Abstract
Streptomyces sp. MWW064 (=NBRC 110611) produces an antitumor cyclic depsipeptide rakicidin D. Here, we report the draft genome sequence of this strain together with features of the organism and generation, annotation and analysis of the genome sequence. The 7.9 Mb genome of Streptomyces sp. MWW064 encoded 7,135 putative ORFs, of which 6,044 were assigned with COG categories. The genome harbored at least three type I polyketide synthase (PKS) gene clusters, seven nonribosomal peptide synthetase (NRPS) gene clusters, and four hybrid PKS/NRPS gene clusters, from which a hybrid PKS/NRPS gene cluster responsible for rakicidin synthesis was successfully identified. We propose the biosynthetic pathway based on bioinformatic analysis, and experimentally proved that the pentadienoyl unit in rakicidins is derived from serine and malonate.
Keywords: Biosynthesis, Nonribosomal peptide synthetase, Polyketide synthase, Rakicidin, Streptomyces
Introduction
Rakicidin D is an inhibitor of tumor cell invasion isolated from the culture broth of an actinomycete strain MWW064 of the genus Streptomyces [1]. To date, five congeners rakicidins A, B, and E from Micromonospora and rakicidins C and D from Streptomyces have been reported [1–4]. Rakicidins share the 15-membered cyclic depsipeptide structure comprising three amino acids and a fatty acid modified with hydroxy and methyl substitutions. The most intriguing part of rakicidins is a rare unusual amino acid, 4-amino-2,4-pentadienoate (APDA), which is present only in a limited range of secondary metabolites of actinomycetes such as BE-43547 [5] and microtermolide [6, 7]. Despite the scarcity of APDA unit in nature, nothing is known about its biosynthesis. Recently, putative biosynthetic genes for rakicidin D were reported [8], but the data is incomplete, no detailed information is shown in the paper, and DNA sequences have not been registered in public databases. Hence, the biosynthesis of rakicidins has been actually unclear yet. In this study, we performed whole genome shotgun sequencing of the strain MWW064 to elucidate the biosynthetic mechanism of rakicidin D. We herein present the draft genome sequence of Streptomyces sp. MWW064, together with the taxonomical identification of the strain, description of its genome properties and annotation of the gene cluster for rakicidin synthesis. We propose the rakicidin-biosynthetic mechanism predicted by bioinformatics analysis and confirmed by precursor-incorporation experiments.
Organism information
Classification and features
In the course of screening for antitumor compounds from actinomycetes, Streptomyces sp. MWW064 was isolated from a marine sediment sample collected in Samut Sakhon province of Thailand and found to produce rakicidin D [1]. The general feature of this strain is shown in Table 1. This strain grew well on ISP 2 and ISP 4 agars. On ISP 5 and ISP 7 agars, the growth was poor. The color of aerial mycelia was white and that of the reverse side was pale red on ISP 2 agar. Diffusible pigments were dark orange on ISP 2 agar medium. Strain MWW064 formed extensively branched- substrate and aerial mycelia. The aerial mycelium formed flexuous spore chains at maturity. The spores were cylindrical, having a smooth surface. A scanning electron micrograph of this strain is shown in Fig. 1. Growth occurred at 15–37 °C (optimum 28 °C) and pH 5–9 (optimum pH 7). Strain MWW064 exhibited growth with 0–3 % (w/v) NaCl (optimum 0 % NaCl). Strain MWW064 utilized glucose and inositol for growth. The gene sequence encoding 16S rRNA was obtained from GenBank/EMBL/DDBJ databases (accession no. GU295447). A phylogenetic tree was reconstructed on the basis of the 16S rRNA gene sequence together with taxonomically close Streptomyces type strains using ClustalX2 [9] and NJPlot [10]. The phylogenetic analysis confirmed that the strain MWW064 belongs to the genus Streptomyces (Fig. 2).
Table 1.
