ABSTRACT
How host–virus co‐evolutionary relationships manifest is one of the most intriguing issues in virology. To address this topic, the mammal–lentivirus relationship can be considered as an interplay of cellular and viral proteins, particularly apolipoprotein B mRNA editing enzyme catalytic polypeptide‐like 3 (APOBEC3) and viral infectivity factor (Vif). APOBEC3s enzymatically restrict lentivirus replication, whereas Vif antagonizes the host anti‐viral action mediated by APOBEC3. In this study, the focus was on the interplay between feline APOBEC3 proteins and two feline immunodeficiency viruses in cats and pumas. To our knowledge, this study provides the first evidence of non‐primate lentiviral Vif being incapable of counteracting a natural host's anti‐viral activity mediated via APOBEC3 protein.
Keywords: apolipoprotein B mRNA editing enzyme catalytic polypeptide‐like 3, feline immunodeficiency virus, lentivirus, viral infectivity factor
List of Abbreviations
- APOBEC3
apolipoprotein B mRNA editing enzyme catalytic polypeptide‐like 3
- FIV
feline immunodeficiency virus
- HIV‐1
human immunodeficiency virus type 1
- OWM
Old World monkey
- PLV
puma lentivirus
- PBMC
peripheral blood mononuclear cells
- SIV
simian immunodeficiency virus
- Vif
viral infectivity factor
Shedding light on the co‐evolutionary history of viruses and hosts is one of the most intriguing topics in the field of virology. However, because viruses have usually become highly diversified through their high evolutionary rates and cross‐species swapping, accessing the co‐evolutionary history of viruses and hosts has been challenging. Focusing on interactions between viral and cellular proteins is a potential strategy for investigating the evolutionary arms race between viruses and hosts, particularly lentiviruses and mammals 1, 2, 3. In particular, the interaction between Vif and APOBEC3 represents a microcosm of the co‐evolutionary relationship 1.
HIV‐1, which causes AIDS in humans, is the most studied lentivirus 4. HIV‐1 replication is robustly restricted by an intrinsic anti‐viral APOBEC3 family protein, APOBEC3G 5. APOBEC3 proteins, including APOBEC3G, are incorporated into nascent viral particles and enzymatically introduce G‐to‐A substitutions in the newly synthesized viral DNA, which results in the abrogation of viral replication 6, 7. To counteract APOBEC3‐mediated anti‐viral action, Vif, an HIV‐1‐encoding protein, induces degradation of APOBEC3 proteins via a ubiquitin/proteasome‐dependent pathway 8, 9.
In contrast to the effects of HIV‐1 infection in humans, it is known that several lineages of OWMs residing in Africa do not develop any AIDS‐like disorders when naturally infected with SIVs, another lineage of lentiviruses 10, 11. Interestingly, the functional interaction between a various lineage of SIV Vif proteins and OWM APOBEC3G proteins has been elucidated 1, 12, 13, possibly providing a clue to understanding the history of co‐evolution and co‐divergence of SIVs and monkeys in the Old World. Furthermore, it has been demonstrated that OWM APOBEC3G can be the barrier that restricts lentiviral cross‐species infection 14, 15.
Another lentivirus, FIV, was first isolated in 1987 from domestic cats (Felis catus) with chronic AIDS‐like disorders 16. Subsequent studies revealed that several lineages of felids are infected with FIV 17, 18, 19.
The puma (Puma concolor; also commonly known as the cougar, panther, or mountain lion) is a felid that resides in the New World 20. An FIV that has been designated FIVpco (also known as PLV), circulates among pumas in the wild 21. The prevalence of FIVpco in wild pumas differs between the geographical areas investigated and/or the epidemiological studies reported 17, 21, 22, 23. Interestingly, because of its lower genetic diversity 22, it has been assumed that FIVpco has not undergone any selective pressures from the host and that wild pumas are naturally infected (i.e., with little or no pathogenicity), as is true for SIV infection in OWMs in Africa 24. However, prominent G‐to‐A substitutions, the mutation signature mediated by APOBEC3, have reportedly been detected in proviral DNA isolated from wild pumas naturally infected with FIVpco 25. Therefore, it is assumed that endogenous puma APOBEC3 protein(s) are resistant to FIVpco Vif‐mediated degradation, which would affect the replication kinetics of FIVpco in pumas.
To elucidate the mechanism of inter‐species viral transmission, domestic cats have been experimentally infected with FIVpco 26, 27, 28, 29. However, PLV1695, a prototypic infectious molecular clone of FIVpco 30, replicates poorly in domestic cats, usually being eradicated around 100 days post‐infection without any treatment 30, 31. FIVpco expands in cultures of PBMCs from pumas in vitro, whereas it only replicates very little in in vitro cultures of domestic cat PBMCs 26. These observations suggest that a barrier inhibits cross‐species transmission of FIVpco into domestic cats. Particularly noteworthy, G‐to‐A hypermutation in proviral DNA, which is the signature of APOBEC3‐mediated mutation, has been observed in cultures of domestic cat PBMCs infected with FIVpco 30.
