Summary
Vitamin K-dependent proteins require carboxylation of certain glutamates for their biological functions. The enzymes involved in the vitamin K-dependent carboxylation include: gamma-glutamyl carboxylase (GGCX), vitamin K epoxide reductase (VKOR) and an as-yet-unidentified vitamin K reductase (VKR). Due to the hydrophobicity of vitamin K, these enzymes are likely to be integral membrane proteins that reside in the endoplasmic reticulum. Therefore, structure-function studies on these enzymes have been challenging, and some of the results are notably controversial. Patients with naturally occurring mutations in these enzymes, who mainly exhibit bleeding disorders or are resistant to oral anticoagulant treatment, provide valuable information for the functional study of the vitamin K cycle enzymes. In this review, we discuss: (i) the discovery of the enzymatic activities and gene identifications of the vitamin K cycle enzymes; (ii) the identification of their functionally important regions and their active site residues; (iii) the membrane topology studies of GGCX and VKOR; and (iv) the controversial issues regarding the structure and function studies of these enzymes, particularly, the membrane topology, the role of the conserved cysteines and the mechanism of active site regeneration of VKOR. We also discuss the possibility that a paralogous protein of VKOR, VKOR-like 1 (VKORL1), is involved in the vitamin K cycle, and the importance of and possible approaches for identifying the unknown VKR. Overall, we describe the accomplishments and the remaining questions in regard to the structure and function studies of the enzymes in the vitamin K cycle.
Keywords: gamma-glutamyl carboxylase, integral membrane proteins, vitamin K, vitamin K reductase, VKORC1 protein
Introduction
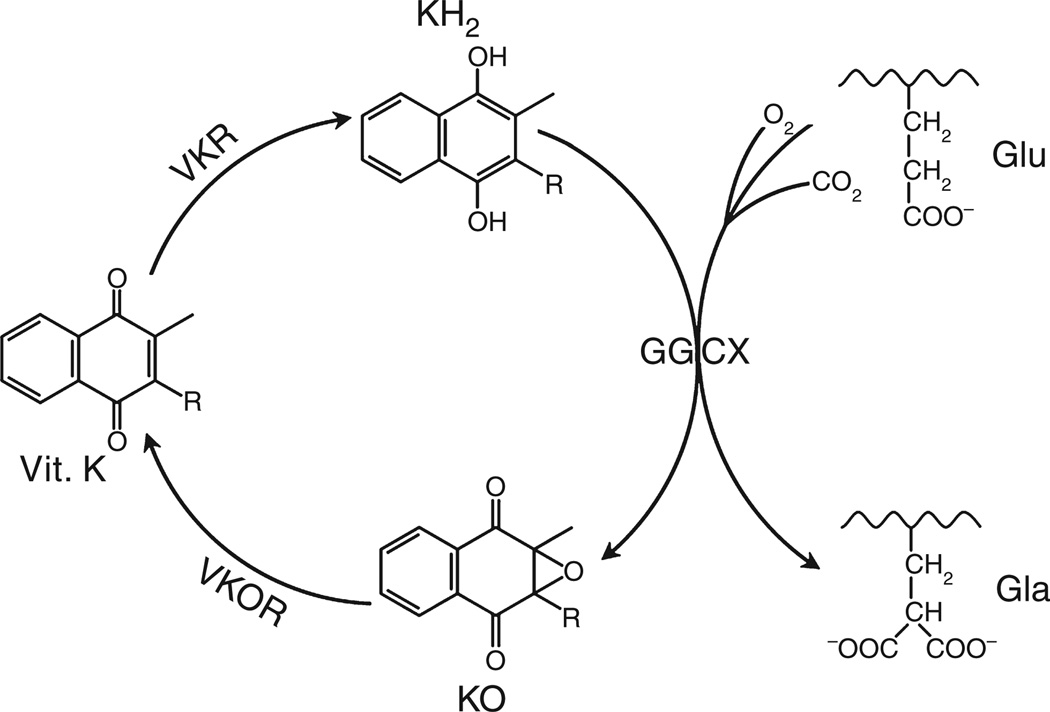
Vitamin K-dependent carboxylation is an essential post-translational modification that converts specific glutamate residues to gamma-carboxyglutamate (Gla) residues in vitamin K-dependent proteins. It is required for the biological function of numerous vitamin K-dependent proteins, including clotting factors (such as factor (F) II, FVII, FIX and FX) and natural anticoagulants (such as protein C, protein S and protein Z). Carboxylation is catalyzed by the enzyme gamma-glutamyl carboxylase (GGCX), which utilizes a reduced form of vitamin K (KH2), carbon dioxide and oxygen as cofactors. Concomitant with each glutamate modification, KH2 is oxidized to vitamin K 2,3-epoxide (KO). KO is converted back to KH2 through a two-step reduction (first to vitamin K, then to KH2) using the enzymes vitamin K epoxide reductase (VKOR) and vitamin K reductase (VKR) in a pathway known as the vitamin K cycle (Fig. 1).
Fig. 1.
Vitamin K cycle. During vitamin K-dependent carboxylation, glutamate (Glu) is converted to gamma-carboxyglutamte (Gla) by gamma-glutamyl carboxylase (GGCX) using a reduced form of vitamin K (KH2), carbon dioxide, and oxygen as cofactors. KH2 is oxidized to vitamin K epoxide (KO). KO is reduced to vitamin K by vitamin K epoxide reductase (VKOR). The reduction of vitamin K to KH2 is carried out by VKOR and an as-yet-unidentified VKR.
Vitamin K-dependent carboxylation was originally observed in clotting factors [1,2]. In all clotting factors, 10 to 12 Gla residues are located in a homologous amino-terminal region referred to as the Gla domain [3]. The multiple Gla residues in this domain adopt a calcium-dependent conformation that promotes clotting factors binding to a membrane surface [4], such as damaged vascular endothelial cells or activated platelets, which allows the localization of clotting factors near the site of vascular injury to either promote or regulate clotting.
With the discovery of new Gla proteins, vitamin K-dependent carboxylation has been implicated in a number of biological functions beyond coagulation. Osteocalcin is a Gla protein produced by osteoblasts and is important for bone formation [5]. Recent studies suggest that osteocalcin also functions as a hormone affecting glucose metabolism in mice [6,7]; whether this is the case in humans still needs to be elucidated [8,9]. The second Gla protein found in bone is matrix Gla protein (MGP), which functions as a strong inhibitor of vascular calcification and connective tissue mineralization [10]. Defects of MGP carboxylation have been associated with cardiovascular diseases and pseudoxanthoma elasticum (PXE) syndrome [11,12]. In addition, MGP has been described as a critical regulator of endothelial cell function, regulating both physiological and tumor-related angiogenesis [13]. Other vitamin K-dependent proteins that are not involved in coagulation include Gas6 (growth arrest-specific protein 6), PRGPs (proline-rich Gla proteins) and TMGs (transmembrane Gla proteins). The metabolic significance of most of these non-coagulation Gla proteins is still poorly understood. A better understanding of the structure-function relationship of the enzymes in the vitamin K cycle will help us to better understand and control a variety of biological processes.
Structure-function studies of GGCX
GGCX protein purification and gene discovery
The enzymatic activity of GGCX was first discovered in the 1970s, in the post-mitochondrial supernatant of hepatocytes [14]. Since then, a number of groups have attempted to purify this integral membrane protein using different approaches; these include salt precipitation, immuno-adsorption and propeptide-based affinity chromatography [15–17]. Knowing that GGCX interacts with the precursor of clotting factors, mainly through an 18-amino acid propeptide, Hubbard et al. [16] attached the propeptide of prothrombin to Sepharose and affinity-purified GGCX from bovine liver microsomes. They found that carboxylation activity in their purified GGCX was 10 000-fold more enriched as compared with the crude microsomes. However, the specific activity of their purified enzyme was still not significantly different to that in the previous partially purified GGCX [18]. A 59-residue peptide containing the propeptide and Gla domain of FIX (FIXQ/S) was later shown to be an excellent affinity ligand for GGCX purification [19]. Using FIXQ/S as the ligand, GGCX was purified up to 90% purity from bovine liver and had an apparent molecular weight of 94 kDa in reduced SDS-PAGE analysis.
With purified bovine GGCX, tryptic peptides were sequenced and used for the PCR amplification of nucleotide fragments in order to screen a bovine liver cDNA library. The longest bovine cDNA insert was used to identify the gene of GGCX from a human liver cDNA library [15]. The identified human GGCX cDNA contains an open reading frame of 2274 nucleotides encoding 758 amino acids. The GGCX gene is located at chromosome 2 at position p12, spans about 13 kb, and contains 15 exons [20,21].
