Abstract
Background
The role of small non-coding microRNAs (miRNAs) in several types of cancer has been evident. However, its expression studies have never been performed in gastric cancer (GC) patients from Saudi population. First time this study was conducted to identify miRNAs that are differentially expressed in GC patients compared with normal controls.
Methods
We investigated the role of miRNAs in GC patients using formalin-fixed paraffin-embedded (FFPE) tissues of 34 samples from GC patients (early stage = 7 and late-stage = 26) and 15 from normal control. We have used miRNA microarray analysis and validated the results by Real-time quantitative PCR (RT-qPCR).
Results
We obtained data of 1082 expressed genes, from cancer tissues and noncancerous tissues (49 samples in total). Where 129 genes were up-regulated (P > 0.05) and 953 genes (P > 0.05) were down-regulated in 49 FFPE tissue samples. Only 33 miRNAs had significant expression in early and late-stage cancer tissues. After candidate miRNAs were selected, RT-qPCR further confirmed that four miRNAs (hsa-miR-200c-3p, hsa-miR-3613, hsa-miR-27b-3p, hsa-miR-4668-5p) were significantly aberrant in GC tissues compared to the normal gastric tissues.
Conclusions
In this study we provide miRNAs profile of GC where many miRNAs showed aberrant expression from normal tissues, suggesting their involvement in the development and progression of gastric cancer. In early and late-stage miR-200c-3p showed significant down regulation as compare to control samples. Many of miRNAs reported in our study showing up-regulation are new and not reported before may be due to population difference. In conclusion, our results suggest that miR-200c-3p had potential to use as diagnostic biomarker for distinguishing GC patients from normal individuals and can be used for diagnosis of cancer at early stage.
Keywords: Gastric cancer, MicroRNAs, Microarray, Real time PCR, Diagnostic biomarker
Background
Gastric Cancer (GC) is the second major cause for cancer-related deaths worldwide mostly prevalent in Asian countries, including Korea, Japan and China [1]. In Saudi Arabia, For all cancer types about 2.9 % is accounted for GC and it is ranked 11th among both male and female population [2]. Multiple factors contribute to the progression of gastric tumors, including diet rich in salted and nitrated food, alcohol consumptions, low consumption of fruits and vegetables, use of tobacco and, especially, Helicobacter pylori infection [3]. Like other malignancies, both genetic and epigenetic factors are involved in the pathogenesis of gastric cancer [4]. Mortality of the disease is potentially reduces if it is diagnosed at early stages in gastric cancer [5].
Tumor markers always remain helpful at early stages in screening of high risks groups. But, the current cancer tumor markers have limited performance in detection of gastric cancer due to low sensitivity and specificity [6]. Therefore, it is required to discover some novel diagnostic biomarkers for the early detection of this malignancy. MicroRNAs (miRNAs) are small group of RNA molecules that regulate expression of different genes by binding to mRNAs. They play an important role in different cellular processes that are necessary to maintain a normal physiological condition. In cancer, particular miRNAs act as tumor supressor or oncogenic hence considered as biomarkers for early diagnosis and accurate prognosis of cancer [7]. They are considered to control a variety of functions in tumor cell including cell proliferation, migration, invasion, differentiation, apoptosis and metabolic processes [8]. Many previous studies suggest that dysregulated expression of miRNAs associated with cancers, and function as tumor inducer and suppressor and their expression may play an important role in cancer progression. The expression of miRNAs in different oncogenic pathways suggests their importance during carcinogenesis [9].
Microarray-based gene expression profiling is a potential technique to study the expression of miRNAs in GC patients [10].
Several previous studies have reported that miRNAs may be used as diagnostic biomarkers in different cancer types [11]. Fang et al. [12] has suggested some oncogenic miRNAs (miR-10b, miR-21, miR-223 and miR-338) and tumor suppressive miRNAs (miR-30a-5p, miR-126 and let-7a) as prognostic signatures in GC patients. Jiang et al. [13] has reported higher expression of miR-421 in early stage GC patients hence suggesting its role as diagnostic biomarker in GC. Using miRNA array in GC patients, abnormal expression of miRNA profiles with up and down-regulated miRNAs has been reported [14].
Although the importance of miRNAs as important prognostic factors in patients with GC is confirmed, but data on the miRNA signature of GC in the Saudi population is missing. Therefore, the present study was undertaken to detect the miRNA expression profile of GC patients and normal gastric FFPE tissue using miRNA 4.0 microarrays. In both early and late GC tumors, 33 miRNAs were found to be differentially expressed and significantly aberrants were validated using RT-qPCR.
Methods
Clinical samples
In this study, 34 gastric cancer tissues samples were collected from King Abdulaziz University (KAU) hospital Jeddah. Informed consent was obtained from patients undergoing a surgical procedure to remove a portion of gastric cancer. This study was approved by medical ethical committee of KAU, Jeddah Saudi Arabia (Reference#174-15). Using the tumor-node-metastasis (TNM) staging of the International Union Against Cancer (1997) [15] all the tumor samples were staged and graded according to the World Health Organization criteria [16]. From these specimens, we have collected 34 FFPE biopsy samples from gastric cancer patients and 15 FFPE gastric biopsy samples from healthy individuals along with detailed clinical history from KAU hospital (Table. 1).
Table 1.
