Preface
Understanding how proteins and other biological macromolecules perform their complicated tasks in the living cell is often aided by knowledge of their three-dimensional structures. Because many tasks involve the cleavage or formation of chemical bonds, structural characterisation at the atomic level is most useful. Recent developments in electron microscopy of frozen hydrated samples (cryo-EM) have provided unprecedented opportunities for the structural characterization of biological macromolecules. This is resulting in a wave of new information about biological processes that were impossible to characterize with previously existing techniques in structural biology.
Introduction
Biological macromolecules adopt complicated three-dimensional structures that are central to their functioning. Some molecules, like enzymes, act alone to provide a chemical environment to favour catalysis of a specific chemical reaction. Other macromolecules form larger complexes with partner proteins, nucleic acids, lipids or sugar molecules. Many of these complexes perform their function through relative movements of individual parts, in a way that resembles how man-made machines work1 Understanding how these complicated structures perform their tasks is a fundamental goal in modern biology.
Macromolecular complexes are too small to see with visible light. Photons with wavelengths that are small enough to visualise atomic-level details are in the X-ray region. X-rays interact so weakly with biological matter that it is difficult to study individual protein complexes. But when multiple copies of a protein are arranged together in a 3D crystal, information about its atomic structure may be obtained from X-ray diffraction experiments. This technique, called X-ray crystallography, has been the most important tool in structural biology for many years (Figure 1). Another technique to characterise protein structure is nuclear magnetic resonance (NMR) spectroscopy, which measures distance-dependent interactions between atoms. NMR may be used to infer the structure of relatively small proteins, and provides unique information about protein dynamics and interactions with other molecules.
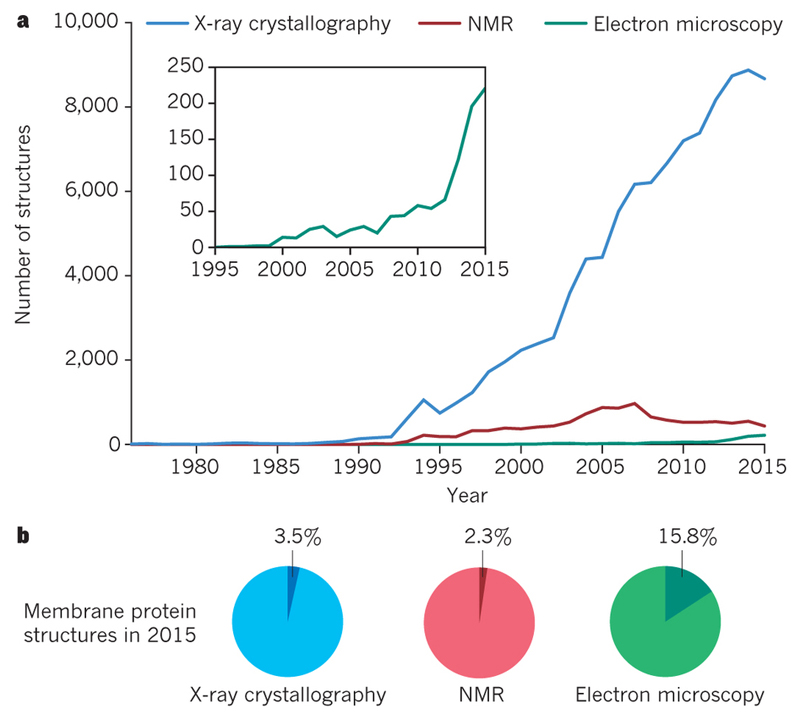
Figure 1. Growth in Structural Biology.
A. Number of structures in the Protein Data Bank as determined by X-ray crystallography (blue), NMR (red) and EM (green). B. Fraction of membrane protein structures in 2015 for each of the three techniques.
One can also use electrons to look at protein structures. Proteins scatter electrons approximately ten thousand times stronger than X-rays, and electrons can be accelerated in electric fields of several hundred thousand Volts to wavelengths that are much smaller than the distances between atoms in protein structures. Moreover, their electric charge makes it relatively easy to focus electrons with electro-magnetic lenses. Therefore, one can build microscopes that use electrons to make images with atomic-level details. For many years, the contributions of electron microscopy (EM) to structural biology were modest in comparison with X-ray crystallography and NMR, but recent trends indicate that this is changing (Figure 1). Earlier this year, the thousandth atomic structure derived from EM images was entered into the main repository for protein structures, the protein data bank (PDB)2. Here, we describe how imaging in an electron microscope can be used to study protein structures, and how rapid progress in EM protein structure determination during the past few years has been heralded as a revolution in structural biology3. We then highlight how the unique characteristics of this technique are already changing structural biology, and identify outstanding opportunities that will continue to transform our understanding of how macromolecules perform their intricate tasks in the cell.
Description of the technique
Because electrons are scattered by molecules in the air, electron microscopes need to be operated at high vacuum. This poses a problem for studying biological samples, most of which naturally occur in an aqueous environment. In addition, biological structures are sensitive to radiation damage. For each electron that contributes to the image formation, there are three electrons that deposit energy in the sample instead. This energy causes cleavage of chemical bonds, and ultimately destroys the structures of interest. By keeping the sample frozen at cryogenic temperatures, cryo-EM allows its preservation in high vacuum and provides some protection against the effects of radiation damage4.
A schematic of a cryo-EM sample inside an electron microscope is shown in Box 1. Typically, a few microliters of a purified protein solution is applied to a metal grid, on top of which lies a thin film of amorphous carbon with holes in it. After blotting away excess liquid with filter paper, the grid is plunged into liquid ethane5. Ideally, this results in a thin layer of non-crystalline, or vitreous ice, in which many copies of the protein adopt a wide range of orientations. Pictures taken through the holes in the carbon film contain 2D projections of individual complexes, which are called particles. Projections from particles in different orientations provide complementary information about the underlying 3D object. Therefore, multiple 2D projections can be combined into a single 3D reconstruction, provided their relative orientations are known. Unfortunately, this information is lost in the experiment, since the individual particles tumble around randomly in solution before the sample is vitrified.
Box 1. Cryo-EM structure determination.
A. A few microliter of sample is applied to a grid and excess liquid is blotted away prior to plunging the grid into liquid ethane. B. The grid is kept at liquid nitrogen temperature and transferred to a transmission electron microscope. Images are taken through the holes of a carbon film on the grid. C. Two-dimensional projection images of many copies of assumedly identical objects in different orientations are combined into a three-dimensional reconstruction. The structure of glutamate dehydrogenase is used as an example34.
