Abstract
Background
Renal cell carcinoma (RCC) is a seventh ranked malignancy with poor prognosis. RCC is lethal at metastatic stage as it does not respond to conventional systemic treatments, and there is an urgent need to find out promising novel biomarkers for effective treatment. The goal of this study was to evaluate the biomarkers that can be potential therapeutic target and predict effective inhibitors to treat the metastatic stage of RCC.
Methods
We conducted transcriptomic profiling to identify differentially expressed genes associated with RCC. Molecular pathway analysis was done to identify the canonical pathways and their role in RCC. Tissue microarrays (TMA) based immunohistochemical stains were used to validate the protein expression of cyclinD1 (CCND1) and were scored semi-quantitatively from 0 to 3+ on the basis of absence or presence of staining intensity in the tumor cell. Statistical analysis determined the association of CCND1 expression with RCC. Molecular docking analyses were performed to check the potential of two natural inhibitors, rutin and curcumin to bind CCND1.
Results
We detected 1490 significantly expressed genes (1034, upregulated and 456, downregulated) in RCC using cutoff fold change 2 and p value < 0.05. Hes-related family bHLH transcription factor with YRPW motif 1 (HEY1), neuropilin 2 (NRP2), lymphoid enhancer-binding factor 1 (LEF1), and histone cluster 1 H3h (HIST1H3H) were most upregulated while aldolase B, fructose-bisphosphate (ALDOB), solute carrier family 12 (SLC12A1), calbindin 1 (CALB1) were the most down regulated genes in our dataset. Functional analysis revealed Wnt/β-catenin signaling as the significantly activated canonical pathway (z score = 2.53) involving cyclin D1 (CCND1). CCND1 was overexpressed in transcriptomic studies (FC = 2.26, p value = 0.0047) and TMA results also showed the positive expression of CCND1 in 53 % (73/139) of RCC cases. The ligands – rutin and curcumin bounded with CCND1 with good affinity.
Conclusion
CCND1 was one of the important upregulated gene identified in microarray and validated by TMA. Docking study showed that CCND1 may act as a potential therapeutic target and its inhibition could focus on the migratory, invasive, and metastatic potential of RCC. Further in vivo and in vitro molecular studies are needed to investigate the therapeutic target potential of CCND1 for RCC treatment.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-016-2775-2) contains supplementary material, which is available to authorized users.
Keywords: Renal cell carcinoma, Cyclin D1, Gene expression profiling, Tissue microarray, Molecular docking, Therapeutic target, Saudi Arabia
Background
Renal Cell carcinoma (RCC) is a major health problem and accounts for approximately 1.5 percent of all cancer deaths [1, 2]. It accounts for about 3 % of all cancers and 2-3 % per year increase in global incidence [1, 3]. For RCC treatment, surgery is the best option at advance stage, however, one third of patients develop metastases even after surgery [4]. At metastatic stage, prognosis is very poor because RCC patients hardly respond to conventional existing systemic treatments and leads to death [5]. RCC treatment is a big challenge without identification of new drug targets and effective remedies. Although previous studies have reported role of gene alterations, their expression and deregulation of molecular signals to be linked with cancer initiation and progression, there still lack of curative therapy for RCC [6–8]. Therefore, identification of a potential drug target and prediction of suitable ligand is crucial for the patients with RCC.
The cyclin D members (D1, D2 and D3) bind to CDKs and are required for the hematopoietic cells proliferation and survival and perform a rate-limiting antiapoptotic function in vivo [9]. Cyclin D1 (CCND1) overexpression is predominantly correlated with early cancer onset, tumor progression, shorter cancer patient survival and increased metastases [10–12]. Induction of vascular endothelial growth factor (VEGF) production by CCND1 promotes oncogenesis by increasing growth and angiogenesis, while downregulation of death receptor, Fas by CCND1 causes chemotherapeutic and apoptosis resistance [13]. Overexpression of CCND1 has been previously reported in many cancers including lung cancers [14], esophageal squamous cell carcinoma [15], head and neck cancer [16], pancreatic cancer [17], pituitary cancer [18], and breast cancer [19].
CCND1 is a proto-oncogene and a good biomarker for tumor progression, found to be deregulated in several cancers, including RCC. CCND1 along with associated cyclins activates cyclin-dependent kinases (CDKs) - CDK4 and CDK6. G1-S phase transition during cell cycle, requires phosphorylation of retinoblastoma (Rb) by CDK4 and CDK6. Hyperphosphorylation of Rb allows expression of genes involved in DNA replication and cell division [20–23]. The ability of CCND1 to exhibit oncogenic property and to regulate a critical G1-S transition checkpoint by activating CDK4/CDK6, makes it a potential therapeutic target of RCC [24–28].
Alternative or synergistic anticancer therapies using natural compounds and their derivatives (polyphenols, flavonoids, alkaloids, saponins, etc.) have been extensively studied [29]. Rutin is a flavonol glycoside found in many plants, including buckwheat; tobacco; asparagus, green tea etc. and contributes to the antibacterial [30], hepatoprotective [31], neuroprotective [32] and antioxidant [33] properties of the plant. It is structurally very similar to quercitrin and has been used therapeutically to decrease capillary fragility, to protect blood capillaries, and as ingredients of multivitamin nutritional supplements and alternative herbal remedies. It can attach to iron ion, thereby averting its binding to H2O2 and free radical generation. In addition, rutin acts as an angiogenesis inhibitor and can stall the VEGF in vitro; also has potential anticancerous and antiproliferative property [34, 35].
Curcumin commonly known as turmeric is a phytopolylphenol pigment isolated from the plant Curcuma longa, and possesses a variety of pharmacologic properties like anti-inflammatory, antineoplastic, antiproliferative, anticancer, apoptosis inducer, chempreventive [36, 37]. It can inhibit the reactive-oxygen species formation, cyclooxygenases (COX) and other metabolic enzymes involved in inflammation; and can disrupt cell signal transduction via inhibition of protein kinase C. It can interact with myriad of biomolecules by covalent and non-covalent binding. The H-bonding and hydrophobic interactions, arising from the aromatic and tautomeric structures in addition to the flexible linker group owe for the non-covalent interactions [38]. Curcumin reportedly suppress cyclin D1 expression by promoting proteolysis and down-regulating its expression and causes inhibition of CDK4-mediated phosphorylation of retinoblastoma protein [39]. It has been reported that curcumin-treated cells show decreased expression of CCND1, resulting in low cell growth rate. This curcumin-induced CCND1 mRNA down-regulation is perhaps mediated by induction of BTG2 as well as inhibition of nuclear translocation of NF-kappaB [40].
In this study, expression profiling of RCC (CEGMR data) identified 1490 significantly differentially expressed genes and molecular pathway analysis predicted alteration in many important cancer related pathways. However, the major finding of this study was identification and tissue microarray based validation of CCND1 as important over-expressed gene/proteins of RCC. Overexpression of CCND1 can trigger cancer by activating many pathways, including Wnt/β-catenin signaling pathway and has been shown to exhibit oncogenic property, making it a potential therapeutic target. We, therefore, attempted docking study to show the therapeutic potential of anticancerous natural ligands (rutin and curcumin) against the identified potential drug target (CCND1).
Methods
Patients and samples
The study was executed on RCC patients from Saudi Arabia and resected tissue samples were collected from collaborating hospitals of Jeddah during the period 2010–2014. For gene expression analysis, fresh surgically resected tumor and normal tissue were collected and stored in RNALater (Invitrogen/Life Technologies, NY, USA) till RNA extraction. All patients included in the present study were Saudi in origin and diagnosed with clear cell or chromophobe renal cell carcinoma without any prior chemotherapy or radiotherapy exposure.
Ethical approval
Local ethical committee has approved this study (08-CEGMR-02-ETH). Patients were included in the present study only after their prior consent.
