Abstract
Methanobactins (Mbns) are a growing family of ribosomally produced, post-translationally modified natural products. Characteristic nitrogen-containing heterocycles and neighboring thioamides allow these compounds to bind copper with high affinity. Genome mining has enabled the identification of Mbn operons in bacterial genomes and the prediction of diverse Mbn structures from operon content and precursor peptide sequence. Here we report the characterization of Mbn from Methylosinus (Ms.) species (sp.) LW4. The peptide backbone is distinct from all previously characterized Mbns, and the post-translational modifications correspond precisely to those predicted on the basis of the Ms. sp. LW4 Mbn operon. Thus, prediction based on genome analysis combined with isolation and structural characterization represents a phylogenetic approach to finding diverse Mbns and elucidating their biosynthetic pathways.
Methane is the sole carbon source for most methanotrophic bacteria, and the primary methane-oxidizing enzyme is the copper enzyme particulate methane monooxygenase (pMMO).1 pMMO can comprise up to a quarter of the cellular protein mass in these bacteria, and their need for copper is correspondingly large. Methanotrophs have thus evolved several different copper acquisition systems, one of which uses siderophore-like natural products (“chalkophores”, from the Greek chalco- “copper”) to import copper.2 These compounds are the ribosomally produced, post-translationally modified natural products (RiPPs)3 known as methanobactins (Mbns).4
Five Mbns have been structurally characterized to date, three via X-ray crystallography,5 one via NMR,6 and one using both techniques (Figure S1).7,8 All five have post-translational modifications consisting of nitrogen-containing heterocycles with neighboring thioamide groups; these moieties comprise the copper-binding ligands. Additional modifications, including N-terminal transamination and threonine sulfonation, are present in some Mbns,5,8 but are not universal.
The first characterized Mbn was that from Methylosinus (Ms.) trichosporium OB3b.7,9 Six years ago, a small ORF proposed to encode a Mbn precursor peptide was identified in the Ms. trichosporium OB3b genome.6 A similar ORF was identified in an unrelated non-methanotrophic species, Azospirillum species (sp.) B510,6 inspiring a genome-mining study that identified putative Mbn precursor peptide-encoding genes, designated mbnA, in 20 operons in 18 species,10 now 51 complete operons in 47 species.11 Based on the conserved genes present in these operons, proteins involved in Mbn transport have been identified11 and specific roles for biosynthesis enzymes have been suggested, but have not yet been experimentally verified. However, if their predicted functions are correct, the precursor peptide sequence and the identities of operon-encoded biosynthesis enzymes enable prediction of candidate structures for the resulting Mbns (Figure S2.)
As proof of principle, we have characterized the Mbn from Ms. sp. LW4. The genome of Ms. sp. LW4 is available, and like other Ms. species, contains an Mbn operon (Figure 1). MbnA precursor peptides consist of a leader and a core sequence, the latter of which contains one or more conserved C(A/G/S)(C/S/T) sequences identifying modifiable cysteines; on the basis of sequence alignments of all known MbnAs, the peptide backbone in the final Ms. sp. LW4 compound is predicted to be MCACNPPWCGTC, in which the two underlined cysteine residues are post-translationally modified to form nitrogen-containing heterocycles and neighboring thioamide groups. This Mbn operon contains only three predicted biosynthesis enzymes: the uncharacterized proteins MbnB and MbnC, which are proposed to mediate oxazolone and thioamide biosynthesis at conserved C(A/G/S)(C/S/T) moieties in the precursor peptide, and MbnN, a PLP-containing transaminase that targets the N-terminal primary amine group.10 Additional enzymes are predicted to mediate other post-translational modifications. For example, MbnF is an FAD-dependent oxidoreductase suggested to be involved in pyrazinedione biosynthesis and MbnS is a PAPS-dependent sulfotransferase that sulfonates threonines; two additional enzymes remain unstudied.10 Because Ms. sp. LW4 lacks these additional enzymes, the heterocycles are predicted to be oxazolones, as in Ms. trichosporium OB3b Mbn. Generation of the oxazolones and thioamides should be accompanied by a transamination reaction mediated by MbnN targeting the N-terminal amine, as well as the formation of a disulfide bond between the two unaltered cysteines (Figure 1) (also present in Ms. trichosporium OB3b, and believed to form post-secretion). These combined modifications yield a predicted mass of 1334.2322 Da for Ms. sp. LW4 CuMbn.