Clinopathological features of GC patients with normal control
| Sample ID | T | N | M | Age | Sex | Tissue type |
|---|---|---|---|---|---|---|
| Normal | ||||||
| S14-0054 | 50 | Female | Normal | |||
| S14-0211 | 40 | Female | Normal | |||
| S14-0267 | 30 | Female | Normal | |||
| S14-0301 | 38 | Female | Normal | |||
| S14-0306 | 31 | Female | Normal | |||
| S12-0959 | 53 | Male | Normal | |||
| S12-0851 | 37 | Male | Normal | |||
| S12-0850 | 31 | Female | Normal | |||
| S12-0710 | 47 | Male | Normal | |||
| S12-0682 | 28 | Female | Normal | |||
| S12-0669 | 36 | Female | Normal | |||
| S12-0447 | 22 | Male | Normal | |||
| S12-0306 | 26 | Female | Normal | |||
| S12-0080 | 32 | Male | Normal | |||
| S12-0049 | 30 | Male | Normal | |||
| Early-stage | ||||||
| S14.4411 | 1 | 0 | 0 | 56 | Female | Cancer |
| S09-1886 | 2 | 0 | 0 | 78 | Male | Cancer |
| S11-4426 | 2 | 0 | 0 | 87 | Male | Cancer |
| S09-2859 | 2 | 0 | 0 | 52 | Male | Cancer |
| S09-3092 | 2 | 0 | 0 | 59 | Female | Cancer |
| S09-3841 | 2 | 0 | 0 | 72 | Male | Cancer |
| S07-2116 | 2 | 0 | 0 | 62 | Male | Cancer |
| Late-stage | ||||||
| S13.0349-1 | 3 | 2 | 0 | 53 | Male | Cancer |
| S13.0214-4E | 3 | 2 | 0 | 47 | Male | Cancer |
| S13.3130-1D | 4 | 1 | 1 | 59 | Male | Cancer |
| S13.3278-1D | 3 | 2 | 0 | 70 | Female | Cancer |
| S13.5248-2 | 4 | 3 | 1 | 55 | Male | Cancer |
| S14.2777 | 2 | 2 | 0 | 58 | Male | Cancer |
| S14.4235 | 4 | 3 | 1 | 18 | Male | Cancer |
| S14.0190-1 | 3 | 2 | 0 | 77 | Male | Cancer |
| S14.1176-1H | 3 | 2 | 0 | 48 | Male | Cancer |
| S09-3254 | 4 | 3 | 1 | 82 | Female | Cancer |
| S09-3678 | 4 | 1 | 0 | 72 | Male | Cancer |
| S09-251 | 3 | 3 | 0 | 57 | Male | Cancer |
| S00-1751 | 2 | 2 | 0 | 55 | Male | Cancer |
| S01-658 | 2 | 2 | 0 | 60 | Male | Cancer |
| S02-3330 | 2 | 2 | 0 | 45 | Male | Cancer |
| S02-3307 | 2 | 2 | 0 | 62 | Male | Cancer |
| S02-423 | 3 | 3 | 1 | 71 | Female | Cancer |
| S03-3123 | 3 | 3 | 0 | 54 | Male | Cancer |
| S06-3929 | 3 | 3 | 0 | 65 | Male | Cancer |
| S06-3390 | 3 | 3 | 0 | 27 | Male | Cancer |
| S06-3357 | 3 | 3 | 0 | 48 | Male | Cancer |
| S06-2460 | 2 | 1 | 0 | 81 | Female | Cancer |
| S06-2397 | 2 | 1 | 0 | 58 | Male | Cancer |
| S06-1949 | 3 | 2 | 0 | 43 | Male | Cancer |
| S06-1939 | 3 | 2 | 0 | 61 | male | Cancer |
| S06-1830 | 2 | 2 | 0 | 43 | Male | Cancer |
| S06-1875 | 2 | 2 | 0 | 60 | Male | Cancer |
RNA extraction and quality analysis
RNeasy FFPE kit was used for extraction of RNA from FFPE tissues according to manufacturer’s instructions. RNA was further purified using DNase I treatment (Ambion, Austin, TX) to eliminate any contaminating DNA. RNA concentrations were calculated using a Nanodrop ND-1000 spectrophotometer (NanoDrop, Wilmington, USA). RNA integrity was evaluated by running electropherograms and RNA integrity number, RIN (a correlative measure that indicates intactness of mRNA) was determined using the RNA 6000 PicoAssay for the Bioanalyzer 2100 (Agilent Technologies, Palo Alto, USA).
Affymetrix miRNA arrays methods
The Affymetrix Genechip miRNA 4.0 array process was carried out according to the manufacturer’s instructions. 1000 ng RNA samples were labeled with the FlashTag™ Biotin using RNA Labeling Kit (Genisphere, Hatfield, PA, USA). The labeled RNA was further quantified, fractionated and hybridized to the miRNA microarray according to the standard protocol. The labeled RNA was then heated at 99 °C and then at 45 °C (for 5 mins at both temp). An Affymetrix® 450 Fluidics Station was used for RNA-array hybridization with agitation at 60 rotations per minute at 48 °C for 16 h. The chips were washed and stained using a Genechip Fluidics Station 450 (Affymetrix, Santa Clara, California, United States). The chips were then scanned with an Affymetrix GCS 3000 canner (Affymetrix, Santa Clara, California, United States). Using the Affymetrix® GeneChip™ Command Console software (AGCC) signal values were computed.
Raw data preparation and statistic analysis
Raw data were extracted automatically in Affymetrix data extraction protocol using the software provided by AGCC. The CEL files import, miRNA level RMA + DABG-All analysis and result export using Affymetrix® Expression Console™ Software. Array data were filtered by probes annotated species. The comparative analysis between GC samples and control samples was carried out using independent T-test and fold-change where the null hypothesis was that no difference exists among two groups. False discovery rate (FDR) was further controlled by the adjustment of p value using Benjamini-Hochberg algorithm. All Statistical tests and visualization of differentially expressed genes were done using R statistical language v. 3.1.2.
MiRNA quantification by real-time quantitative PCR (RT-qPCR)
For miRNA quantification SYBR Green RT-qPCR assay was used. Using miScript Reverse Transcription kit (Qiagen) one microgram of RNA was reverse-transcribed to cDNA. Further using miScript SYBR Green PCR kit (Qiagen) along with Universal primer and the miRNA-specific primers qPCR is performed in ABI 7900 Real-time PCR system (Applied Biosystems). The miRNA-specific primer sequences were designed using miRNA sequences obtained from the miRBase database. Each reaction mixture contained 10 μl of 2× Fast SYBR Green Master Mix (Life Technologies, Grand Island, NY), 0.5 μl of dye (ROX) II (50x), 1 μl of forward primer, 1 μl of reverse primer, 10 μL of distilled water, and 1 μl of cDNA template. Each sample was run in duplicates and U6 miRNA was used as control to normalize the expression levels of miRNAs. Mean expression values of each miRNAs (dublicate) relative to U6 RNA were calculated using the 2-∆CT method [17], wherein ∆Ct = Ct(tumor)-Ct(control) and control is the sample without treatment.