RNA extraction and array processing
Qiagen RNeasy Mini Kit (Qiagen, Hilden, Germany) was used to extract total RNA from fresh kidney tissue, Nano Drop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) was used for concentration determination and RNA quality was checked with Bioanalyzer (Agilent Technologies, CA, USA). Out of 20 specimen, only 7 tumor and 5 control samples passed the selection criteria of RNA integrity number (RIN) >5 and were judged fit to be used for array expression analysis. We used Human Gene 1.0 ST GeneChip arrays (Affymetrix, Santa Clara, CA, USA) for transcriptomics studies (Life Technology, Grand Island, NY), interrogating 764,885 probes and 36,079 annotated reference sequences (NCBI build 36). We processed 250 ng RNA of 12 samples using the Ambion WT Expression Kit (Life Technologies, Austin, TX), GeneChip Hybridization, Wash and Stain Kit (Affymetrix, Santa Clara, CA) and GeneChip WT Terminal Labeling and Controls Kit (Affymetrix, Santa Clara, CA). The hybridization of 5500 ng of cDNA was done in a hybridization oven at 45 °C under rotation (60 rpm) for 17 h. After complete processing, the arrays were scanned in the GeneChip Scanner 3000 7G and GeneChip Command Console Software (AGCC) were used to generate probe cell intensity data (CEL files).
Gene expression analysis
We carried transcriptomic profiling of 12 samples, seven RCC and five normal kidney tissues. To gain confidence with our limited number samples, we performed a comparative analysis with independent expression datasets from NCBI’s GEO database (GSE781, n = 34; GSE7023, n = 47; and GSE6344, n = 40) for confirmation. Affymetrix. CEL files were imported and analyzed using Partek Genomics Suite version 6.6 (Partek Inc., MO, USA). Default settings robust multi-chip averaged (RMA) was used to log-transform data set and for normalization. Analysis of Variance (ANOVA) was applied, and differentially expressed genes (DEGs) were identified with cut off fold change > 2 and p value <0.05. Principal component analysis (PCA) was performed to assess overall expression pattern among sample groups, similar samples were grouped together.
Tissue microarray and immunohistochemistry
Tissue microarrays (TMA) were designed and constructed for 139 primary RCC and 34 normal kidney tissue as previously described [41]. Experienced pathologist reviewed hematoxylin and eosin (HE) slides of RCC and normal kidney tissue. 1.5 mm tissue cores from areas of interest were chosen from donor block(s) and transferred to recipient paraffin block of TMA Master 1.14 SP3 (3D Histech Ltd, Budapest, Hungary). HE staining of TMA slides was repeated to assess basic morphology of slide construction.
Immunohistochemical studies were performed on positive-charged leica plus slides (Leica Microsystems, Wetzler, Germany) mounted with 4 μm of TMA paraffin blocks. Deparaffinisation of sections was done using xylene, followed by rehydration in an automated BenchMark XT immunostainer (Ventana® Medical systems Inc., Tucson, AZ, USA) and pretreatment in prediluted cell conditioning 1 (CC1) solution for an hour. Immunostaining of TMA slides was done by incubating anti-CCND1 antibody at 37 °C for 16 min, followed by washing, counterstaining (with Mayer’s hematoxylin) and mounting using Ventana® Ultraview Universal DAB detection kit. For analysis and interpretation both negative (with tris-buffered saline only) and positive (with primary antibody) control slides were used. Sections were evaluated independently by the pathologist without knowing the clinicopathological characteristics of RCC patients. Immunostainings were scored semiquantitatively from 0 to 4 + .
Functional and pathway analysis
We performed pathway analyses and Gene ontology (GO) studies for differentially regulated genes in RCC to find associated biological networks and molecular processes, using Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems, Redwood City, CA). Significantly expressed genes with Affymetrix ID, expression level and p-value were uploaded into IPA software to identify the most significant altered biological functions and networks. Fisher’s exact test was used to calculate the significance of association between trancriptomic data and canonical pathways of RCC.
Molecular docking studies
The 3-D crystal structure of cyclin D1 was retrieved from RCSB’s Protein Data Bank (PDB) – PDB id: 2w96: Chain A. Structure visualization and illustration was done using PyMol (DeLano Scientific) (Fig. 1). The molecular structure of rutin and curcumin were retrieved from NCBI’s PubChem compound database with CID 5280805 and 969516 respectively (Fig. 2).
Fig. 1.
Molecular structure of rutin and curcumin retrieved from NCBI’s PubChem compound database with CID 5280805 and 969516
Fig. 2.
Overall cyclin D1 structure depicted as ribbon diagram (PDB: 2 W96)
Molecular docking was performed using Molecular Docking Server on [42]. The MMFF94 force field geometry optimization method was used for energy minimization of ligand molecule: rutin and curcumin using DockingServer. Gasteiger partial charges were added to the ligand atoms at pH 7.0. Non-polar hydrogen atoms were merged, and rotatable bonds were defined. Rest methodology was followed in sequential manner as previously described [2, 6, 43].
Supporting data availability
Data series (Accession No. GSE781, GSE7023, GSE6344) used in present study are available at NCBI’s Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/).
Results
This study focused on utilizing transcriptomic profiling to identify biomarkers associated with RCC and conducting molecular docking analysis to assess the interactions between potential target and drugs. We identified CCND1 as important overexpressed gene/proteins of RCC and demonstrated its potential as possible anticancer drug target.
Identification of differentially expressed genes
Three-dimensional scatter plot of PCA demonstrated that RCC and control tissues are distinctly clustered (Fig. 3). We did genome-wide transcription profiling of fresh RCC specimens and identified 1490 differentially expressed genes; 1034 up-regulated and 456 down-regulated using unadjusted p value < 0.05 (Additional file 1). Number of differentially expressed genes reduced to 141 (22 up-regulated and 119 down regulated) on applying the stringent condition of false discovery rate with p value < 0.05 while keeping all other above parameter same (Fig. 4, Table 1). Hes-related family bHLH transcription factor with YRPW motif 1 (HEY1), neuropilin 2 (NRP2), lymphoid enhancer-binding factor 1 (LEF1), and histone cluster 1 H3h (HIST1H3H) were the most upregulated ones while aldolase B, fructose-bisphosphate (ALDOB), solute carrier family 12 (SLC12A1), calbindin 1 (CALB1) were the most down regulated genes in our dataset. We compared our identified differentially expressed genes list with re-analyzed GEO data series (GSE781, GSE6344 and GSE7023) and identified over-expression of CCND1 in all dataset, thus supporting our result (Table 2).
Fig. 3.
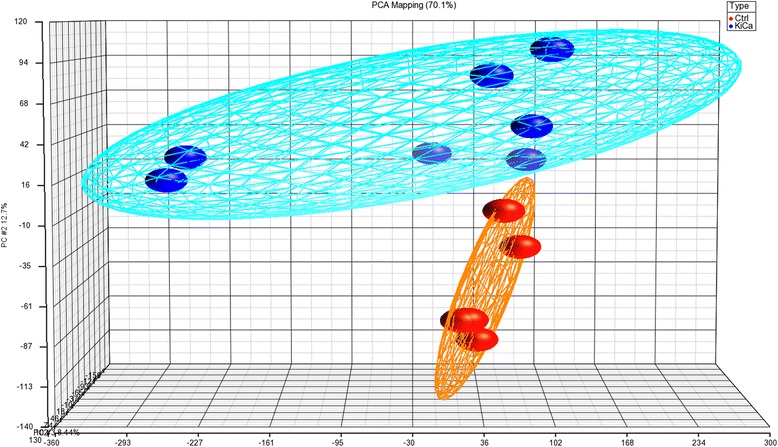
Scatter plot of PCA show grouping of similar type based on genome-wide expression values, as represented as eclipse, where each ball represents one sample. Blue and red is representing RCC and normal kidney tissue
Fig. 4.
