings in 17 patients. Neurology. 2013;81:1697–1703. doi: 10.1212/01.wnl.0000435296.72400.a1. [DOI] [PMC free article] [PubMed] [Google Scholar]