The fact that human APOBEC3G robustly limits HIV‐1 replication in the absence of Vif both in vitro 5, 8 and in vivo 32, 33, 34, is convincing evidence that human APOBEC3G confers intrinsic immunity restricting HIV‐1 replication. Similar to what occurs in HIV‐1 infection in humans, certain APOBEC3 proteins from domestic cats exhibit strong anti‐FIV activity, whereas this anti‐viral action is counteracted by the Vif protein of FIV in domestic cats (FIVfca) 35, 36, 37. The presence of the vif gene in FIV strongly suggests that there was co‐evolutionary interplay between feline APOBEC3 and FIV Vif proteins. In fact, as described above, one study has suggested that feline APOBEC3 is the barrier that inhibits cross‐species transmission of FIVpco (strain PLV1695) into domestic cats 30. However, the counteracting activity of FIVpco Vif against APOBEC3 proteins from both domestic cats and pumas has not been elucidated. To address these issues, we herein focused on the role of FIVpco Vif in antagonizing APOBEC3 proteins from domestic cats and pumas.
We set out to compare the amino acid sequences of APOBEC3s from domestic cats and pumas and those of Vifs from FIVfca and FIVpco. Human APOBEC3s and HIV‐1 Vif, respectively, were used as the putative outgroups. It should be noted that domestic cats and pumas express five different APOBEC3 proteins; namely APOBEC3Z2a, APOBEC3Z2b, APOBEC3Z2c, APOBEC3Z3 and APOBEC3Z2Z3 35, 36, 37, 38, whereas humans encode seven APOBEC3 proteins (APOBEC3A, B, C, D, F, G and H) 7, 8. As shown in Figure 1, we found that puma APOBEC3Z2 (Fig. 1a) and APOBEC3Z3 (Fig. 1b) are similar to cat orthologs rather than human orthologs. On the other hand, although FIVpco Vif are more closely related to FIVfca Vif than to HIV‐1 Vif (Fig. 1c), the values for identity and similarity of FIVpco Vif to FIVfca Vif are relatively low compared to those for puma APOBEC3s to cat APOBEC3s (Fig. 1a,b). These results indicate that lentiviral vif is relatively diversified in comparison with host APOBEC3 proteins. Moreover, we compared the FIV Vif sequences from various felids; namely FIVfca in cats, FIVpco in pumas, FIVple in lions (Panthera leo) and FIVoma in Pallas' cats (Otocolobus manul). As shown in Figure 1d, FIV vif is highly diversified but forms a cluster in the same lineage. This figure also shows that the sequence of PLV1695 vif is close to that of the vif of the other FIVpco from wild pumas (Fig. 1d), suggesting that the use of PLV1695 Vif in our subsequent experiments is biologically relevant.
Figure 1.
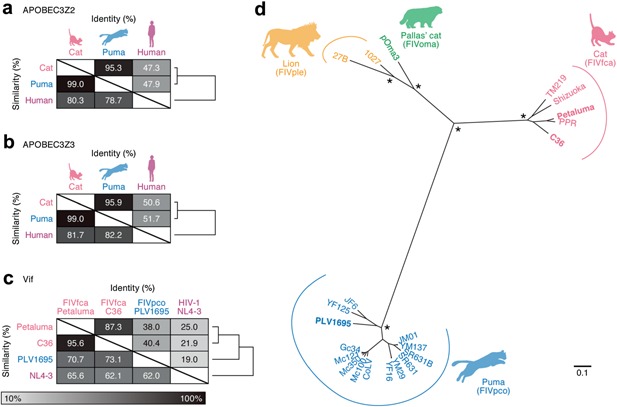
Comparison of APOBEC3 and Vif sequences. The identity and similarity of amino acid sequences of (a) APOBEC3Z2, (b) APOBEC3Z3 and (c) Vif. Unrooted phylogenetic trees are shown on the right of the respective matrix. The sequences of human APOBEC3 (a, b) and HIV‐1 Vif (c) were used as the outgroups. The identity and similarity of amino acid sequences were analyzed using GENETYX v 9.0. The following sequences were used in this analysis; GenBank accession numbers are indicated in parentheses: domestic cat, APOBEC3Z2a (EU109281), APOBEC3Z3 (EU011792); puma, APOBEC3Z2 (GU097659), APOBEC3Z3 (GU097659); human, APOBEC3Z2 (NM_014508), APOBEC3Z3 (NM_001166002); FIVfca Vif strain Petaluma (M25381), strain C36 (AY600517); FIVpco strain PLV1695 (DQ192583) and HIV‐1 strain NL4‐3 (M19921). Note that human APOBEC3Z2 and APOBEC3Z3 are identical to APOBEC3C and APOBEC3H, respectively 55. A3Z2, APOBEC3Z2; A3Z3, APOBEC3Z3. (d) Unrooted phylogenetic tree of FIV vif reconstructed by the maximum likelihood method. The common names of the hosts of each FIV are provided below each illustration. The names of the cognitive FIV lineage of each host are shown in parentheses. The FIV strains used in the experiments, the results of which are shown in Figures 2 and 3, are shown in bold. Note that the bootstrap values on the nodes of respective FIV lineages (FIVfca, FIVpco, FIVoma, and FIVple) are more than 80 (indicated with asterisks). The scale bar indicates an evolutionary distance of 0.1 nucleotide substitutions per site. We used the following 23 FIV strains to construct this phylogenetic tree (GenBank accession numbers in parentheses): FIVfca, Petaluma (M25381), TM219 (M59418), C36 (AY600517), Shizuoka (LC079040), PPR (M36968), BM3070 (AF474246); FIVoma, pOma3 (AY713445); FIVpco, PLV1695 (DQ192583), Gc34 (EF455603), Mc350 (EF455604), Mc100 (EF455605), Mc121 (EF455606), YM29 (EF455607), YF16 (EF455608), JM01 (EF455609), JF6 (EF455610), YM137 (EF455611), YF125 (EF455612), SR631 (EF455613), SR631B (EF455614), CoLV (EF455615); FIVple, 27B (EU117991) and 1027 (EU117992).