Classification and general features of Streptomyces sp. MWW064 [13]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [24] | |
| Phylum Actinobacteria | TAS [25] | ||
| Class Actinobacteria | TAS [26] | ||
| Order Actinomycetales | TAS [26–29] | ||
| Suborder Streptomycineae | TAS [26, 29] | ||
| Family Streptomycetaceae | TAS [26, 28–31] | ||
| Genus Streptomyces | TAS [28, 31–33] | ||
| Species undetermined | - | ||
| strain: MWW064 | TAS [1] | ||
| Gram stain | Gram-positive | NAS | |
| Cell shape | Branched mycelia | IDA | |
| Motility | Not reported | ||
| Sporulation | Sporulating | IDA | |
| Temperature range | 15 °C to 37 °C | IDA | |
| Optimum temperature | 28 °C | IDA | |
| pH range; Optimum | 5 to 9; 7 | IDA | |
| Carbon source | D-glucose, inositol | IDA | |
| MIGS-6 | Habitat | Marine sediment | TAS [1] |
| MIGS-6.3 | Salinity | 0 % to 3 % NaCl | IDA |
| MIGS-22 | Oxygen requirement | Aerobic | IDA |
| MIGS-15 | Biotic relationship | Free-living | IDA |
| MIGS-14 | Pathogenicity | Not reported | |
| MIGS-4 | Geographic location | Samut Sakhon province, Thailand | TAS [1] |
| MIGS-5 | Sample collection | February 2, 2008 | NAS |
| MIGS-4.1 | Latitude | 13° 32’ 55” N | NAS |
| MIGS-4.2 | Longitude | 100° 16’ 39” E | NAS |
| MIGS-4.4 | Altitude | 8.6 m. above sea level | NAS |
a Evidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [34]
Fig. 1.
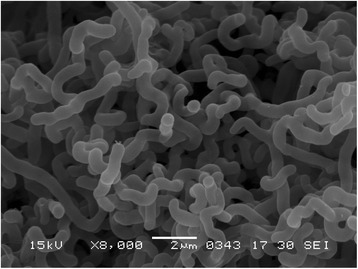
Scanning electron micrograph of Streptomyces sp. MWW064 grown on 1/2 ISP 2 agar for 7 days at 28 °C. Bar, 2 μm
Fig. 2.
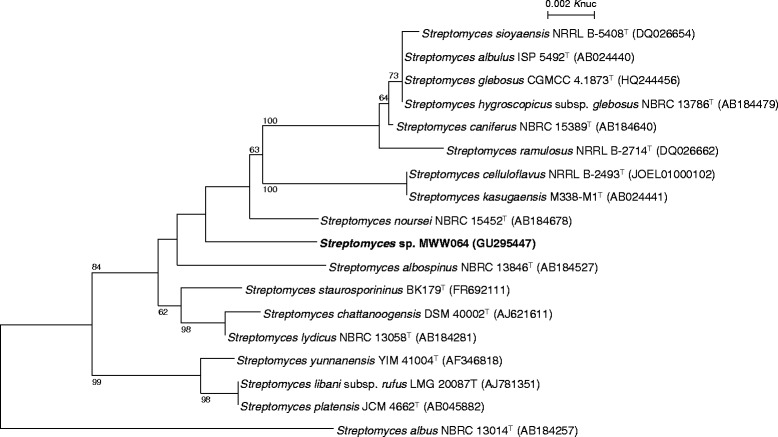
Phylogenetic tree of Streptomyces sp. MWW064 and phylogenetically close type strains, showing over 98.5 % similarity, based on 16S rRNA gene sequences. The accession numbers for 16S rRNA genes are shown in parentheses. The tree uses sequences aligned by ClustalX2 [9], and constructed by the neighbor-joining method [35]. All positions containing gaps were eliminated. The building of the tree also involves a bootstrapping process repeated 1,000 times to generate a majority consensus tree, and only bootstrap values above 50 % are shown at branching points. Streptomyces albus NBRC 13014T was used as an outgroup
Chemotaxonomic data
The isomer of diaminopimelic acid in the whole-cell hydrolysate was analyzed according to the method described by Hasegawa et al. [11]. Isoprenoid quinones and cellular fatty acids were analyzed as described previously [12]. The whole-cell hydrolysate of strain MWW064 contained ll-diaminopimelic acid as its diagnostic peptidoglycan diamino acid. The predominant menaquinones were identified as MK-9(H2), MK-9(H4) and MK-9(H6); MK-10(H2), MK-10(H4) and MK-10(H6) were also detected as minor components. The major cellular fatty acids were found to be anteiso-C15:0, iso-C15:0, C16:0, anteiso-C17:0, iso-C17:0 and iso-C16:0.
Genome sequencing information
Genome project history
In collaboration between Toyama Prefectural University and NBRC, the organism was selected for genome sequencing to elucidate the rakicidin biosynthetic pathway. We successfully accomplished the genome project of Streptomyces sp. MWW064 as reported in this paper. The draft genome sequences have been deposited in the INSDC database under the accession number BBUY01000001-BBUY01000099. The project information and its association with MIGS version 2.0 compliance are summarized in Table 2 [13].
