Abstract
OBJECTIVE
To determine the prevalence of HOXB13 G84E mutation in a population of prostate cancer patients of Ashkenazi Jewish heritage, an ethnic group common to the New York City area.
MATERIALS AND METHODS
A retrospective analysis of germline DNA samples from Ashkenazi Jewish men and a comparison group of non-Ashkenazi men treated for prostate cancer at our institution. Patients were genotyped for G84E using a TaqMan assay (Applied Biosystems). Positive cases were confirmed using Sanger sequencing.
RESULTS
Median age at prostate cancer diagnosis was 68 years for 889 Ashkenazi Jewish patients, 64 years for 920 non-Ashkenazi Jewish patients. The median follow up was 9 years for Ashkenazi Jewish patients and 8.8 years for non-Ashkenazi Jewish patients. Only 4 patients were found to be heterozygous carriers of G84E. They were all of non-Ashkenazi Jewish ancestry and were diagnosed at 70, 66, 78, and 49 years of age. Two of them presented with high-risk prostate cancer. The prevalence of G84E in the non-Ashkenazi sample was 0.4%.
CONCLUSIONS
HOXB13 G84E mutation was not observed in prostate cancer patients of Ashkenazi Jewish ancestry treated at our institution. Screening for G84E, therefore, may be unnecessary in Ashkenazi Jewish men if these results are validated by other studies.
Keywords: Prostatic neoplasms, germline mutation, risk
Introduction
Prostate cancer has a proven hereditary basis; almost one-quarter of prostate cancer cases occur in familial clusters, and 9% can be attributed to cancer traits that segregate in an autosomal dominant Mendelian inheritance pattern (1, 2). Men with first-degree relatives with prostate cancer have a 2–4 times higher risk of developing prostate cancer over the course of their lives.(3) Furthermore, African Americans are at an increased risk of developing prostate cancer independent of socioeconomic factors, indicating that genetic variants may determine the risk of prostate cancer in this group (4).
In a recent study, Ewing et al, sequenced 202 genes in the 17q21–22 region, which has been implicated in prostate cancer susceptibility, in germline DNA of 94 unrelated familial prostate cancer patients selected for linkage to the candidate region (5). Probands from four families were heterozygous for a rare missense mutation (G84E) in HOXB13. The mutation was also found in all 18 men in these four families who had prostate cancer and had DNA available for analysis. Overall, the mutation was found in 72 (1.4%) of the 5,083 men with prostate cancer but only 1 (0.1%) of the 1,401 control subjects (0.1%) (P = 8.5×10−7). Furthermore, the mutation was significantly more prevalent in men with early-onset, familial prostate cancer (3.1%) than in those with late–onset disease and no family history of prostate cancer (0.6%). Subsequent studies by other groups have also reported the prevalence of G84E mutation in familial prostate cancer patients. In a study cohort consisting of 928 familial prostate cancer patients and 930 controls, Breyer et al. found the G84E mutation in 1.9% of probands, with a 2.7% occurrence in patients with more than 3 affected family members (6). Akbari et al. performed a case-control study, sequencing germline DNA from peripheral leukocytes of 1,843 men diagnosed with prostate cancer and 2,225 controls to identify possible mutations in HOXB13. The mutation was more prevalent in Caucasian men than in ethnically matched controls (0.7% vs. 0.1%, P = 0.01) (7). Finally, Karlsson et al. genotyped samples from two population-based, Swedish, case-control studies and found the prevalence of the G84E mutation to be more than 1% in the general Swedish population, which is higher than we and others have reported for controls (around 0.1%) (8).
The aim of this study is to determine the prevalence of HOXB13 G84E mutation in men of Ashkenazi Jewish heritage treated for prostate cancer at Memorial Sloan-Kettering Cancer Center.