Functional regions and critical residues in GGCX
Since the purification of GGCX and the cloning of its gene, significant progress has been made in understanding how GGCX interacts with its substrates and achieves catalysis [22]. GGCX is a dual-function enzyme with multiple substrates. It recognizes its protein substrate through a relatively tight binding to the propeptide of vitamin K-dependent proteins, which tethers the substrate to the enzyme [23]. During the process of carboxylation, the γ-hydrogen of the glutamates of the substrate is abstracted, followed by the addition of CO2 [24]. Simultaneously, GGCX oxidizes KH2 to KO to provide the energy required for the carboxylation [25]. Therefore, the functional regions and critical residues in GGCX include a propeptide binding site, a glutamate binding site, a vitamin K binding site, a carboxylation active site, an epoxidation active site and, possibly, a CO2 binding site.
The binding of the propeptide to GGCX initiates a structural reorientation of GGCX by which the Gla domain of the substrate is positioned at the catalytic site of GGCX [26,27]. Because GGCX is an integral membrane protein with limited structural information, identification of its functional regions has been very challenging and confusing. Using peptide-based affinity labeling and site-directed mutagenesis analysis, the propeptide binding site and the carboxylation active site were located within the N-terminal of GGCX between residues 1 and 225 [28,29]. However, results from limited trypsin digestion and site-directed mutagenesis studies suggest that the propeptide binding site was located at the C-terminal of GGCX, between residues 495 and 513 [30,31]. Characterization of the naturally occurring W501S GGCX mutation suggests that the major effect of this mutation is to decrease the affinity of GGCX for the propeptide binding [32]. Recently, GGCX was incorporated as single molecules into nanodiscs, which provide a native-like membrane environment; the assembled GGCX complexes were structurally stable and catalytically active [26]. Hydrogen/deuterium exchange mass spectrometry was used to characterize specific regions of GGCX that exhibited structural rearrangements upon binding to propeptides. One of the most pronounced enhancements in the deuterium exchange was observed in peptides 491–507. Together, these results suggest that the propeptide binding region is located in the C-terminal of GGCX.
Identification of GGCX’s glutamate binding site came primarily from the characterization of naturally occurring GGCX mutations and their neighboring conserved residues. Patients with homozygous mutations of L394R in GGCX have a severe bleeding disorder due to decreased biological activities of all clotting factors [33]. In vitro characterization of the purified enzyme shows that the Ki of the competitive inhibitor for the pentapeptide FLEEL carboxylation is 110-fold higher for L394R than that of the wild-type enzyme [34]. Mutations of L394 and of neighboring residues, Y395 and W399, resulted in defective glutamate binding and significantly decreased carboxylation efficiency [35]. These results suggest that glutamate recognition is the primary function of the highly conserved region between residues 393 and 404 in GGCX. This glutamate-binding region of GGCX has been confirmed further by the nanodisc-hydrogen/deuterium exchange mass spectrometry study [26].
Identifying GGCX’s catalytic residues for carboxylation and epoxidation has been less successful. Numerous studies have suggested that free cysteine residues in GGCX are important for both carboxylation and epoxidation reactions [36–38]. Based on these observations, Dowd et al. [39] used a chemical model to develop a ‘base strength amplification mechanism’ for vitamin K-dependent carboxylation. They proposed that two free cysteines were involved in GGCX’s active site. One cysteine served as a weak base to deprotonate KH2, forming a strong base to abstract the γ-hydrogen of the glutamate, and the other cysteine provided the binding site of CO2. Further support for this hypothesis was provided by Pudota et al. [40], who suggested that C99 and C450 are the two active site cysteines required for both carboxylation and epoxidation reactions. However, results from site-directed mutagenesis, chemical modification and peptide fingerprinting suggest that cysteine residues are not involved in GGCX catalysis. Furthermore, C99 and C450 form the only disulfide bond in GGCX, which is important for GGCX stability [41,42]. That GGCX’s catalytic residues are not cysteines has been further confirmed by Rishavy et al. [43], who suggest that the catalytic base, which deprotonates KH2, is not a cysteine but an activated amine.
According to the ‘base strength amplification mechanism’, a weak base in GGCX is required for the conversion of KH2 to a strong base [39]. A quantum chemical study suggests that the alkoxide of vitamin K is most likely the strong base that abstracts a proton from the γ-carbon of glutamates [44,45]. After the initial step (the removal of a proton from KH2), the energetics of the reaction are thermodynamically favorable. Cysteines have been excluded as GGCX’s weak base [41–43]; the candidate base residues were searched using sequence alignment of the Leptospira GGCX homolog with mammalian orthologs [46]. Three conserved histidines and one conserved lysine were identified. Mutating these candidate residues to alanine showed that only the K218A mutant was completely inactive for both the carboxylation and the epoxidation reactions. In addition, the activity of the K218A mutant could be rescued with small amines. Based on this study, the authors concluded that K218 (together with K217) serves as the weak base to deprotonate KH2 and initiate the carboxylation reaction.
Membrane topology of GGCX
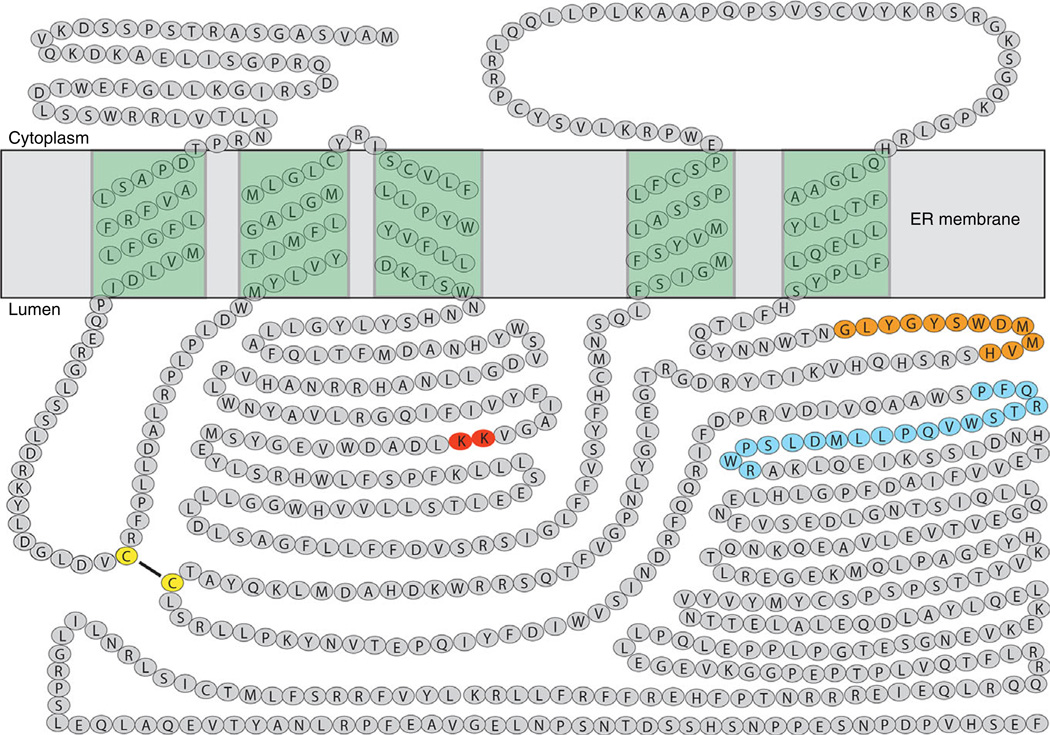
To better understand the structure and function relationships of integral membrane proteins, it is necessary to know their membrane topologies, especially when their three-dimensional structure is unavailable. To determine the membrane topology of GGCX, Tie et al. [47] used in vitro translation/cotranslocation, combined with an N-linked glycosylation reporter tag, to map the topological orientation of the predicted transmembrane domain (TMD) in the GGCX sequence. In addition, they determined the orientations of the amino and carboxyl termini of full-length GGCX by expressing the tagged proteins in HEK293 cells. Results from this study suggest that GGCX spans the endoplasmic reticulum (ER) membrane five times, with its amino terminus located in the cytoplasm and its carboxyl terminus located in the ER lumen (Fig. 2). This topology model places the identified propeptide binding site, the glutamate binding site and the vitamin K deprotonate active site in the ER lumen, which is where vitamin K-dependent carboxylation occurs.
Fig. 2.
Proposed membrane topology of gamma-glutamyl carboxylase (GGCX). GGCX spans the endoplasmic reticulum (ER) membrane five times with its N-terminus located in the cytoplasm and C-terminus located in ER lumen. The glutamate binding site is shown as orange filled circles; the propeptide binding site is shown as blue filled circles; the proposed vitamin K deprotonation residues are shown as red filled circles; and the disulfide linked cysteines are shown as yellow filled circles.
This topology, however, has been called into question by placing a vitamin K-dependent protein binding sequence (VKS, residues 343–355) in the cytoplasm [48,49]. To clarify this issue, microsomes from insect cells expressing GGCX were treated with proteinase K, and the luminal protected fragment was purified [41]. The N-terminal sequence of the purified fragment corresponds to the sequence of GGCX, starting from residue 352, confirming the cytoplasmic location of the VKS region. In addition, results from the functional expression of a two-chain GGCX suggest that the fifth TMD in the proposed topology model is the last and only TMD in the C-terminal peptide of the two-chain GGCX [50], a finding that further supports the proposed topology model of GGCX.