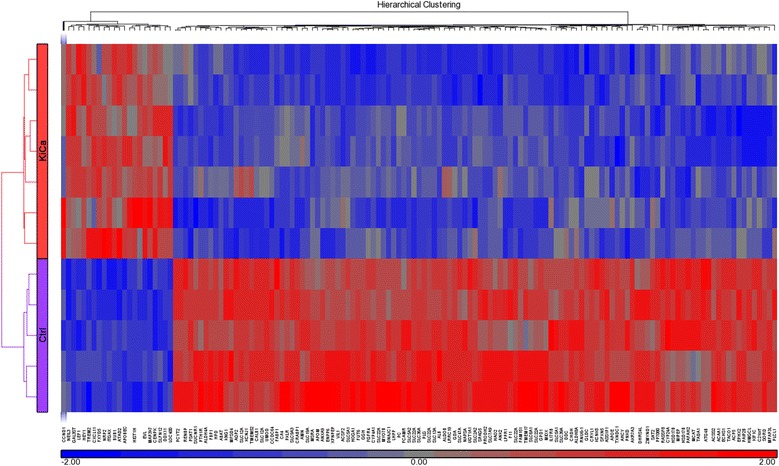
Hierarchical clustering and functional analysis of significantly differentially expressed genes in kidney cancer using Affymetrix Human ST 1.0 array and Partek Genomics suite (ver 6.6)
Table 1.
Differentially expressed significant genes in RCC
| Gene symbol | Gene name | RefSeq | p-value | Fold-change |
|---|---|---|---|---|
| HEY1 | hes-related family bHLH transcription factor with YRPW motif 1 | NM_001040708 | 7.88E-06 | 3.64128 |
| NRP2 | neuropilin 2 | ENST00000272849 | 0.00017 | 3.63215 |
| LEF1 | lymphoid enhancer-binding factor 1 | NM_001130713 | 4.04E-05 | 3.54448 |
| HIST1H3H | histone cluster 1, H3h | NM_003536 | 3.29E-05 | 2.87948 |
| ITGAX | integrin, alpha X (complement component 3 receptor 4 subunit) | NM_000887 | 3.23E-05 | 2.69367 |
| BUB1 | BUB1 mitotic checkpoint serine | NM_001278616 | 3.35E-05 | 2.62163 |
| MAP3K7CL | MAP3K7 C-terminal like | NM_001286620 | 2.81E-05 | 2.54523 |
| FXYD5 | FXYD domain containing ion transport regulator 5 | NM_001164605 | 0.000158 | 2.45618 |
| HIST1H2AI | histone cluster 1, H2ai | NM_003509 | 0.000108 | 2.27918 |
| CENPK | centromere protein K | NM_001267038 | 6.01E-05 | 2.27502 |
| CCND1 | cyclin D1 | NM_053056 | 0.004789 | 2.25898 |
| DDX11 | DEAD | NM_001257144 | 0.000178 | 2.24119 |
| APOBEC3D | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-li | NM_152426 | 2.61E-06 | 2.23905 |
| ATAD2 | ATPase family, AAA domain containing 2 | NM_014109 | 0.000129 | 2.18905 |
| HIST1H3F | histone cluster 1, H3f | NM_021018 | 0.00016 | 2.14196 |
| TREM2 | triggering receptor expressed on myeloid cells 2 | NM_018965 | 0.00021 | 2.13092 |
| GAL3ST4 | galactose-3-O-sulfotransferase 4 | NM_024637 | 1.50E-05 | 2.11736 |
| LOC400464 | uncharacterized LOC400464 | AK127420 | 9.33E-05 | 2.11281 |
| CXCL11 | chemokine (C-X-C motif) ligand 11 | NM_005409 | 0.0002 | 2.07422 |
| NEIL3 | nei endonuclease VIII-like 3 (E. coli) | NM_018248 | 2.73E-05 | 2.06874 |
| EVL | Enah | ENST00000553771 | 5.57E-05 | 2.06506 |
| SLFN12L | schlafen family member 12-like | ENST00000361112 | 9.07E-05 | 2.02832 |
| ALDH4A1 | aldehyde dehydrogenase 4 family, member A1 | NM_001161504 | 4.94E-07 | −16.1809 |
| SLC22A12 | solute carrier family 22 (organic anion | NM_001276326 | 9.50E-06 | −16.4997 |
| SLC47A2 | solute carrier family 47 (multidrug and toxin extrusion), me | NM_001099646 | 5.60E-06 | −17.2214 |
| HAO2 | hydroxyacid oxidase 2 (long chain) | NM_001005783 | 0.000192 | −17.3322 |
| SLC6A19 | solute carrier family 6 (neutral amino acid transporter), me | NM_001003841 | 0.000195 | −18.4585 |
| XPNPEP2 | X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound | NM_003399 | 0.000131 | −18.9386 |
| CYP4A11 | cytochrome P450, family 4, subfamily A, polypeptide 11 | XR_246241 | 7.59E-05 | −19.2237 |
| SLC22A6 | solute carrier family 22 (organic anion transporter), member 6 | NM_004790 | 5.33E-06 | −20.6522 |
| KCNJ1 | potassium inwardly-rectifying channel, subfamily J, member 1 | NM_000220 | 8.79E-05 | −22.4359 |
| TMEM52B | transmembrane protein 52B | NM_001079815 | 0.000112 | −23.1374 |
| SLC12A3 | solute carrier family 12 (sodium | NM_000339 | 1.14E-06 | −27.7638 |
| HPD | 4-hydroxyphenylpyruvate dioxygenase | NM_001171993 | 1.88E-05 | −29.7949 |
| SLC5A12 | solute carrier family 5 (sodium | XM_006718157 | 0.000155 | −30.9613 |
| KNG1 | kininogen 1 | NM_000893 | 4.28E-06 | −34.7144 |
| SLC13A3 | solute carrier family 13 (sodium-dependent dicarboxylate tra | NM_001011554 | 1.93E-06 | −37.6483 |
| SLC36A2 | solute carrier family 36 (proton | NM_181776 | 4.06E-07 | −37.8957 |
| PLG | plasminogen | NM_000301 | 4.87E-07 | −43.0535 |
| SLC22A8 | solute carrier family 22 (organic anion transporter), member | NM_001184732 | 4.50E-06 | −45.8474 |
| UMOD | uromodulin | NM_001008389 | 2.44E-06 | −68.9599 |
| CALB1 | calbindin 1, 28 kDa | NM_004929 | 2.29E-06 | −78.3947 |
| SLC12A1 | solute carrier family 12 | ENST00000330289 | 0.000135 | −79.6698 |
| ALDOB | aldolase B, fructose-bisphosphate | NM_000035 | 2.56E-05 | −87.9122 |
Negative fold change value indicates the downregulation
*bold data shows CCND1 (Cyclin D1) was overexpressed (fold change = 2.258) and statistically significant (p-value = 0.00478)
Table 2.
Expression of CCND1 in Saudi RCC patients (CEGMR dataset) and GEO dataset
Validation of CCND1
Transcriptomic profiling revealed distinct CCND1 overexpression (FC = 2.26, p value = 0.0047). Validation study based on TMA-immunohistochemistry staining showed the positive expression of CCND1 in 53 % (73/139) of RCC cases (Fig. 5).
Fig. 5.
