Abstract
Cell-cycle checkpoints are generally global in nature: one unattached kinetochore prevents the segregation of all chromosomes; stalled replication forks inhibit late origin firing throughout the genome. A potential exception to this rule is the regulation of replication fork progression by the S-phase DNA damage checkpoint. In this case, it is possible that that the checkpoint is global, and that it slows all replication forks in the genome. However, it is also possible that the checkpoint acts locally at sites of DNA damage, and only slows those forks that encounter DNA damage. Whether the checkpoint regulates forks globally or locally has important mechanistic implications for how replication forks deal with damaged DNA during S phase.
Keywords: DNA Replication, Replication Forks, S-Phase DNA Damage Checkpoint, Intra-S Checkpoint
Introduction
“The dream of every cell is to become two cells” is the basic tenet of cell cycle, as stated by Francois Jacob in 1965. In order to achieve this dream, cells guard their most valuable capital, the genetic information, from endogenous and exogenous damaging agents [1,2]. However, surveying the genome for insults is a complex challenge. Proteins involved in sensing damage have to do so amidst large excess of undamaged template. The situation is more complicated during S phase when potential competing structures are available [3].
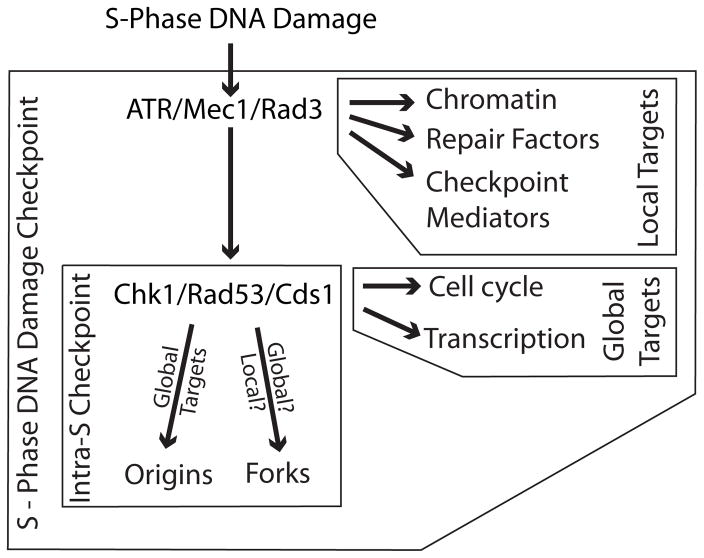
Central mechanisms for responding to DNA damage and other potentially lethal insults are the quality-control checkpoints that arrest the cell cycle and allow time for problems to be resolved [4,5]. These checkpoints, whether responding to DNA damage during G2 or unattached kinetochores during mitosis, are intrinsically global in nature. They respond to trouble in one part of the cell and regulate diverse and distant targets. A potential exception to this rule is the intra-S-phase branch of the S-phase DNA damage checkpoint, which slows DNA replication in response to DNA damage (Figure 1) [6]. It is possible that the effect of this checkpoint—the slowing of replication forks in response to DNA damage—only affects forks that actually encounter DNA damage, instead of globally regulating the progression of all replication forks. This review will discuss the possibility that the regulation of replication forks is a local phenomenon and the mechanistic implications of such a possibility.
Figure 1.
A Schematic Diagram of the S-Phase DNA Damage Checkpoint
DNA damage that block polymerases during S-phase activates the ATR/Mec1/Rad3 checkpoint kinase, which activates the S-phase DNA damage checkpoint. ATR/Mec1/Rad3 is activated locally at sites of DNA damage, therefore its targets are local to sites of damage. However, one of its major targets is activation of the S-phase checkpoint effector kinase Chk1/Rad53/Cds1, which is freely diffusible and triggers a global checkpoint response. A subset of Chk1/Rad53/Cds1 targets, those that constitute the intra-S-phase branch of the checkpoint, regulate replication dynamics by inhibiting origin firing and slowing fork progression. Although the regulation of origin firing is an intrinsically global effect, it is unclear if forks are regulated globally, with all forks being slowed, or locally, with slowing reflecting checkpoint-dependent pausing of forks that encounter damage. The many protein targets of these kinases have been reported recently [15,63–65].