Materials and Methods
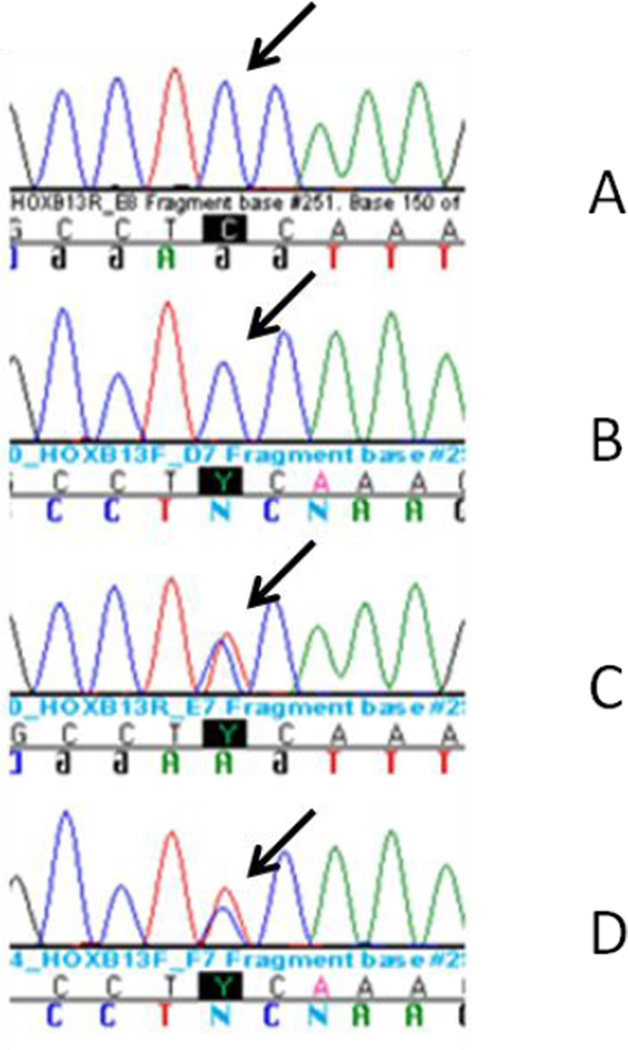
Germline DNA was extracted from blood samples of 889 Ashkenazi Jewish and a comparison group of 920 non-Ashkenazi Jewish men treated for prostate cancer at Memorial Sloan-Kettering Cancer Center between 1990 and 2006. Ancestry was self-reported. DNA was collected through IRB-approved protocol. A TaqMan assay (Applied Biosystems) specific to G84E was designed, and the genotyping was performed by real-time PCR according to the manufacturer’s instructions. Detection was performed on the ABI Prism 7900 using the Sequence Detection Software (Applied Biosystems). Suspected carriers of the SNP were verified through Sanger sequencing (Figure 1). Primer3 software v 0.4.0 (available at http://frodo.wi.mit.edu/) was used to design PCR primers for Sanger validation of samples with the variant, as indicated by the TaqMan assay. PCR was performed using Platinum Taq DNA Polymerase (Invitrogen), and PCR products were cleaned-up using Exo/Sap IT (USB) and sequenced at the MSKCC Genomics Core Laboratory. Sequencher v. 4.8 (Gene Codes) was used for assembling and aligning the reads to the reference sequence (Human genome build 19) obtained from Ensembl (http://www.ensembl.org).
Figure 1.
DNA Sequence Chromatogram Obtained from Germline DNA of Men with Prostate Cancer Carrying Wild Type (A: forward stand, B: reverse strand) or Mutated HOXB13 (C: forward stand, D: reverse strand)
Results
The median age at diagnosis of prostate cancer was 68 and 64 years for Ashkenazi Jewish and non-Ashkenazi Jewish patients, respectively. Of a total of 1,809 prostate cancer patients included in the study, 4 patients were found to be heterozygous for the G84E mutation. None of the patients carrying the mutation were of Ashkenazi Jewish descent, and none had family history of the disease. Those patients were white males 70, 66, 78, and 49 years old at diagnosis. Two presented with high-risk prostate cancer and both died, with one having prostate cancer documented as the cause of death (Table 2). Thus, the prevalence of this mutation is 0% in the Ashkenazi Jewish prostate cancer patients and 0.4% in the non-Ashkenazi Jewish prostate cancer patients in our study.