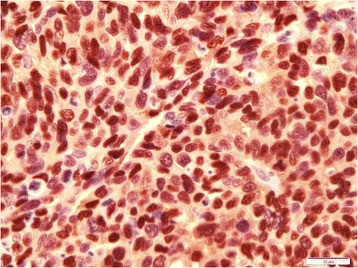
Immunohistochemistry stain for CCND1 show positive staining in RCC (original magnification × 60)
Pathways and networks underlying RCC
Pathway analysis of identified DEGs revealed the biofunctions, molecular network and canonical pathways association with RCC (Table 3). Most significantly inhibited pathways were synaptic long term potentiation (z-score = −2.33), NRF2-mediated oxidative stress response (z-score = −2.33), production of nitric oxide and reactive oxygen species in macrophages (z-score = −2.324), and renin-angiotensin signaling (z-score = −2.121). Wnt/β-catenin signaling was significantly activated pathway (z-score = 2.53) involving following genes; cyclin D1 (CCND1, FC = 2.26), CD44 molecule (CD44, FC = 2.31), v-myc avian myelocytomatosis viral oncogene homolog (c-Myc, FC = 2.31), HNF1 homeobox A (TCF1, FC = −2.26), secreted frizzled-related protein 1 (SFRP1, FC = −4.45) (Fig. 6). We found over expression of CCND1 playing important role in regulation of Wnt/β-catenin signaling along with other cancer related pathways like Acute Myeloid Leukemia Signaling, Non-Small Cell Lung Cancer Signaling, PTEN Signaling, Regulation of Cellular Mechanics by Calpain Protease, ErbB2-ErbB3 Signaling, HER-2 Signaling in Breast Cancer, HER-2 Signaling in Breast Cancer, Thyroid Cancer Signaling, Endometrial Cancer Signaling etc. Further extensive molecular pathway analysis may help to better understand the mechanism of RCC initiation, invasion and metastasis.
Table 3.
Canonical pathways predicted by Ingenuity Pathway Analysis for significant genes differentially expressed in kidney cancer
| Ingenuity canonical pathways | -log (p-value) | z-score | Molecules |
|---|---|---|---|
| Wnt/β-catenin Signaling | 0.271 | 2.530 | CSNK1E,MYC,PPP2R4,TGFBR3,CD44,LEF1,SFRP1,UBC,CCND1,HNF1A,ACVR2A,LRP1 |
| Synaptic Long Term Potentiation | 0.481 | −2.333 | PLCB4,PPP1R1A,PPP1R3C,PPP3R1,PRKAR2A,CACNA1C,PLCL1,PLCD4,PRKCZ,PRKCA |
| NRF2-mediated Oxidative Stress Response | 1.5 | −2.333 | GSTA3,AKR7A2,AKR7A3,GSTM1,GSTM3,NQO2,ABCC2,NQO1,DNAJC19,SOD1,PRKCZ,DNAJC11,AKR1A1,SCARB1,FMO1,GSTA1,AOX1,TXN,PRKCA,EPHX1 |
| Production of Nitric Oxide and Reactive Oxygen Species in Macrophages | 0.512 | −2.324 | PPARA,MAP3K15,APOE,APOM,PPP1R3C,PRKCZ,APOL1,ALB,PPP2R4,CYBA,APOC1,CHUK,APOD,RBP4,PRKCA |
| Sperm Motility | 1.19 | −2.309 | PLA2G16,SLC16A10,PLCB4,PLA2R1,PRKAR2A,PNPLA3,PLCL1,PLA2G12B,PDE1A,PLCD4,PLA2G7,PRKCZ,PRKCA |
| Renin-Angiotensin Signaling | 0.279 | −2.121 | ADCY9,GRB2,REN,PRKAR2A,CCL5,PRKCZ,AGT,PRKCA |
| Nitric Oxide Signaling in the Cardiovascular System | 0.27 | −1.890 | KNG1,CAV1,PRKAR2A,CACNA1C,PDE1A,PRKCZ,PRKCA |
| Antioxidant Action of Vitamin C | 3.01 | 1.897 | PLA2G16,NAPEPLD,PLA2R1,SLC23A3,PLA2G7,GLRX,SLC2A3,PLCB4,SLC23A1,SLC2A2,PNPLA3,CHUK,TXN,PLA2G12B,PLCL1,PLCD4 |
| Aldosterone Signaling in Epithelial Cells | 1.34 | −1.897 | DNAJC12,DNAJC19,PDPK1,HSPD1,HSPA2,PRKCZ,HSPA12A,DNAJC11,PLCB4,SCNN1G,SLC12A1,CRYAA/LOC102724652,SCNN1B,PLCL1,PLCD4,PRKCA,AHCY |
| Valine Degradation I | 10.11 | NaN | ECHS1,ABAT,ACADSB,BCKDHB,BCAT1,HIBCH,HIBADH,AUH,DLD,DBT,EHHADH,HADHA,ALDH6A1 |
| Ethanol Degradation II | 9.4 | NaN | HSD17B10,ADH6,ALDH1B1,ALDH4A1,ACSS1,ALDH9A1,ADH5,ALDH2,AKR1A1,ALDH3A2,ACSS2,ADHFE1,ACSL1,ALDH7A1,DHRS4 |
| Fatty Acid β-oxidation I | 9.4 | NaN | HSD17B10,ECHS1,SLC27A2,ACAA1,ACAA2,SCP2,ECI2,AUH,ACSL4,IVD,EHHADH,ACADM,HADHA,ACSL1,HADH |
| FXR/RXR Activation | 9.17 | NaN | PPARA,KNG1,APOE,PKLR,APOH,ABCC2,SLC22A7,HNF1A,CYP8B1,MTTP,PCK2,SCARB1,SLC10A2,FGFR4,LPL,GC,AGT,APOM,SDC1,UGT2B4,CYP27A1,SERPINF2,APOL1,ALB,FABP6,APOC1,FBP1,G6PC,SLC51B,RBP4,APOD |
| Serotonin Degradation | 8.46 | NaN | ADH6,HSD17B10,ALDH4A1,ALDH1B1,UGT3A1,UGT2B4,UGT2B7,UGT1A1,ALDH9A1,ADH5,ALDH2,AKR1A1,SMOX,ALDH3A2,ADHFE1,DHRS4,ALDH7A1,MAOA |
| Noradrenaline and Adrenaline Degradation | 7.86 | NaN | ADH6,HSD17B10,ALDH4A1,ALDH1B1,ALDH9A1,ADH5,ALDH2,AKR1A1,SMOX,ALDH3A2,ADHFE1,DHRS4,ALDH7A1,MAOA |
| Tryptophan Degradation | 7.34 | NaN | ALDH4A1,ALDH1B1,ALDH2,AKR1A1,SMOX,ALDH3A2,DDC,ALDH9A1,ALDH7A1,MAOA |
| PXR/RXR Activation | 4.04 | NaN | PPARA,SCD,GSTM1,ABCB1,ABCC2,PRKAR2A,CES2,HMGCS2,UGT1A1,PCK2,ALDH3A2,GSTA1,G6PC,CYP2B6 |
| Acute Myeloid Leukemia Signaling | 0.491 | 0.816 | RUNX1,MYC,GRB2,LEF1,CCND1,HNF1A,IDH1 |
| Non-Small Cell Lung Cancer Signaling | 0.481 | −0.816 | GRB2,PDPK1,EGF,ERBB2,CCND1,PRKCA |
| PTEN Signaling | 0.662 | 0.905 | FGFR3,GRB2,FGFR4,TGFBR3,PREX2,ITGA5,FGFR2,PDPK1,CHUK,CCND1,PRKCZ |
| Regulation of Cellular Mechanics by Calpain Protease | 0.453 | −1.000 | GRB2,ITGA5,EGF,CCND1,ACTN1 |
| ErbB2-ErbB3 Signaling | 0.967 | −1.890 | MYC,GRB2,NRG3,PDPK1,ERBB3,ERBB2,CCND1 |
| Aryl Hydrocarbon Receptor Signaling | 2.07 | NaN | GSTA3,GSTM1,ALDH4A1,ALDH1B1,GSTM3,NQO2,NQO1,ALDH8A1,CCND1,ALDH9A1,MYC,ALDH1L1,ALDH1L2,ALDH3A2,GSTA1,ALDH5A1,ALDH6A1,ALDH7A1 |
| HER-2 Signaling in Breast Cancer | 1.01 | NaN | GRB2,PARD6B,EGF,ERBB3,ITGB8,ERBB2,CCND1,PRKCZ,PRKCA |
| Thyroid Cancer Signaling | 0.828 | NaN | CXCL10,MYC,LEF1,CCND1,HNF1A |
| Endometrial Cancer Signaling | 0.765 | NaN | MYC,GRB2,PDPK1,LEF1,ERBB2,CCND1 |
| Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis | 0.615 | NaN | CXCL8,FN1,IL1RL1,CXCL12,CCL5,HNF1A,CCND1,FCGR1A,PRKCZ,C5,MYC,PLCB4,F2RL1,PPP3R1,LEF1,CHUK,SFRP1,PLCL1,PLCD4,FCGR3A/FCGR3B,TNFSF13B,LRP1,PRKCA,ADAMTS4 |
| Estrogen-mediated S-phase Entry | 0.278 | NaN | MYC,CCND1 |
*bold data shows presence and importance of CCND1 among identified canonical pathways
Fig. 6.