It is possible to determine the relative orientations of individual particles a posteriori by processing the experimental 2D projection images in a computer6. This process is known as single-particle analysis. Single-particle cryo-EM suffers from low contrast in the images because proteins scatter electrons only by about 30% more than the surrounding ice. This in itself would not be a problem if one could use many electrons to accurately determine small differences in image intensities. However, to reduce the effects of radiation damage, one needs to carefully limit the number of electrons used for imaging, which results in extremely noisy images. This noise impedes the accurate assignment of the particle orientations, which becomes the major bottleneck.
Recent progress
Larger complexes and complexes containing oligonucleotides give rise to images with higher signal-to-noise ratios, and internal symmetry is also helpful. This explains why cryo-EM structures of ribosomes and icosahedral virus capsids have long been at the forefront of the field. By 2008, several virus structures had been solved to near-atomic resolution7–9. Around the same time, ribosome structures could be calculated to 6 Å, e.g.10–12. These successes built on many important developments in instrumentation, experimental procedures and image processing during the preceding forty years. For an overview of these contributions, we refer to reviews by others13–15. In what follows, we focus on the more recent developments that have changed the scope of cryo-EM structure determination in the past few years.
By 2010, the quality of the available structures contrasted with theoretical considerations about radiation damage that predicted cryo-EM structure determination should be possible to atomic resolution for protein complexes with molecular weights down to 100 kDa16. Part of the difference between prediction and observation could be explained by the inefficient detection of electrons. Images were originally recorded on photographic film, while in the early 2000s the development of digital charge-coupled device (CCD) cameras17 had opened the way to higher throughput and automated data acquisition18. Photographic film only detects about a third of all incoming electrons. CCDs are even less efficient, as an additional conversion from electrons into photons leads to an overall detection rate below one fifth of the incoming electrons19. This affects cryo-EM structure determination right at its bottleneck: where low numbers of electrons to limit radiation damage result in images that are too noisy for reliable orientation determination.
Around 2012, three different commercial companies had produced prototypes of a new generation of digital electron detectors. The new chips were sufficiently resistant to radiation damage to allow direct detection of electrons, which improved their efficiency to approximately half of the incoming electrons20. Moreover, modern electronics around these direct-electron detectors enabled fast image capture, much like burst mode in modern photographic cameras. Counting of individual electron events on the fastest camera led to even better electron detection21. Fast image capture also allowed addressing an outstanding problem of imaging frozen hydrated samples: energy deposited by the incoming electrons causes movement inside the ice layer, which blurs the resulting images. Processing of movies recorded during electron exposure effectively removed the blurring effects22,23 which facilitated structure determination to unprecedented resolution and from much less data than before21,24.
Meanwhile, another major impediment to high-resolution cryo-EM structure determination had been solved. Because many macromolecular machines adopt a range of different conformations in solution, and purification or complex formation are seldom perfect, cryo-EM samples often contain a variety of structures. When such mixtures are subjected to single-particle analysis, the 2D projections of different 3D structures need to be separated. A generally applicable solution to the mixture problem was first provided by 3D maximum likelihood classification algorithms25,26. Maximum likelihood methods use a statistical description of the data, which makes them more robust to noise than methods that were previously in common use27. Alternative classification approaches soon followed, and the new methods turned the mixture problem into an opportunity. Whereas until then the presence of multiple structures had blurred cryo-EM structures, valuable insights into protein dynamics could now be obtained from a single experiment, e.g. see28.
When in 2013 the new electron detectors became commercially available, the field was poised for a revolution. The unprecedented image quality from the new detectors allowed more accurate orientational assignments and better separation of particles from distinct structural states. Early structures that heralded a new era included the coenzyme F420-reducing hydrogenase29, the mammalian ion channel TRPV130 and the large subunit of the mitochondrial ribosome from yeast31. The extension of maximum likelihood methods into an empirical Bayesian approach made image processing more accessible to non-experts, as crucial parameters no longer needed tuning but were estimated from the data themselves32. Moreover, the automated data acquisition procedures that were originally developed for CCDs facilitated the recording of large data sets, from which the best particles could be selected using image classification. As a result, in the past three years many different groups have solved cryo-EM structures to atomic resolution for a wide range of specimens. Complexes with molecular weights below 200 kDa have become feasible targets33,34, and achievable resolution limits now extend beyond 3 Å2,35–37, and may even surpass 2 Å in favourable cases34.
Current opportunities
Many of the new structures are of proteins that are naturally embedded in the membrane (Fig. 2). Membrane proteins are hard to purify in solution, as one needs to stabilise their hydrophobic membrane-spanning domains with detergents. Because detergents also make crystallisation notoriously difficult, membrane proteins have been a blind spot in structural biology for many years (Fig. 1). This is unfortunate, as approximately half of all known small molecule drugs bind to membrane proteins. Cryo-EM structure determination does not require crystallisation. Instead, membrane proteins can be imaged directly when solubilised in detergents or amphipols38, or stabilised in a lipid environment using nano-discs that are formed within a scaffold of an amphipathic protein belt39 or the recently developed saposin-lipoprotein system40. The first membrane protein structure that was solved using the new cryo-EM technology was that of the transient receptor potential cation channel subfamily V member 1 (TRPV1)30. This protein is responsible for the burning sensation from chilli peppers and is an important drug target for pain. Its structure was first solved in an empty state that was solubilised in amphipols. A complex with a spider toxin and a small-molecule compound that is similar to capsaicin, the active component in chilli peppers, was also determined41. A recent, improved TRPV1 structure inside the more natural environment of a nano-disc also allowed visualisation of lipid substrates and provided more detailed mechanistic insights42. The initial TRPV1 structure was followed by many other medically relevant ion channels, such as the voltage-dependent calcium channel CaV1.143, a sodium-potassium channel44, the glycine receptor45. Combined, these structures have yielded a wealth of new information on how cells regulate ion transport across the membrane.