Checkpoint Activation
During S phase, the replicative polymerases encounter DNA lesions leading to activation of the S-phase damage checkpoint [3]. The primary damage sensor, conserved from yeast to mammals, is the ATR kinase (ATM and Rad3 related kinase, the homologs of which are Mec1 in budding yeast and Rad3 in fission yeast) [7,8]. ATR recognizes a wide range of lesions caused by different agents such as methyl-methane sulfonate (MMS), ultra-violet radiation (UV), aphidicolin, cis-platin, and hydroxyurea (HU) [1,3,6,7]. It does so by identifying a common substrate generated in response to damage in S phase – ssDNA coated with replication protein A (RPA) [8–12]. In response to all of these treatments, replicative polymerases stall, while the replicative helicase continues, leading to uncoupling [9,13]. This uncoupling leads to generation of ssDNA, which gets coated with RPA. Using this structure as a substrate allows ATR, in collaboration with a host of other proteins, to respond to diverse lesions [7]. Hence, presence of lesions alone is not sufficient for activation of ATR. Replication across damaged template leading to formation of stalled polymerases is necessary.
Once ATR is activated, it relays the signal to an effector kinase—Chk1 in mammals or Cds1/Rad53 in fission/budding yeast—through various adaptor and mediator proteins [7]. The amount of Chk1 activated is proportional to the amount of ssDNA generated, accounting for the dose dependent activation of the checkpoint [9,14]. While ATR is localized at the scene of crime, targeting fork associated and chromatin factors, the effector kinases act as transducers, spreading the signal throughout the cell (Figure 1)[15,16]. They target a wide variety of proteins that regulate cell cycle, DNA repair, gene expression and cellular metabolism [16–21].
Global Regulation of Origin Firing by the Checkpoint
One branch of the S-phase DNA damage, often referred to as the intra-S phase checkpoint, slows replication of the damaged template (Figure 1) [22]. The intra-S-phase branch itself slows replication in two separate ways: inhibiting origin firing and slowing fork progression [23–29].
The regulation of origin firing is an intrinsically global response, since unfired origins that are distant from the damage are blocked from firing. Consistent with this global nature of origin inhibition, the lowest level of DNA damage sufficient to trigger checkpoint activation appears to be sufficient to fully inhibit further origin firing [25,28]. For instance, low doses of UV in human cells cause a 5-fold reduction in origin firing and increasing the UV dose does not increase the amount of origin inhibition [28]. Treatment with ionizing radiation (IR) reduced origin firing in a similar fashion. A two-fold reduction in origin firing was observed at the lowest doses that trigger checkpoint activation and no additional inhibition was seen at higher doses [25]. This switch-like behavior, in which origin firing is maximally inhibited as soon as the checkpoint is activated, fits with the classic checkpoint paradigm, in which damage at one place in the cell activates a global cellular response that inhibits cell-cycle progression, in this case by inhibiting further origin firing.
The molecular details of origin inhibition are becoming clear in both mammals and budding yeast. In mammals, Chk1 regulates origin firing by inhibiting both cyclin-dependent kinase (CDK) and Dbf4-dependent kinase (DDK), the two major replication kinases. Chk1 inhibits CDK by phosphorylating Cdc25A and thereby targeting it for degradation [30–32]. Cdc25A is a phosphatase required for removing the inhibitory phosphorylation from the Cdk2-CyclinE/A isoforms of CDK, which are required for origin firing. Chk1 further phosphorylates and inhibits DDK, which is also required for origin initiation [33]. In budding yeast Rad53 hyper-phosphorylates the Dbf4 subunit of DDK and as well as the origin initiation factor Sld3 in response to DNA damage and thereby prevents origin firing [34,35].