GGCX genotype and the clinical phenotype
Since the identification of the GGCX gene, over 30 naturally occurring GGCX mutations have been discovered in patients with vitamin K-related disorders [51]. Characterization of some of these mutations provides invaluable information regarding the function of GGCX. Defects of vitamin K-dependent carboxylation have long been known to cause bleeding disorders, referred to as vitamin K-dependent coagulation factors deficiency (VKCFD) [52]. Recently, GGCX mutations have also been linked to non-bleeding clinical phenotypes, mainly the PXE-like syndrome [53]. The PXE-like syndrome is characterized by aberrant mineralization of soft connective tissue resulting in fragmentation of elastic fibers, involving primarily the skin, eyes and cardiovascular system [54]. GGCX mutations in PXE-like patients resulted in a lower level of carboxylated MGP – a strong inhibitor of vascular calcification and connective tissue mineralization. Some PXE-like patients have comorbid VKCFD syndrome [53], while others have normal blood coagulation [55]. It is not clear why some GGCX mutations result in bleeding disorders while others cause non-bleeding syndromes. This is mainly because our current knowledge of GGCX’s function was obtained from in vitro experimentation under artificial conditions, which has limited usefulness in understanding the clinical consequences of these mutations. To better address this issue, a GGCX-deficient mouse strain was generated by gene targeting [56]. However, due to embryonic lethality, all homozygous GGCX-deficient mice succumbed to massive intra-abdominal hemorrhages shortly after birth. While liver-specific GGCX-deficient mice have been created to again attempt to examine GGCX function in vivo [57], it appears that manipulating GGCX variants and GGCX substrates in mice is impracticable. Recently, CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas9 was used to create a GGCX-deficient cell line that contains vitamin K-dependent reporter proteins, which appears to be a very useful tool for studying the functional consequences of GGCX mutations in their native milieu [58].
Structure-function studies of VKOR
VKOR protein purification and gene discovery
Since the discovery of the enzymatic activity of VKOR in 1970 [59], numerous attempts to purify VKOR from hepatic microsomes have been unsuccessful [60–63]. Purification of VKOR is exceptionally difficult because it is rendered inactive by any detergent that achieves its solubilization. Due to the loss of activity that accompanied purification, it was postulated that VKOR was a multi-enzyme complex.
The first partial purification of VKOR was reported in 1985 [63]. Sodium cholate-solubilized rat hepatic microsomes were separated by a discontinuous sucrose gradient to isolate a 200S microsomal sub-fraction that contained VKOR activity. However, attempts to further purify VKOR from the 200S fraction resulted in loss of enzymatic activity. A ~17 kDa warfarin-sensitive protein was identified by chemical modification of the free cysteines using radioactive N -[3H]ethylmaleimide ([3H]NEM). Both KO and warfarin can effectively block the incorporation of [3H]NEM into the reduced protein. Therefore, it has been suggested that the [3H]NEM-labeled component is a single polypeptide containing one disulfide bond that exhibits VKOR activity. As VKOR was generally thought to be a multi-enzyme complex, the authors concluded that it was unlikely that a single peptide of relatively low molecular weight could catalyze these complex reactions [63]. In fact, the results of Lee et al. [64] agree well with the fact that VKOR is an 18 kDa enzyme that employs a CXXC redox center as its active site.
Still mistakenly assuming that VKOR was a multi-enzyme complex, VKOR purification then focused on searching for the potential component(s). Cain et al. [62] subjected CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate)-solubilized rat liver microsomes to a gel-filtration column. Enzymatic activity was lost, but some activity could be regained by combining the flow-through and the retained fractions. The flow-through was further separated by ion-exchange chromatography; the unbound fraction also restored VKOR activity. Characterization of the unbound proteins suggested that microsomal epoxide hydrolase (mEH), an integral membrane protein, was the second component of the assumed VKOR complex [61,62]. However, it was later shown that a mouse knockout of mEH had no defects in vitamin K metabolism [65], which indicated that mEH was not involved in KO reduction. Therefore, it is very unlikely that VKOR is actually a multi-enzyme complex.
Traditional biochemical approaches to protein identification did not contribute to VKOR identification, even after several decades of efforts. It was not until 2004 that the gene encoding VKOR was identified independently by two laboratories using alternative approaches: siRNA functional screening and interspecies genetic linkage analysis [66,67]. The putative VKOR gene was found to encode for a 163-amino acid protein and was confirmed by overexpression in insect cells and HEK293 cells. Recombinant VKOR has been purified from insect cells [64], and this purified single-peptide enzyme can accomplish both the conversion of KO to vitamin K and vitamin K to KH2 [64].
Functional regions and critical residues in VKOR
The cloning of the VKOR gene made it possible to study VKOR functions at the molecular level. Shortly after the identification of the VKOR gene, a protein sequence alignment of widely divergent species showed that four cysteines (C43, C51, C132 and C135 of human VKOR) are absolutely conserved; these cysteines are proposed to be VKOR’s active site residues [68] (Fig. 3). The C132XXC135 motif was hypothesized to perform the nucleophilic attack on vitamin K epoxide, as proposed by a chemical model study [69]. This hypothesis was confirmed by mutagenesis studies showing that mutating either of the cysteines renders VKOR inactive [70–72].
Fig. 3.
Multiple sequence alignment of vitamin K epoxide reductase (VKOR) near the conserved cysteine residues. The amino acid sequences of VKOR from widely divergent species were aligned by CLUSTAL W. Completely conserved cysteine residues are indicated by numbers according to the position of amino acid residues in the human VKOR sequence.
While there is no doubt that the CXXC motif is VKOR’s active site, the role of the other two conserved cysteines (C43 and C51, the loop cysteines) remains controversial. Based on studies of bacterial VKOR homologues (VKORHs), the loop cysteines of human VKOR were proposed to shuttle electrons to the active site cysteines [73,74]. It is clear that the loop cysteines in bacterial VKORHs accept electrons from their Trx-like domain or from DsbA; these electrons are then transferred to the active site cysteines [75]. Mutating either of the loop cysteines blocks the electron transfer pathway and renders the enzyme inactive. However, the mechanism in human VKOR appears to be different. Although the C43S mutation significantly decreased VKOR activity, VKOR molecules with C51 mutated to serine or with both C43 and C51 mutated to serine retained almost full activity [70,72]. This suggests that the loop cysteines are not required for VKOR activity. A caveat to this conclusion is that these studies used dithiothreitol as a reductant to reduce the active site disulfide in vitro, which may bypass the function of the loop cysteines.
Recently, a cell-based assay was established for the functional study of vitamin K cycle enzymes in mammalian cells using a chimeric vitamin K-dependent reporter protein [76]. The reporter protein in this assay is protein C (PC) with its Gla domain replaced by that of FIX (FIXgla). The rationale for using FIXgla-PC as the reporter protein is that the uncarboxylated reporter protein should be selectively degraded by the cells [77] to decrease the background; FIXgla enables the ready detection of the carboxylated reporter protein by a conformational specific monoclonal antibody that recognizes only carboxylated FIXgla. For functional studies of VKOR, transcription activator-like effector nucleases (TALENs)-mediated genome editing was used to knockout the endogenous VKOR and VKORL1 genes in the reporter cells [78]. A VKOR molecule with both C43 and C51 mutated to alanine retains ~90% activity in cell-based assays compared with the wild-type enzyme. Additionally, the entire loop between the two cysteines (including C43 and C51) can be deleted with only a minor effect on VKOR’s activity. These results strongly suggest that transfer of electrons from the loop cysteines to the active site cysteines cannot be the major mechanism in human VKOR.
VKOR is the target of warfarin – the most widely used oral anticoagulant. Naturally occurring VKOR mutations have been detected, and these mutations result in warfarin resistance in rats, mice and humans. As these mutations are widely dispersed throughout the VKOR sequence, and not all the mutations are resistant to warfarin inhibition in a cell-based assay [78], it seems unlikely that all of these residues are involved in warfarin binding. The most frequent mutation residue, Y139, together with its neighboring residues (TY139A), has been proposed to be part of the warfarin-binding site [79]. The Y139F VKOR mutant is resistant to warfarin inhibition; because the only difference between tyrosine and phenylalanine is the hydroxyl group, this hydroxyl group is therefore proposed to play an essential role in warfarin binding [72]. Interestingly, in a functional study of the consequences of warfarin-resistant VKOR mutants on the recycling of vitamin K, Matagrin et al. [80] reported that all functional VKOR mutants at residue Y139 produce 3-hydroxyvitamin K, suggesting that the hydroxyl group of Y139 is involved in the catalytic mechanism corresponding to the dehydration of KO. These results together suggest that the TYA and CXXC motifs form a binding pocket for both the substrate (KO) and inhibitor (warfarin) binding.
In addition to affecting warfarin sensitivity, one naturally occurring mutation in VKOR (R98W) has been associated with VKCFD [66]. Characterization of the R98W mutant suggests that this mutation disrupts a diarginine ER retention motif, resulting in mislocalization of the mutated protein to the ER, and exits the ER membrane by cellular quality control systems [81].