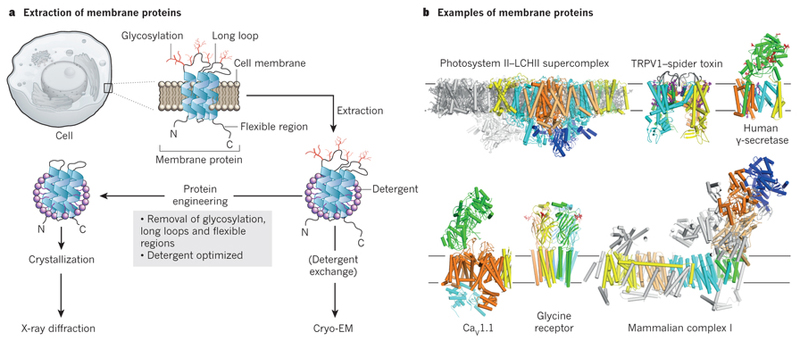
Figure 2. Membrane protein structural biology.
Membrane proteins need to be extracted from their membranes using detergent solubilisation. Membrane protein crystallisation is difficult and often requires protein engineering to remove flexible loops and sugar. Cryo-EM structure determination allows direct imaging of membrane proteins in detergents or in more natural environments like nano-discs. The presence of sugars or very large membrane complexes does not preclude cryo-EM structure determination. The structures of the photosystem II - light harvesting supercomplex60, TRPV1 in complex with a spider toxin42, gamma-secretase33, the voltage-gated calcium channel CaV1.143, the glycine receptor45, and bovine complex-I59 are shown as examples.
Another advantage of cryo-EM is that flexible regions do not impede structure determination. Whereas flexible loops or sugars are typically removed using complicated protein engineering approaches to facilitate crystallisation46, fully glycosylated wild-type proteins may be used for cryo-EM. For example, the structure of the human gamma-secretase complex was solved despite the presence of at least eleven sugar chains and a long disordered loop33. This membrane-embedded protease generates the amyloid-beta peptides that aggregate in the brains of Alzheimer's patients. With an ordered mass of approximately 130 kDa, this is currently the smallest cryo-EM structure with a resolution beyond 3.5 Å. Another example is the heavily glycosylated structure of the human immuno-deficiency virus type I (HIV1) envelope glycoprotein trimer. This protein recognises receptors on the surface of immune cells and mediates viral entry. Studying the structure of the fully glycosylated native protein provided key information that may help vaccine development47,48. Likewise, recent structures of the viral glycoprotein GP1 from Ebola bound to neutralising antibodies49 and the Niemann-Pick C1 transporter50 provide clues on human immunity against Ebola and how this virus fuses its membrane with that of the host.
Because wild-type proteins can be used and only micrograms of purified protein are required also means that complexes purified from native sources may be suitable for cryo-EM structure determination. This has allowed structure determination of various large membrane protein complexes that had resisted crystallographic studies for many years, including the ryanodine receptor51–53, the inositol triphosphate receptor54, glutamate receptors55–58 mammalian complex-I59, photosystem II – light harvesting supercomplex (PSII-LHCII)60, and ATP synthases61,62.
The possibility to determine structures of large complexes from native sources has also had a huge impact on the characterisation of soluble macromolecular machines (Fig. 3). More than fifty high-resolution ribosome structures have appeared in the past two years, yielding a wealth of new information on the control of protein biosynthesis. For example, studies on both mammalian63,64 and yeast31 mitochondrial ribosomes have revealed how coevolution with the mitochondrial genome has effected their structure. Other macromolecular machines have become amenable to structural studies for the first time in history, as exemplified by the structures that are now emerging of the inflammasome65,66, the spliceosome e.g.67–69, the signalosome70,71, the exosome72, the anaphase promoting complex73,74, the 26S proteasome75,76 the SNARE complex77,78, the dynein:dynactin complex79,80, the chaperone heat-shock protein 9081, and target-of-rapamycin (TOR)82,83. All of these machines change conformation, rearrange subunits and bind different partners and substrates during their assembly and working cycles. Cryo-EM enables the study of the often short-lived conformations and interactions that are difficult to isolate or stabilise biochemically.
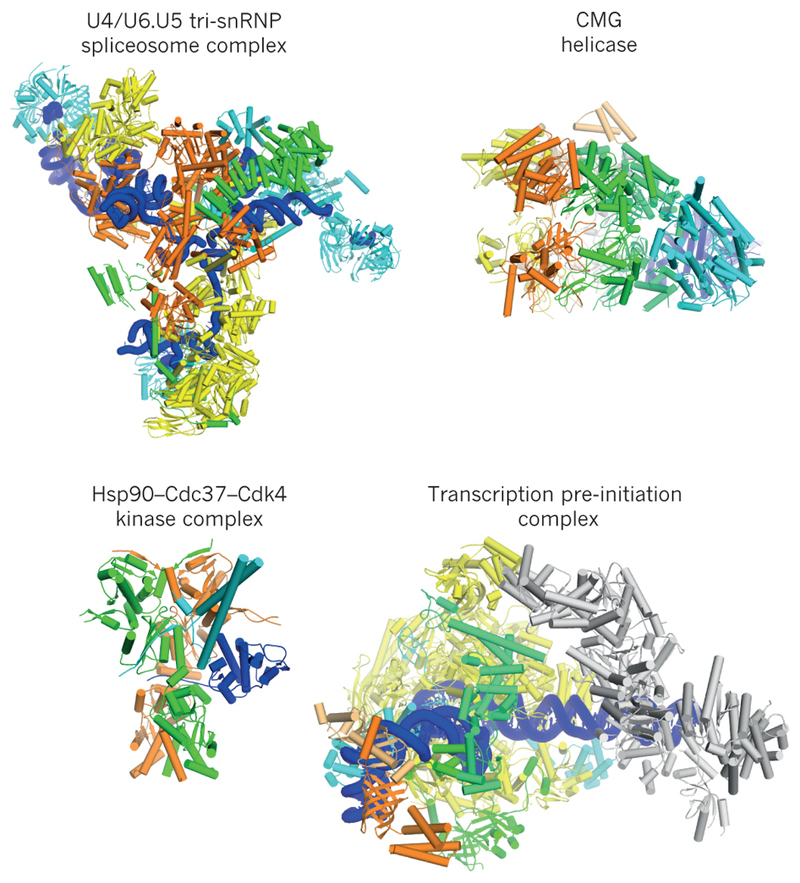
Figure 3. Soluble macromolecular machines.
Examples of cryo-EM structures are shown for the U4/U6.U5 tri-snRNP spliceosomal complex from yeast67; the eukaryotic replicative CMG helicase86; the human Hsp90-Cdc37-Cdk4 kinase complex81; and the human transcription pre-initiation complex91.