Next, we used an in vitro cell culture system to evaluate whether FIVpco Vif can degrade feline APOBEC3 proteins. For this assay, we obtained the FLAG‐tagged codon‐optimized open reading frames of FIVfca Vif strain Petaluma 39, FIVfca strain C36 40 and FIVpco strain PLV1695 26 from GeneArt Gene Synthesis service (Life Technologies, Carlsbad, CA, USA) (the GenBank accession numbers are presented in the legend of Figure 1, and the codon‐optimized sequences are available on request). The obtained DNA fragments were inserted the BamHI‐SalI site of pDON‐AI plasmid (Takara, Kyoto, Japan). The expression plasmids for HA‐tagged APOBEC3Z3 and APOBEC3Z2Z3 from domestic cats 35 and pumas 37 were kindly provided by Dr. Carsten Münk (Heinrich Heine University, Düsseldorf, Germany). To produce FIV‐based lentivirus vector, pFP93 (pFIVgagpolΔvif; we used a replication‐incompetent vif‐deficient FIV packaging construct derived from clone FIV 34TF10 (GenBank accession no. M25381; 430 ng) 41, pTiger‐luc (pFIVΨ‐luc) (430 ng; Addgene, Cambridge, MA, USA), and pMD.G (pVSVg, a VSVg expression plasmid; 150 ng). We co‐transfected these plasmids into HEK293T cells by using PEI Max (Polysciences, Warrington, PA, USA) and harvested the cells and viral particles from culture supernatants at 48 hr post‐transfection. The harvested samples were used for SDS‐PAGE/western blotting or lentiviral reporter assays as previously described 32, 39, 42, 43. Briefly, we used the following antibodies for western blotting: an anti‐FLAG polyclonal antibody (Sigma, St Louis, MO, USA), an anti‐HA antibody (3F10; Roche, Indianapolis, IN, USA), an anti‐FIV p27Capsid antibody (PAK3‐2C1; Santa Cruz Biotechnology, Santa Cruz, CA, USA); an anti‐α‐tubulin antibody (DM1A; Sigma) and an anti‐VSVg antibody (P5DA; Roche). For FIV reporter assays, we used HEK293T cells for the target cells 44.
It is known that feline (i.e., both cats and pumas) APOBEC3Z3 and APOBEC3Z2Z3 can impair FIV infection 36, 37, 39. Consistent with these previous observations, we incorporated domestic cat APOBEC3Z3 (Fig. 2a) and APOBEC3Z2Z3 (Fig. 2b) into the released virions and found that the packaged APOBEC3 proteins decreased FIV infectivity in a dose‐dependent manner (Fig. 2c,d). Also, FIVfca Vif (strains Petaluma and C36) significantly augmented viral infectivity by impairing the incorporation of these proteins into released virions (Fig. 2).
Figure 2.