Results and discussion
Expression profiles of miRNA in gastric cancer
We used miRNA 4.0 array gene chips, to evaluate miRNA expression profiles between FFPE GC and normal tissues. By adjusting average change >2-fold and value <0.05 as a cut-off level, 1082 expressed genes were differentially expressed between the cancerous and normal control. Among them, one-way ANOVA showed that only 33 miRNAs had significant expression, where 8 (has-mir-193a-5p, has-mir-200c-3p, has-mir-1227-5p, has-mir-1909-3p, has-mir-1378, has-mir-3613-3p, has-mir-5196-5p and has-mir-7704) were apparently expressed in early stage cancer tissues (p < 0.05) and 25 (hsa-miR-22-3p, hsa-miR-28-3p, hsa-miR-29a-3p, hsa-miR-99a-5p, hsa-miR-27b-3p, hsa-miR-143-3p, hsa-miR-145-5p, hsa-miR-1251-5p, hsa-miR-126-3p, hsa-miR-185-5p, hsa-miR-200c-3p, hsa-miR-130b-3p, hsa-miR-378a-3p, hsa-miR-877-5p, hsa-miR-128, hsa-miR-378c, hsa-miR-3613-3p, hsa-miR-4532, hsa-miR-4668-5p, hsa-miR-4800-5p, hsa-miR-6124, hsa-miR-6812-5p, hsa-miR-7150, hsa-miR-7704, hsa-miR-7847-3p) were in advanced stage cancer samples as compare to normal control (p < 0.05) (Table. 2). As we used 2-fold as cut off value so some aberrant miRNAs in the chip analysis may be neglected. Differentially expressed genes were related to signal transduction, metabolism, angiogenesis, cell structure and cycle, and gene protein expression were mostly down-regulated. We found 13 new among 33 aberrant miRNAs that have not been reported before in GC studies. These miRNAs named as hsa-miR-1227-5p, hsa-miR-1909-3p, hsa-miR-3178, hsa-miR-7704 (in early-stage GC) and hsa-miR-1251-5p, hsa-miR-877-5p, hsa-mir-1281, hsa-miR-4532, hsa-miR-6124, hsa-miR-6812-5p, hsa-mir-7150, hsa-mir-7704 and hsa-mir-7847-3p (in late-stage GC) (Table. 2). There are many concerns about the integrity of miRNA from FFPE tissues and their suitability for microarray as RNA might be modified in chemical reaction and become fragmented in FFPE tissues [18] but due to very small size of miRNA (about 19-22 nucleotides) it’s not degraded in FFPE preparation. Many previous studies of miRNA microarray supporting a correlation in miRNA expression results from fresh and FFPE cancer tissues samples [19–21]. We found in all samples good value of RNA integrity and optical density (data not shown). In order to assess miRNA data after filtering we built box plot to visualize the distribution of data. The distributions of log2 ratios are almost same among miRNA chips. The probe-level data for the box plots are distributed from about 2–14 on the log scale (Fig. 1a). To check variation between cancer and normal tissues samples we assess the data using scatter-plot. Around 70–80 % miRNA seems to be same in cancerous and normal tissues used in this study (Fig. 1b).
Table 2.
Expression profile of up and down-regulated miRNAs in GC patients
| Transcript ID(Array Design) | Accession | Fold Change (linear) (GC vs. Control) | FDR p-value (GC vs. Control) |
|---|---|---|---|
| Detected in early stage (I and II) | |||
| hsa-miR-193a-5p | MIMAT0004614 | −2.06796 | 0.77052133 |
| hsa-miR-200c-3p | MIMAT0000617 | −22.552013 | 0.77052133 |
| hsa-miR-1227-5p | MIMAT0022941 | −1.717431 | 0.77052133 |
| hsa-miR-1909-3p | MIMAT0007883 | −1.951977 | 0.77052133 |
| hsa-miR-3178 | MIMAT0015055 | −1.826664 | 0.72999652 |
| hsa-miR-3613-3p | MIMAT0017991 | 2.981591 | 0.72999652 |
| hsa-miR-5196-5p | MIMAT0021128 | −1.959981 | 0.77052133 |
| hsa-miR-7704 | MIMAT0030019 | −1.893422 | 0.72999652 |
| U79 | _ | 1.738033 | 0.74726754 |
| Detected in early stage (III and IV) | |||
| hsa-miR-22-3p | MIMAT0000077 | −1.90434 | 0.10052633 |
| hsa-miR-28-3p | MIMAT0004502 | −1.87632 | 0.145678142 |
| hsa-miR-29a-3p | MIMAT0000427 | −1.96579 | 0.196109756 |
| hsa-miR-99a-5p | MIMAT0000097 | −1.97584 | 0.21331947 |
| hsa-miR-27b-3p | MIMAT0000419 | −2.29128 | 0.275381924 |
| hsa-miR-143-3p | MIMAT0000435 | −2.20679 | 0.132397944 |
| hsa-miR-145-5p | MIMAT0000437 | −2.20593 | 0.271934938 |
| hsa-miR-1251-5p | MIMAT0028700 | −2.70147 | 0.014814394 |
| hsa-miR-126-3p | MIMAT0005903 | −1.9555 | 0.292936143 |
| hsa-miR-185-5p | MIMAT0001339 | −1.55828 | 0.212836342 |
| hsa-miR-200c-3p | MIMAT0000455 | −2.18027 | 0.113424849 |
| hsa-miR-130b-3p | MIMAT0000691 | −2.41458 | 0.126769698 |
| hsa-miR-378a-3p | MIMAT0000732 | −1.78021 | 0.074619747 |
| hsa-miR-877-5p | MIMAT0004949 | 1.696113 | 0.064059796 |
| hsa-miR-1281 | MIMAT0005939 | −1.97312 | 0.064954696 |
| hsa-miR-378c | MIMAT0016847 | −2.51734 | 0.016444786 |
| hsa-miR-3613-3p | MIMAT0017991 | 2.507303 | 0.212235198 |
| hsa-miR-4532 | MIMAT0019071 | −2.05911 | 0.046570249 |
| hsa-miR-4668-5p | MIMAT0019745 | 2.097354 | 0.268773791 |
| hsa-miR-4800-5p | MIMAT0019978 | 1.510005 | 0.257957349 |
| hsa-miR-6124 | MIMAT0024597 | 2.156251 | 0.088004364 |
| hsa-miR-6812-5p | MIMAT0027524 | 1.583428 | 0.250692071 |
| hsa-miR-7150 | MIMAT0028211 | 1.628161 | 0.277892706 |
| hsa-miR-7704 | MIMAT0030019 | −1.64036 | 0.016444786 |
| hsa-miR-7847-3p | MIMAT0030422 | 1.59577 | 0.244713443 |
| U3 | _ | 1.780994 | 0.060093686 |
| U63 | _ | −1.69859 | 0.116804752 |
| U3-2B | _ | 1.761599 | 0.070969781 |
| U3-2 | _ | 1.761599 | 0.070969781 |
| U3-3 | _ | 1.761599 | 0.070969781 |
| U3-4 | _ | 1.761599 | 0.070969781 |
Fig. 1.