Nuclear complexes that act on DNA and RNA form another group of molecular machines that has traditionally been difficult to analyse. Both the proteins and the oligonucleotide substrates in these complexes are typically highly dynamic molecules that engage in transient interactions with each other. Consequently, many large nuclear complexes have proven challenging to make or purify. Showcasing the potential of the technique to study highly dynamic complexes, various cryo-EM structures have started to shed light on the molecular details of some of the most fundamental processes of life. Structures of the DNA replication machinery have unveiled molecular details of how genomes are copied84–87, while multiple structures of RNA polymerases have led to new insights into how DNA is transcribed into RNA88–92. RNA polymerases have even been studied in situ, inside the capsid of dsRNA viruses93,94. Other examples include a structure that provides new insights into genetic recombination95, structures that show how the retroviral recombination machinery engages with the host nucleosome to insert its DNA96,97, a structure of bacterial group II intron showing how this bacterial DNA mobile element catalyses self-splicing in conjunction with a small intron-encoded protein98, and structures that have aided our understanding of the mechanisms of type II and type III CRISPR systems99,100.
In almost all of the above-mentioned studies, image classification played a crucial role in selecting structurally homogeneous subsets of particles for structure determination. For many structures, a strikingly small fraction of the particles in the initial data set is selected for calculation of the final map. Apparently, the majority of particles in cryo-EM data sets are somehow unsuitable for high-resolution structure determination and image classification allows selecting only the very best ones. Moreover, much like man-made machines, macromolecular machines employ movements of parts relative to each other in their functioning. Whereas one would typically need to trap dynamic complexes in a single state to facilitate crystallisation, the new image classification algorithms offer the unique opportunity to visualise the full conformational freedom of these complexes in a single experiment (Fig. 4). A striking example of this is the membrane-embedded ATP synthase, which acts as a molecular turbine, converting a proton flux into rotation to synthesise the energy currency for the cell in the form of highly energetic adenosine triphosphate (ATP) molecules. Image classification revealed the presence of three different rotated states of this machine from a single cryo-EM data set61. In another example, imaging of actively translating human polysomes, i.e. multiple ribosomes bound to a single mRNA molecule, followed by extensive image classification led to structural snapshots of the entire translation cycle101.
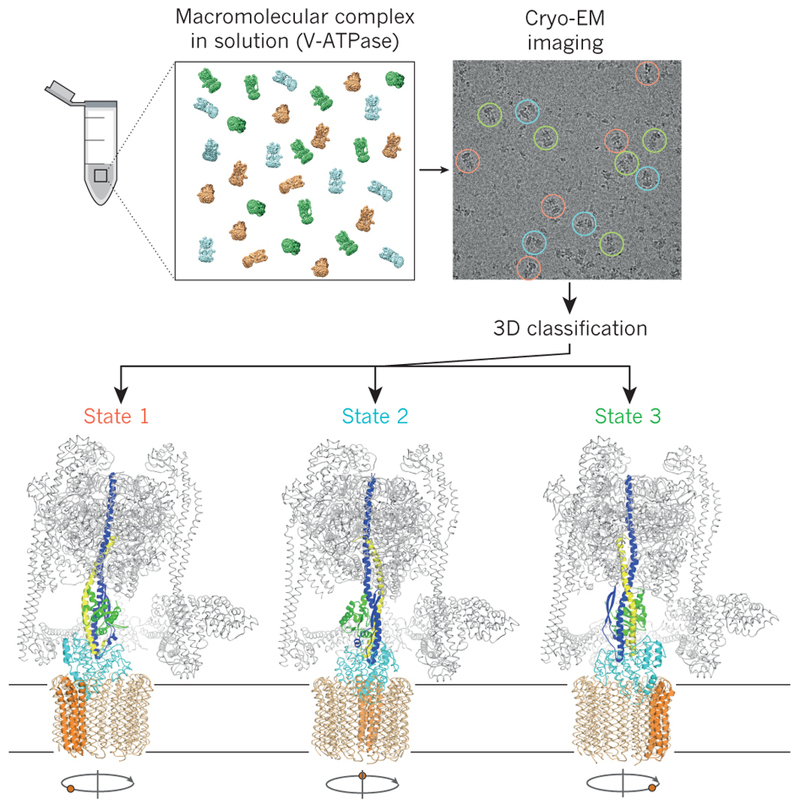
Figure 4. Image classification allows studying macromolecular dynamics.
Mixtures of macromolecular complexes in distinct conformational or compositional states may be imaged directly, and image classification may be used to obtain structures for each of the states. Thereby, image classification allows characterization of the functional cycles of dynamic molecular machines from a single experiment. Different functional states of the eukaryotic V-type ATPase61 are shown as an example.
Looking ahead
Several challenges remain to be overcome if cryo-EM structure determination is to continue its transformative growth in structural biology. Its elevated cost is an immediate concern. The 300kV microscopes that are best for high-resolution structure determination cost in excess of $5 million, and their maintenance contracts are consistently expensive. Microscopes that operate at 200kV are cheaper and can also be used to produce atomic-resolution structures102,103, but their cost still runs in the millions. To facilitate broad access to high-end microscopes, regional or national facilities are quickly gaining in popularity104. Nevertheless, sample optimisation requires access to an electron microscope on a regular basis, preferably weekly if not daily, which is not practical through centralised facilities. Although expensive high-voltage machines are in principle not needed for screening, currently available cheaper microscopes, which often operate at 100-120 kV, lack the coherence (i.e. they can't generate electrons that are all in-phase) to visualise particles with molecular weights below several hundred thousand daltons. Therefore, an urgent need exists for the development of affordable screening microscopes with more coherent electron sources.
The costs of storing and processing large volumes of data also form a problem. Automated data acquisition on a high-end microscope can yield several terabytes of images every day, and processing times may run into hundred thousands of computing core hours per data set. To avoid buying and maintaining costly high-performance computing infrastructure, alternative solutions such as cloud computing105 and implementation of image processing algorithms on cheaper graphical processing units (GPUs) are being explored106,107.
Besides reducing costs and expanding capacity to increase accessibility, there is still ample scope for improving the available technology itself. Much of recent progress was achieved by improving electron detection efficiency from approximately 30% on film to 50% on direct-electron detectors. The latter could be made even better. An important aspect of efficient electron detection is the ability to count individual electrons as they hit the detector, which requires fast read-outs21,108. Of the three commercially available detectors, only one detector is currently fast enough to allow counting, but one still needs to lower the intensity of the electron beam to avoid flooding the chip with too many events. Through the use of more modern technology, with even faster electronics and better chip design, detectors with up to 90% efficiency could in principle be made.