Although the inhibition of late origin firing is widely conserved, the biological significance of this response is unclear. Inhibition of origin firing in response to DNA damage seems most practical as a means to prevent additional forks encountering damage. However, early origins fire irrespective of the presence of damage [25,27,36]. Therefore forks will continue to encounter damage, albeit less frequently, which may provide more time for repair.
Direct evidence for a less important role for the origin-inhibition branch of the intra-S-phase checkpoint comes from budding yeast. mec1-100, a hypomorphic allele of mec1, which fails to block origin firing but maintains fork regulation, is not sensitive to HU or MMS [37]. This result suggests that fork regulation is the crucial function of the checkpoint. It may be best to think of origin regulation as precautionary—it may be helpful in reducing the frequency of damage encounters, but it does nothing to directly deal with damage.
Regulation of Fork Progression by the Checkpoint
In addition to inhibiting origin firing, the intra-S-phase branch of the S-phase DNA damage checkpoint actively regulates forks by slowing them in response to damage [25,38–40]. The bulky lesions induced by UV, MMS and 4-nitroquinoline 1-oxide (4NQO), treatments commonly used to induce the checkpoint, are believed to block the replicative polymerases [41–43]. Nonetheless, replication forks are able to bypass these lesions, presumably by recruiting translesion polymerases, by recombinational template switching, or by downstream repriming [44–46]. Bypass of damage does not seem to inevitably slow forks because, in checkpoint mutants, damaged DNA is replicated with little or no slowing of bulk DNA synthesis [22,47–49]. Rather, the checkpoint actively slows replication on damaged templates, presumably to allow a more robust response to the damage.
In mammals, the intra-S-phase branch of the checkpoint regulates forks in response to DNA damage. Depletion of Tipin, a factor required for Chk1 activation, prevents slowing of fork progression in response to UV damage, suggesting that the checkpoint actively regulates forks in response to UV damage [40]. However, the role of Tipin, and its partner Timeless, at forks encountering damage is complex, since their depletion compromises Chk1 activation itself [40,50]. Camptothecin (CPT) treatment, which impedes polymerases during S phase, also shows fork slowing in a Chk1 dependent manner [29]. The Hus1 checkpoint clamp subunit and the Werner’s Syndrome helicase are also required for Chk1-dependent fork slowing in response to CPT treatment [51,52]. In addition, Chk1 is indirectly required for maintaining normal fork speeds in the absence of DNA damage via its regulation of origin firing [53]. However, in this case, Chk1 acts to increase fork rates by limiting origin firing, and thus does not appear to have any relation to damage-induced checkpoint-dependent slowing of replication forks.
There is contradictory data about the checkpoint dependence of fork regulation in budding yeast. The Diffley lab has shown that wild-type cells and checkpoint mutants both slow replication fork progression in response to MMS [54]. Hence, fork slowing seems to be a passive response to polymerase stalling lesions independent of the checkpoint. However, the Aparicio lab has shown that Rad53 actively slows down forks in the presence of MMS and fork restart requires its inactivation [38]. Moreover mec1-100, which fails to block origin firing, is not sensitive to HU or MMS unlike mec1Δ [37]. Thus, the checkpoint seems to actively regulate forks.
It is widely believed that the main role of the intra-S-phase checkpoint at forks is to maintain its fork stability [55]. However, much of the evidence for this claim comes from studies using HU arrest to stall forks. Restart of forks from an HU arrest is used to measure the role of checkpoint proteins at the forks. Checkpoint mutants often fail to restart replication after the removal of HU [56–58]. However, it is important to realize that the mechanism of slowing induced by polymerase-blocking DNA damage cannot be the same as the mechanism of arrest induced by HU, which arrests the entire fork. During HU arrest, forks are stalled for prolonged duration in the range of hours, and in the absence of the checkpoint forks are unable to recover from these arrests. In contrast, when a replicative polymerase stalls across a MMS or UV lesion, the fork pausing is transient in nature, and in the absence of the checkpoint, the fork appears to be able to continue replication [40]. Hence, the role of the checkpoint at forks appears to be more than simply maintaining the stability of the replisome components at the site of damage. Likewise, in budding and fission yeast, rad53Δ and cds1Δ cells manage to complete replication without much slowing even in the presence of damage caused by MMS, demonstrating that the checkpoint is not required for fork stability in response to DNA damage [22,47–49]. Recent work from the Labib lab shows that the replisome components associated with the fork in checkpoint mutants and wild-type cells are similar in response to damage [59]. Hence the checkpoint may have different roles at the fork depending on whether the fork stalls for an extended time or simply pauses briefly to negotiate damage.