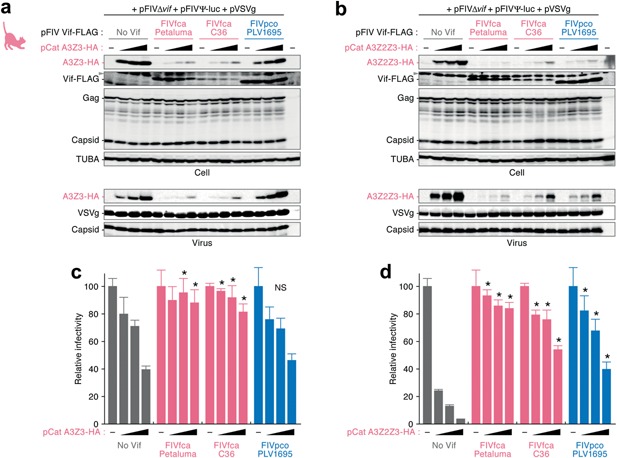
Resistance of domestic cat APOBEC3Z3 to FIVpco Vif. The plasmids expressing FLAG‐tagged FIV Vif (800 ng) and HA‐tagged domestic cat APOBEC3Z3 or APOBEC3Z2Z3 (0, 50, 100 and 290 ng) were co‐transfected with the plasmids for producing FIV‐based lentiviral vector (see the main text for detail) into HEK293T cells. The cell extracts and viral particles from culture supernatants were analyzed by (a, b) western blotting and (c, d) FIV‐based reporter assay. The assays were performed in triplicate. Representative results of western blotting are shown in (a) and (b). In panels (a) and (b); gray arrowheads indicate non‐specific bands. (c) and (d) show the percentages of viral infectivity compared to the value without APOBEC3 expression. The error bars indicate SDs. *, P < 0.05 versus “No Vif” by Student's t‐test. A3Z3, APOBEC3Z3; A3Z2Z3, APOBEC3Z2Z3; NS, no statistically significant difference compared to the values for “No Vif”.
We then assessed the ability of FIVpco Vif to degrade feline APOBEC3 proteins. Although FIVpco Vif degraded domestic cat APOBEC3Z2Z3, as in the case of FIVfca Vif (Fig. 2b), it was of interest that FIVpco Vif was unable to degrade domestic cat APOBEC3Z3 (Fig. 2a). The undegraded cat APOBEC3Z3 proteins were efficiently incorporated into the released viral particles (Fig. 2a,b, bottom panels) and suppressed viral infectivity (Fig. 2c,d). These findings suggest that domestic cat APOBEC3Z3 is resistant to FIVpco Vif‐mediated degradation.
Next, we analyzed the functional relationship between puma APOBEC3 and FIV Vif proteins. As in the case of domestic cat APOBEC3 proteins, puma APOBEC3Z3 and APOBEC3Z2Z3 proteins suppressed viral infectivity in a dose‐dependent manner (Fig. 3). We also found that the anti‐viral action of puma APOBEC3 proteins is significantly antagonized by FIVfca Vif (Fig. 3). However, although puma APOBEC3Z2Z3 is counteracted by FIVpco Vif (Fig. 3b,d), we found that FIVpco Vif is unable to degrade the APOBEC3Z3 protein of the puma, its natural host (Fig. 3a), and that viral infectivity is suppressed by the packaged APOBEC3Z3 protein as in the case of an absence of Vif expression (Fig. 3c).
Figure 3.
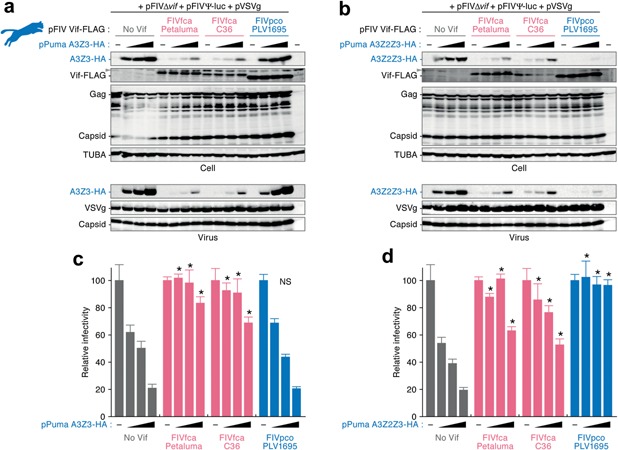
Resistance of puma APOBEC3Z3 to FIVpco Vif. The plasmids expressing FLAG‐tagged FIV Vif (800 ng) and HA‐tagged puma APOBEC3Z3 or APOBEC3Z2Z3 (0, 50, 100 and 290 ng) were co‐transfected with the plasmids for producing FIV‐based lentiviral vector (see the main text for detail) into HEK293T cells. The cell extracts and viral particles from culture supernatants were analyzed by (a, b) western blotting and (c, d) FIV‐based reporter assay. The assays were performed in triplicate. Representative results of western blotting are shown in (a) and (b); gray arrowheads indicate non‐specific bands. (c) and (d) show the percentages of viral infectivity compared to the value without APOBEC3 expression. The error bars indicate SDs. *, P < 0.05 versus “No Vif” by Student's t‐test. A3Z3, APOBEC3Z3; A3Z2Z3, APOBEC3Z2Z3; NS, no statistically significant difference compared to the values of “No Vif”.
In this study, we found that FIVpco Vif is unable to degrade domestic cat APOBEC3Z3. This finding raises the possibility that anti‐FIV activity of feline APOBEC3Z3 is not crucial for FIV replication and that the antagonizing ability of FIV Vif against feline APOBEC3Z3 is dispensable for FIV. However, here we also demonstrated that feline APOBEC3Z3 proteins significantly attenuate FIV infectivity (Fig. 2). Moreover, it has been reported that Vif is a prerequisite for FIV replication in both in vitro cell culture systems 45, 46 and in vivo 47. Taken together, these observations argue against the possibility described in the second sentence of this paragraph.