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31 | Finishing quality | Improved-high-quality draft |
| MIGS-28 | Libraries used | 454 shotgun library, Illumina paired-end library |
| MIGS 29 | Sequencing platforms | 454 GS FLX+, Illumina HiSeq1000 |
| MIGS 31.2 | Fold coverage | 8.9 ×, 93.5 ×, respectively |
| MIGS 30 | Assemblers | Newbler v2.8, GenoFinisher |
| MIGS 32 | Gene calling method | Progidal |
| Locus Tag | SSP35 | |
| GenBank ID | BBUY00000000 | |
| GenBank Date of Release | February 20, 2016 | |
| GOLD ID | Not registered | |
| BIOPROJECT | PRJDB3538 | |
| MIGS 13 | Source Material Identifier | NBRC 110611 |
| Project relevance | Industrial |
Growth conditions and genomic DNA preparation
Streptomyces sp. MWW064 was deposited in the NBRC culture collection with the registration number of NBRC 110611. Its monoisolate was grown on polycarbonate membrane filter (Advantec) on double diluted ISP 2 agar medium (0.2 % yeast extract, 0.5 % malt extract, 0.2 % glucose, 2 % agar, pH 7.3) at 28 °C. High quality genomic DNA for sequencing was isolated from the mycelia with an EZ1 DNA Tissue Kit and a Bio Robot EZ1 (Qiagen) according to the protocol for extraction of nucleic acid from Gram-positive bacteria. The size, purity, and double-strand DNA concentration of the genomic DNA were measured by pulsed-field gel electrophoresis, ratio of absorbance values at 260 nm and 280 nm, and Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies), respectively, to assess the quality of genomic DNA.
Genome sequencing and assembly
Shotgun and paired-end libraries were prepared and subsequently sequenced using 454 pyrosequencing technology and HiSeq1000 (Illumina) paired-end technology, respectively (Table 2). The 70 Mb shotgun sequences and 739 Mb paired-end sequences were assembled using Newbler v2.8 and subsequently finished using GenoFinisher [14] to yield 99 scaffolds larger than 500 bp.
Genome annotation
Coding sequences were predicted by Prodigal [15] and tRNA-scanSE [16]. The gene functions were annotated using an in-house genome annotation pipeline, and PKS- and NRPS-related domains were searched using the SMART and PFAM domain databases. PKS and NRPS gene clusters and their domain organizations were determined as reported previously [17]. Substrates of adenylation (A) and acyltransferase (AT) domains were predicted using antiSMASH [18]. BLASTP search against the NCBI nr databases were also used for predicting function of proteins encoded in the rakicidin biosynthetic gene cluster.
Genome properties
The total size of the genome is 7,870,697 bp and the GC content is 71.1 % (Table 3), similar to other genome-sequenced Streptomyces members. Of the total 7,206 genes, 7,135 are protein-coding genes and 71 are RNA genes. The classification of genes into COGs functional categories is shown in Table 4. As for secondary metabolite pathways by modular PKSs and NRPSs, Streptomyces sp. MWW064 has at least four hybrid PKS/NRPS gene clusters, three type I PKS gene clusters, and seven NRPS gene clusters. According to the assembly line mechanism [19], we predicted the chemical backbones that each cluster will synthesize (Table 5), suggesting the potential of Streptomyces sp. MWW064 to produce diverse polyketide- and nonribosomal peptide-compounds as the secondary metabolites.
Table 3.
Genome statistics
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 7,904,619 | 100.0 |
| DNA coding (bp) | 6,855,885 | 86.7 |
| DNA G + C (bp) | 5,597,799 | 70.8 |
| DNA scaffolds | 99 | - |
| Total genes | 7,206 | - |
| Protein coding genes | 7,135 | 99.0 |
| RNA genes | 71 | 0.99 |
| Pseudogenes | - | - |
| Genes in internal clusters | 2,610 | 36.2 |
| Genes with function prediction | 4,515 | 62.7 |
| Genes assigned to COGs | 6,044 | 83.9 |
| Genes with Pfam domains | 4,870 | 67.6 |
| Genes with signal peptides | 559 | 7.8 |
| Genes with transmembrane helices | 1,550 | 21.5 |
| CRISPR repeats | 1 | - |
Table 4.
Number of genes associated with general COG functional categories
| Code | Value | % age | Description |
|---|---|---|---|
| J | 279 | 4.6 | Translation, ribosomal structure and biogenesis |
| A | 4 | 0.1 | RNA processing and modification |
| K | 696 | 11.5 | Transcription |
| L | 452 | 7.5 | Replication, recombination and repair |
| B | 6 | 0.1 | Chromatin structure and dynamics |
| D | 55 | 0.9 | Cell cycle control, Cell division, chromosome partitioning |
| V | 132 | 2.2 | Defense mechanisms |
| T | 432 | 7.1 | Signal transduction mechanisms |
| M | 294 | 4.9 | Cell wall/membrane biogenesis |
| N | 33 | 0.5 | Cell motility |
| U | 95 | 1.6 | Intracellular trafficking and secretion |
| O | 223 | 3.7 | Posttranslational modification, protein turnover, chaperones |
| C | 386 | 6.4 | Energy production and conversion |
| G | 474 | 7.8 | Carbohydrate transport and metabolism |
| E | 651 | 10.8 | Amino acid transport and metabolism |
| F | 134 | 2.2 | Nucleotide transport and metabolism |
| H | 253 | 4.2 | Coenzyme transport and metabolism |
| I | 323 | 5.3 | Lipid transport and metabolism |
| P | 404 | 6.7 | Inorganic ion transport and metabolism |
| Q | 385 | 6.4 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 1,032 | 17.1 | General function prediction only |
| S | 440 | 7.3 | Function unknown |
| - | 1,091 | 18.1 | Not in COGs |
The total is based on the total number of protein coding genes in the genome
Table 5.