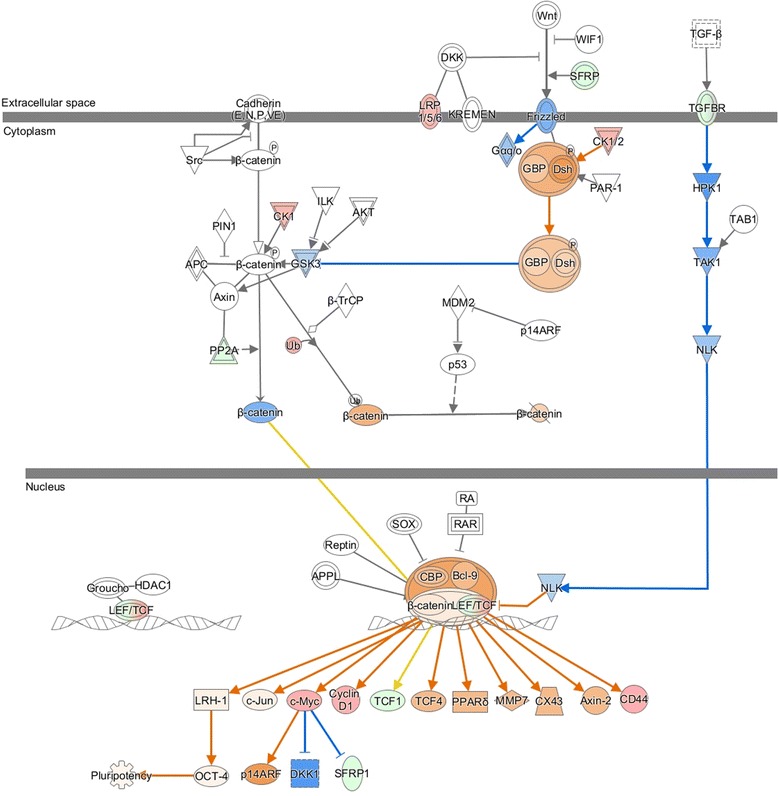
Wnt-β catenin signaling was significantly activated (z-score = 2.53) in RCC based on following differentially expressed genes: cyclin D1 (CCND1, FC = 2.25898), CD44 molecule (CD44, FC = 2.31667), v-myc avian myelocytomatosis viral oncogene homolog (c-Myc, FC = 2.31324), HNF1 homeobox A (TCF1, FC = −2.26661), secreted frizzled-related protein 1 (SFRP1, FC = −4.45838). Red represents overexpression and green underexpression
Docking studies
We made a structural attempt to study possible binding of two natural famed ligands with the potential therapeutic drug target, Cyclin D1 for cancer therapeutics. CCND1 protein has a classical double cyclin box domain fold, comprising of 11 alpha-helices [44].
Molecular docking studies predicted good interactions between three dimensional structure of drug target (CCND1, PDBID: 2w96) and selected ligands; rutin and curcumin. Molecular docking revealed that both the compounds are able to bind in the ligand binding domain. In silico docking studies revealed interaction of two active compounds with the common vital ligand binding site residues (Leu91, Lys149, Asn151) of cyclin D1. Both rutin and curcumin docked at a common ligand binding site of CCND1 slightly varied intensity as estimated by their size, structure, stereochemistry (Figs. 7 and 8; Table 4). We also examined their complete interaction profile including hydrogen bonds, HB plot, polar, hydrophobic, pi-pi and cation-pi interactions. The estimated free energy of binding with Cyclin D1 for rutin was −4.26 kcal/mol and for curcumin was −4.67 kcal/mol which is very similar, however, the estimated inhibition constant (Ki) was 757.57 μM and 380.02 μM respectively.
Fig. 7.
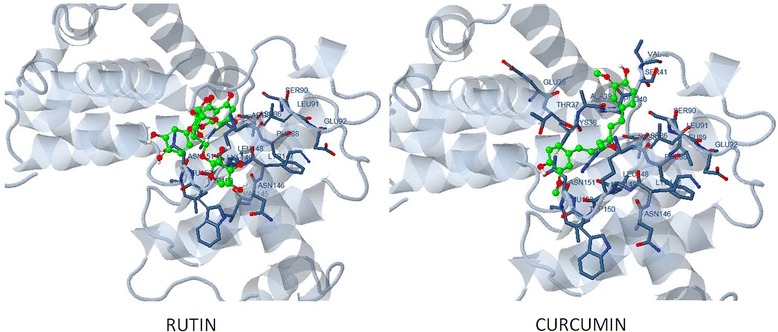
Molecular docking conformation and interactions of rutin and curcumin with Cyclin D1
Fig. 8.
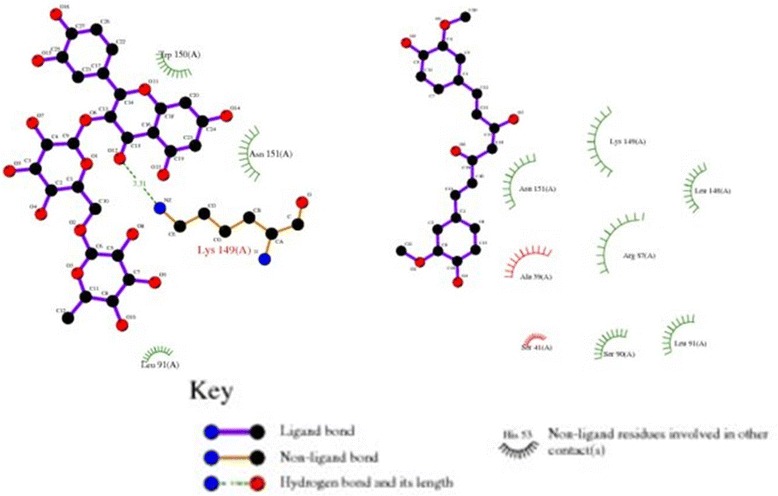
Two-dimensional plot showing the primary interacting residues of Cyclin D1
Table 4.
Docking features and values for rutin and curcumin
| Features | RUTIN | CURCUMIN |
|---|---|---|
| Est. Free energy of binding | −4.26 kcal/mol | −4.67 kcal/mol |
| Est. Inhibition Constant, Ki | 757.57 μM | 380.02 μM |
| vdW + Hbond + desolv Energy | −5.43 kcal/mol | −6.37 kcal/mol |
| Electrostatic energy | −0.01 kcal/mol | −0.12 kcal/mol |
| Total intermolecular energy | −5.44 kcal/mol | −6.49 kcal/mol |
| Interaction surface | 653.668 | 684.416 |
Discussion
RCC is a complex heterogeneous tumors involving altered genes and proteins. We performed a transcriptional profiling and functional analysis of RCC to understand the role of identified significant genes in regulation of physiological processes through biological pathways/networks. We found CCND1 as one of the significantly expressed gene and potential biomarker RCC.