Table 2.
Patient and tumor characteristics of HOXB13 mutation (rs138213197) heterozygous carriers in 920 non-Ashkenazi sporadic prostate cancer patients treated at Memorial Sloan Kettering-Cancer Center
| Age at diagnosis |
PSA at diagnosis |
Gleason score |
TNM stage | Metastasis | Death from Prostate Cancer |
|---|---|---|---|---|---|
| 70 | < 10 | 4+5 | T3cN0MX | Yes | Unknown |
| 66 | < 10 | 3+3 | T1c | Unknown | Unknown |
| 78 | Unknown | 5+5 | T3aN1M1b | Yes | Yes |
| 49 | >10 | 3+4 | T3aN1M0 | No | No |
Discussion
Prostate cancer has well known hereditary components; however, no high-penetrance susceptibility gene conferring increased risk for this disease has been reported until a rare SNP, G84E in HOXB13, was found to be associated with a 20-fold increase in the risk of prostate cancer among over 5,000 men of European descent.(5) This finding has been validated in several studies.(6)
The Ashkenazi Jewish population, compromising about 80% of Jews worldwide, is known to be a relatively homogeneous genetic group with well-known ancestry-associated mutations. For example, 2.3 percent (120 out of 5,318) of Ashkenazi Jewish individuals were found to carry one of three founder mutations in BRCA1 and BRCA2 (9), five times higher than of the general population (10). Ashkenazi Jews are present in high numbers in the New York City area (about 12% of the population) and well represented in our DNA repository. The availability of samples from this unique population, and the possibility of G84E being a founder mutation in Ashkenazi Jewish men, stimulated this effort to genotype HOXB13 G84E variant in DNA to determine this variant’s prevalence in Ashkenazi Jewish men with prostate cancer. None of the patients genotyped, however, tested positive for G84E.
Previously published results suggest a potential role for genetic testing for G84E in individuals with early-onset prostate cancer or a family history of the disease. It is notable that a single mutation was found, simplifying the development and deployment of a clinical test. However, before testing for G84E is offered outside research studies, critical questions remain to be answered, including: What is the actual risk associated with this mutation? What screening would be recommended and how will it impact clinical care of men with prostate cancer? If these questions are answered, and if it was determined that testing for G84E could change disease management, then Ashkenazi Jewish patients could be saved the anxiety and the expense of testing if our findings are validated by other studies in the same population.
The statistical power to find differences in prevalence is always an issue in small studies like ours. However, considering an approximate prevalence of G84E of 0.6% in prostate cancer patients and 0.1% in controls, we would have had more than 90% power to find carriers of G84E in our study cohort. Another limitation of this study is that genotyped individuals were all from a single referral center in New York City and may not be representative of the population of affected individuals in general. Therefore, further studies will be needed to validate these findings.
Table 1.
Patient and tumor characteristics of 889 Ashkenazi Jewish and 920 non-Ashkenazi sporadic prostate cancer patients treated at Memorial Sloan Kettering-Cancer Center
| Variable | Non-Ashkenazi (%) | Ashkenazi (%) |
|---|---|---|
| Median age on diagnosis | 64 | 68 |
| Race | ||
| White | 93.7 | - |
| Asian | 2.8 | - |
| Black | 1.0 | - |
| Other | 2.5 | - |
| PSA | ||
| <10 | 63.9 | 69.1 |
| 10–20 | 19.0 | 18.6 |
| >20 | 17.1 | 12.3 |
| Gleason Score | ||
| <=6 | 35.3 | 38.4 |
| 7 | 42.1 | 38.7 |
| 8–10 | 22.6 | 22.9 |
| T stage | ||
| T1 | 44.4 | 45.7 |
| T2 | 38.4 | 38.0 |
| T3 | 17.2 | 16.3 |
| Median follow up (Months) | 105 | 108 |
| Death due to prostate cancer | 10.3 | 13.5 |
Acknowledgments
Research funding
Supported by Sabin Family Research Fund and the Sharon Corzine Research Initiative.