Modular PKS and NRPS gene clusters in Streptomyces sp. MWW064
| Gene cluster | Encoded in | No. of modular PKS and NRPS genes | No. of modules | Backbone of predicted product |
|---|---|---|---|---|
| pks/nrps-1 (rak) | scaffold 9 | 6 | 7 | R-C3-C3-Ser-C2-Gly-X |
| pks/nrps-2 | scaffold 5 | 6 | 14 | C2-C2-C2-C2-C2-Gly-C2-C2-C2-C2-C2-C2-C2-C2 |
| pks/nrps-3 | scaffold 2 | 4 | 3 | C?-C?-X |
| pks/nrps-4 | scaffold 11 | 1 | 2 | X-C2 |
| pks-1 | scaffold 18 | 5 | 5 | C?-C3-C2-C2-C? |
| pks-2 | scaffold 23 | 1 | 1 | C? |
| other pks a | scaffolds 11, 39, 45 | >3 | >10 | C2-C2-C3-C2, C2-C2, C2-C2-C2, C2 |
| nrps-1 | scaffold 11 | 4 | 4 | X-X-Val-X |
| nrps-2 | scaffold 18 | 3 | 3 | R-Val-X |
| nrps-3 | scaffold 9 | 2 | 3 | R-Cys-mCys |
| nrps-4 | scaffold 13 | 3 | 4 | Val-Gly-Ser-Pro |
| nrps-5 | scaffold 2 | 1 | 1 | Ser |
| nrps-6 | scaffold 12 | 1 | 1 | Thr |
| other nrps a | scaffolds 3, 5 | >2 | >6 | X-X-X-X-X, Cys |
anot completely sequenced. R, starter unit; C3, C3 unit derived from methylmalonyl-CoA; C2, C2 unit derived from malonyl-CoA; X, unpredictable amino acid; C?, unpredictable carbon unit derived from acyl-CoA; mCys, methylated cysteine
Insights from the genome sequence
Rakicidin biosynthetic pathway in Streptomyces sp. MWW064
The chemical structure of rakicidin D suggested that it is synthesized by a hybrid PKS/NRPS pathway. Among the four hybrid PKS/NRPS gene clusters present in Streptomyces sp. MWW064 (Table 5), pks/nrps-1 is most likely responsible for rakicidin synthesis because the carbon backbone of the predicted product (R-C3-C3-Ser-C2-Gly-X) is in good accordance with that of rakicidin D. Genes in pks/nrps-1 (Table 6) encode enzymes necessary for rakicidin biosynthesis (Fig. 3). This cluster contains three PKS genes (SSP35_09_01910, SSP35_09_01900, SSP35_09_01880) and three NRPS genes (SSP35_09_01890, SSP35_09_01870, SSP35_09_01860), corresponding to rakAB, rakC, rakEF, rakD, rakG, and rakH [8], respectively. Based on the collinearity rule of modular PKS/NRPS pathways, it is deduced that RakAB loads a starter molecule (‘R’ in Fig. 3), and subsequently RakAB and RakC add a diketide chain to the starter by condensation of two methylmalonyl-CoA molecules, since the substrates of their AT domains are likely methylmalonyl-CoA (‘ATm’ in Fig. 3). An NRPS RakD and the remaining PKS RakEF are most likely involved in the APDA supply: the A domain of RakD has signature amino acid residues for serine, and RakEF contains a set of domains (AT, KR, DH) for malonate incorporation, ketoreduction, and dehydration to provide a double bond between C9 and C10. In addition, the DH domain in RakEF is also proposed to be responsible for the dehydration of the primary hydroxy group of the incorporated serine molecule on the basis of the following reasons although experimental evidences are required. First, no dehydratase gene is present near the rakicidin cluster. In the biosynthesis of dehydroalanine in bacterial peptides such as lantibiotics, a dehydratase catalyzes the exo-methylene formation from serine [20, 21]. Second, the order of KR and DH domains in RakEF is unusual: among the three hundred type I PKS genes for eighty actinomycete polyketides, the order of two domains is exclusively DH-KR [22]. The only exception can be seen in the PKS genes for enediynes in which the chain elongation is iteratively catalyzed as similar to type II PKS [23]. The unusual order of KR-DH may render an undescribed function to the DH domain of RakEF. After formation of APDA moiety, RakG is likely responsible for the condensation of glycine and the following N-methylation, and RakH for asparagine condensation. Hydroxylation of asparagine would be catalyzed by asparagine hydroxylase encoded by rakO in the downstream of the cluster, to yield rakicidin D. On the basis of the above-mentioned bioinfomatic evidences, we here propose the biosynthetic pathway of rakicidin D as shown Fig. 3.