HEY1, an upregulated gene, has been reported to be mediator of notch signaling, showing pro-oncogenic function and promotes cancer progression [45, 46]. Neuropilin-2 (Nrp2) is a well known receptor for the vascular endothelial growth factor-C (VEGF-C) and activates lymph nodes as well as promotes tumor metastasis by lymphangiogenesis [47, 48]. LEF1 interacts with β-catenin and plays critical role in proliferation of RCC by activating downstream target genes [49, 50]. Wnt/β-catenin signaling, found activated, regulates embryonic development and is involved in many diseases including cancer, polycystic kidney disease [51–54]. WNT signal and its paracrine mode to growth of cancer cells makes it clinically important to understand the metastasis of tumor cells [53, 55, 56]. HIST1H3H is frequently altered chromatin factors in many cancers [57, 58]. Aldolase, a family member of glycolysis enzymes, was found to be significantly affecting RCC. Aldolase-A was reportedly upregulated while aldolase-B was downregulated in RCC and human primary hepatocellular carcinoma [59–62]. SLC12 family members are involved in regulation of cell volume, blood pressure and chloride concentration, and play a critical role in diseases like cancer, epilepsy and osteoporosis [63]. In the present study, SLC12 was down regulated that is in accordance to other findings [64]. CALB1 is reported to be altered in RCC and found to be negatively stained compared to normal tissue [61, 65].
CCND1 was overexpressed in our as well as other transcriptomics studies [66–69]. We validated CCND1 overexpression by using tissue microarray platform and in silico docking analysis was done to check its therapeutic potential as it plays a key role in G1-S phase transition of cell cycle. There are reports of anti-proliferative, apoptosis inducing and chemopreventive effects of natural bioactive flavonoids like baicalein, catechin, genistein, quercetin, and rutin. Docking analysis showed that rutin and curcumin binds to CCND1 and can potentially inhibit downstream CCND1/CDK4/CDK6 complex formation, required for G1-S phase transition. Our finding demonstrate the anticancer drug targets potential of CCND1 and rutin and curcumin as potential inhibitors, however, this in silico docking study has to be validated further.
Conclusion
Our microarray and immunohistochemistry results suggest significantly high levels of cyclin D1 expression in RCC. Distinct transcriptomic signatures identified for RCC needs verification at larger dataset and additional significant genes need to be further validated for identification of novel biomarkers. The critical role of CCND1 in RCC metastasis by activating G1-S transition of cell cycle has drawn our attention to examine its potential as anticancer drug target. Our in silico docking study shown CCND1 protein as an attractive anticancer target and natural flavanoids rutin and curcumin as potential anticancer drug of RCC and they may be promising in the prevention of kidney cancer too. Quantitative structure-activity relationship studies, ligand binding, efficacy and toxicity should be further investigated before clinical trials. Clinical and therapeutic applications of these natural ligands were initially limited by their low solubility and bioavailability but combination with adjuvant and nano-technology based delivery vehicles can immensely improve their potential. Moreover, these are reported to act in synergism with several other natural compounds or synthetic agents routinely used in chemotherapy and can assist in cancer prevention and treatment when used alone or in combination with other conventional treatments.
Acknowledgment
This project and publication was supported by the NSTIP strategic technologies program in the Kingdom of Saudi Arabia – Project No (10-BIO1258-03, 09-BIO1073-03, 08-MED120-03). The authors also acknowledge with thanks Science and Technology Unit, King Abdulaziz University for technical support. Authors would like to acknowledge the Deanship of Scientific Studies, King Abdulaziz University, Jeddah, Saudi Arabia for funding the research (HiCi-1434-117-2). We also thank the patients, physicians, nurses, and pathologists of the King Abdulaziz University Hospital, and King Faisal Specialist Hospital and Research Center, Jeddah, Saudi Arabia.
Declaration
This article has been published as part of BMC Cancer Volume 16 Supplement 2, 2016: Proceedings of the 3rd International Genomic Medicine Conference: cancer. The full contents of the supplement are available online at http://bmccancer.biomedcentral.com/articles/supplements/volume-16-supplement-2.
Availability of data and material
Data series (Accession No. GSE781, GSE7023, GSE6344) used in present study are available atNCBI’s Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/).
Authors’ contributions
SK, JM and HF participated in the study design. HJS, AAA, FA and MS performed data collection, DNA extraction and microarray studies. AB and JM did tissue microarray, immunohistochemistry and pathological studies. SK, ZM and HN analyzed data, interpreted the results and drafted the manuscript. KA, AMA, AC, AS and MHQ participated in critical review, editing and finalization of manuscript. All authors have read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Ethical approval
Local ethical committee has approved this study (08-CEGMR-02-ETH). Patients were included in the present study only after their prior consent.
Ethical approval information is included in the materials and methods section. Local ethical committee has approved this study under the approval number 08-CEGMR-02-ETH.
Consent for publication
Patients’ consent information are included in the material and methods section. Patients were only included in the present study after their prior consent.
Additional file
Differentially expressed genes of RCC from Saudi patients on comparing RCC with normal kidney tissue. Transcriptomics profiling revealed 1490 differentially expressed genes, 1034 up-regulated and 456 down-regulated, with fold change ≥ 2 and p-value < 0.05. (XLSX 209 kb)
Contributor Information
Sajjad Karim, Phone: +966 12 6401000, Email: skarim1@kau.edu.sa, Email: sajjad_k_2000@yahoo.com.
Mohammed H. Al-Qahtani, Phone: +966 12 6401000, Email: mhalqahtani@kau.edu.sa
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Mirza Z, Schulten HJ, Farsi HM, Al-Maghrabi JA, Gari MA, Chaudhary AG, Abuzenadah AM, Al-Qahtani MH, Karim S. Molecular interaction of a kinase inhibitor midostaurin with anticancer drug targets, S100A8 and EGFR: transcriptional profiling and molecular docking study for kidney cancer therapeutics. PLoS One. 2015;10(3):e0119765. doi: 10.1371/journal.pone.0119765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatt JR, Finelli A. Landmarks in the diagnosis and treatment of renal cell carcinoma. Nat Rev Urol. 2014;11(9):517–25. doi: 10.1038/nrurol.2014.194. [DOI] [PubMed] [Google Scholar]
- 4.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373(9669):1119–32. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 5.Xue S, Li QW, Che JP, Guo Y, Yang FQ, Zheng JH. Decreased expression of long non-coding RNA NBAT-1 is associated with poor prognosis in patients with clear cell renal cell carcinoma. Int J Clin Exp Pathol. 2015;8(4):3765–74. [PMC free article] [PubMed] [Google Scholar]
- 6.Mirza Z, Schulten HJ, Farsi HM, Al-Maghrabi JA, Gari MA, Chaudhary AG, Abuzenadah AM, Al-Qahtani MH, Karim S. Impact of S100A8 expression on kidney cancer progression and molecular docking studies for kidney cancer therapeutics. Anticancer Res. 2014;34(4):1873–84. [PubMed] [Google Scholar]