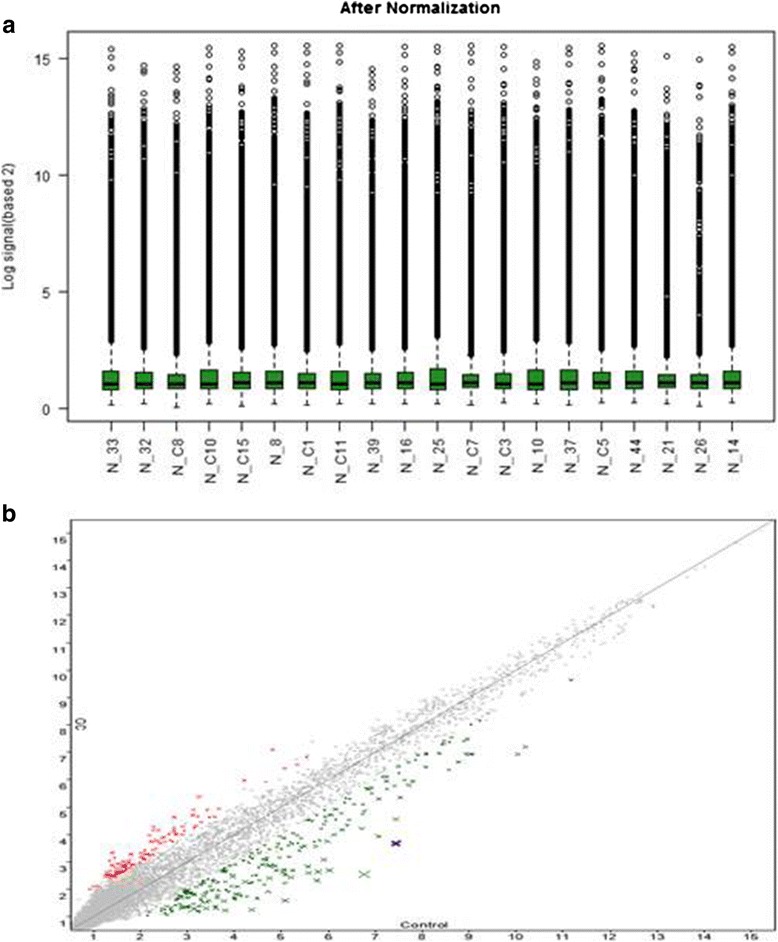
(a) Comparison of sample distribution using box plot (b) and scatter plot to show difference in miRNAs expression between cancerous and normal control samples
Identification of differentially expressed miRNAs in early vs late-stage GC
We analyzed the expression of miRNAs significantly expressed from cancerous (early and last stage) and normal tissues using Volcano plot filtering. For up and down-regulated miRNAs genes we used ≥ 2.0 fold change and value P value (<0.05). In this plot, differentially expressed genes are statistically significant and are shown in red and green points (Fig. 2). We identified 1082 differently expressed miRNA and only 33 had significant expression, where 9 genes showed up-regulation and 24 genes showed down-regulation of expression in GC (early and late-stage GC) as compare to normal gastric tissues. Further hierarchical clustering analyses (Fig. 3) were performed on the basis of differentially expressed miRNAs from cancerous versus normal tissues. Based on this hierarchical clustering, we have divided miRNAs expression in three groups: early-stage, late-stage and normal tissues. These results showed change in cancerous and control tissue samples. We have identified many new miRNAs from this study from Saudi population that have never been reported in studies related to cancer as well as in GC. Among 33 aberrant miRNAs (early and late-stage GC) 13 were new and rest of the miRNAs were reported before in cancer studies. Similarly, from remaining 20 miRNAs, only 9 were reported in GC studies before. Most of the miRNAs were down-regulated only 9 were up-regulated. In early-stage GC tissue samples, 8 miRNAs had aberrant expression as compare to normal control. Among them 7 was up-regulated and only 1 showed down-regulation (Table. 2). Some of them were not reported before in GC except has-mir-200c-3p. Some miRNAs identified in early-stage cancer tissues samples, including hsa-miR-193a-5p, hsa-miR-3613 and hsa-miR-5196-5p were reported already in cervical cancer and acute lymphoblastic leukemia [22, 23]. In our study, significant down-regulation of has-mir-200c-3p has been seen in early and late-stage GC tissue samples.