Is Regulation of Fork Progression by the Checkpoint Global or Local?
Unlike the global regulation of origin firing by the checkpoint, the regulation of replication forks could be either global or local. Global regulation of fork progression would entail the slowing of all replication forks in the cell, regardless of whether or not they encounter DNA damage. As such, it would be analogous to the global regulation of origin firing, in that such regulation would be a precautionary mechanism that would regulate forks in case they might encounter damage. Alternatively, local regulation of forks would only slow forks that were actually replicating damaged DNA.
The difference between global and local regulation of replication forks has important mechanistic implications. If forks are globally slowed, then every fork presumably moves uniformly slowly across undamaged DNA. Such slow replication would provide more time for repair and might also be associated with a mode of replication that is better able to deal with DNA damage when it encounters it. However, this mode of fork slowing would be largely prophylactic and not involved in directly dealing with DNA damage. Alternatively, if forks are only locally slowed at sites of DNA damage, then forks presumably move at normal speeds between lesions and pause transiently, in a checkpoint dependent manner, at sites of damage. As such, local regulation of forks does not so much slow fork progression as cause numerous brief pauses, which, when averaged over time, gives the appearance of slow fork progression. Transient pauses at sites of damage could allow for error-free bypass of the lesion by replication-coupled recombination or other mechanisms of lesion bypass [60]. In any case, local regulation of replication forks implies an intimate interaction between the checkpoint and the replication of the damage template. Because available fork-rate measurements, even single-molecule measurements, actually measure the average fork rate over many kilobases, and the doses of UV and MMS damage used cause lesions about every 500 bp [61,62], multiple transient pauses would have a similar effect on average fork rate as uniform slowing. Therefore, the available data does not distinguish between uniform slowing and multiple transient pauses. Distinguishing between these two possibilities will probably require the monitoring of replication forks moving in real-time.
One indication that fork regulation may be local is that the extent of fork slowing is dependent on the dose of damage. If activation of the checkpoint slowed all forks, one might expect to see the sort of threshold response seen in the inhibition of origin firing, that checkpoint activation, even at low doses, would suffice to trigger fork slowing, and that additional damage would not lead to additional slowing. On the contrary, the extent of fork slowing is dose dependent in a number of systems [24,25,39]. Interestingly, in both mammalian cells and fission yeast, IR, which causes double- and single-strand breaks, does not slow fork progression, even though it activates the checkpoint to a comparable extent to that of the doses of MMS that do slow forks [25,48,49]. It is possible that, because the doses of IR used cause only 10s to 100s of double-strand breaks, even if each break stalls two forks, it would still not affect the overall rate of DNA replication. This situation contrasts with that of UV or MMS damage, in which the millions of lesions caused would lead to slowing even if each lesion causes only a very brief checkpoint-dependent pause.
When considering whether the checkpoint is global or local, a subtle distinction arises between the location of checkpoint action and the extent of checkpoint signaling. The issue we are most concerned with here is where the checkpoint effects are manifest: at all forks or only at forks that encounter damage. However, there is also a potential difference as to whether a fork encountering damage can trigger local checkpoint activation, in which the checkpoint effector kinases are only active in the vicinity of the fork, or whether checkpoint activation requires global activation of the checkpoint effector kinases. If local checkpoint activation is possible, a single fork encountering single lesion could be subject to checkpoint-dependent regulation without triggering a cell-wide checkpoint response. Such events would be cryptic; they would be observable neither by checkpoint kinases assays, nor by slowing of replication, because so few forks would be affected. Cryptic checkpoint regulation would allow for checkpoint functions in many currently unrecognized situations. Alternatively, it is possible that in order to function, the checkpoint effector kinases must be globally activated. If so, it is possible that they phosphorylate all forks, as a precaution against encountering damage, but that the consequence of that phosphorylation is only manifest as slowing when forks encounter damage. Whether checkpoint signaling is an obligate global phenomena, or can sometimes occur locally, is an interesting and important question; however it has less significant implications for the mechanism of the checkpoint that the question of whether forks are slowed globally or only at sites of damage.