Membrane topology of VKOR
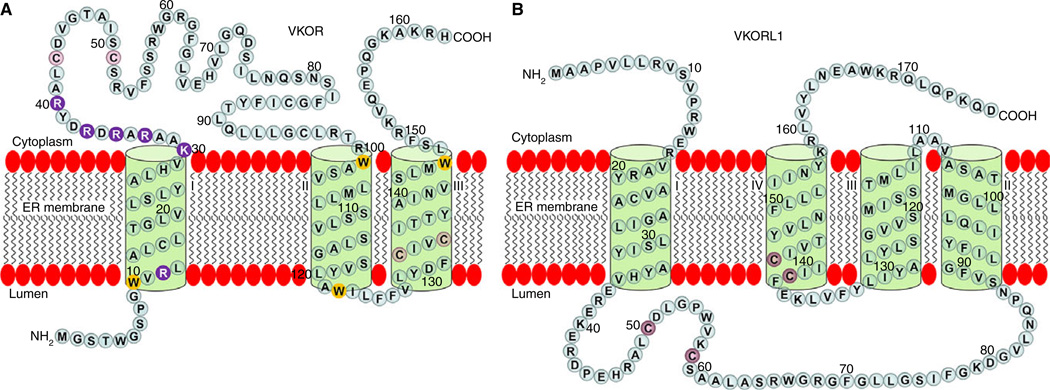
The membrane topology of VKOR has been a major controversy in the community of VKOR researchers. Both three- and four-TMD topology models have been proposed for VKOR; these are based on biochemical approaches [82,83] and on the crystal structure of a bacterial (Synechococcus) VKORH (Syn-VKORH) [74]. Although the membrane orientations of the last two TMDs of VKOR are not in doubt, the orientation of the first TMD is a source of contention. The three-TMD model suggests a Nout/Cin orientation for TMD1; a Nin/Cout orientation is suggested by the four-TMD model. One of the main factors that affects the orientation of the TMD is the distribution of the charged residues flanking the hydrophobic transmembrane segment, called the ‘positive inside rule,’ which applies to both bacterial and mammalian membrane proteins [84,85]. As human VKOR and Syn-VKORH have different charged-residue distributions flanking the first TMD, the ‘positive inside rule’ predicts a three-TMD model for human VKOR and a four-TMD model for Syn-VKORH. Additionally, human VKOR and Syn-VKORH only share ~20% sequence identity, so deducting the membrane topology of human VKOR from Syn-VKORH may not be appropriate. Furthermore, the three-TMD model is based on an examination of the potential of each predicted TMD of human VKOR to function as an authentic stop-transfer sequence and the detection of the termini locations of full-length VKOR in both intact microsomes and in live cells in situ [82,83]. This makes it more likely that human VKOR is a three-TMD protein (Fig. 4A).
Fig. 4.
Proposed membrane topology of vitamin K epoxide reductase (VKOR) and VKORL1. (A) A three-transmembrane domain (TMD) topology model of VKOR is illustrated. The N-terminus of VKOR is located in the endoplasmic reticulum (ER) lumen and the C-terminus located in the cytoplasm. The positively charged residues flanking TMD1 are shown as purple filled circles; the membrane interfacial tryptophans are shown as yellow filled circles; and conserved active site cysteines and loop cysteines are shown as red filled circles. (B) A four-TMD topology model of VKORL1 is illustrated. Both the N-terminus and C-terminus of VKORL1 are located in the cytoplasm. Conserved active site cysteines and loop cysteines are shown as red filled circles.
In the three-TMD model, the CXXC active site of VKOR is located within the last TMD, close to the ER lumen. The hydrophobic environment of the CXXC redox center is consistent with the hydrophobicity of the vitamin K substrate. In addition, four of the six tryptophan residues in VKOR are located in the membrane’s bilayer interface. Tryptophan residues have been shown to exhibit a strong tendency to remain within the interfacial region, serving as anchoring residues that fix the transmembrane helix within the lipid bilayer [86]. Therefore, the positively charged residues and the tryptophan residues in VKOR both seem to lock VKOR into a three-TMD configuration.
Clarifying the topological structure of VKOR is important because the two existing topology models place the functionally disputed conserved loop cysteines on different sides of the ER membrane. The three-TMD model places the conserved loop cysteines in the cytoplasm, the opposite side of the ER membrane from the CXXC active site. It has been argued that the three-TMD model is unlikely because the loop cysteines transfer electrons to the active site. It is worth noting that VKOR can be changed from a three-TMD molecule to a fully active four-TMD molecule by mutating the positively charged residues flanking the first TMD [82]. However, even when the loop cysteines are located on the same side of the ER membrane as the active site cysteines, they do not appear to be involved in electron transfer [82]. Interestingly, Mycobacterium tuberculosis VKORH (Mt-VKOR) has four TMDs in its VKOR domain and has its loop cysteines located on the same side of the membrane as the active site cysteines. These loop cysteines are clearly required for Mt-VKORH’s activity in E. coli [87] but are not essential for vitamin K reduction in mammalian cells [88]. Results from these studies suggest that the conserved loop cysteines in VKOR are not involved in active site regeneration, and that VKOR’s active site is directly reduced by an as-yet-unknown physiological reductant.
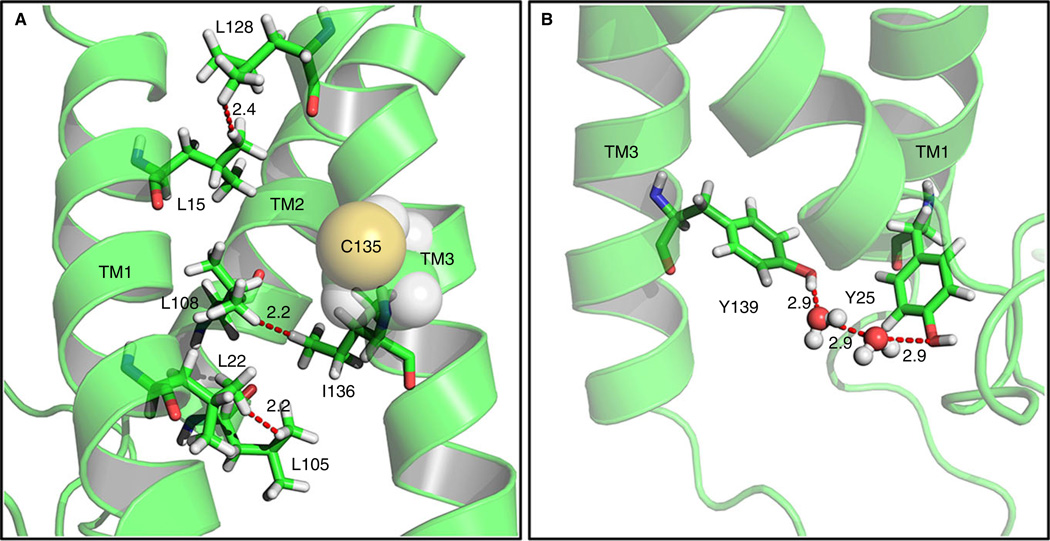
Further support for the three-TMD topology model of VKOR comes from a molecular dynamics simulation of VKOR in a palmitoyl-oleoyl-phosphatidylethanolamine lipid bilayer [89]. This study shows that the three-TMD topology model of VKOR is more stable than the four-TMD model, as the hydrophobic interactions between leucine residues in different transmembrane helices only occur in the three-TMD model (Fig. 5A). Additionally, in the four-TMD model, residue Y139 of the warfarin-binding motif faces to the outside of the core of the TMDs, whereas in the three-TMD model, Y139 interacts with Y25 through a H2O molecule to form a substrate/warfarin binding pocket (Fig. 5B).
Fig. 5.
Molecular dynamics simulations of vitamin K epoxide reductase (VKOR) as a three-transmembrane domain (TMD) molecule. (A) Five hydrophobic interactions between leucine and/or isoleucine residue pairs within TMDs stabilize the three-TMD topology model of VKOR. (B) The proposed warfarin binding residue Y139 (in the TYA motif) is stabilized by interaction with a tyrosine in TMD1 (Y25) via a water network.
Structure-function study of VKORL1
During the identification of VKOR, homologous searches with VKOR detected a paralogous protein called VKOR-like 1 (VKORL1) [66]. VKORL1 appears to have a different membrane topology (Fig. 4B) and a different mechanism for its active site regeneration as compared with VKOR [90]. Although the structure-function relationships of VKOR have been extensively studied, the functional study of VKORL1 has not been reported until recently [90–93].