Fig. 2.
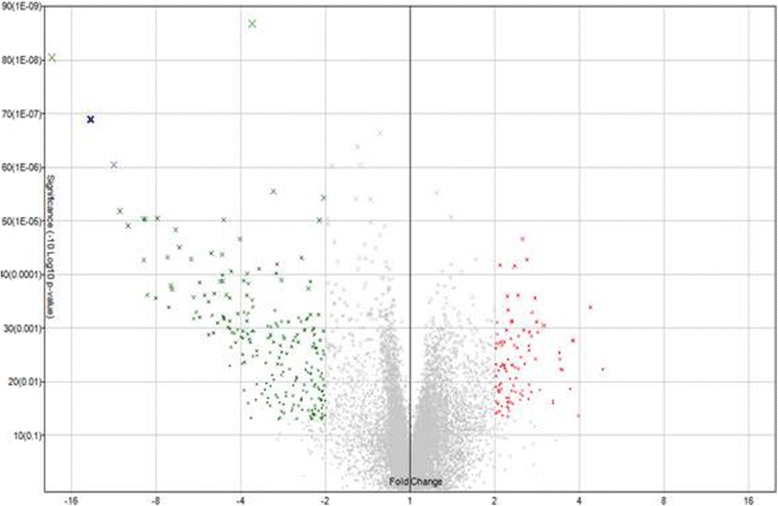
Volcano plot showing the relative expression of miRNA genes. The vertical lines correspond to 2.0-fold up and down, respectively, and the horizontal line represents a value of 0.05. The red point in the plot represents the differentially expressed genes with statistical significance
Fig. 3.
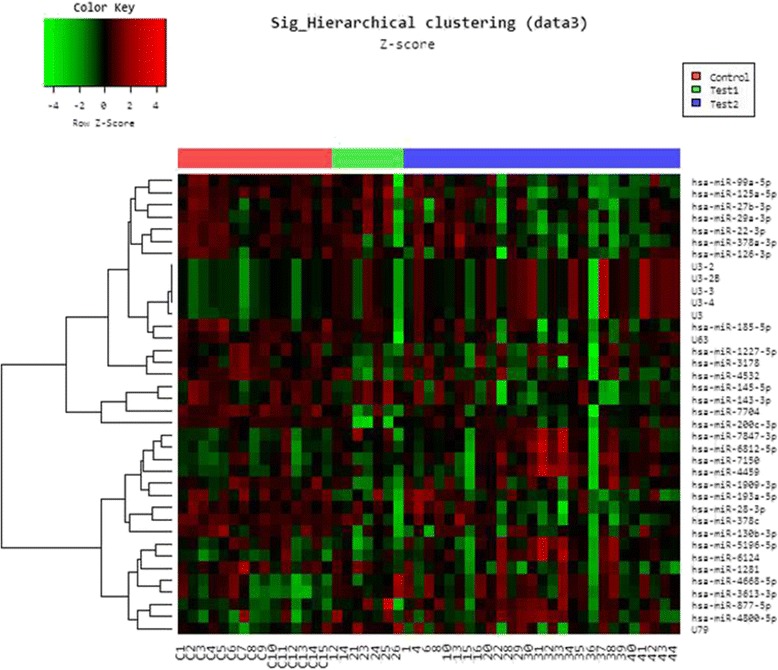
Hierarchical clustering for differentially expressed miRNAs in cancer versus normal pass volcano plot. Red indicates high relative expression and green indicates low relative expression
The miR-200 family consists of five members and is encoded by two genes on chromosomes 12 and function as tumor regulator [24, 25]. It plays an important role in different types of cancer by inhibition, invasion, migration, proliferation and drug resistance. Previous studies have shown down-regulation of miR-200c in GC that is consistent with our study [26, 27]. The miR-200c has been reported in various types of cancers, including colorectal [28], breast [29], cholangiocarcinoma [30], colorectal [31], hepatocellular [32], lung adenocarcinoma [33], ovarian [34] and renal cell carcinoma [35]. The miR-200 family inhibits the epithelial-mesenchymal transition and metastasis by down-regulating ZEB1 and ZEB2 (Zinc-finger E-box Binding homeobox 1 and 2). The miR-200 inhibits angiogenesis by regulating interleukin-8 and CXCL1 secreted by cancer cells. Marked reduction of angiogenesis has been observed after delivering miR-200 members into the tumour endothelium [36]. Another study reported that extracellular matrix proteins and peptidases are targeted by miR-200 and alters the tumor microenvironment to inhibit metastasis [37]. H. pylori induced infection is another risk factor for progression of GC. Matsushima et al. [38] have characterized that decreased expression of tumor suppressor family miR-200 has been seen in H.pylori positive GC patients. Few other studies reported down-regulation of miR-200 in H. pylori-infected GC and increased expression of anti-apoptotic proteins, Bcl-2 and XIAP hence inhibiting apoptosis [39–41].