The noise in the images may be further decreased through the use of energy filters, which are optical devices to remove electrons that have lost part of their energy in the sample and therefore no longer contribute constructively to the image. Energy filters were originally considered most useful for relatively thick samples, e.g. whole cells, where more electrons loose energy while passing through the sample. However, some of the highest resolution and smallest structures currently available were done using energy-filtered imaging, indicating that removal of these electrons is also beneficial for thinner samples, e.g. see2,33,34,36.
Another promising hardware development is that of phase plates. Phase plates are optical devices that produce a difference in the phase of the scattered and the unscattered electron waves. This generates up to an order of magnitude better contrast in the images at low resolution, which will be of particular interest for imaging complexes that are too small to yield enough contrast with existing optics109. Other optical devices that may yield better images, in particular at higher resolutions, are aberration correctors. Using complicated combinations of lenses, these devices counteract the effects of imperfections in the optical system of the microscope. Although currently not needed to reach resolution beyond 3 Å, these correctors might help when resolutions around 2 Å are to be achieved110.
Sample preparation methods provide another important opportunity for improvement. Although movie-processing has come a long way to correct for beam-induced sample motions, large movements during the early stages of electron exposure are often too fast to correct for21,29,111. This forms an important constraint on high-resolution structure determination, as most of the high-resolution information is destroyed by radiation damage during these early stages112. Developments in cryo-EM sample preparation aim at reducing or stopping this motion, for example by replacing both the copper grid and the amorphous holey carbon film by gold113, or by using graphene or graphene oxide films114,115.
Also the requirements on quantity and purity of the sample may be improved. Currently, several microliters of a purified protein sample, typically at concentrations between 0.1-5 μM, are needed to prepare a single cryo-EM grid. Purification on the grid itself, through the use of specifically adhered affinity tags116, may relax demands on sample concentration and purity. Moreover, because less than 0.1% of this volume will remain on the grid after blotting and only a fraction of the grid surface is typically used for data acquisition, savings in required sample volumes of multiple orders of magnitude are in principle possible. Current investigations in this direction include spraying pico-liter sized droplets onto cryo-EM grids117. When multiple components are mixed together in a precisely timed manner, spraying samples onto grids and quickly freezing them can also be used for time-resolved studies118. Combined with modern microfluidics and the capability to tackle mixtures through image classification, this may allow following reactions on a millisecond timescale119.
In software development, better image formation models may be necessary to calculate higher resolution structures. In addition, better algorithms are needed to deal with complicated, many-component mixtures and to identify minority states. In particular, the presence of multiple, continuously flexing domains still represents a challenge. Alternative procedures that describe the entire ensemble of structures instead of dividing the data into subsets have the potential to reveal the conformational complexity of highly dynamic molecular machines67,120.
Prospects
Each of the on-going developments mentioned above has the potential to further enlarge the scope of cryo-EM single-particle analysis. Better detectors, more widespread use of energy filters, phase-plates, stopping beam-induced motions, and possibly correctors of optical aberrations in the microscope will all result in images with higher signal-to-noise ratios. Combined with improved image processing algorithms, this will yield higher resolution structures of smaller complexes and from more complicated mixtures than currently possible.
Being able to visualise complexes with molecular weights around 100 kDa will widen the applicability of the technique to many more drug targets. For example, G-protein coupled receptors (GPCRs) are highly sought-after targets for pharmaceutical industry. In complex with conformation specific antibodies or (hetero-trimeric) G-proteins, these should soon become amenable to cryo-EM structure determination. Combined with the possibility to achieve resolutions well beyond 3 Å34 on complexes bound to drug candidates121, this will lead to routine cryo-EM structure-based drug design on targets that are currently extremely difficult to characterise using alternative techniques.
The ability to obtain structures from small sub-populations of the data through image processing, possibly in combination with sample preparation methods on the nanoliter scale, will also expand the applicability of the technique to low-abundant complexes in the cell. Combined with time-resolved methods, even transient complexes and intermediate conformational states may be sampled, which will provide unprecedented insights into the functioning of large macromolecular machines. For example, structural characterisation at the molecular or even atomic level of extremely large and flexible machines like the nuclear pore complex122, or the characterisation of complexes involved in the organisation of chromatin123 are now on the horizon.
Yet, inside the cell macromolecules do not act in isolation. The cell is densely packed with many different molecules that form highly intricate interaction networks that are finely tuned to keep the cell alive. Therefore, the detailed structural information about individual complexes obtained by single-particle analysis needs to be understood in the context of their cellular environment. Excitingly, imaging with electrons may provide unique opportunities for this task too. Electron cryo-tomography can be used to calculate 3D reconstructions of frozen cells by taking multiple images of the same sample at different tilt angles inside the microscope124. Again, radiation damage is a major limitation, but resolutions may be increased by averaging over repeated sub-structures within the cell, e.g. see125. The tomographic approaches will benefit from the same hardware and software developments that are also driving single-particle analysis forwards. Combined, these techniques may bridge the gap between biophysics and cell biology, and provide a roadmap towards our understanding of the living cell at a molecular level.
Acknowledgements
We thank Yifan Cheng, David Barford and Richard Henderson for comments on an early version of this manuscript. S.H.W.S is funded by the UK Medical Research Council (MC_UP_A025_1013).