- 7.Merdad A, Karim S, Schulten HJ, Jayapal M, Dallol A, Buhmeida A, Al-Thubaity F, Gari IM, Chaudhary AG, Abuzenadah AM, et al. Transcriptomics profiling study of breast cancer from kingdom of Saudi Arabia revealed altered expression of adiponectin and fatty acid binding Protein4: is lipid metabolism associated with breast cancer? BMC Genomics. 2015;16 Suppl 1:S11. doi: 10.1186/1471-2164-16-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Choi YJ, Saez B, Anders L, Hydbring P, Stefano J, Bacon NA, Cook C, Kalaszczynska I, Signoretti S, Young RA, et al. D-cyclins repress apoptosis in hematopoietic cells by controlling death receptor Fas and its ligand FasL. Dev Cell. 2014;30(3):255–67. doi: 10.1016/j.devcel.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diehl JA. Cycling to cancer with cyclin D1. Cancer Biol Ther. 2002;1(3):226–31. doi: 10.4161/cbt.72. [DOI] [PubMed] [Google Scholar]
- 11.Jares P, Colomer D, Campo E. Genetic and molecular pathogenesis of mantle cell lymphoma: perspectives for new targeted therapeutics. Nat Rev Cancer. 2007;7(10):750–62. doi: 10.1038/nrc2230. [DOI] [PubMed] [Google Scholar]
- 12.Thomas GR, Nadiminti H, Regalado J. Molecular predictors of clinical outcome in patients with head and neck squamous cell carcinoma. Int J Exp Pathol. 2005;86(6):347–63. doi: 10.1111/j.0959-9673.2005.00447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shintani M, Okazaki A, Masuda T, Kawada M, Ishizuka M, Doki Y, Weinstein IB, Imoto M. Overexpression of cyclin DI contributes to malignant properties of esophageal tumor cells by increasing VEGF production and decreasing Fas expression. Anticancer Res. 2002;22(2A):639–47. [PubMed] [Google Scholar]
- 14.Yamanouchi H, Furihata M, Fujita J, Murakami H, Yoshinouchi T, Takahara J, Ohtsuki Y. Expression of cyclin E and cyclin D1 in non-small cell lung cancers. Lung Cancer (Amsterdam, Netherlands) 2001;31(1):3–8. doi: 10.1016/S0169-5002(00)00160-4. [DOI] [PubMed] [Google Scholar]
- 15.Ikeguchi M, Sakatani T, Ueta T, Kaibara N. Cyclin D1 expression and retinoblastoma gene protein (pRB) expression in esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2001;127(9):531–6. doi: 10.1007/s004320100265. [DOI] [PubMed] [Google Scholar]
- 16.Izzo JG, Papadimitrakopoulou VA, Li XQ, Ibarguen H, Lee JS, Ro JY, El-Naggar A, Hong WK, Hittelman WN. Dysregulated cyclin D1 expression early in head and neck tumorigenesis: in vivo evidence for an association with subsequent gene amplification. Oncogene. 1998;17(18):2313–22. doi: 10.1038/sj.onc.1202153. [DOI] [PubMed] [Google Scholar]
- 17.Gansauge S, Gansauge F, Ramadani M, Stobbe H, Rau B, Harada N, Beger HG. Overexpression of cyclin D1 in human pancreatic carcinoma is associated with poor prognosis. Cancer Res. 1997;57(9):1634–7. [PubMed] [Google Scholar]
- 18.Simpson DJ, Frost SJ, Bicknell JE, Broome JC, McNicol AM, Clayton RN, Farrell WE. Aberrant expression of G(1)/S regulators is a frequent event in sporadic pituitary adenomas. Carcinogenesis. 2001;22(8):1149–54. doi: 10.1093/carcin/22.8.1149. [DOI] [PubMed] [Google Scholar]
- 19.Arnold A, Papanikolaou A. Cyclin D1 in breast cancer pathogenesis. J Clin Oncol. 2005;23(18):4215–24. doi: 10.1200/JCO.2005.05.064. [DOI] [PubMed] [Google Scholar]
- 20.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3(1):11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 21.Mundle SD, Saberwal G. Evolving intricacies and implications of E2F1 regulation. FASEB J. 2003;17(6):569–74. doi: 10.1096/fj.02-0431rev. [DOI] [PubMed] [Google Scholar]
- 22.Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10(7):699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 23.Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14(19):2393–409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 24.Ho A, Dowdy SF. Regulation of G(1) cell-cycle progression by oncogenes and tumor suppressor genes. Curr Opin Genet Dev. 2002;12(1):47–52. doi: 10.1016/S0959-437X(01)00263-5. [DOI] [PubMed] [Google Scholar]
- 25.Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol. 2006;24(11):1770–83. doi: 10.1200/JCO.2005.03.7689. [DOI] [PubMed] [Google Scholar]
- 26.Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov. 2009;8(7):547–66. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- 27.Amin HM, McDonnell TJ, Medeiros LJ, Rassidakis GZ, Leventaki V, O’Connor SL, Keating MJ, Lai R. Characterization of 4 mantle cell lymphoma cell lines. Arch Pathol Lab Med. 2003;127(4):424–31. doi: 10.5858/2003-127-0424-COMCLC. [DOI] [PubMed] [Google Scholar]
- 28.Musgrove EA. Cyclins: roles in mitogenic signaling and oncogenic transformation. Growth Factors (Chur, Switzerland) 2006;24(1):13–9. doi: 10.1080/08977190500361812. [DOI] [PubMed] [Google Scholar]
- 29.Ali R, Mirza Z, Ashraf GM, Kamal MA, Ansari SA, Damanhouri GA, Abuzenadah AM, Chaudhary AG, Sheikh IA. New anticancer agents: recent developments in tumor therapy. Anticancer Res. 2012;32(7):2999–3005. [PubMed] [Google Scholar]
- 30.van der Watt E, Pretorius JC. Purification and identification of active antibacterial components in Carpobrotus edulis L. J Ethnopharmacol. 2001;76(1):87–91. doi: 10.1016/S0378-8741(01)00197-0. [DOI] [PubMed] [Google Scholar]
- 31.Domitrović R, Jakovac H, Vasiljev Marchesi V, Vladimir-Knežević S, Cvijanović O, Tadić Ž, Romić Ž, Rahelić D. Differential hepatoprotective mechanisms of rutin and quercetin in CCl(4)-intoxicated BALB/cN mice. Acta Pharmacol Sin. 2012;33(10):1260–70. doi: 10.1038/aps.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hao G, Dong Y, Huo R, Wen K, Zhang Y, Liang G. Rutin inhibits neuroinflammation and provides neuroprotection in an experimental rat model of subarachnoid hemorrhage, possibly through suppressing the RAGE-NF-kappaB inflammatory signaling pathway. Neurochem Res. 2016. [DOI] [PubMed]
- 33.Shahid A, Ali R, Ali N, Kazim Hasan S, Rashid S, Majed F, Sultana S. Attenuation of genotoxicity, oxidative stress, apoptosis and inflammation by rutin in benzo(a)pyrene exposed lungs of mice: plausible role of NF-kappaB, TNF-alpha and Bcl-2. Journal of Complementary & Integrative Medicine. 2016. [DOI] [PubMed]
- 34.Luo H, Jiang BH, King SM, Chen YC. Inhibition of cell growth and VEGF expression in ovarian cancer cells by flavonoids. Nutr Cancer. 2008;60(6):800–9. doi: 10.1080/01635580802100851. [DOI] [PubMed] [Google Scholar]
- 35.Yu SH, Yu JM, Yoo HJ, Lee SJ, Kang DH, Cho YJ, Kim DM. Anti-proliferative effects of rutin on OLETF Rat vascular smooth muscle cells stimulated by glucose variability. Yonsei Med J. 2016;57(2):373–81. doi: 10.3349/ymj.2016.57.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng AL, Hsu CH, Lin JK, Hsu MM, Ho YF, Shen TS, Ko JY, Lin JT, Lin BR, Ming-Shiang W, et al. Phase I clinical trial of curcumin, a chemopreventive agent, in patients with high-risk or pre-malignant lesions. Anticancer Res. 2001;21(4B):2895–900. [PubMed] [Google Scholar]
- 37.Goel A, Kunnumakkara AB, Aggarwal BB. Curcumin as “Curecumin”: from kitchen to clinic. Biochem Pharmacol. 2008;75(4):787–809. doi: 10.1016/j.bcp.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 38.Priyadarsini KI. Chemical and structural features influencing the biological activity of curcumin. Curr Pharm Des. 2013;19(11):2093–100. doi: 10.2174/138161213805289228. [DOI] [PubMed] [Google Scholar]
- 39.Mukhopadhyay A, Banerjee S, Stafford LJ, Xia C, Liu M, Aggarwal BB. Curcumin-induced suppression of cell proliferation correlates with down-regulation of cyclin D1 expression and CDK4-mediated retinoblastoma protein phosphorylation. Oncogene. 2002;21(57):8852–61. doi: 10.1038/sj.onc.1206048. [DOI] [PubMed] [Google Scholar]