Table 6.
ORFs in the rakicidin-biosynthetic gene cluster of Streptomyces sp. MWW064
| ORF (locus tag) | Size (aa) | Deduced function | Protein homolog [origin] | Identity/similarity (%) | Accession number |
|---|---|---|---|---|---|
| SSP35_09_01970 | 243 | unknown | hypothetical protein [Streptomyces natalensis] | 63/73 | WP_030067339 |
| SSP35_09_01960 | 331 | transcriptional regulator | hypothetical protein DT87_01625 [Streptomyces sp. NTK 937] | 59/69 | KDQ65969 |
| SSP35_09_01950 | 358 | 3-oxoacyl-ACP synthase | 3-oxoacyl-ACP synthase [Streptomyces sp. NRRL S-920] | 77/87 | WP_030791445 |
| SSP35_09_01940 | 79 | ACP | phosphopantetheine-binding protein [Streptomyces bingchenggensis BCW-1] | 68/79 | ADI05068 |
| SSP35_09_01930 | 406 | ketosynthase | 3-oxoacyl-ACP synthase [Streptomyces sp. NRRL S-15] | 81/89 | WP_031089521 |
| SSP35_09_01920 | 146 | unknown | methylmalonyl-CoA epimerase [Salinispora pacifica] | 80/87 | WP_018222873 |
| SSP35_09_01910 (RakAB) | 2,902 | PKS | hypothetical protein [Streptomyces vitaminophilus] | 63/73 | WP_018385948 |
| SSP35_09_01900 (RakC) | 1,624 | PKS | non-ribosomal peptide synthetase [Micromonospora sp. M42] | 60/70 | EWM63000 |
| SSP35_09_01890 (RakD) | 1,126 | NRPS | hypothetical protein [Streptomyces vitaminophilus] | 68/78 | WP_018385946 |
| SSP35_09_01880 (RakEF) | 1,950 | PKS | hypothetical protein [Streptomyces vitaminophilus] | 64/73 | WP_018385945 |
| SSP35_09_01870 (RakG) | 1,556 | NRPS | hypothetical protein, partial [Micromonospora purpureochromogenes] | 64/74 | WP_030498976 |
| SSP35_09_01860 (RakH) | 1,565 | NRPS | amino acid adenylation domain protein [Nostoc punctiforme PCC 73102] | 38/55 | ACC80782 |
| SSP35_09_01850 | 563 | ABC transporter | hypothetical protein [Micromonospora purpureochromogenes] | 62/73 | WP_030498978 |
| SSP35_09_01840 (RakL) | 251 | type-II thioesterase | hypothetical protein [Streptomyces vitaminophilus] | 64/73 | WP_018385940 |
| SSP35_09_01830 (RakN) | 1,013 | NRPS | non-ribosomal peptide synthetase [Micromonospora sp. M42] | 55/63 | EWM63010 |
| SSP35_09_01820 (RakO) | 331 | asparagine oxygenase | clavaminate synthase [Streptomyces sp. LaPpAH-202] | 64/75 | WP_026235187 |
| SSP35_09_01810 | 809 | unknown | penicillin amidase [Amycolatopsis nigrescens] | 63/74 | WP_026359955 |
| SSP35_09_01800 | 205 | transcriptional regulator | putative LuxR family transcriptional regulator [Streptomyces glaucescens] | 71/81 | AIR96926 |
SSP35_09_01910, SSP35_09_01900, SSP35_09_01890, SSP35_09_01880, SSP35_09_01870, SSP35_09_01860, SSP35_09_01840, SSP35_09_01830, and SSP35_09_01820 are corresponding to RakA plus RakB (RakAB), RakC, RakD, RakE plus RakF (RakEF), RakG, RakH, RakL, RakN, and RakO, previously reported in the reference [8], respectively, and SSP35_09_01940 may possibly be corresponding to RakI
Fig. 3.