In late-stage GC tissue samples, 8 miRNAs were up-regulated and 17 showed down-regulation with 10 new miRNAs (has-mir-1251-5p, has-mir-877-5p, has-mir-1281, has-mir-378c, has-mir-4532, has-mir-6124, has-mir-6812-5p, has-mir-7150, has-mir-7704 and has-mir-7847-3p) not reported before (Table. 2). Many miRNAs involve in tumorigenesis and altered in variety of cancers are detected in our study such as tumor-suppressor (has-mir-22-3p, has-mir-28-3p, has-mir-99a-5p, has-mir-130b-3p, has-mir-378a-3p, has-mir-3613-3p, has-mir-4668-5p and has-mir-4800-5p) are reported before in colorectal [42], cervical [22] and breast cancer [43]. Some of the miRNAs (has-mir-29a-3p, has-mir-27b-3p, has-mir-143-3p, has-mir-145-5p, has-mir-126-3p, has-mir-185-5p, has-mir-200c-3p) differential expressed in late-stage GC samples had similar kind of expression in previously published GC studies. The down-regulation of has-miR-29a-3p in our study is consistent with previous study of GC where decreased expression of has-miR-29a-3p promotes cell proliferation by suppressing the expression of cell cycles regulators [44, 45]. The receptor tyrosine kinase like orphan receptor 1 (ROR1) protein is an oncogenic protein. The miR-27b-3p suppress the expression of ROR1 in GC hence function as tumor suppressor [46]. Similar expression of has-mir-27b-3p has been detected in our study. Furthermore, other GC related miRNAs such as has-mir-143-3p, has-mir-145-5p, has-mir-126-3p, has-mir-185-5p have found to be down-regulated in our study have shown similar expression in other studies and reported as tumor-suppressors [47–49]. In addition to miRNAs, we found seven small nucleolar RNA (snoRNAs) differentially expressed in both early and late-stage tumor samples. In our study, only 1 snoRNA (U63) showed down-regulation and others (U79, U3, U3-2b, U3-2, U3-3 and U3-4) were up-regulated and are related to human C/D box group (Table. 2). Several recent studies have highlighted the role of snoRNAs as miRNAs precursors and have similar function like miRNAs [50, 51]. Another study on Epstein-Barr virus genome, suggests their role in infection [52]. What role these snoRNAs may play in GC is interesting to explore.
The present study has limitations due to small number of tumor samples (n = 34), especially samples of early stages. Therefore, results of the present study need further validation using larger group of GC patients in future. Our results showed that significant down-regulation of has-mir-200c-3p is markedly observed in early-stage GC as compare to late-stage GC samples.
Validation of dysregulated miRNAs by quantitative PCR analysis
Further using real-time quantitative PCR analysis, results from expression array were validated. We selected four significantly aberrant miRNAs with 2-fold change including hsa-miR-200c-3p, hsa-miR-3613, hsa-miR-27b-3p, hsa-miR-3613 from both stages to test in cancerous versus normal tissues and miRNA U6 was used as control. The miRNA expression analysis of 2 oncogenic hsa-miR-200c and hsa-miR-27b-3p was significantly down-regulated while hsa-miR-3613 and hsa-miR-3613 showed up-regulation in comparison with normal control (Fig. 4a and b) that is consistent with miRNAs array results. Significantly down-regulation of hsa-miR-200c has been reported previously in many studies of cancer indicating the role of hsa-miR-200c as tumor suppressor [24–34]. hsa-miR-200c had multiple roles in regulating tumor cell growth by inhibiting the metastasis and invasion of hepatocellular carcinoma and gastric carcinoma [35, 36]. At last, accordant down-regulation of hsa-miR-200c in GC tissues was detected, in comparison with the normal tissues. We also found aberrant expression of some new miRNAs in GC patients, which have no report of aberrant expression in any other studies.
Fig. 4.

Quantification of the highly expressed miRNAs in the human GC versus normal control in four selected upregulated miRNA. (a) Early-stage and (b) Late-stage miRNAs
Conclusions
In conclusion, we explored the miRNAs expression of FFPE gastric tissues from GC patients and normal control. Many miRNAs showed aberrant expression in cancerous versus normal control. As it is evident from many previous studies and also the current results strongly suggest that hsa-miR-200c acts as a tumor suppressor miRNA that plays a potential role in the oncogenesis in humans. Hence highlighting its functions as a tumor-suppressive miRNA and prognostic marker in GC patients in Saudi population. The significance and role of aberrant miRNAs expression of GC patients in Saudi population will be better understood as more miRNAs will be identified. Our study may be helpful in future to identify potential prognostic biomarkers for GC.
Acknowledgments
Declarations
This article has been published as part of BMC Genomics Volume 17 Supplement 9, 2016: Proceedings of the 3rd International Genomic Medicine Conference: genomics. The full contents of the supplement are available online at http://bmcgenomics.biomedcentral.com/articles/supplements/volume-17-supplement-9.Publication fees for this article are paid by the Center of Excellence in Genomic Medicine Research (CEGMR), King Abdulaziz University, Jeddah, Kingdom of Saudi Arabia.
Acknowledgements
This project was funded by the National Plan for Science, Technology and Innovation (MAARIFAH)–King Abdulaziz City for Science and Technology-the Kingdom of Saudi Arabia-award number (12-BIO-2725-03). The authors also, acknowledge with thanks Science and Technology Unit, King Abdulaziz University for technical support.
Authors’ contributions
FB and MIN made substantial contributions to design the study. SA, MY, AMA, MHA and EIA were involved in expression arrays and data interpretation. AJF and AS were responsible for sample collection and of clinical databases. FB and MIN drafted the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study was approved by the Research Committee of the Biomedical Ethics Unit, Faculty of Medicine, King Abdulaziz University, Jeddah, Saudi Arabia.