References
- 1.Alberts B. The Cell as a Collection of Protein Machines: Preparing the Next Generation of Molecular Biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee S, et al. 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016;351:871–875. doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuhlbrandt W. The Resolution Revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- 4.Taylor KA, Glaeser RM. Electron Diffraction of Frozen, Hydrated Protein Crystals. Science. 1974;186:1036–1037. doi: 10.1126/science.186.4168.1036. [DOI] [PubMed] [Google Scholar]
- 5.Dubochet J, Chang JJ, Freeman R, Lepault J, McDowall AW. Frozen aqueous suspensions. Ultramicroscopy. 1982;10:55–61. [Google Scholar]
- 6.Frank J, Verschoor A, Boublik M. Computer averaging of electron micrographs of 40S ribosomal subunits. Science. 1981;214:1353–1355. doi: 10.1126/science.7313694. [DOI] [PubMed] [Google Scholar]
- 7.Jiang W, et al. Backbone structure of the infectious ε15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- 8.Yu X, Jin L, Zhou ZH. 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, et al. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci USA. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villa E, et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci USA. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuette J-C, et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. The EMBO journal. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seidelt B, et al. Structural Insight into Nascent Polypeptide Chain-Mediated Translational Stalling. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agard D, Cheng Y, Glaeser RM, Subramaniam S. Chapter Two - Single-Particle Cryo-Electron Microscopy (Cryo-EM): Progress, Challenges, and Perspectives for Further Improvement. Advances in Imaging and Electron Physics. 2014;185:113–137. [Google Scholar]
- 14.Frank J. Generalized single-particle cryo-EM--a historical perspective. Microscopy. 2016;65:3–8. doi: 10.1093/jmicro/dfv358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vinothkumar KR, Henderson R. Single particle electron cryomicroscopy: trends, issues and future perspective. Quarterly Reviews of Biophysics. 2016;49:251–276. doi: 10.1017/S0033583516000068. [DOI] [PubMed] [Google Scholar]
- 16.Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Quarterly Reviews of Biophysics. 1995;28:171–193. doi: 10.1017/s003358350000305x. [DOI] [PubMed] [Google Scholar]
- 17.Krivanek OL, Mooney PE. Applications of slow-scan CCD cameras in transmission electron microscopy. Ultramicroscopy. 1993;49:95–108. [Google Scholar]
- 18.Potter CS, et al. Leginon: a system for fully automated acquisition of 1000 electron micrographs a day. Ultramicroscopy. 1999;77:153–161. doi: 10.1016/s0304-3991(99)00043-1. [DOI] [PubMed] [Google Scholar]
- 19.McMullan G, Chen S, Henderson R, Faruqi AR. Detective quantum efficiency of electron area detectors in electron microscopy. Ultramicroscopy. 2009;109:1126–1143. doi: 10.1016/j.ultramic.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMullan G, Faruqi AR, Clare D, Henderson R. Comparison of optimal performance at 300keV of three direct electron detectors for use in low dose electron microscopy. Ultramicroscopy. 2014;147:156–163. doi: 10.1016/j.ultramic.2014.08.002. [A comparison of the three commercially available direct electron detectors illustrates they are all better than photographic film.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Meth. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brilot AF, et al. Beam-induced motion of vitrified specimen on holey carbon film. J Struct Biol. 2012;177:630–637. doi: 10.1016/j.jsb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell MG, et al. Movies of Ice-Embedded Particles Enhance Resolution in Electron Cryo-Microscopy. Structure. 2012;20:1823–1828. doi: 10.1016/j.str.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bai X-C, Fernández IS, McMullan G, Scheres SHW. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2:e00461–e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheres SHW, et al. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nature Meth. 2006;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 26.Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. J Struct Biol. 2013;183:377–388. doi: 10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sigworth FJ. A Maximum-Likelihood Approach to Single-Particle Image Refinement. J Struct Biol. 1998;122:328–339. doi: 10.1006/jsbi.1998.4014. [DOI] [PubMed] [Google Scholar]
- 28.Clare DK, et al. ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the GroEL Chaperonin. Cell. 2012;149:113–123. doi: 10.1016/j.cell.2012.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allegretti M, Mills DJ, McMullan G, Kühlbrandt W, Vonck J. Atomic model of the F 420-reducing [NiFe] hydrogenase by electron cryo-microscopy using a direct electron detector. eLife. 2014;3:7237. doi: 10.7554/eLife.01963. [One of the first three high-resolution structures heralding the resolution revolution in cryo-EM structure determination.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [One of the first three high-resolution structures heralding the resolution revolution in cryo-EM structure determination.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amunts A, et al. Structure of the Yeast Mitochondrial Large Ribosomal Subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [One of the first three high-resolution structures heralding the resolution revolution in cryo-EM structure determination.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheres SHW. A Bayesian View on Cryo-EM Structure Determination. J Mol Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [Introduction of a statistical framework for classification and high-resolution refinement with reduced needs for expert supervision.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai X-C, et al. An atomic structure of human γ-secretase. Nature. 2015;525:212–217. doi: 10.1038/nature14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merk A, et al. Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery. Cell. 2016;165:1698–1707. doi: 10.1016/j.cell.2016.05.040. [The highest reported resolution (1.8 Å) of a single-particle reconstruction thus far.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campbell MG, Veesler D, Cheng A, Potter CS, Carragher B. 2.8 Å resolution reconstruction of the Thermoplasma acidophilum20S proteasome using cryo-electron microscopy. eLife. 2015;4:e01963. doi: 10.7554/eLife.06380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartesaghi A, et al. 2.2 A resolution cryo-EM structure of -galactosidase in complex with a cell-permeant inhibitor. Science. 2015;348:1147–1151. doi: 10.1126/science.aab1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife. 2015;4:e06980–e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Althoff T, Mills DJ, Popot JL, Kühlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I 1III 2IV 1. The EMBO journal. 2011;30:4652–4664. doi: 10.1038/emboj.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2009;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frauenfeld J, et al. A saposin-lipoprotein nanoparticle system for membrane proteins. Nature Meth. 2016;13:345–351. doi: 10.1038/nmeth.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504:113–118. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Y, Cao E, Julius D, Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu J, et al. Structure of the voltage-gated calcium channel Cav1.1 complex. Science. 2015;350:aad2395–aad2395. doi: 10.1126/science.aad2395. [DOI] [PubMed] [Google Scholar]