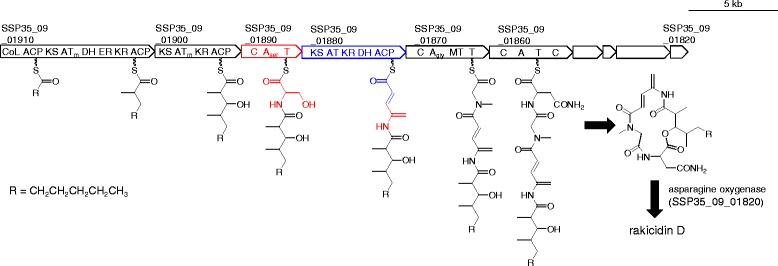
Genetic map of rakicidin biosynthetic gene cluster of Streptomyces sp. MWW064 and the biosynthetic mechanism of rakicidin D
Identification of biosynthetic precursors of the APDA moiety
To verify the predicted biosynthetic origin of the APDA unit, feeding experiments using 13C-labeled precursors were carried out. Inoculation, cultivation, extraction, and purification were performed in the same manner as previously reported [1]. Addition of sodium [2-13C]acetate or [1-13C]-L-serine (20 mg/100 ml medium/flask, 10 flasks for [2-13C]acetate, 3 flasks for [1-13C]-L-serine) was initiated at 48 h after inoculation and periodically carried out every 24 h for four times. After further incubation for 24 h, the whole culture broths were extracted with 1-butanol and several steps of purification yielded 2.5 mg and 1.7 mg of 13C-labeled rakicidin D, respectively. The 13C NMR spectrum of these labeled rakicidin D is shown in Table 7. Feeding of sodium [2-13C]acetate gave enrichments at C9 of the APDA unit and three carbons C18, C20, and C22 in the aliphatic chain of the fatty acid moiety. [1-13C]-L-serine feeding enriched C10 of the APDA unit and the carbonyl carbon of Gly (C5). These results unambiguously indicated that the APDA unit is derived from an acetate and a serine (Fig. 4). Labeling of C5 by serine-feeding can be explained by the interconversion between glycine and serine by transformylase in primary metabolism for amino acid supply.
Table 7.
Incorporation of 13C-labeled precursors into rakicidin D
| Position | δC | Relative enrichmentsa | |
|---|---|---|---|
| [2-13C]acetate | [1-13C]-L-serine | ||
| 1 | 169.2 | 0.89 | 1.58 |
| 2 | 54.9 | 1.14 | 1.19 |
| 3 | 72.5 | 1.57 | 1.07 |
| 4 | 172.7 | 1.31 | 1.95 |
| 5 | 167.6 | 0.68 | 6.61 |
| 6 | 52.5 | 0.77 | 1.15 |
| 7 | 36.5 | 1.00 | 1.00 |
| 8 | 166.0 | 0.77 | 1.43 |
| 9 | 118.8 | 3.46 | 1.54 |
| 10 | 138.4 | 1.03 | 13.97 |
| 11 | 137.9 | 0.95 | 0.67 |
| 12 | 117.1 | 1.02 | 1.33 |
| 13 | 172.5 | 1.61 | 1.26 |
| 14 | 41.7 | 1.86 | 1.36 |
| 15 | 78.1 | 1.25 | 1.21 |
| 16 | 33.9 | 1.91 | 1.39 |
| 17 | 32.8 | 1.30 | 1.55 |
| 18 | 27.0 | 2.64 | 1.26 |
| 19 | 28.9 | 1.15 | 1.58 |
| 20 | 31.3 | 3.37 | 1.43 |
| 21 | 22.1 | 1.05 | 1.28 |
| 22 | 14.0 | 3.37 | 1.61 |
| 23 | 15.4 | 1.51 | 1.07 |
| 24 | 13.3 | 1.75 | 1.32 |
a 13C signal intensity of each peak in the labeled 1 divided by that of the corresponding signal in the unlabeled 1, respectively, normalized to give an enrichment ratio of 1 for the unenriched peak of C7. The numbers in bold type indicate 13C-enriched atoms from 13C-labeled precursors
Fig. 4.
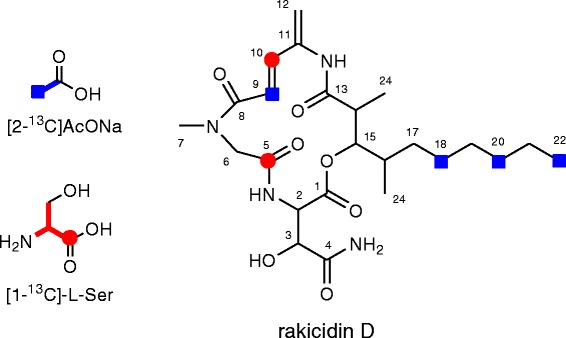
Incorporation of 13C-labeled precursors into rakicidin D
Conclusions
The 7.9 Mb draft genome of Streptomyces sp. MWW064, a producer of rakicidin D isolated from marine segment, has been deposited at GenBank/ENA/DDBJ under the accession number BBUY00000000. We successfully identified the PKS/NRPS hybrid gene cluster for rakicidin synthesis and proposed the plausible biosynthetic pathway. Labeled precursor incorporation experiments showed the APDA moiety is synthesized from serine and malonate. These finding will open up possibilities of genetic engineering to synthesize more potential rakicidin-based antitumor compounds and discovering new bioactive compounds possessing APDA units.