Abbreviations
- GC
Gastric cancer
- miRNAs
microRNAs
- FFPE
Formalin-fixed paraffin-embedded
- qPCR
Real-time quantitative PCR
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Saudi Cancer Registry . Cancer incidence report in Saudi Arabia 2010. Riyadh (KSA): Saudi Cancer Registry; 2014. [Google Scholar]
- 3.Mínguez B, Lachenmayer A. Diagnostic and prognostic molecular markers in hepatocellular carcinoma. Dis Markers. 2011;31(3):181–190. doi: 10.3233/DMA-2011-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009;374(9688):477–490. doi: 10.1016/S0140-6736(09)60617-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gotoda T, Yamamoto H, Soetikno RM. Endoscopic submucosal dissection of early gastric cancer. J Gastroenterol. 2006;41(10):929–942. doi: 10.1007/s00535-006-1954-3. [DOI] [PubMed] [Google Scholar]
- 6.Li C, Jian Fang L, Qu C, Qing Qing Q, Min Y, Bing Ya L, Zheng Gang Z. MiRNA-199a-3p: a potential circulating diagnostic biomarker for early gastric cancer. J Surg Oncol. 2013;108(2):89–92. doi: 10.1002/jso.23358. [DOI] [PubMed] [Google Scholar]
- 7.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, Dirbas FM. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138(3):592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni L, Zhang WG, Nan KJ, Song TS, Huang C. MicroRNA profiling of human gastric cancer. Mol Med Report. 2009;2(6):963–970. doi: 10.3892/mmr_00000199. [DOI] [PubMed] [Google Scholar]
- 10.Jiang HB, Yang TJ, Lu P, Ma YJ. Gene expression profiling of gastric cancer. Eur Rev Med Pharmacol Sci. 2014;18(15):2109–2115. [PubMed] [Google Scholar]
- 11.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 12.Fang Y, Shen H, Li H, Cao Y, Qin R, Long L, Zhu X, Xie C, Xu W. miR-106a confers cisplatin resistance by regulating PTEN/Akt pathway in gastric cancer cells. Acta Biochim Biophys Sin. 2013;45(11):963–72. doi: 10.1093/abbs/gmt106. [DOI] [PubMed] [Google Scholar]
- 13.Jiang Z, Guo J, Xiao B, Miao Y, Huang R, Li D, Zhang Y. Increased expression of miR-421 in human gastric carcinoma and its clinical association. J Gastroenterol. 2010;45(1):17–23. doi: 10.1007/s00535-009-0135-6. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Luo F, Li Q, Xu M, Feng D, Zhang G, Wu W. Identification of new aberrantly expressed miRNAs in intestinal-type gastric cancer and its clinical significance. Oncol Rep. 2011;26(6):1431–1439. doi: 10.3892/or.2011.1437. [DOI] [PubMed] [Google Scholar]
- 15.Sobin LH, Wittekind CH. TNM classification of malignant tumors. 5. New York: Wiley; 1997. pp. 59–62. [Google Scholar]
- 16.Solcia E, Capella C. Endocrine tumours of the gastrointestinal tract. In: Solcia E, Kloppel G, Sobin LH, editors. Histological typing of endocrine tumours. 2. Berlin: Springer; 2000. pp. 61–8. [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Liu A, Xu X. MicroRNA isolation from formalin-fixed, paraffin-embedded tissues. Methods Mol Biol. 2011;724:259–267. doi: 10.1007/978-1-61779-055-3_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Smyth P, Flavin R, Cahill S, Denning K, Aherne S, Guenther SM, O’Leary JJ, Sheils O. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007;7:36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glud M, Klausen M, Gniadecki R, Rossing M, Hastrup N, Nielsen FC, Drzewiecki KT. MicroRNA expression in melanocytic nevi: the usefulness of formalin-fixed, paraffin-embedded material for miRNA microarray profiling. J Invest Dermatol. 2009;129(5):1219–1224. doi: 10.1038/jid.2008.347. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Chen J, Radcliffe T, LeBrun DP, Tron VA, Feilotter H. An array-based analysis of microRNA expression comparing matched frozen and formalin-fixed paraffin-embedded human tissue samples. J Mol Diagn. 2008;10(6):513–519. doi: 10.2353/jmoldx.2008.080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67(13):6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 23.Schotte D, Moqadam FA, Lange-Turenhout EAM, Chen C, Van IJcken WFJ, Pieters R, Den Boer ML. Discovery of new microRNAs by small RNAome deep sequencing in childhood acute lymphoblastic leukemia. Leukemia. 2011;25(9):1389–1399. doi: 10.1038/leu.2011.105. [DOI] [PubMed] [Google Scholar]
- 24.Tang H, Deng M, Tang Y, Xie X, Guo J, Kong Y, Ye F, Su Q, Xie X. miR-200b and miR-200c as prognostic factors and mediators of gastric cancer cell progression. Clin Cancer Res. 2013;19(20):5602–5612. doi: 10.1158/1078-0432.CCR-13-1326. [DOI] [PubMed] [Google Scholar]
- 25.Humphries B, Yang C. The microRNA-200 family: small molecules with novel roles in cancer development, progression and therapy. Oncotarget. 2015;6(9):6472. doi: 10.18632/oncotarget.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valladares-Ayerbes M, Reboredo M, Medina-Villaamil V, Iglesias-Díaz P, Lorenzo-Patiño MJ, Haz M, Santamarina I, Blanco M, Fernández-Tajes J, Quindós M, Carral A. Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J Transl Med. 2012;10(1):186. doi: 10.1186/1479-5876-10-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng X, Wang Z, Fillmore R, Xi Y. MiR-200, a new star miRNA in human cancer. Cancer Lett. 2014;344(2):166–173. doi: 10.1016/j.canlet.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu L, Qiu M, Tan G, Liang Z, Qin Y, Chen L, Chen H, Liu J. miR-200c Inhibits invasion, migration and proliferation of bladder cancer cells through down-regulation of BMI-1 and E2F3. J Transl Med. 2014;12(1):305. doi: 10.1186/s12967-014-0305-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song C, Liu LZ, Pei XQ, Liu X, Yang L, Ye F, Xie X, Chen J, Tang H. miR-200c inhibits breast cancer proliferation by targeting KRAS. Oncotarget. 2015;6(33):34968–34978. doi: 10.18632/oncotarget.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng F, Jiang J, Yu Y, Tian R, Guo X, Li X, Shen M, Xu M, Zhu F, Shi C, Hu J. Direct targeting of SUZ12/ROCK2 by miR-200b/c inhibits cholangiocarcinoma tumourigenesis and metastasis. Br J Cancer. 2013;109(12):3092–3104. doi: 10.1038/bjc.2013.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun J, Ding W, Zh, J, Chen W. MiR-200 suppresses metastases of colorectal cancer through ZEB1. Tumour Biol. 2015; 1-7 [DOI] [PubMed]