Westhofen et al. [93] expressed VKORL1 in HEK293 cells, then examined VKORL1’s ability to reduce KO and vitamin K from crude cell membranes. This study showed that VKORL1 reduces KO 18 054-fold slower than VKOR. Based on these results, the authors hypothesized that the primary physiological function of VKORL1 is to reduce vitamin K to KH2. As KH2 has been reported to be an effective antioxidant, helping to prevent cell death caused by oxidative stress, these authors further concluded that VKORL1 was responsible for driving vitamin K-mediated intracellular antioxidation pathways that were critical to cell survival. It should be noted, however, that this conclusion is based on comparing the specific enzymatic activity of VKORL1 in crude cell membranes (total proteins) in the presence of detergent with that of purified VKOR reconstituted in intact membranes [64]. As cell-based studies show that VKORL1 has a substantial role in reducing KO to support vitamin K-dependent carboxylation, and HEK293 cells with both VKOR and VKORL1 ablated grow and function normally [78,90], it is premature to conclude that the primary physiological function of VKORL1 is to prevent oxidative stress in cells.
A recent study suggests that VKORL1 might rescue VKOR’s activity in extrahepatic tissues during anticoagulation therapy [92]. In this study, VKORL1 and VKOR were expressed in Pichia pastoris and their susceptibility to vitamin K antagonists (VKAs) was compared using a DTT-driven microsomal activity assay. Results from this study show that the catalytic efficiency of VKORL1 in reducing KO is two-fold higher than that of VKOR. Compared with VKOR, VKORL1 is ~50-fold more resistant to VKA inhibition. VKOR’s and VKORL1’s mRNA level and KO reductase activity were also compared in mouse and rat tissues. In liver, the mRNA level of VKORL1 is ~10-fold lower than that of VKOR, while in extrahepatic tissues the mRNA level of VKORL1 is systematically higher than that of VKOR. Results from activity assays of rat tissues suggest that VKOR contributes more than 90% of the KO reductase activity in liver, while VKORL1 contributes ~55% of the KO reductase activity in testis and ~22% in lung. Based on these results, the authors suggest that VKORL1 supports vitamin K-dependent carboxylation mainly in extrahepatic tissues and rescues VKOR’s activity during anticoagulation therapy.
Although VKORL1 can efficiently reduce KO to support vitamin K-dependent carboxylation in the DTT-driven and cell-based assays [78,90,92], it appears unlikely that VKORL1 plays a key role in the vitamin K cycle under physiological conditions. If so, the murine knockout of VKOR would be viable [94] and no significant decrease of KO reductase activity in the VKOR knockout cells would have been observed [78].
Functional studies of VKR
To date, we know very little about the enzymes that reduce vitamin K, but there are several lines of evidence that demonstrate VKR’s importance. Most notably, patients that are overdosed with warfarin can be rescued by large doses of vitamin K through the action of a warfarin-resistant enzyme that reduces vitamin K, designated as the ‘antidotal enzyme’. The antidotal effect of vitamin K was first discovered in 1966 [95] and was recently demonstrated in the VKOR-knockout mouse [94] and in a cell-based vitamin K cycle enzyme study [76]. However, despite decades of effort [96–98], the identity of VKR is still unknown.
It has been proposed that vitamin K is reduced to KH2 via two pathways [97]: a warfarin-sensitive pathway (accomplished by VKOR) and a warfarin-resistant pathway (catalyzed by a NAD(P)H-dependent antidotal enzyme). As VKOR is capable of reducing vitamin K to KH2 [64, 70, 99], it has been proposed that VKOR could form a dimer with one molecule reducing KO to vitamin K and the other molecule reducing vitamin K to KH2 [99]. However, studies using the purified enzyme suggest that VKOR reduces KO to vitamin K ~50-fold faster than it reduces vitamin K to KH2 [64]. When VKOR is inactivated by warfarin or knocked out by gene targeting in HEK293 cells, those cells can still efficiently reduce vitamin K to support vitamin K-dependent carboxylation [76,78]. Additionally, VKOR-deficient mice died due to extensive intracerebral hemorrhage; however, this lethal phenotype could be rescued by the oral administration of vitamin K [94]. Furthermore, VKOR was unable to convert KO directly to KH2 to support vitamin K-dependent carboxylation in a cell-based assay [76]. Therefore, it is unlikely that VKOR is the major physiological contributor to vitamin K reduction.
NQO1 (NAD(P)H quinone oxidoreductase 1) has long been known as the warfarin-resistant antidotal enzyme for vitamin K reduction [98]. Rat liver microsomes depleted of NQO1 displayed reduced vitamin K-dependent carboxylation activity, and this activity could be restored by adding purified NQO1 [97]. However, dicoumarol, a strong inhibitor of NQO1, was unable to inhibit vitamin K reduction in HEK293 cells [76], and NQO1-deficient mice survived at the same frequency as wild-type mice when poisoned with warfarin [96]. In addition, an enzyme kinetic study of membrane fractions from mouse liver microsomes shows the existence of NAD(P)H-dependent VKR activity in both wild-type and NQO1-deficient mice [96]. These results suggest that NQO1 is not the antidotal enzyme for reducing vitamin K to KH2 under physiological conditions.
VKR, like GGCX and VKOR, is likely to be an integral membrane protein residing in the ER [96]. Traditional biochemical approaches to protein identification are often unsuccessful for membrane protein identification, as illustrated by the history of efforts to identify VKOR. The identification of VKOR spanned several decades and was ultimately achieved, not by biochemical approaches, but by alternative approaches employing interspecies genetic linkage analysis [66] and siRNA functional screening [67]. We believe that a similar non-traditional approach to protein identification may be the key to identifying VKR. One promising avenue is the recently developed genome-scale CRISPR-Cas9 knockout library screening [100]. Detailed insights into the characteristics of warfarin-resistant and warfarin-sensitive VKRs will not only extend our understanding of the mechanism of vitamin K reduction, but will also help us to develop new ways of regulating coagulation and treating thrombosis.
Acknowledgments
This work was supported by NIH grant HL077740 (to both authors).
Footnotes
Disclosure of Conflict of Interests
The authors hold a patent on the use of VKOR for producing vitamin K-dependent proteins licensed to Emergent.
References
- 1.Stenflo J, Fernlund P, Egan W, Roepstorff P. Vitamin K dependent modifications of glutamic acid residues in prothrombin. Proc Natl Acad Sci USA. 1974;71:2730–2733. doi: 10.1073/pnas.71.7.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelsestuen GL, Zytkovicz TH, Howard JB. The mode of action of vitamin K. Identification of gamma-carboxyglutamic acid as a component of prothrombin. J Biol Chem. 1974;249:6347–6350. [PubMed] [Google Scholar]
- 3.Vermeer C. Gamma-carboxyglutamate-containing proteins and the vitamin K-dependent carboxylase. Biochem J. 1990;266:625–636. doi: 10.1042/bj2660625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelsestuen GL. Enhancement of vitamin-K-dependent protein function by modification of the gamma-carboxyglutamic acid domain: studies of protein C and factor VII. Trends Cardiovasc Med. 1999;9:162–167. doi: 10.1016/s1050-1738(99)00024-9. [DOI] [PubMed] [Google Scholar]
- 5.Ducy P, Desbois C, Boyce B, Pinero G, Story B, Dunstan C, Smith E, Bonadio J, Goldstein S, Gundberg C, Bradley A, Karsenty G. Increased bone formation in osteocalcin-deficient mice. Nature. 1996;382:448–452. doi: 10.1038/382448a0. [DOI] [PubMed] [Google Scholar]