Acknowledgements
This research was supported by a Grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, and Technology of Japan to Y.I. We are grateful to Dr. S. Kitani at Osaka University for thoughtful discussion on the biosynthesis of rakicidins. We also thank Ms. Yuko Kitahashi, Ms. Satomi Saitou and Ms. Chiyo Shibata for assistance to this study.
Authors’ contributions
HK elucidated rakicidin-biosynthetic pathway and drafted the manuscript. AI carried out feeding experiments using labeled precursors. NI annotated the genome sequences. AH sequenced the genome. MH performed chemotaxonomic experiments. EH examined the features of the strain. TN coordinated collaboration between Japan and Thailand. WP isolated the strain. YI designed this study and edited the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- A
Adenylation
- ABC
ATP-binding cassette
- ACP
Acyl carrier protein
- APDA
4-amino-2,4-pentadienoate
- AT
Acyltransferase
- C
Condensation
- CoA
Coenzyme A
- CoL
CoA ligase
- DDBJ
DNA Data Bank of Japan
- DH
Dehydratase
- ER
Enoylreductase
- ISP
International Streptomyces project
- KR
Ketoreductase
- KS
Ketosynthase
- NBRC
Biological Resource Center, National Institute of Technology and Evaluation
- NMR
Nuclear magnetic resonance
- NRPS
Nonribosomal peptide synthetase
- PKS
Polyketide synthase
- T
Thiolation
References
- 1.Igarashi Y, Shimasaki R, Miyanaga S, Oku N, Onaka H, Sakurai H, Saiki I, Kitani S, Nihira T, Wimonsiravude W, et al. Rakicidin D, an inhibitor of tumor cell invasion from marine-derived Streptomyces sp. J Antibiot. 2010;63(9):563–5. doi: 10.1038/ja.2010.70. [DOI] [PubMed] [Google Scholar]
- 2.McBrien KD, Berry RL, Lowe SE, Neddermann KM, Bursuker I, Huang S, Klohr SE, Leet JE. Rakicidins, new cytotoxic lipopeptides from Micromonospora sp. fermentation, isolation and characterization. J Antibiot. 1995;48(12):1446–52. doi: 10.7164/antibiotics.48.1446. [DOI] [PubMed] [Google Scholar]
- 3.Oku N, Matoba S, Yamazaki YM, Shimasaki R, Miyanaga S, Igarashi Y. Complete stereochemistry and preliminary structure-activity relationship of rakicidin A, a hypoxia-selective cytotoxin from Micromonospora sp. J Nat Prod. 2014;77(11):2561–5. doi: 10.1021/np500276c. [DOI] [PubMed] [Google Scholar]
- 4.Hu J, Wunderlich D, Sattler I, Feng X, Grabley S, Thiericke R. Rakicidin C, a new cyclic depsipeptide from Streptomyces sp. Eur. J. Org. Chem. 2000;2000(19):3353–3356
- 5.Nishioka H, Nakajima S, Nagashima M, Kojiri K, Suda H. BE-43547 series substances, their manufacture with Streptomyces species, and their use as antitumor agents. Japan Patent 1998:JP 10147594 A 19980602.