Figure 1.
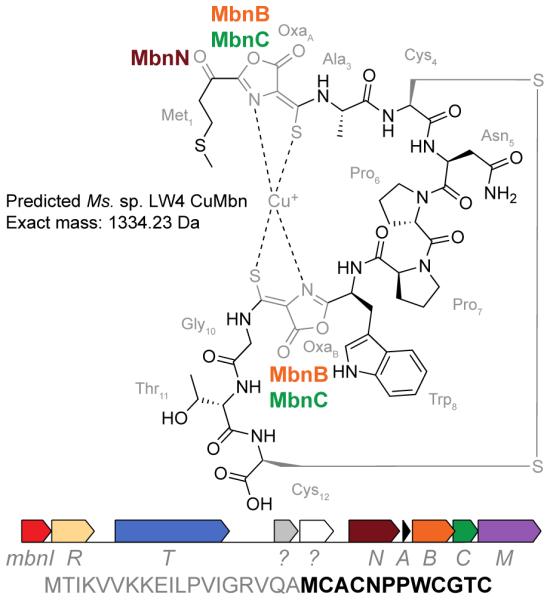
Predicted structure for Mbn from Ms. sp. LW4 (top) along with its Mbn operon (middle) and MbnA sequence (bottom, leader peptide in gray and core peptide in black). Biosynthesis genes encode the precursor peptide (MbnA), the uncharacterized proteins MbnB and MbnC, and the aminotransferase MbnN. Other genes are related to regulation (MbnI, MbnR) or transport (MbnT, MbnM).
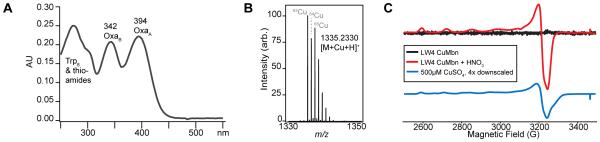
When grown under conditions of copper starvation, Ms. sp. LW4 cultures take on a yellowish cast. Initial purification of mildly hydrophobic compounds from copper-stabilized spent medium using a Diaion HP-20 column12 results in the elution of a yellow-brown compound. The UV-visible spectrum (Figure 2A) of the apo compound is very similar to those of previously isolated Mbns.5-7 Major features can be assigned to the tryptophan and thioamides (overlapping, center at 273 nm), oxazolones B (342 nm) and A (395 nm).
Figure 2.
Spectroscopic characterization of Ms. sp. LW4 Mbn. (A) UV-vis spectrum of ~45μM apo Ms. sp. LW4 Mbn. Peaks at 342 and 394 nm correspond to the two oxazolone rings; enhanced absorbance below 300 nm is likely due to the presence of a tryptophan. (B) Positive ion mode LC-MS spectrum of Ms. sp. LW4 CuMbn. The copper isotopic signature is visible. (C) EPR spectrum of 222 μM Ms. sp. LW4 CuMbn (black). After nitric acid treatment (red), the signal (g∥= 2.400, g⊥= 2.08, and A∥ = 132 G) resembles type 2 copper (blue) and is consistent with copper release and oxidation by nitric acid.