- 6.Ferron M, Lacombe J. Regulation of energy metabolism by the skeleton: osteocalcin and beyond. Arch Biochem Biophys. 2014;561:137–146. doi: 10.1016/j.abb.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 7.Lee NK, Sowa H, Hinoi E, Ferron M, Ahn JD, Confavreux C, Dacquin R, Mee PJ, McKee MD, Jung DY, Zhang Z, Kim JK, Mauvais-Jarvis F, Ducy P, Karsenty G. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130:456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Booth SL, Centi A, Smith SR, Gundberg C. The role of osteocalcin in human glucose metabolism: marker or mediator? Nat Rev Endocrinol. 2013;9:43–55. doi: 10.1038/nrendo.2012.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gundberg CM, Lian JB, Booth SL. Vitamin K-dependent carboxylation of osteocalcin: friend or foe? Adv Nutr. 2012;3:149–157. doi: 10.3945/an.112.001834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schurgers LJ, Cranenburg EC, Vermeer C. Matrix Gla-protein: the calcification inhibitor in need of vitamin K. Thromb Haemost. 2008;100:593–603. [PubMed] [Google Scholar]
- 11.Willems BA, Vermeer C, Reutelingsperger CP, Schurgers LJ. The realm of vitamin K dependent proteins: shifting from coagulation toward calcification. Mol Nutr Food Res. 2014;58:1620–1635. doi: 10.1002/mnfr.201300743. [DOI] [PubMed] [Google Scholar]
- 12.Vanakker OM, Martin L, Schurgers LJ, Quaglino D, Costrop L, Vermeer C, Pasquali-Ronchetti I, Coucke PJ, De Paepe A. Low serum vitamin K in PXE results in defective carboxylation of mineralization inhibitors similar to the GGCX mutations in the PXE-like syndrome. Lab Invest. 2010;90:895–905. doi: 10.1038/labinvest.2010.68. [DOI] [PubMed] [Google Scholar]
- 13.Sharma B, Albig AR. Matrix Gla protein reinforces angiogenic resolution. Microvasc Res. 2013;85:24–33. doi: 10.1016/j.mvr.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah DV, Suttie JW. The vitamin K dependent, in vitro production of prothrombin. Biochem Biophys Res Commun. 1974;60:1397–1402. doi: 10.1016/0006-291x(74)90353-2. [DOI] [PubMed] [Google Scholar]
- 15.Wu SM, Cheung WF, Frazier D, Stafford DW. Cloning and expression of the cDNA for human gamma-glutamyl carboxylase. Science. 1991;254:1634–1636. doi: 10.1126/science.1749935. [DOI] [PubMed] [Google Scholar]
- 16.Hubbard BR, Ulrich MM, Jacobs M, Vermeer C, Walsh C, Furie B, Furie BC. Vitamin K-dependent carboxylase: affinity purification from bovine liver by using a synthetic propeptide containing the gamma-carboxylation recognition site. Proc Natl Acad Sci USA. 1989;86:6893–6897. doi: 10.1073/pnas.86.18.6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Metz M, Vermeer C, Soute BA, van Scharrenburg GJM, Slotboom AJ, Hemker HC. Partial purification of bovine liver vitamin K-dependent carboxylase by immunospecific adsorption onto antifactor X. FEBS Lett. 1981;123:215–218. doi: 10.1016/0014-5793(81)80290-6. [DOI] [PubMed] [Google Scholar]
- 18.Girardot JM. Vitamin K-dependent carboxylase. Partial purification and properties of the enzyme-substrate complex. J Biol Chem. 1982;257:15008–15011. [PubMed] [Google Scholar]
- 19.Wu SM, Morris DP, Stafford DW. Identification and purification to near homogeneity of the vitamin K-dependent carboxylase. Proc Natl Acad Sci USA. 1991;88:2236–2240. doi: 10.1073/pnas.88.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu SM, Stafford DW, Frazier LD, Fu YY, High KA, Chu K, Sanchez-Vega B, Solera J. Genomic sequence and transcription start site for the human gamma-glutamyl carboxylase. Blood. 1997;89:4058–4062. [PubMed] [Google Scholar]
- 21.Kuo WL, Stafford DW, Cruces J, Gray J, Solera J. Chromosomal localization of the gamma-glutamyl carboxylase gene at 2p12. Genomics. 1995;25:746–748. doi: 10.1016/0888-7543(95)80024-g. [DOI] [PubMed] [Google Scholar]
- 22.Stafford DW, Hebling CM. Vitamin K cycles and gamma-carboxylation of coagulation factors. Recent Adv Thromb Hemostasis. 2008;2008:27–44. [Google Scholar]
- 23.Pan LC, Price PA. The propeptide of rat bone gamma-carboxyglutamic acid protein shares homology with other vitamin K-dependent protein precursors. Proc Natl Acad Sci USA. 1985;82:6109–6113. doi: 10.1073/pnas.82.18.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman PA, Shia MA, Gallop PM, Griep AE. Vitamin K-dependent gamma-carbon-hydrogen bond cleavage and nonmandatory concurrent carboxylation of peptide-bound glutamic acid residues. Proc Natl Acad Sci USA. 1979;76:3126–3129. doi: 10.1073/pnas.76.7.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larson AE, Friedman PA, Suttie JW. Vitamin K-dependent carboxylase. Stoichiometry of carboxylation and vitamin K 2,3-epoxide formation. J Biol Chem. 1981;256:11032–11035. [PubMed] [Google Scholar]
- 26.Parker CH, Morgan CR, Rand KD, Engen JR, Jorgenson JW, Stafford DW. A conformational investigation of propeptide binding to the integral membrane protein gamma-glutamyl carboxylase using nanodisc hydrogen exchange mass spectrometry. Biochemistry. 2014;53:1511–1520. doi: 10.1021/bi401536m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins-Gruber SL, Mutucumarana VP, Lin PJ, Jorgenson JW, Stafford DW, Straight DL. Effect of vitamin K-dependent protein precursor propeptide, vitamin K hydroquinone, and glutamate substrate binding on the structure and function of {gamma}-glutamyl carboxylase. J Biol Chem. 2010;285:31502–31508. doi: 10.1074/jbc.M110.143297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugiura I, Furie B, Walsh CT, Furie BC. Profactor IX propeptide and glutamate substrate binding sites on the vitamin K-dependent carboxylase identified by site-directed mutagenesis. J Biol Chem. 1996;271:17837–17844. doi: 10.1074/jbc.271.30.17837. [DOI] [PubMed] [Google Scholar]
- 29.Yamada M, Kuliopulos A, Nelson NP, Roth DA, Furie B, Furie BC, Walsh CT. Localization of the factor IX propeptide binding site on recombinant vitamin K dependent carboxylase using benzoylphenylalanine photoaffinity peptide inactivators. Biochemistry. 1995;34:481–489. doi: 10.1021/bi00002a012. [DOI] [PubMed] [Google Scholar]
- 30.Lin PJ, Jin DY, Tie JK, Presnell SR, Straight DL, Stafford DW. The putative vitamin K-dependent gamma-glutamyl carboxylase internal propeptide appears to be the propeptide binding site. J Biol Chem. 2002;277:28584–28591. doi: 10.1074/jbc.M202292200. [DOI] [PubMed] [Google Scholar]
- 31.Wu SM, Mutucumarana VP, Geromanos S, Stafford DW. The propeptide binding site of the bovine gamma-glutamyl carboxylase. J Biol Chem. 1997;272:11718–11722. doi: 10.1074/jbc.272.18.11718. [DOI] [PubMed] [Google Scholar]
- 32.Soute BA, Jin DY, Spronk HM, Mutucumarana VP, Lin PJ, Hackeng TM, Stafford DW, Vermeer C. Characteristics of recombinant W501S mutated human gamma-glutamyl carboxylase. J Thromb Haemost. 2004;2:597–604. doi: 10.1111/j.1538-7836.2004.00686.x. [DOI] [PubMed] [Google Scholar]
- 33.Brenner B, Sanchez-Vega B, Wu SM, Lanir N, Stafford DW, Solera J. A missense mutation in gamma-glutamyl carboxylase gene causes combined deficiency of all vitamin K-dependent blood coagulation factors. Blood. 1998;92:4554–4559. [PubMed] [Google Scholar]
- 34.Mutucumarana VP, Stafford DW, Stanley TB, Jin DY, Solera J, Brenner B, Azerad R, Wu SM. Expression and characterization of the naturally occurring mutation L394R in human gamma-glutamyl carboxylase. J Biol Chem. 2000;275:32572–32577. doi: 10.1074/jbc.M006808200. [DOI] [PubMed] [Google Scholar]
- 35.Mutucumarana VP, Acher F, Straight DL, Jin DY, Stafford DW. A conserved region of human vitamin K-dependent carboxylase between residues 393 and 404 is important for its interaction with the glutamate substrate. J Biol Chem. 2003;278:46488–46493. doi: 10.1074/jbc.M307707200. [DOI] [PubMed] [Google Scholar]
- 36.Canfield LM, Sinsky TA, Suttie JW. Vitamin K-dependent carboxylase: purification of the rat liver microsomal enzyme. Arch Biochem Biophys. 1980;202:515–524. doi: 10.1016/0003-9861(80)90457-9. [DOI] [PubMed] [Google Scholar]
- 37.Mack DO, Suen ET, Girardot JM, Miller JA, Delaney R, Johnson BC. Soluble enzyme system for vitamin K-dependent carboxylation. J Biol Chem. 1976;251:3269–3276. [PubMed] [Google Scholar]
- 38.Friedman PA, Shia M. Some characteristics of a vitamin K-dependent carboxylating system from rat liver microsomes. Biochem Biophys Res Commun. 1976;70:647–654. doi: 10.1016/0006-291x(76)91096-2. [DOI] [PubMed] [Google Scholar]
- 39.Dowd P, Hershline R, Ham SW, Naganathan S. Vitamin K and energy transduction: a base strength amplification mechanism. Science. 1995;269:1684–1691. doi: 10.1126/science.7569894. [DOI] [PubMed] [Google Scholar]