Rather, we found that FIVpco Vif is incapable of counteracting the APOBEC3Z3 protein of pumas, its natural host (Fig. 3a,c). Even in the case of SIVs naturally infected with OWMs in Africa, SIV Vif proteins counteract the APOBEC3G proteins of their natural hosts 13. Therefore, to the best of our knowledge, this is the first report demonstrating that a non‐primate lentiviral Vif protein is unable to degrade an anti‐viral APOBEC3 protein of its natural host (Fig. 3a,c). FIVpco strain PLV1695, which we used this study and which is the only infectious clone of FIVpco reported so far, is able to replicate in puma PBMCs 26, and pumas in the wild are naturally infected with FIVpco 17, 21, 22, 23. However, as described above, a previous study has demonstrated that G‐to‐A mutations, which are presumably mediated by endogenous APOBEC3, are detectable in the proviral DNA of wild pumas naturally infected with FIVpco 25. Therefore, taken together with our findings, puma APOBEC3Z3 may partially suppress, but not completely prevent, FIVpco replication.
As described above, several lentiviruses have been identified in mammals and most of them encode Vif 48, 49. This fact strongly suggests that there are various patterns of cross‐species transmissions and evolutionary histories between lentiviruses and their hosts. For instance, Etienne and colleagues have recently reported that chimpanzee APOBEC3 proteins play a pivotal role as the barrier that protects interspecies transmission of OWM lentiviruses 50. In addition, D'arc and colleagues have reported that gorilla APOBEC3G is resistant to the degradation mediated by the Vif protein of SIV in chimpanzee 51. In contrast, here we found that FIVfca Vif degrades anti‐viral APOBEC3 proteins (i.e., APOBEC3Z3 and APOBEC3Z2Z3) of both cat and puma (Figs 2,3). These results suggest that FIVfca Vif has a potent and broader ability to counteract the anti‐viral actions of various feline APOBEC3 proteins. In this regard, it is known that a predator–prey system is one of the triggers leading to the interspecies transmission of viruses 52. In fact, previous studies have reported that pumas and domestic cats are sympatric in North America and share certain pathogens such as feline leukemia virus 53, 54, strongly supporting the possibility that FIVfca‐infected domestic cats can be puma's prey and that FIVfca can be transmitted to puma.
In summary, our observations suggest that the evolutionary interplay between viruses and hosts, particularly Vif and APOBEC3, is more complicated than expected. Clarifying the interplay between lentiviral Vif and mammalian APOBEC3 may a provide clues to elucidating the evolutionary bottleneck of emerging/re‐emerging viruses, including lentiviruses.
DISCLOSURE
All authors have no interests to declare.
ACKNOWLEDGMENTS
We would like to thank Drs. Eric M. Poeschla (Mayo Clinic, Rochester, MN, USA) and Carsten Münk (Heinrich Heine University, Germany) for providing pFP93 and HA‐tagged feline APOBEC3 expression plasmids, respectively. We also thank Dr. Kenta Matsuda (National Institute of Health, Bethesda, MD, USA) for crucial comments and Ms. Kotubu Misawa (Institute for Virus Research, Kyoto University, Japan) for her dedicated support. This study was supported in‐part by CREST, JST (to KS); Health Labour Sciences Research Grant 26361601 from MHLW (to KS); Sumitomo Foundation Research Grant (to KS); Imai Memorial Trust (to KS); Ichiro Kanehara Foundation (to KS); Kanae Foundation (to KS); Suzuken Memorial Foundation (to KS); Uehara Memorial Foundation (to KS); Takeda Science Foundation (to KS); JSPS Research Fellowship PD 15J06242 (to RY); Scientific Research C 15K07166 (to KS) from JSPS; a Grant‐in‐Aid for Scientific Research on Innovative Areas 24115008 from MEXT (to YK); Research on HIV/AIDS from Japan Agency for Medical Research and Development (AMED) 15Afk0410013h0001 (to YK); JSPS Core‐to‐Core Program, A. Advanced Research Networks (to YK); and the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Dynamic Approaches to Living System) from AMED (to EY).