Positive mode mass spectrometry (MS) analysis indicates the presence of a compound with a single charge and m/z 1335.2330. The detected ion species has an m/z within 1 ppm of the predicted [M+H+Cu]+ ion and a distinct isotopic distribution consistent with the presence of copper, as observed for other Mbns (Figure 2B).5-7 Present in the tandem MS spectrum of this ion species is a short sequence tag representing a diproline moiety (Figure S3A) as well as several fragment ions associated with tryptophan-containing peptides (m/z 130.6, 159.09, and 170.60; Figure S3B). Both a diproline moiety and a tryptophan are present in the predicted Ms. sp LW4 Mbn structure (Figures 1, S2, S3). Thus, the isolated compound has a mass that exactly matches the predicted mass, and contains elements of the predicted Ms. sp LW4 Mbn backbone. The presence of copper was confirmed by ICP-MS, and no other metal ions were detected above background.
To assess the oxidation state of bound copper, CuMbn samples were analyzed via electron paramagnetic resonance (EPR) spectroscopy (Figure 2C). No signal is initially observed, but after incubation with 10% HNO3 a Cu(II) signal appears (Figure 2C). Spin quantitation (244 μM EPR-active copper) compared to ICP-MS analysis (222 μM total copper) indicates quantitative release and oxidation of bound Cu(I), consistent with Mbn binding a single Cu(I) ion as previously observed.13 The MS and EPR results strongly suggest that the compound isolated from Ms. sp. LW4 is indeed consistent with the structure predicted based on MbnA sequence and operon content (Figure 1).
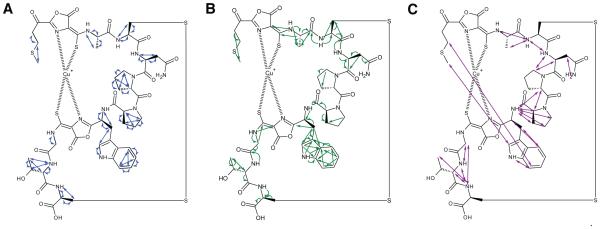
We thus pursued NMR characterization using samples without isotopic enrichment. [1H-1H] TOCSY, [1H-1H] ROESY, [1H-13C] HMBC, [1H-13C] HSQC, and [1H-15N] HSQC (Table S1) were performed in 10% D2O in 10 mM phosphate buffer pH 6.5; all but [1H-15N] HSQC were also performed in 100% D2O. The results are consistent with the proposed structure (Figures 3, S4, Table S1). The peaks are sharp, ruling out paramagnetic effects that would be caused by Cu(II). In the 1H-15N HSQC experiment, the asparagine primary amine protons and eight additional proton signals were detected, accounting for seven backbone amides and the secondary amine in the tryptophan ring. No amide proton signals are detectable for five residues, including Pro6 and Pro7 (as expected), as well as Met1 (predicted to lose the N-terminal primary amine due to the aminotransferase MbnN), Cys2, and Cys9 (the modified cysteines). This is consistent with the presence of oxazolone rings8 but not imidazolones and not some pyrazinedione tautomers, in which an extra amine is present and visible (Figure S1).6 There are proton and carbon signals consistent with side chains for all residues except for the modified Cys2 and Cys9 (Figure 3A). TOCSY correlations were observed between neighboring residues, and were interrupted by the proton-poor oxazolone/thioamide moieties; longer-range NOE interactions measured via ROESY support a three-dimensional structure with Mbn wrapped around the copper ion, like that observed for other CuMbns5,7 (Figure 3B). In the 1H-13C HMBC experiments, longer-range carbon-proton interactions were also observed for most residues as well as several carbons present in the oxazolone/thioamide moieties.
Figure 3.
NMR support for the proposed structure of Ms. sp. LW4 CuMbn. (A) Proton-proton correlations observed via [1H-1H] TOCSY. (B) Multibond proton-carbon coupling observed via [1H-13C]-HMBC. (C) Inter-residue Nuclear Overhauser Effect (NOE) interactions observed via [1H-1H]-ROESY.