- 40.Pudota BN, Miyagi M, Hallgren KW, West KA, Crabb JW, Misono KS, Berkner KL. Identification of the vitamin K-dependent carboxylase active site: Cys-99 and Cys-450 are required for both epoxidation and carboxylation. Proc Natl Acad Sci USA. 2000;97:13033–13038. doi: 10.1073/pnas.97.24.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tie JK, Jin DY, Loiselle DR, Pope RM, Straight DL, Stafford DW. Chemical modification of cysteine residues is a misleading indicator of their status as active site residues in the vitamin K-dependent gamma-glutamyl carboxylation reaction. J Biol Chem. 2004;279:54079–54087. doi: 10.1074/jbc.M408945200. [DOI] [PubMed] [Google Scholar]
- 42.Tie JK, Mutucumarana VP, Straight DL, Carrick KL, Pope RM, Stafford DW. Determination of disulfide bond assignment of human vitamin K-dependent gamma-glutamyl carboxylase by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Biol Chem. 2003;278:45468–45475. doi: 10.1074/jbc.M309164200. [DOI] [PubMed] [Google Scholar]
- 43.Rishavy MA, Pudota BN, Hallgren KW, Qian W, Yakubenko AV, Song JH, Runge KW, Berkner KL. A new model for vitamin K-dependent carboxylation: the catalytic base that deprotonates vitamin K hydroquinone is not Cys but an activated amine. Proc Natl Acad Sci USA. 2004;101:13732–13737. doi: 10.1073/pnas.0404989101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu S, Liu S, Davis CH, Stafford DW, Pedersen LG. Quantum chemical study of the mechanism of action of vitamin k carboxylase in solvent. Int J Quantum Chem. 2010;110:2744–2751. doi: 10.1002/qua.22740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis CH, Ii DD, Stafford DW, Pedersen LG. Quantum chemical study of the mechanism of action of vitamin K carboxylase (VKC). IV. Intermediates and transition states. J Phys Chem A. 2007;111:7257–7261. doi: 10.1021/jp068564y. [DOI] [PubMed] [Google Scholar]
- 46.Rishavy MA, Hallgren KW, Yakubenko AV, Shtofman RL, Runge KW, Berkner KL. Bronsted analysis reveals Lys218 as the carboxylase active site base that deprotonates vitamin K hydroquinone to initiate vitamin K-dependent protein carboxylation. Biochemistry. 2006;45:13239–13248. doi: 10.1021/bi0609523. [DOI] [PubMed] [Google Scholar]
- 47.Tie J, Wu SM, Jin D, Nicchitta CV, Stafford DW. A topological study of the human gamma-glutamyl carboxylase. Blood. 2000;96:973–978. [PubMed] [Google Scholar]
- 48.Pudota BN, Hommema EL, Hallgren KW, McNally BA, Lee S, Berkner KL. Identification of sequences within the gamma-carboxylase that represent a novel contact site with vitamin K-dependent proteins and that are required for activity. J Biol Chem. 2001;276:46878–46886. doi: 10.1074/jbc.M108696200. [DOI] [PubMed] [Google Scholar]
- 49.Rishavy MA, Berkner KL. Vitamin K oxygenation, glutamate carboxylation, and processivity: defining the three critical facets of catalysis by the vitamin K-dependent carboxylase. Adv Nutr. 2012;3:135–148. doi: 10.3945/an.111.001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tie JK, Zheng MY, Hsiao KL, Perera L, Stafford DW, Straight DL. Transmembrane domain interactions and residue proline 378 are essential for proper structure, especially disulfide bond formation, in the human vitamin K-dependent gamma-glutamyl carboxylase. Biochemistry. 2008;47:6301–6310. doi: 10.1021/bi800235r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watzka M, Geisen C, Scheer M, Wieland R, Wiegering V, Dorner T, Laws HJ, Gumruk F, Hanalioglu S, Unal S, Albayrak D, Oldenburg J. Bleeding and non-bleeding phenotypes in patients with GGCX gene mutations. Thromb Res. 2014;134:856–865. doi: 10.1016/j.thromres.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 52.Napolitano M, Mariani G, Lapecorella M. Hereditary combined deficiency of the vitamin K-dependent clotting factors. Orphanet J Rare Dis. 2010;5:21. doi: 10.1186/1750-1172-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanakker OM, Martin L, Gheduzzi D, Leroy BP, Loeys BL, Guerci VI, Matthys D, Terry SF, Coucke PJ, Pasquali-Ronchetti I, De Paepe A. Pseudoxanthoma elasticum-like phenotype with cutis laxa and multiple coagulation factor deficiency represents a separate genetic entity. J Invest Dermatol. 2007;127:581–587. doi: 10.1038/sj.jid.5700610. [DOI] [PubMed] [Google Scholar]
- 54.Marconi B, Bobyr I, Campanati A, Molinelli E, Consales V, Brisigotti V, Scarpelli M, Racchini S, Offidani A. Pseudoxanthoma elasticum and skin: clinical manifestations, histopathology, pathomechanism, perspectives of treatment. Intractable Rare Dis Res. 2015;4:113–122. doi: 10.5582/irdr.2015.01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kariminejad A, Bozorgmehr B, Najafi A, Khoshaeen A, Ghalandari M, Najmabadi H, Kariminejad MH, Vanakker OM, Hosen MJ, Malfait F, Quaglino D, Florijn RJ, Bergen AA, Hennekam RC. Retinitis pigmentosa, cutis laxa, and pseudoxanthoma elasticum-like skin manifestations associated with GGCX mutations. J Invest Dermatol. 2014;134:2331–2338. doi: 10.1038/jid.2014.191. [DOI] [PubMed] [Google Scholar]
- 56.Zhu A, Sun H, Raymond RM, Jr, Furie BC, Furie B, Bronstein M, Kaufman RJ, Westrick R, Ginsburg D. Fatal hemorrhage in mice lacking gamma-glutamyl carboxylase. Blood. 2007;109:5270–5275. doi: 10.1182/blood-2006-12-064188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Azuma K, Tsukui T, Ikeda K, Shiba S, Nakagawa K, Okano T, Urano T, Horie-Inoue K, Ouchi Y, Ikawa M, Inoue S. Liver-specific gamma-glutamyl carboxylase-deficient mice display bleeding diathesis and short life span. PLoS ONE. 2014;9:e88643. doi: 10.1371/journal.pone.0088643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tie JK, Carneiro JDA, Jin DY, Martinhago CD, Vermeer C, Stafford DW. Characterization of vitamin K-dependent carboxylase mutations that cause bleeding and non-bleeding disorders. Blood. 2016 doi: 10.1182/blood-2015-10-677633. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bell RG, Matschiner JT. Vitamin K activity of phylloquinone oxide. Arch Biochem Biophys. 1970;141:473–476. doi: 10.1016/0003-9861(70)90164-5. [DOI] [PubMed] [Google Scholar]
- 60.Begent LA, Hill AP, Steventon GB, Hutt AJ, Pallister CJ, Cowell DC. Characterization and purification of the vitamin K1 2,3 epoxide reductases system from rat liver. J Pharm Pharmacol. 2001;53:481–486. doi: 10.1211/0022357011775776. [DOI] [PubMed] [Google Scholar]
- 61.Guenthner TM, Cai D, Wallin R. Co-purification of microsomal epoxide hydrolase with the warfarin-sensitive vitamin K1 oxide reductase of the vitamin K cycle. Biochem Pharmacol. 1998;55:169–175. doi: 10.1016/s0006-2952(97)00431-0. [DOI] [PubMed] [Google Scholar]
- 62.Cain D, Hutson SM, Wallin R. Assembly of the warfarin-sensitive vitamin K 2,3-epoxide reductase enzyme complex in the endoplasmic reticulum membrane. J Biol Chem. 1997;272:29068–29075. doi: 10.1074/jbc.272.46.29068. [DOI] [PubMed] [Google Scholar]
- 63.Lee JJ, Principe LM, Fasco MJ. Identification of a warfarin-sensitive protein component in a 200S rat liver microsomal fraction catalyzing vitamin K and vitamin K 2,3-epoxide reduction. Biochemistry. 1985;24:7063–7070. doi: 10.1021/bi00346a007. [DOI] [PubMed] [Google Scholar]
- 64.Chu PH, Huang TY, Williams J, Stafford DW. Purified vitamin K epoxide reductase alone is sufficient for conversion of vitamin K epoxide to vitamin K and vitamin K to vitamin KH2. Proc Natl Acad Sci U S A. 2006;103:19308–19313. doi: 10.1073/pnas.0609401103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miyata M, Kudo G, Lee YH, Yang TJ, Gelboin HV, Fernandez-Salguero P, Kimura S, Gonzalez FJ. Targeted disruption of the microsomal epoxide hydrolase gene. Microsomal epoxide hydrolase is required for the carcinogenic activity of 7,12-dimethylbenz[a]anthracene. J Biol Chem. 1999;274:23963–23968. doi: 10.1074/jbc.274.34.23963. [DOI] [PubMed] [Google Scholar]
- 66.Rost S, Fregin A, Ivaskevicius V, Conzelmann E, Hortnagel K, Pelz HJ, Lappegard K, Seifried E, Scharrer I, Tuddenham EG, Muller CR, Strom TM, Oldenburg J. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 67.Li T, Chang CY, Jin DY, Lin PJ, Khvorova A, Stafford DW. Identification of the gene for vitamin K epoxide reductase. Nature. 2004;427:541–544. doi: 10.1038/nature02254. [DOI] [PubMed] [Google Scholar]
- 68.Goodstadt L, Ponting CP. Vitamin K epoxide reductase: homology, active site and catalytic mechanism. Trends Biochem Sci. 2004;29:289–292. doi: 10.1016/j.tibs.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 69.Silverman RB. Chemical model studies for the mechanism of vitamin K epoxide reductase. J Am Chem Soc. 1981;103:5939–5941. [Google Scholar]