- 6.Carr G, Poulsen M, Klassen JL, Hou Y, Wyche TP, Bugni TS, Currie CR, Clardy J. Microtermolides A and B from termite-associated Streptomyces sp. and structural revision of vinylamycin. Org Lett. 2012;14(11):2822–5. doi: 10.1021/ol301043p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Igarashi M, Shida T, Sasaki Y, Kinoshita N, Naganawa H, Hamada M, Takeuchi T. Vinylamycin, a new depsipeptide antibiotic, from Streptomyces sp. J Antibiot. 1999;52(10):873–9. doi: 10.7164/antibiotics.52.873. [DOI] [PubMed] [Google Scholar]
- 8.Albright JC, Goering AW, Doroghazi JR, Metcalf WW, Kelleher NL. Strain-specific proteogenomics accelerates the discovery of natural products via their biosynthetic pathways. J Ind Microbiol Biotechnol. 2014;41(2):451–9. doi: 10.1007/s10295-013-1373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 10.Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78(5):364–9. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 11.Hasegawa T, Takizawa M, Tanida S. A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol. 1983;29(4):319–322. doi: 10.2323/jgam.29.319. [DOI] [Google Scholar]
- 12.Hamada M, Yamamura H, Komukai C, Tamura T, Suzuki K, Hayakawa M. Luteimicrobium album sp. nov., a novel actinobacterium isolated from a lichen collected in Japan, and emended description of the genus Luteimicrobium. J Antibiot. 2012;65(8):427–31. doi: 10.1038/ja.2012.45. [DOI] [PubMed] [Google Scholar]
- 13.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26(5):541–7. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohtsubo Y, Maruyama F, Mitsui H, Nagata Y, Tsuda M. Complete genome sequence of Acidovorax sp. strain KKS102, a polychlorinated-biphenyl degrader. J Bacteriol. 2012;194(24):6970–1. doi: 10.1128/JB.01848-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–64. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komaki H, Ichikawa N, Hosoyama A, Fujita N, Igarashi Y. Draft genome sequence of marine-derived Streptomyces sp. TP-A0598, a producer of anti-MRSA antibiotic lydicamycins. Stand Genomic Sci. 2015;10:58. doi: 10.1186/s40793-015-0046-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. antiSMASH 2.0--a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41(Web Server issue):W204–12. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106(8):3468–96. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 20.Bierbaum G, Gotz F, Peschel A, Kupke T, van de Kamp M, Sahl HG. The biosynthesis of the lantibiotics epidermin, gallidermin, Pep5 and epilancin K7. Antonie Van Leeuwenhoek. 1996;69(2):119–127. doi: 10.1007/BF00399417. [DOI] [PubMed] [Google Scholar]
- 21.Okesli A, Cooper LE, Fogle EJ, van der Donk WA. Nine post-translational modifications during the biosynthesis of cinnamycin. J Am Chem Soc. 2011;133(34):13753–60. doi: 10.1021/ja205783f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichikawa N, Sasagawa M, Yamamoto M, Komaki H, Yoshida Y, Yamazaki S, Fujita N. DoBISCUIT: a database of secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2013;41(Database issue):D408–14. doi: 10.1093/nar/gks1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Lanen SG, Shen B. Biosynthesis of enediyne antitumor antibiotics. Curr Top Med Chem. 2008;8(6):448–59. doi: 10.2174/156802608783955656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87(12):4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garrity GM, Holt JG. The road map to the manual. Second. New York: Springer; 2001. pp. 119–169. [Google Scholar]
- 26.Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol. 1997;47:479–491. doi: 10.1099/00207713-47-2-479. [DOI] [Google Scholar]
- 27.Buchanan RE. Studies in the Nomenclature and Classification of the Bacteria: II. The Primary Subdivisions of the Schizomycetes. J Bacteriol. 1917;2(2):155–64. doi: 10.1128/jb.2.2.155-164.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol. 1980;30:225–420. doi: 10.1099/00207713-30-1-225. [DOI] [PubMed] [Google Scholar]
- 29.Zhi XY, Li WJ, Stackebrandt E. An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int J Syst Evol Microbiol. 2009;59(Pt 3):589–608. doi: 10.1099/ijs.0.65780-0. [DOI] [PubMed] [Google Scholar]
- 30.Kim SB, Lonsdale J, Seong CN, Goodfellow M. Streptacidiphilus gen. nov., acidophilic actinomycetes with wall chemotype I and emendation of the family Streptomycetaceae (Waksman and Henrici (1943)AL) emend. Rainey et al. 1997. Antonie Van Leeuwenhoek. 2003;83(2):107–16. doi: 10.1023/A:1023397724023. [DOI] [PubMed] [Google Scholar]
- 31.Wellington EM, Stackebrandt E, Sanders D, Wolstrup J, Jorgensen NO. Taxonomic status of Kitasatosporia, and proposed unification with Streptomyces on the basis of phenotypic and 16S rRNA analysis and emendation of Streptomyces Waksman and Henrici 1943, 339AL. Int J Syst Bacteriol. 1992;42(1):156–60. doi: 10.1099/00207713-42-1-156. [DOI] [PubMed] [Google Scholar]
- 32.Waksman SA, Henrici AT. The nomenclature and classification of the Actinomycetes. J Bacteriol. 1943;46(4):337–41. doi: 10.1128/jb.46.4.337-341.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Witt D, Stackebrandt E. Unification of the genera Streptoverticillium and Streptomyces, and amendation of Streptomyces Waksman and Henrici 1943, 339 AL. Syst Appl Microbiol. 1990;13:361–371. doi: 10.1016/S0723-2020(11)80234-1. [DOI] [Google Scholar]
- 34.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]