REFERENCES
- 1. Compton A.A., Malik H.S., Emerman M. ( 2013) Host gene evolution traces the evolutionary history of ancient primate lentiviruses. Philos Trans R Soc Lond B Biol Sci 368: 20120496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sato K., Gee P., Koyanagi Y. ( 2012) Vpu and BST2: Still not there yet? Front Microbiol 3: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sauter D. ( 2014) Counteraction of the multifunctional restriction factor tetherin. Front Microbiol 5: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barre‐Sinoussi F., Chermann J.C., Rey F., Nugeyre M.T., Chamaret S., Gruest J., Dauguet C., Axler‐Blin C., Vezinet‐Brun F., Rouzioux C., Rozenbaum W., Montagnier L. ( 1983) Isolation of a T‐lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220: 868–71. [DOI] [PubMed] [Google Scholar]
- 5. Sheehy A.M., Gaddis N.C., Choi J.D., Malim M.H. ( 2002) Isolation of a human gene that inhibits HIV‐1 infection and is suppressed by the viral Vif protein. Nature 418: 646–50. [DOI] [PubMed] [Google Scholar]
- 6. Albin J.S., Harris R.S. ( 2010) Interactions of host APOBEC3 restriction factors with HIV‐1 in vivo: Implications for therapeutics. Expert Rev Mol Med 12: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kitamura S., Ode H., Iwatani Y. ( 2011) Structural features of antiviral APOBEC3 proteins are linked to their functional activities. Front Microbiol 2: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Desimmie B.A., Delviks‐Frankenberrry K.A., Burdick R.C., Qi D., Izumi T., Pathak V.K. ( 2014) Multiple APOBEC3 restriction factors for HIV‐1 and one Vif to rule them all. J Mol Biol 426: 1220–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Refsland E.W., Harris R.S. ( 2013) The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol 371: 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klatt N.R., Silvestri G., Hirsch V. ( 2012) Nonpathogenic simian immunodeficiency virus infections. Cold Spring Harb Perspect Med 2: a007153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Locatelli S., Peeters M. ( 2012) Cross‐species transmission of simian retroviruses: How and why they could lead to the emergence of new diseases in the human population. AIDS 26: 659–73. [DOI] [PubMed] [Google Scholar]
- 12. Compton A.A., Hirsch V.M., Emerman M. ( 2012) The host restriction factor APOBEC3G and retroviral Vif protein coevolve due to ongoing genetic conflict. Cell Host Microbe 11: 91–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Compton A.A., Emerman M. ( 2013) Convergence and divergence in the evolution of the APOBEC3G‐Vif interaction reveal ancient origins of simian immunodeficiency viruses. PLoS Pathog 9: e1003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hatziioannou T., Princiotta M., Piatak M., Jr. , Yuan F., Zhang F., Lifson J.D., Bieniasz P.D. ( 2006) Generation of simian‐tropic HIV‐1 by restriction factor evasion. Science 314: 95. [DOI] [PubMed] [Google Scholar]
- 15. Mariani R., Chen D., Schrofelbauer B., Navarro F., Konig R., Bollman B., Munk C., Nymark‐Mcmahon H., Landau N.R. ( 2003) Species‐specific exclusion of APOBEC3G from HIV‐1 virions by Vif. Cell 114: 21–31. [DOI] [PubMed] [Google Scholar]
- 16. Pedersen N.C., Ho E.W., Brown M.L., Yamamoto J.K. ( 1987) Isolation of a T‐lymphotropic virus from domestic cats with an immunodeficiency‐like syndrome. Science 235: 790–3. [DOI] [PubMed] [Google Scholar]
- 17. Troyer J.L., Pecon‐Slattery J., Roelke M.E., Johnson W., Vandewoude S., Vazquez‐Salat N., Brown M., Frank L., Woodroffe R., Winterbach C., Winterbach H., Hemson G., Bush M., Alexander K.A., Revilla E., O'Brien S.J. ( 2005) Seroprevalence and genomic divergence of circulating strains of feline immunodeficiency virus among Felidae and Hyaenidae species. J Virol 79: 8282–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pecon‐Slattery J., Troyer J.L., Johnson W.E., O'Brien S.J. ( 2008) Evolution of feline immunodeficiency virus in Felidae: Implications for human health and wildlife ecology. Vet Immunol Immunopathol 123: 32–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vandewoude S., Troyer J., Poss M. ( 2010) Restrictions to cross‐species transmission of lentiviral infection gleaned from studies of FIV. Vet Immunol Immunopathol 134: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Caso A., Lopez‐Gonzalez C., Payan E., Eizirik E., De Oliveira T., Leite‐Pitman R., Kelly M., Valderrama C., Lucherini M. ( 2008) Puma concolor. The IUCN red list of threatened species Version 20152 [Cited 17 July 2015.] Available from URL: http://wwwiucnredlistorg