- 70.Jin DY, Tie JK, Stafford DW. The conversion of vitamin K epoxide to vitamin K quinone and vitamin K quinone to vitamin K hydroquinone uses the same active site cysteines. Biochemistry. 2007;46:7279–7283. doi: 10.1021/bi700527j. [DOI] [PubMed] [Google Scholar]
- 71.Wajih N, Sane DC, Hutson SM, Wallin R. Engineering of a recombinant vitamin K-dependent gamma-carboxylation system with enhanced gamma-carboxyglutamic acid forming capacity: evidence for a functional CXXC redox center in the system. J Biol Chem. 2005;280:10540–10547. doi: 10.1074/jbc.M413982200. [DOI] [PubMed] [Google Scholar]
- 72.Rost S, Fregin A, Hunerberg M, Bevans CG, Muller CR, Oldenburg J. Site-directed mutagenesis of coumarin-type anticoagulant-sensitive VKORC1: evidence that highly conserved amino acids define structural requirements for enzymatic activity and inhibition by warfarin. Thromb Haemost. 2005;94:780–786. doi: 10.1160/TH05-02-0082. [DOI] [PubMed] [Google Scholar]
- 73.Schulman S, Wang B, Li W, Rapoport TA. Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc Natl Acad Sci U S A. 2010;107:15027–15032. doi: 10.1073/pnas.1009972107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li W, Schulman S, Dutton RJ, Boyd D, Beckwith J, Rapoport TA. Structure of a bacterial homologue of vitamin K epoxide reductase. Nature. 2010;463:507–512. doi: 10.1038/nature08720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hatahet F, Boyd D, Beckwith J. Disulfide bond formation in prokaryotes: history, diversity and design. Biochim Biophys Acta. 2014;1844:1402–1414. doi: 10.1016/j.bbapap.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tie JK, Jin DY, Straight DL, Stafford DW. Functional study of the vitamin K cycle in mammalian cells. Blood. 2011;117:2967–2974. doi: 10.1182/blood-2010-08-304303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tokunaga F, Takeuchi S, Omura S, Arvan P, Koide T. Secretion, gamma-carboxylation, and endoplasmic reticulum-associated degradation of chimeras with mutually exchanged Gla domain between human protein C and prothrombin. Thromb Res. 2000;99:511–521. doi: 10.1016/s0049-3848(00)00258-9. [DOI] [PubMed] [Google Scholar]
- 78.Tie JK, Jin DY, Tie K, Stafford DW. Evaluation of warfarin resistance using transcription activator-like effector nucleases-mediated vitamin K epoxide reductase knockout HEK293 cells. J Thromb Haemost. 2013;11:1556–1564. doi: 10.1111/jth.12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oldenburg J, Bevans CG, Muller CR, Watzka M. Vitamin K epoxide reductase complex subunit 1 (VKORC1): the key protein of the vitamin K cycle. Antioxid Redox Signal. 2006;8:347–353. doi: 10.1089/ars.2006.8.347. [DOI] [PubMed] [Google Scholar]
- 80.Matagrin B, Hodroge A, Montagut-Romans A, Andru J, Fourel I, Besse S, Benoit E, Lattard V. New insights into the catalytic mechanism of vitamin K epoxide reductase (VKORC1) - The catalytic properties of the major mutations of rVKORC1 explain the biological cost associated to mutations. FEBS Open Biol. 2013;3:144–150. doi: 10.1016/j.fob.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Czogalla KJ, Biswas A, Rost S, Watzka M, Oldenburg J. The Arg98Trp mutation in human VKORC1 causing VKCFD2 disrupts a di-arginine-based ER retention motif. Blood. 2014;124:1354–1362. doi: 10.1182/blood-2013-12-545988. [DOI] [PubMed] [Google Scholar]
- 82.Tie JK, Jin DY, Stafford DW. Human vitamin K epoxide reductase and its bacterial homologue have different membrane topologies and reaction mechanisms. J Biol Chem. 2012;287:33945–33955. doi: 10.1074/jbc.M112.402941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tie JK, Nicchitta C, von Heijne G, Stafford DW. Membrane topology mapping of vitamin K epoxide reductase by in vitro translation/cotranslocation. J Biol Chem. 2005;280:16410–16416. doi: 10.1074/jbc.M500765200. [DOI] [PubMed] [Google Scholar]
- 84.von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- 85.Hartmann E, Rapoport TA, Lodish HF. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci U S A. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.de Planque MR, Bonev BB, Demmers JA, Greathouse DV, Koeppe RE, 2nd, Separovic F, Watts A, Killian JA. Interfacial anchor properties of tryptophan residues in transmembrane peptides can dominate over hydrophobic matching effects in peptide-lipid interactions. Biochemistry. 2003;42:5341–5348. doi: 10.1021/bi027000r. [DOI] [PubMed] [Google Scholar]
- 87.Wang X, Dutton RJ, Beckwith J, Boyd D. Membrane topology and mutational analysis of Mycobacterium tuberculosis VKOR, a protein involved in disulfide bond formation and a homologue of human vitamin K epoxide reductase. Antioxid Redox Signal. 2011;14:1413–1420. doi: 10.1089/ars.2010.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tie JK, Jin DY, Stafford DW. Mycobacterium tuberculosis vitamin K epoxide reductase homologue supports vitamin K-dependent carboxylation in mammalian cells. Antioxid Redox Signal. 2012;16:329–338. doi: 10.1089/ars.2011.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu S, Tie JK, Stafford DW, Pedersen LG. Membrane topology for human vitamin K epoxide reductase. J Thromb Haemost. 2014;12:112–114. doi: 10.1111/jth.12450. [DOI] [PubMed] [Google Scholar]
- 90.Tie JK, Jin DY, Stafford DW. Conserved loop cysteines of vitamin K epoxide reductase complex subunit 1-like 1 (VKORC1L1) are involved in its active site regeneration. J Biol Chem. 2014;289:9396–9407. doi: 10.1074/jbc.M113.534446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Caspers M, Czogalla KJ, Liphardt K, Muller J, Westhofen P, Watzka M, Oldenburg J. Two enzymes catalyze vitamin K 2,3-epoxide reductase activity in mouse: VKORC1 is highly expressed in exocrine tissues while VKORC1L1 is highly expressed in brain. Thromb Res. 2015;135:977–983. doi: 10.1016/j.thromres.2015.01.025. [DOI] [PubMed] [Google Scholar]
- 92.Hammed A, Matagrin B, Spohn G, Prouillac C, Benoit E, Lattard V. VKORC1L1, an enzyme rescuing the vitamin K 2,3-epoxide reductase activity in some extrahepatic tissues during anticoagulation therapy. J Biol Chem. 2013;288:28733–28742. doi: 10.1074/jbc.M113.457119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Westhofen P, Watzka M, Marinova M, Hass M, Kirfel G, Muller J, Bevans CG, Muller CR, Oldenburg J. Human vitamin K 2,3-epoxide reductase complex subunit 1-like 1 (VKORC1L1) mediates vitamin K-dependent intracellular antioxidant function. J Biol Chem. 2011;286:15085–15094. doi: 10.1074/jbc.M110.210971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Spohn G, Kleinridders A, Wunderlich FT, Watzka M, Zaucke F, Blumbach K, Geisen C, Seifried E, Muller C, Paulsson M, Bruning JC, Oldenburg J. VKORC1 deficiency in mice causes early postnatal lethality due to severe bleeding. Thromb Haemost. 2009;101:1044–1050. [PubMed] [Google Scholar]
- 95.O’Reilly RA, Aggeler PM. Surreptitious ingestion of coumarin anticoagulant drugs. Ann Intern Med. 1966;64:1034–1041. doi: 10.7326/0003-4819-64-5-1034. [DOI] [PubMed] [Google Scholar]
- 96.Ingram BO, Turbyfill JL, Bledsoe PJ, Jaiswal AK, Stafford DW. Assessment of the contribution of NAD(P)H-dependent quinone oxidoreductase 1 (NQO1) to the reduction of vitamin K in wild-type and NQO1-deficient mice. Biochem J. 2013;456:47–54. doi: 10.1042/BJ20130639. [DOI] [PubMed] [Google Scholar]
- 97.Wallin R. Vitamin K antagonism of coumarin anticoagulation. A dehydrogenase pathway in rat liver is responsible for the antagonistic effect. Biochem J. 1986;236:685–693. doi: 10.1042/bj2360685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wallin R, Gebhardt O, Prydz H. NAD(P)H dehydrogenase and its role in the vitamin K (2-methyl-3-phytyl-1,4-naphthaquinone)- dependent carboxylation reaction. Biochem J. 1978;169:95–101. doi: 10.1042/bj1690095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rishavy MA, Hallgren KW, Wilson LA, Usubalieva A, Runge KW, Berkner KL. The vitamin K oxidoreductase is a multimer that efficiently reduces vitamin K epoxide to hydroquinone to allow vitamin K-dependent protein carboxylation. J Biol Chem. 2013;288:31556–31566. doi: 10.1074/jbc.M113.497297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]