- 40.Kwon YK, Jun JM, Shin SW, Cho JW, Suh SI. Curcumin decreases cell proliferation rates through BTG2-mediated cyclin D1 down-regulation in U937 cells. Int J Oncol. 2005;26(6):1597–603. [PubMed] [Google Scholar]
- 41.Gomaa W, Ke Y, Fujii H, Helliwell T. Tissue microarray of head and neck squamous carcinoma: validation of the methodology for the study of cutaneous fatty acid-binding protein, vascular endothelial growth factor, involucrin and Ki-67. Virchows Arch. 2005;447(4):701–9. doi: 10.1007/s00428-005-0002-7. [DOI] [PubMed] [Google Scholar]
- 42.Bikadi Z, Hazai E. Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J Cheminformatics. 2009;1:15. doi: 10.1186/1758-2946-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gueguen N, Desquiret-Dumas V, Leman G, Chupin S, Baron S, Nivet-Antoine V, Vessières E, Ayer A, Henrion D, Lenaers G, et al. Resveratrol directly binds to mitochondrial complex I and increases oxidative stress in brain mitochondria of aged mice. PLoS One. 2015;10(12):e0144290. doi: 10.1371/journal.pone.0144290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Day PJ, Cleasby A, Tickle IJ, O’Reilly M, Coyle JE, Holding FP, McMenamin RL, Yon J, Chopra R, Lengauer C, et al. Crystal structure of human CDK4 in complex with a D-type cyclin. Proc Natl Acad Sci U S A. 2009;106(11):4166–70. doi: 10.1073/pnas.0809645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alana L, Sese M, Canovas V, Punyal Y, Fernandez Y, Abasolo I, de Torres I, Ruiz C, Espinosa L, Bigas A, et al. Prostate tumor OVerexpressed-1 (PTOV1) down-regulates HES1 and HEY1 notch targets genes and promotes prostate cancer progression. Mol Cancer. 2014;13:74. doi: 10.1186/1476-4598-13-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belandia B, Powell SM, Garcia-Pedrero JM, Walker MM, Bevan CL, Parker MG. Hey1, a mediator of notch signaling, is an androgen receptor corepressor. Mol Cell Biol. 2005;25(4):1425–36. doi: 10.1128/MCB.25.4.1425-1436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yasuoka H, Kodama R, Tsujimoto M, Yoshidome K, Akamatsu H, Nakahara M, Inagaki M, Sanke T, Nakamura Y. Neuropilin-2 expression in breast cancer: correlation with lymph node metastasis, poor prognosis, and regulation of CXCR4 expression. BMC Cancer. 2009;9:220. doi: 10.1186/1471-2407-9-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caunt M, Mak J, Liang WC, Stawicki S, Pan Q, Tong RK, Kowalski J, Ho C, Reslan HB, Ross J, et al. Blocking neuropilin-2 function inhibits tumor cell metastasis. Cancer Cell. 2008;13(4):331–42. doi: 10.1016/j.ccr.2008.01.029. [DOI] [PubMed] [Google Scholar]
- 49.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25(57):7469–81. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 50.Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res. 2007;13(14):4042–5. doi: 10.1158/1078-0432.CCR-06-2316. [DOI] [PubMed] [Google Scholar]
- 51.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149(6):1192–205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 52.Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, Levy DB, Smith KJ, Preisinger AC, Hedge P, McKechnie D, et al. Identification of FAP locus genes from chromosome 5q21. Science (New York, NY) 1991;253(5020):661–5. doi: 10.1126/science.1651562. [DOI] [PubMed] [Google Scholar]
- 53.Rodova M, Islam MR, Maser RL, Calvet JP. The polycystic kidney disease-1 promoter is a target of the beta-catenin/T-cell factor pathway. J Biol Chem. 2002;277(33):29577–83. doi: 10.1074/jbc.M203570200. [DOI] [PubMed] [Google Scholar]
- 54.Rubinfeld B, Souza B, Albert I, Muller O, Chamberlain SH, Masiarz FR, Munemitsu S, Polakis P. Association of the APC gene product with beta-catenin. Science (New York, NY) 1993;262(5140):1731–4. doi: 10.1126/science.8259518. [DOI] [PubMed] [Google Scholar]
- 55.Malanchi I, Huelsken J. Cancer stem cells: never Wnt away from the niche. Curr Opin Oncol. 2009;21(1):41–6. doi: 10.1097/CCO.0b013e32831d1faf. [DOI] [PubMed] [Google Scholar]
- 56.Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, Gerald WL, Massague J. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138(1):51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coulombe-Huntington J, Lam KC, Dias C, Majewski J. Fine-scale variation and genetic determinants of alternative splicing across individuals. PLoS Genet. 2009;5(12):e1000766. doi: 10.1371/journal.pgen.1000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L, Sun L, Huang J, Jiang M. Cyclin-dependent kinase inhibitor 3 (CDKN3) novel cell cycle computational network between human non-malignancy associated hepatitis/cirrhosis and hepatocellular carcinoma (HCC) transformation. Cell Prolif. 2011;44(3):291–9. doi: 10.1111/j.1365-2184.2011.00752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Unwin RD, Craven RA, Harnden P, Hanrahan S, Totty N, Knowles M, Eardley I, Selby PJ, Banks RE. Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics. 2003;3(8):1620–32. doi: 10.1002/pmic.200300464. [DOI] [PubMed] [Google Scholar]
- 60.Mathupala SP, Rempel A, Pedersen PL. Aberrant glycolytic metabolism of cancer cells: a remarkable coordination of genetic, transcriptional, post-translational, and mutational events that lead to a critical role for type II hexokinase. J Bioenerg Biomembr. 1997;29(4):339–43. doi: 10.1023/A:1022494613613. [DOI] [PubMed] [Google Scholar]
- 61.Perroud B, Lee J, Valkova N, Dhirapong A, Lin PY, Fiehn O, Kultz D, Weiss RH. Pathway analysis of kidney cancer using proteomics and metabolic profiling. Mol Cancer. 2006;5:64. doi: 10.1186/1476-4598-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song H, Xia SL, Liao C, Li YL, Wang YF, Li TP, Zhao MJ. Genes encoding Pir51, Beclin 1, RbAp48 and aldolase b are up or down-regulated in human primary hepatocellular carcinoma. World J Gastroenterol. 2004;10(4):509–13. doi: 10.3748/wjg.v10.i4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arroyo JP, Kahle KT, Gamba G. The SLC12 family of electroneutral cation-coupled chloride cotransporters. Mol Aspects Med. 2013;34(2–3):288–98. doi: 10.1016/j.mam.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 64.Skubitz KM, Skubitz AP. Differential gene expression in renal-cell cancer. J Lab Clin Med. 2002;140(1):52–64. doi: 10.1067/mlc.2002.125213. [DOI] [PubMed] [Google Scholar]
- 65.Cross SS, Hamdy FC, Deloulme JC, Rehman I. Expression of S100 proteins in normal human tissues and common cancers using tissue microarrays: S100A6, S100A8, S100A9 and S100A11 are all overexpressed in common cancers. Histopathology. 2005;46(3):256–69. doi: 10.1111/j.1365-2559.2005.02097.x. [DOI] [PubMed] [Google Scholar]
- 66.Lenburg ME, Liou LS, Gerry NP, Frampton GM, Cohen HT, Christman MF. Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data. BMC Cancer. 2003;3:31. doi: 10.1186/1471-2407-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13(16):4740–9. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 68.Tun HW, Marlow LA, von Roemeling CA, Cooper SJ, Kreinest P, Wu K, Luxon BA, Sinha M, Anastasiadis PZ, Copland JA. Pathway signature and cellular differentiation in clear cell renal cell carcinoma. PLoS One. 2010;5(5):e10696. doi: 10.1371/journal.pone.0010696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Furge KA, Chen J, Koeman J, Swiatek P, Dykema K, Lucin K, Kahnoski R, Yang XJ, Teh BT. Detection of DNA copy number changes and oncogenic signaling abnormalities from gene expression data reveals MYC activation in high-grade papillary renal cell carcinoma. Cancer Res. 2007;67(7):3171–6. doi: 10.1158/0008-5472.CAN-06-4571. [DOI] [PubMed] [Google Scholar]