- 21. Olmsted R.A., Langley R., Roelke M.E., Goeken R.M., Adger‐Johnson D., Goff J.P., Albert J.P., Packer C., Laurenson M.K., Caro T.M. ( 1992) Worldwide prevalence of lentivirus infection in wild feline species: epidemiologic and phylogenetic aspects. J Virol 66: 6008–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Biek R., Rodrigo A.G., Holley D., Drummond A., Anderson C.R., Jr. , Ross H.A., Poss M. ( 2003) Epidemiology, genetic diversity, and evolution of endemic feline immunodeficiency virus in a population of wild cougars. J Virol 77: 9578–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carpenter M.A., Brown E.W., Culver M., Johnson W.E., Pecon‐Slattery J., Brousset D., O'Brien S.J. ( 1996) Genetic and phylogenetic divergence of feline immunodeficiency virus in the puma (Puma concolor). J Virol 70: 6682–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vandewoude S., Apetrei C. ( 2006) Going wild: lessons from naturally occurring T‐lymphotropic lentiviruses. Clin Microbiol Rev 19: 728–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Franklin S.P., Troyer J.L., Terwee J.A., Lyren L.M., Boyce W.M., Riley S.P., Roelke M.E., Crooks K.R., Vandewoude S. ( 2007) Frequent transmission of immunodeficiency viruses among bobcats and pumas. J Virol 81: 10961–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vandewoude S., O'Brien S.J., Langelier K., Hardy W.D., Slattery J.P., Zuckerman E.E., Hoover E.A. ( 1997) Growth of lion and puma lentiviruses in domestic cat cells and comparisons with FIV. Virology 233: 185–92. [DOI] [PubMed] [Google Scholar]
- 27. Vandewoude S., O'Brien S.J., Hoover E.A. ( 1997) Infectivity of lion and puma lentiviruses for domestic cats. J Gen Virol 78 (Pt 4): 795–800. [DOI] [PubMed] [Google Scholar]
- 28. Vandewoude S., Hageman C.A., O'Brien S.J., Hoover E.A. ( 2002) Nonpathogenic lion and puma lentiviruses impart resistance to superinfection by virulent feline immunodeficiency virus. J Acquir Immune Defic Syndr 29: 1–10. [DOI] [PubMed] [Google Scholar]
- 29. Vandewoude S., Hageman C.L., Hoover E.A. ( 2003) Domestic cats infected with lion or puma lentivirus develop anti‐feline immunodeficiency virus immune responses. J Acquir Immune Defic Syndr 34: 20–31. [DOI] [PubMed] [Google Scholar]
- 30. Poss M., Ross H.A., Painter S.L., Holley D.C., Terwee J.A., Vandewoude S., Rodrigo A. ( 2006) Feline lentivirus evolution in cross‐species infection reveals extensive G‐to‐A mutation and selection on key residues in the viral polymerase. J Virol 80: 2728–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Terwee J.A., Yactor J.K., Sondgeroth K.S., Vandewoude S. ( 2005) Puma lentivirus is controlled in domestic cats after mucosal exposure in the absence of conventional indicators of immunity. J Virol 79: 2797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sato K., Takeuchi J.S., Misawa N., Izumi T., Kobayashi T., Kimura Y., Iwami S., Takaori‐Kondo A., Hu W.S., Aihara K., Ito M., An D.S., Pathak V.K., Koyanagi Y. ( 2014) APOBEC3D and APOBEC3F potently promote HIV‐1 diversification and evolution in humanized mouse model. PLoS Pathog 10: e1004453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sato K., Izumi T., Misawa N., Kobayashi T., Yamashita Y., Ohmichi M., Ito M., Takaori‐Kondo A., Koyanagi Y. ( 2010) Remarkable lethal G‐to‐A mutations in vif‐proficient HIV‐1 provirus by individual APOBEC3 proteins in humanized mice. J Virol 84: 9546–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krisko J.F., Martinez‐Torres F., Foster J.L., Garcia J.V. ( 2013) HIV restriction by APOBEC3 in humanized mice. PLoS Pathog 9: e1003242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Münk C., Beck T., Zielonka J., Hotz‐Wagenblatt A., Chareza S., Battenberg M., Thielebein J., Cichutek K., Bravo I.G., O'Brien S.J., Löchelt M., Yuhki N. ( 2008) Functions, structure, and read‐through alternative splicing of feline APOBEC3 genes. Genome Biol 9: R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Larue R.S., Lengyel J., Jonsson S.R., Andresdottir V., Harris R.S. ( 2010) Lentiviral Vif degrades the APOBEC3Z3/APOBEC3H protein of its mammalian host and is capable of cross‐species activity. J Virol 84: 8193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zielonka J., Marino D., Hofmann H., Yuhki N., Löchelt M., Münk C. ( 2010) Vif of feline immunodeficiency virus from domestic cats protects against APOBEC3 restriction factors from many felids. J Virol 84: 7312–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stern M.A., Hu C., Saenz D.T., Fadel H.J., Sims O., Peretz M., Poeschla E.M. ( 2010) Productive replication of Vif‐chimeric HIV‐1 in feline cells. J Virol 84: 7378–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yoshikawa R., Takeuchi J.S., Yamada E., Nakano Y., Ren F., Tanaka H., Munk C., Harris R.S., Miyazawa T., Koyanagi Y., Sato K. ( 2015) Vif determines the requirement for CBF‐beta in APOBEC3 degradation. J Gen Virol 96: 887–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. De Rozieres S., Mathiason C.K., Rolston M.R., Chatterji U., Hoover E.A., Elder J.H. ( 2004) Characterization of a highly pathogenic molecular clone of feline immunodeficiency virus clade C. J Virol 78: 8971–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saenz D.T., Barraza R., Loewen N., Teo W., Poeschla E.M. ( 2012) Feline immunodeficiency virus‐based lentiviral vectors. Cold Spring Harb Protoc 2012: 71–6. [DOI] [PubMed] [Google Scholar]