- 32.Yeh TS, Wang F, Chen TC, Yeh CN, Yu MC, Jan YY, Chen MF. Expression profile of microRNA-200 family in hepatocellular carcinoma with bile duct tumor thrombus. Ann Surg. 2014;259(2):346–354. doi: 10.1097/SLA.0000000000000223. [DOI] [PubMed] [Google Scholar]
- 33.Roybal JD, Zang Y, Ahn YH, Yang Y, Gibbons DL, Baird BN, Alvarez C, Thilaganathan N, Liu DD, Saintigny P, Heymach JV. miR-200 inhibits lung adenocarcinoma cell invasion and metastasis by targeting Flt1/VEGFR1. Mol Cancer Res. 2011;9(1):25–35. doi: 10.1158/1541-7786.MCR-10-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu X, Macdonald DM, Huettner PC, Feng Z, El Naqa IM, Schwarz JK, Mutch DG, Grigsby PW, Powel SN, Wang X. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol. 2009;114(3):457–464. doi: 10.1016/j.ygyno.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 35.Nakada C, Matsuura K, Tsukamoto Y, Tanigawa M, Yoshimoto T, Narimatsu T, Nguyen LT, Hijiya N, Uchida T, Sato F, Mimata H. Genome‐wide microRNA expression profiling in renal cell carcinoma: significant down‐regulation of miR‐141 and miR‐200c. J Pathol. 2008;216(4):418–427. doi: 10.1002/path.2437. [DOI] [PubMed] [Google Scholar]
- 36.Pecot CV, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C, Wu S, Han HD, Shah MY, Rodriguez-Aguayo C, Bottsford-Miller J. Tumour angiogenesis regulation by the miR-200 family. Nat Commun. 2013;4:2427. doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schliekelman MJ, Gibbons DL, Faca VM, Creighton CJ, Rizvi ZH, Zhang Q, Wong CH, Wang H, Ungewiss C, Ahn YH, Shin DH. Targets of the tumor suppressor miR-200 in regulation of the epithelial-mesenchymal transition in cancer. Cancer Res. 2011;71(24):7670–7682. doi: 10.1158/0008-5472.CAN-11-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsushima K, Isomoto H, Inoue N, Nakayama T, Hayashi T, Nakayama M, Nakao K, Hirayama T, Kohno S. MicroRNA signatures in helicobacter pylori-infected gastric mucosa. Int J Cancer. 2011;128(2):361–70. doi: 10.1002/ijc.25348. [DOI] [PubMed] [Google Scholar]
- 39.Furukawa-Hibi Y, Yoshida-Araki K, Ohta T, Ikeda K, Motoyama N. FOXO forkhead transcription factors induce G(2)-M checkpoint in response to oxidative stress. J Biol Chem. 2002;277:26729–26732. doi: 10.1074/jbc.C200256200. [DOI] [PubMed] [Google Scholar]
- 40.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284:23204–23216. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu W, Xu H, Zhu D, Zhi H, Wang T, Wang J, Jiang B, Shu Y, Liu P. miR-200bc/429 cluster modulates multidrug resistance of human cancer cell lines by targeting BCL2 and XIAP. Cancer Chemother Pharmacol. 2012;69:723–731. doi: 10.1007/s00280-011-1752-3. [DOI] [PubMed] [Google Scholar]
- 42.Almeida MI, Nicoloso MS, Zeng L, Ivan C, Spizzo R, Gafà R, Xiao L, Zhang X, Vannini I, Fanini F, Fabbri M. Strand-specific miR-28-5p and miR-28-3p have distinct effects in colorectal cancer cells. Gastroenterology. 2012;142(4):886–896. doi: 10.1053/j.gastro.2011.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Persson H, Kvist A, Rego N, Staaf J, Vallon-Christersson J, Luts L, Loman N, Jonsson G, Naya H, Hoglund M, Borg A. Identification of new microRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene. Cancer Res. 2011;71(1):78–86. doi: 10.1158/0008-5472.CAN-10-1869. [DOI] [PubMed] [Google Scholar]
- 44.Zhao Z, Wang L, Song W, Cui H, Chen G, Qiao F, Hu J, Zhou R, Fan H. Reduced miR-29a-3p expression is linked to the cell proliferation and cell migration in gastric cancer. World J. Surg. Oncol. 2015;13(1):1. doi: 10.1186/s12957-015-0513-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui Y, Su WY, Xing J, Wang YC, Wang P, Chen XY, Shen ZY, Cao H, Lu YY, Fang JY. MiR-29a inhibits cell proliferation and induces cell cycle arrest through the downregulation of p42.3 in human gastric cancer. PLoS One. 2011;6(10):e25872. doi: 10.1371/journal.pone.0025872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tao J, Zhi X, Zhang X, Fu M, Huang H, Fan Y, Guan W, Zou C. miR-27b-3p suppresses cell proliferation through targeting receptor tyrosine kinase like orphan receptor 1 in gastric cancer. J Exp Clin Cancer Res. 2015;34(1):1. doi: 10.1186/s13046-015-0253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xing X, Li Z. Expression of miR-143 and miR-145 and their functional study in gastric carcinoma. Chinese J Gastrointest. Surg. 2015;18(1):50–3. [PubMed] [Google Scholar]
- 48.Feng R, Chen X, Yu Y, Su L, Yu B, Li J, Cai Q, Yan M, Liu B, Zhu Z. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298(1):50–63. doi: 10.1016/j.canlet.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 49.Tan Z, Jiang H, Wu Y, Xie L, Dai W, Tang H, Tang S. miR-185 is an independent prognosis factor and suppresses tumor metastasis in gastric cancer. Mol Cell Biochem. 2014;386(1-2):223–31. doi: 10.1007/s11010-013-1860-y. [DOI] [PubMed] [Google Scholar]
- 50.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–686. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hutzinger R, Feederle R, Mrazek J, Schiefermeier N, Balwierz PJ, et al. Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome. PLoS Pathog. 2009;5:e1000547. doi: 10.1371/journal.ppat.1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]