Supported in part by funds provided by David H. Koch through the Prostate Cancer Foundation.
Supported by the Sidney Kimmel Center for Prostate and Urologic Cancers.
References
- 1.Carter BS, Bova GS, Beaty TH, Steinberg GD, Childs B, Isaacs WB, et al. Hereditary prostate cancer: epidemiologic and clinical features. J Urol. 1993 Sep;150(3):797–802. doi: 10.1016/s0022-5347(17)35617-3. PubMed PMID: 8345587. Epub 1993/09/01. eng. [DOI] [PubMed] [Google Scholar]
- 2.Gronberg H, Isaacs SD, Smith JR, Carpten JD, Bova GS, Freije D, et al. Characteristics of prostate cancer in families potentially linked to the hereditary prostate cancer 1 (HPC1) locus. JAMA. 1997 Oct 15;278(15):1251–1255. doi: 10.1001/jama.1997.03550150055035. PubMed PMID: 9333266. Epub 1997/10/23. eng. [DOI] [PubMed] [Google Scholar]
- 3.Zeegers MP, Jellema A, Ostrer H. Empiric risk of prostate carcinoma for relatives of patients with prostate carcinoma: a meta-analysis. Cancer. 2003 Apr 15;97(8):1894–903. doi: 10.1002/cncr.11262. PubMed PMID: 12673715. Epub 2003/04/04. eng. [DOI] [PubMed] [Google Scholar]
- 4.Haas GP, Sakr WA. Epidemiology of prostate cancer. CA Cancer J Clin. 1997 Sep-Oct;47(5):273–287. doi: 10.3322/canjclin.47.5.273. PubMed PMID: 9314822. Epub 1997/10/07. eng. [DOI] [PubMed] [Google Scholar]
- 5.Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, et al. Germline mutations in HOXB13 and prostate-cancer risk. The New England journal of medicine. 2012 Jan 12;366(2):141–149. doi: 10.1056/NEJMoa1110000. PubMed PMID: 22236224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breyer JP, Avritt TG, McReynolds KM, Dupont WD, Smith JR. Confirmation of the HOXB13 G84E germline mutation in familial prostate cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012 Aug;21(8):1348–1353. doi: 10.1158/1055-9965.EPI-12-0495. PubMed PMID: 22714738. Pubmed Central PMCID: PMC3415588. Epub 2012/06/21. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akbari MR, Trachtenberg J, Lee J, Tam S, Bristow R, Loblaw A, et al. Association Between Germline HOXB13 G84E Mutation and Risk of Prostate Cancer. Journal of the National Cancer Institute. 2012 Aug 1;104(16):1260–1262. doi: 10.1093/jnci/djs288. PubMed PMID: 22781434. Epub 2012/07/12. eng. [DOI] [PubMed] [Google Scholar]
- 8.Karlsson R, Aly M, Clements M, Zheng L, Adolfsson J, Xu J, et al. A Population-based Assessment of Germline HOXB13 G84E Mutation and Prostate Cancer Risk. European urology. 2012 Jul 20; doi: 10.1016/j.eururo.2012.07.027. PubMed PMID: 22841674. Epub 2012/07/31. Eng. [DOI] [PubMed] [Google Scholar]
- 9.Struewing JP, Hartge P, Wacholder S, Baker SM, Berlin M, McAdams M, et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med. 1997 May 15;336(20):1401–1408. doi: 10.1056/NEJM199705153362001. PubMed PMID: 9145676. Epub 1997/05/15. eng. [DOI] [PubMed] [Google Scholar]
- 10.Warner E, Foulkes W, Goodwin P, Meschino W, Blondal J, Paterson C, et al. Prevalence and penetrance of BRCA1 and BRCA2 gene mutations in unselected Ashkenazi Jewish women with breast cancer. J Natl Cancer Inst. 1999 Jul 21;91(14):1241–1247. doi: 10.1093/jnci/91.14.1241. PubMed PMID: 10413426. Epub 1999/07/21. eng. [DOI] [PubMed] [Google Scholar]