- 42. Kobayashi T., Takeuchi J.S., Ren F., Matsuda K., Sato K., Kimura Y., Misawa N., Yoshikawa R., Nakano Y., Yamada E., Tanaka H., Hirsch V.M., Koyanagi Y. ( 2014) Characterization of red‐capped mangabey tetherin: implication for the co‐evolution of primates and their lentiviruses. Sci Rep 4: 5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakano Y., Matsuda K., Yoshikawa R., Yamada E., Misawa N., Hirsch V.M., Koyanagi Y., Sato K. ( 2015) Down‐modulation of primate lentiviral receptors by Nef proteins of SIVcpz and related SIVs: Implication for the evolutionary event on the emergence of SIVcpz. J Gen Virol 96: 2867–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yoshikawa R., Izumi T., Yamada E., Nakano Y., Misawa N., Ren F., Carpenter M.A., Ikeda T., Munk C., Harris R.S., Miyazawa T., Koyanagi Y., Sato K. ( 2015) A naturally occurring domestic cat APOBEC3 variant confers resistance to FIV infection. J Virol 90: 474–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Inoshima Y., Kohmoto M., Ikeda Y., Yamada H., Kawaguchi Y., Tomonaga K., Miyazawa T., Kai C., Umemura T., Mikami T. ( 1996) Roles of the auxiliary genes and AP‐1 binding site in the long terminal repeat of feline immunodeficiency virus in the early stage of infection in cats. J Virol 70: 8518–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lockridge K.M., Himathongkham S., Sawai E.T., Chienand M., Sparger E.E. ( 1999) The feline immunodeficiency virus vif gene is required for productive infection of feline peripheral blood mononuclear cells and monocyte‐derived macrophages. Virology 261: 25–30. [DOI] [PubMed] [Google Scholar]
- 47. Shen X., Leutenegger C.M., Stefano Cole K., Pedersen N.C., Sparger E.E. ( 2007) A feline immunodeficiency virus vif‐deletion mutant remains attenuated upon infection of newborn kittens. J Gen Virol 88: 2793–9. [DOI] [PubMed] [Google Scholar]
- 48. Desrosiers R.C. ( 2007) Nonhuman Lentiviruses In: Knipe D.M., Howley P.M., eds. Fields Virology, 5th edn Philadelphia: Lippincott Williams & Wilkins. [Google Scholar]
- 49. Gifford R.J. ( 2012) Viral evolution in deep time: lentiviruses and mammals. Trends Genet 28: 89–100. [DOI] [PubMed] [Google Scholar]
- 50. Etienne L., Bibollet‐Ruche F., Sudmant P.H., Wu L.I., Hahn B.H., Emerman M. ( 2015) The role of the antiviral APOBEC3 gene family in protecting chimpanzees against lentiviruses from monkeys. PLoS Pathog 11: e1005149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. D'arc M., Ayouba A., Esteban A., Learn G.H., Boue V., Liegeois F., Etienne L., Tagg N., Leendertz F.H., Boesch C., Madinda N.F., Robbins M.M., Gray M., Cournil A., Ooms M., Letko M., Simon V.A., Sharp P.M., Hahn B.H., Delaporte E., Mpoudi Ngole E., Peeters M. ( 2015) Origin of the HIV‐1 group O epidemic in western lowland gorillas. Proc Natl Acad Sci U S A 112: E1343–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leendertz F.H., Zirkel F., Couacy‐Hymann E., Ellerbrok H., Morozov V.A., Pauli G., Hedemann C., Formenty P., Jensen S.A., Boesch C., Junglen S. ( 2008) Interspecies transmission of simian foamy virus in a natural predator‐prey system. J Virol 82: 7741–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brown, M.A. , Cunningham, M.W. , Roca, A.L. , Troyer, J.L. , Johnson, W.E. O'Brien, S.J. ( 2008) Genetic characterization of feline leukemia virus from Florida panthers. Emerg Infect Dis, 14: 252–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bevins S.N., Carver S., Boydston E.E., Lyren L.M., Alldredge M., Logan K.A., Riley S.P., Fisher R.N., Vickers T.W., Boyce W., Salman M., Lappin M.R., Crooks K.R., Vandewoude S. ( 2012) Three pathogens in sympatric populations of pumas, bobcats, and domestic cats: Implications for infectious disease transmission. PLoS ONE 7: e31403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Larue R.S., Andresdottir V., Blanchard Y., Conticello S.G., Derse D., Emerman M., Greene W.C., Jonsson S.R., Landau N.R., Lochelt M., Malik H.S., Malim M.H., Munk C., O'Brien S.J., Pathak V.K., Strebel K., Wain‐Hobson S., Yu X.F., Yuhki N., Harris R.S. ( 2009) Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol 83: 494–7. [DOI] [PMC free article] [PubMed] [Google Scholar]