While these data are consistent with the presence of two oxazolone/thioamide moieties, the modifications are proton-poor, which hinders conclusive heterocycle identification by NMR. However, acid hydrolysis monitored by UV-Vis spectroscopy strongly supports this conclusion. Exposure of Ms. sp. LW4 Mbn to 100 mM HCl results in hydrolysis of first the C-terminal oxazolone (OxaB) and then the N-terminal oxazolone ring (OxaA) on a timescale similar to that observed for Ms. trichosporium OB3b Mbn (Figures 4, S5).6 Notably, barring oxazolines,14 acid sensitivity is significantly less in other common RiPP heterocycles, including those in Methylocystis (Mc.) Mbns,6 and the oxazoles/thiazoles in microcins and lantibiotics.
Figure 4.
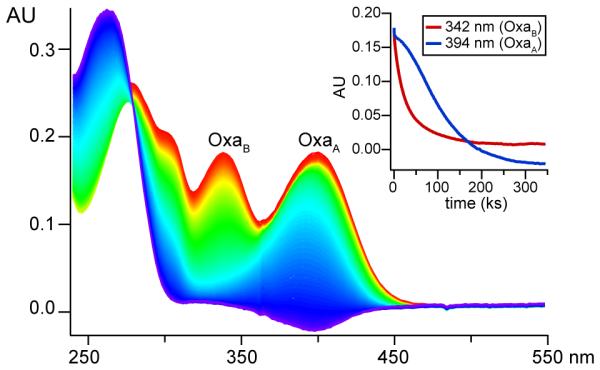
Acid hydrolysis of ~40 μM Ms. sp. LW4 Mbn. Inset: decrease in OxaB at 342nm (red) and OxaA at 395nm (blue) over 96 h.
Structural characterization of CuMbns is not always straightforward. The crystal structure of the original CuMbn from Ms. trichosporium OB3b was revised via NMR to correctly identify the heterocycles and other products of post-translational modifications.7,8 In addition, the NMR structure of CuMbn from Mc. sp. SB2 may be incorrect; the first heterocycle is proposed to be a 5-membered imidazolone ring,6 but the crystal structures of CuMbn from closely related Mc. species have a 6-membered pyrazinedione in that location.5 Mc. rosea SV97 and Mc. sp. SB2 have identical MbnAs, and MbnB and MbnC sequences that differ by 3 and 1 residues, respectively; MbnB and MbnC are proposed to form the initial heterocycles.10 Two closely related Mc. species, Mc. hirsuta CSC1 and Mc. sp. M also produce Mbns with a pyrazinedione and not an imidazolone at that position.5 Other Mc. Mbn biosynthesis genes are also well-conserved, including the FAD-dependent monooxygenase MbnF (proposed to be involved in pyrazinedione biosynthesis) and the sulfotransferase MbnS (86-99% and 93-99%).10 Genomic and bioinformatic data thus strongly suggest that Mbns produced by the two species should be identical.
One potential explanation for this discrepancy would be the presence of the hydroxy-oxo tautomer and a protonated secondary amine rather than the untautomerized pyrazinedione ring, which is doubly hydroxylated and has a tertiary amine (Figure S6). Importantly, the secondary amine present in this tautomer could produce [1H-13C-HMBC] and [1H-15N]-HMBC NMR signals quite similar to those observed for the ring assigned as an imidazolone in Mc. sp. SB2 CuMbn. The presence of this tautomer is in fact suggested by the Mc. sp. M CuMbn electron density map,5 in which the amine appears protonated (Figure S6).
For Ms. sp. LW4 CuMbn, several lines of evidence support the correct assignment of both rings as oxazolones, including the susceptibility of the nitrogen-containing heterocycles to acid hydrolysis, a mass shift consistent with glycine formation after acid hydrolysis (Figures S5, S7), and the lack of additional nitrogen signals in the 1H-15N HSQC spectra. In addition, the Ms. sp. LW4 Mbn operon contains only the uncharacterized biosynthesis proteins MbnB and MbnC and the transaminase MbnN, with no other biosynthesis genes.