- 44.Hite RK, et al. Cryo-electron microscopy structure of the Slo2.2 Na+-activated K+ channel. Nature. 2015;527:198–203. doi: 10.1038/nature14958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du J, Lü W, Wu S, Cheng Y, Gouaux E. Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature. 2015;526:224–229. doi: 10.1038/nature14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baker M. Making membrane proteins for structures: a trillion tiny tweaks. Nature Meth. 2010;7:429–434. doi: 10.1038/nmeth0610-429. [DOI] [PubMed] [Google Scholar]
- 47.Lyumkis D, et al. Cryo-EM Structure of a Fully Glycosylated Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Misasi J, et al. Structural and molecular basis for Ebola virus neutralization by protective human antibodies. Science. 2016;351:1343–1346. doi: 10.1126/science.aad6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gong X, et al. Structural Insights into the Niemann-Pick C1 (NPC1)-Mediated Cholesterol Transfer and Ebola Infection. Cell. 2016;165:1467–1478. doi: 10.1016/j.cell.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan Z, et al. Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature. 2014;517:50–55. doi: 10.1038/nature14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Efremov RG, Leitner A, Aebersold R, Raunser S. Architecture and conformational switch mechanism of the ryanodine receptor. Nature. 2014;517:39–43. doi: 10.1038/nature13916. [DOI] [PubMed] [Google Scholar]
- 53.Zalk R, et al. Structure of a mammalian ryanodine receptor. Nature. 2014;517:44–49. doi: 10.1038/nature13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fan G, et al. Gating machinery of InsP3R channels revealed by electron cryomicroscopy. Nature. 2015;527:336–341. doi: 10.1038/nature15249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tajima N, et al. Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature. 2016;534:63–68. doi: 10.1038/nature17679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu S, et al. Mechanism of NMDA Receptor Inhibition and Activation. Cell. 2016;165:704–714. doi: 10.1016/j.cell.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herguedas B, et al. Structure and organization of heteromeric AMPA-type glutamate receptors. Science. 2016;352:aad3873–aad3873. doi: 10.1126/science.aad3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meyerson JR, et al. Structural mechanism of glutamate receptor activation and desensitization. Nature. 2014;514:328–334. doi: 10.1038/nature13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vinothkumar KR, Zhu J, Hirst J. Architecture of mammalian respiratory complex I. Nature. 2014;515:80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei X, et al. Structure of spinach photosystem II–LHCII supercomplex at 3.2 Å resolution. Nature. 2016;534:69–74. doi: 10.1038/nature18020. [DOI] [PubMed] [Google Scholar]
- 61.Zhao J, Benlekbir S, Rubinstein JL. Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature. 2015;521:241–245. doi: 10.1038/nature14365. [Example how image classification can reveal multiple functional states of dynamic molecular machines from a single experiment.] [DOI] [PubMed] [Google Scholar]
- 62.Allegretti M, et al. Horizontal membrane-intrinsic α-helices in the stator a-subunit of an F-type ATP synthase. Nature. 2015;521:237–240. doi: 10.1038/nature14185. [DOI] [PubMed] [Google Scholar]
- 63.Amunts A, Brown A, Toots J, Scheres SHW, Ramakrishnan V. The structure of the human mitochondrial ribosome. Science. 2015;348:95–98. doi: 10.1126/science.aaa1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greber BJ, et al. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 2015;348:303–308. doi: 10.1126/science.aaa3872. [DOI] [PubMed] [Google Scholar]
- 65.Zhang L, et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 2015;350:404–409. doi: 10.1126/science.aac5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu Z, et al. Structural and biochemical basis for induced self-propagation of NLRC4. Science. 2015;350:399–404. doi: 10.1126/science.aac5489. [DOI] [PubMed] [Google Scholar]
- 67.Nguyen THD, et al. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature. 2016;530:298–302. doi: 10.1038/nature16940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Agafonov DE, et al. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science. 2016;351:1416–1420. doi: 10.1126/science.aad2085. [DOI] [PubMed] [Google Scholar]
- 69.Wan R, et al. The 3.8 A structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science. 2016;351:466–475. doi: 10.1126/science.aad6466. [DOI] [PubMed] [Google Scholar]
- 70.Mosadeghi R, et al. Structural and kinetic analysis of the COP9-Signalosome activation and the cullin-RING ubiquitin ligase deneddylation cycle. eLife. 2016;5 doi: 10.7554/eLife.12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cavadini S, et al. Cullin–RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature. 2016;531:598–603. doi: 10.1038/nature17416. [DOI] [PubMed] [Google Scholar]
- 72.Liu J-J, et al. CryoEM structure of yeast cytoplasmic exosome complex. Cell Res. 2016;26:822–837. doi: 10.1038/cr.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature. 2015;522:450–454. doi: 10.1038/nature14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang S, et al. Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature. 2016;533:260–264. doi: 10.1038/nature17973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schweitzer A, et al. Structure of the human 26S proteasome at a resolution of 3.9 Å. Proc Natl Acad Sci USA. 2016;113:7816–7821. doi: 10.1073/pnas.1608050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dambacher CM, Worden EJ, Herzik MA, Martin A, Lander GC. Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. eLife. 2016;5:e13027. doi: 10.7554/eLife.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou Q, et al. Cryo-EM structure of SNAP-SNARE assembly in 20S particle. Cell Res. 2015;25:551–560. doi: 10.1038/cr.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao M, et al. Mechanistic insights into the recycling machine of the SNARE complex. Nature. 2015;518:61–67. doi: 10.1038/nature14148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chowdhury S, Ketcham SA, Schroer TA, Lander GC. Structural organization of the dynein–dynactin complex bound to microtubules. Nat Struct Mol Biol. 2015;22:345–347. doi: 10.1038/nsmb.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Urnavicius L, et al. The structure of the dynactin complex and its interaction with dynein. Science. 2015;347:1441–1446. doi: 10.1126/science.aaa4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Verba KA, et al. Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science. 2016;352:1542–1547. doi: 10.1126/science.aaf5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aylett CHS, et al. Architecture of human mTOR complex 1. Science. 2015;351:48–52. doi: 10.1126/science.aaa3870. [DOI] [PubMed] [Google Scholar]
- 83.Baretić D, Berndt A, Ohashi Y, Johnson CM, Williams RL. Tor forms a dimer through an N-terminal helical solenoid with a complex topology. Nature Communications. 2016;7:11016. doi: 10.1038/ncomms11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fernández-Leiro R, Conrad J, Scheres SH, Lamers MH. cryo-EM structures of the E. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease and τ. eLife. 2015;4:459. doi: 10.7554/eLife.11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li N, et al. Structure of the eukaryotic MCM complex at 3.8 Å. Nature. 2015;524:186–191. doi: 10.1038/nature14685. [DOI] [PubMed] [Google Scholar]