Taken together, these data identify a novel Mbn structurally distinct from both the originally characterized Ms. trichosporium OB3b Mbn and the group of related Mc. Mbns. Most important, the proposed Ms. sp. LW4 Mbn structure perfectly matches the predicted structure and is completely consistent with the assignment of biosynthetic roles to the proteins MbnB, MbnC, and MbnN. Such correlations between MbnA sequences, operon contents, and experimentally determined structures provide a complementary and alternative strategy towards characterization of previously uninvestigated protein families involved in Mbn biosynthesis, and can be extended to uncharacterized Mbns and Mbn-like natural products from a range of bacteria.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Marina G. Kalyuzhnaya for Ms. sp. LW4 and Dr. Yongbo Zhang for NMR assistance. IMSERC is supported by NIH grants 1S10OD012016-01/1S10RR019071-01A1, Northwestern University/the state of Illinois, the IIN and the NSF under NNCI-1542205. High-resolution mass spectral analyses were partially supported by the NRTDP (GM108569).
Funding Sources
Funding for this work was provided by the NSF (MCB0842366 to A.C.R.), the NIH (GM070473 and GM118035 to A.C.R., GM111097 to B.M.H., AT009143 to N.L.K., 5T32GM008382 to M.O.R.), and the AHA (14PRE20460104 to G.E.K.).
Footnotes
Supporting Information. An attached PDF contains supplemental experimental procedures and data.
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Sirajuddin S, Rosenzweig AC. Biochemistry. 2015;54:2283. doi: 10.1021/acs.biochem.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Semrau JD, DiSpirito AA, Yoon S. FEMS Microbiol. Rev. 2010;34:496. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- (3).Velásquez JE, van der Donk WA. Curr. Opin. Chem. Biol. 2011;15:11. doi: 10.1016/j.cbpa.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Kenney GE, Rosenzweig AC. ACS Chem. Biol. 2012;7:260. doi: 10.1021/cb2003913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Ghazouani, el A, Basle A, Gray J, Graham DW, Firbank SJ, Dennison C. Proc. Nat. Acad. Sci. USA. 2012;109:8400. doi: 10.1073/pnas.1112921109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Krentz BD, Mulheron HJ, Semrau JD, DiSpirito AA, Bandow NL, Haft DH, Vuilleumier S, Murrell JC, McEllistrem MT, Hartsel SC, Gallagher WH. Biochemistry. 2010;49:10117. doi: 10.1021/bi1014375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kim HJ, Graham DW, DiSpirito AA, Alterman MA, Galeva N, Larive CK, Asunskis D, Sherwood PMA. Science. 2004;305:1612. doi: 10.1126/science.1098322. [DOI] [PubMed] [Google Scholar]
- (8).Behling LA, Hartsel SC, Lewis DE, DiSpirito AA, Choi DW, Masterson LR, Veglia G, Gallagher WH. J. Am. Chem. Soc. 2008;130:12604. doi: 10.1021/ja804747d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).DiSpirito AA, Zahn JA, Graham DW, Kim HJ, Larive CK, Derrick TS, Cox CD, Taylor A. J. Bacteriol. 1998;180:3606. doi: 10.1128/jb.180.14.3606-3613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kenney GE, Rosenzweig AC. BMC Biol. 2013;11:17. doi: 10.1186/1741-7007-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Dassama LMK, Kenney GE, Ro SY, Zielazinski EL, Rosenzweig AC. In review [Google Scholar]
- (12).Bandow NL, Gallagher WH, Behling LA, Choi DW, Semrau JD, Hartsel SC, Gilles VS, DiSpirito AA. Methods Enzymol. 2011;495:259. doi: 10.1016/B978-0-12-386905-0.00017-6. [DOI] [PubMed] [Google Scholar]
- (13).Hakemian AS, Tinberg CE, Kondapalli KC, Telser J, Hoffman BM, Stemmler TL, Rosenzweig AC. J. Am. Chem. Soc. 2005;127:17142. doi: 10.1021/ja0558140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Lee J, Hao Y, Blair PM, Melby JO, Agarwal V, Burkhart BJ, Nair SK, Mitchell DA. Proc. Nat. Acad. Sci. USA. 2013;110:12954. doi: 10.1073/pnas.1306101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.