- 86.Yuan Z, et al. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol. 2016;23:217–224. doi: 10.1038/nsmb.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abid Ali F, et al. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nature Communications. 2016;7:10708. doi: 10.1038/ncomms10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hoffmann NA, et al. Molecular structures of unbound and transcribing RNA polymerase III. Nature. 2015;528:231–236. doi: 10.1038/nature16143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Murakami K, et al. Structure of an RNA polymerase II preinitiation complex. Proc Natl Acad Sci USA. 2015;112:13543–13548. doi: 10.1073/pnas.1518255112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bernecky C, Herzog F, Baumeister W, Plitzko JM, Cramer P. Structure of transcribing mammalian RNA polymerase II. Nature. 2016;529:551–554. doi: 10.1038/nature16482. [DOI] [PubMed] [Google Scholar]
- 91.He Y, et al. Near-atomic resolution visualization of human transcription promoter opening. Nature. 2016;533:359–365. doi: 10.1038/nature17970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Plaschka C, et al. Transcription initiation complex structures elucidate DNA opening. Nature. 2016;533:353–358. doi: 10.1038/nature17990. [DOI] [PubMed] [Google Scholar]
- 93.Zhang X, et al. In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus. Nature. 2015;527:531–534. doi: 10.1038/nature15767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu H, Cheng L. Cryo-EM shows the polymerase structures and a nonspooled genome within a dsRNA virus. Science. 2015;349:1347–1350. doi: 10.1126/science.aaa4938. [DOI] [PubMed] [Google Scholar]
- 95.Ru H, et al. Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures. Cell. 2015;163:1138–1152. doi: 10.1016/j.cell.2015.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Maskell DP, et al. Structural basis for retroviral integration into nucleosomes. Nature. 2015;523:366–369. doi: 10.1038/nature14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ballandras-Colas A, et al. Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function. Nature. 2016;530:358–361. doi: 10.1038/nature16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qu G, et al. Structure of a group II intron in complex with its reverse transcriptase. Nat Struct Mol Biol. 2016;23:549–557. doi: 10.1038/nsmb.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jiang F, et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351:867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Taylor DW, et al. Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science. 2015;348:581–585. doi: 10.1126/science.aaa4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Behrmann E, et al. Structural Snapshots of Actively Translating Human Ribosomes. Cell. 2015;161:845–857. doi: 10.1016/j.cell.2015.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Campbell MG, et al. Near-atomic resolution reconstructions using a mid-range electron microscope operated at 200kV. J Struct Biol. 2014;188:183–187. doi: 10.1016/j.jsb.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liang B, et al. Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy. Cell. 2015;162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Saibil HR, Grünewald K, Stuart DI. A national facility for biological cryo-electron microscopy. Acta Crystallogr D Biol Crystallogr. 2015;71:127–135. doi: 10.1107/S1399004714025280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cianfrocco MA, Leschziner AE. Low cost, high performance processing of single particle cryo-electron microscopy data in the cloud. eLife. 2015;4:483. doi: 10.7554/eLife.06664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schmeisser M, et al. Parallel, distributed and GPU computing technologies in single-particle electron microscopy. Acta Crystallogr D Biol Crystallogr. 2009;65:659–671. doi: 10.1107/S0907444909011433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kimanius D, Forsberg BO, Scheres S, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. bioRxiv. 2016 doi: 10.1101/059717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McMullan G, Clark AT, Turchetta R, Faruqi AR. Enhanced imaging in low dose electron microscopy using electron counting. Ultramicroscopy. 2009;109:1411–1416. doi: 10.1016/j.ultramic.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Danev R, Baumeister W. Cryo-EM single particle analysis with the Volta phase plate. eLife. 2016;5 doi: 10.7554/eLife.13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fischer N, et al. Structure of the E. coli ribosome–EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM. Nature. 2015;520:567–570. doi: 10.1038/nature14275. [DOI] [PubMed] [Google Scholar]
- 111.Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. eLife. 2014;3:e01963. doi: 10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stark H, Zemlin F, Boettcher C. Electron radiation damage to protein crystals of bacteriorhodopsin at different temperatures. Ultramicroscopy. 1996;63:75–79. [Google Scholar]
- 113.Russo CJ, Passmore LA. Ultrastable gold substrates for electron cryomicroscopy. Science. 2014;346:1377–1380. doi: 10.1126/science.1259530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Russo CJ, Passmore LA. Controlling protein adsorption on graphene for cryo-EM using low-energy hydrogen plasmas. Nature Meth. 2014;11:649–652. doi: 10.1038/nmeth.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pantelic RS, Meyer JC, Kaiser U, Baumeister W, Plitzko JM. Graphene oxide: A substrate for optimizing preparations of frozen-hydrated samples. J Struct Biol. 2010;170:152–156. doi: 10.1016/j.jsb.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 116.Kelly DF, Dukovski D, Walz T. New Applications for Affinity Grids in Preparing EM Specimens. Microsc Microanal. 2010;16:840–841. [Google Scholar]
- 117.Jain T, Sheehan P, Crum J, Carragher B, Potter CS. Spotiton: A prototype for an integrated inkjet dispense and vitrification system for cryo-TEM. J Struct Biol. 2012;179:68–75. doi: 10.1016/j.jsb.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Unwin N. Acetylcholine receptor channel imaged in the open state. Nature. 1995;373:37–43. doi: 10.1038/373037a0. [DOI] [PubMed] [Google Scholar]
- 119.Chen B, et al. Structural Dynamics of Ribosome Subunit Association Studied by Mixing-Spraying Time-Resolved Cryogenic Electron Microscopy. Structure. 2015;23:1097–1105. doi: 10.1016/j.str.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dashti A, et al. Trajectories of the ribosome as a Brownian nanomachine. Proc Natl Acad Sci USA. 2014;111:17492–17497. doi: 10.1073/pnas.1419276111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li H, et al. Structure- and function-based design of Plasmodium-selective proteasome inhibitors. Nature. 2016;530:233–236. doi: 10.1038/nature16936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kosinski J, et al. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science. 2016;352:363–365. doi: 10.1126/science.aaf0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Song F, et al. Cryo-EM Study of the Chromatin Fiber Reveals a Double Helix Twisted by Tetranucleosomal Units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- 124.Baumeister W. Electron tomography: towards visualizing the molecular organization of the cytoplasm. Curr Opin Struct Biol. 2002;12:679–684. doi: 10.1016/s0959-440x(02)00378-0. [DOI] [PubMed] [Google Scholar]
- 125.Mahamid J, et al. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 2016;351:969–972. doi: 10.1126/science.aad8857. [This work shows an exciting vision how electron tomography and subtomogram averaging can bridge the gap between structural biology of isolated complexes and cell biology.] [DOI] [PubMed] [Google Scholar]