Conclusion
The intra-S-phase branch of the S phase checkpoint slows replication forks in response to damage. This replication fork slowing is presumably important for replicating the damaged template. Nonetheless, the mechanistic role of the slowing is still unclear. In fact, it is not even clear if the checkpoint slows all forks or only those that encounter damage. Understanding this basic question of how the checkpoint regulates replication dynamics is a crucial step towards understanding how the checkpoint functions to protect the genome.
Acknowledgments
This work was supported by a National Institutes of Health grant to NR [GM069957].
Abbreviations
- ATR
ATM and Rad3 related kinase
- MMS
Methyl-methane sulfonate
- UV
Ultra-violet radiation
- HU
Hydroxyurea
- RPA
Replication protein A
- IR
Ionizing radiation
- CPT
Camptothecin
- CDK
Cyclin-dependent kinase
- DDK
Dbf4-dependent kinase
References
- 1.Zhou BS, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 2.Labib K, De Piccoli G. Surviving chromosome replication: the many roles of the S-phase checkpoint pathway. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 2011;366:3554–61. doi: 10.1098/rstb.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annual Review of Biochemistry. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 4.Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–22. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 5.Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–34. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 6.Bartek J, Lukas C, Lukas J. Checking on DNA Damage in S phase. Nature. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 7.Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Current Opinion in Cell Biology. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- 8.Cimprich KA, Cortez D. ATR: An Essential Regulator of Genome Integrity. Nature Review Molecular Cell Biology. 2009;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byun TS, Pacek M, Yee M, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes & Development. 2005;19:1040–52. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–8. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 11.Kim SM, Kumagai A, Lee J, Dunphy WG. Phosphorylation of Chk1 by ATM- and Rad3-related (ATR) in Xenopus egg extracts requires binding of ATRIP to ATR but not the stable DNA-binding or coiled-coil domains of ATRIP. The Journal of Biological Chemistry. 2005;280:38355–64. doi: 10.1074/jbc.M508673200. [DOI] [PubMed] [Google Scholar]
- 12.Ball HL, Ehrhardt MR, Mordes DA, Glick GG, Chazin WJ, Cortez D. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Molecular and Cellular Biology. 2007;27:3367–77. doi: 10.1128/MCB.02238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veaute X, Sarasin A. UV lesions located on the leading strand inhibit DNA Replication but do not inhibit SV40 T-antigen helicase activity. DNA Repair. 2000;459:19–28. doi: 10.1016/s0921-8777(99)00052-x. [DOI] [PubMed] [Google Scholar]
- 14.Shimada K, Pasero P, Gasser SM. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes & Development. 2002;16:3236–52. doi: 10.1101/gad.239802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smolka MB, Albuquerque CP, Chen S, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10364–9. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nature Cell Biology. 2003;5:255–260. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- 17.Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, Cocito A, Doksani Y, Klein H, Gómez-González B, Aguilera A, Katou Y, Shirahige K, Foiani M. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell. 2011;146:233–46. doi: 10.1016/j.cell.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bermejo R, Kumar A, Foiani M. Preserving the genome by regulating chromatin association with the nuclear envelope. Trends in Cell Biology. 2012;22:465–73. doi: 10.1016/j.tcb.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Bashkirov VI, Bashkirova EV, Haghnazari E, Heyer W. Direct Kinase-to-Kinase Signaling Mediated by the FHA Phosphoprotein Recognition Domain of the Dun1 DNA Damage Checkpoint Kinase. Molecular and Cellular Biology. 2003;23:1441–1452. doi: 10.1128/MCB.23.4.1441-1452.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smolka MB, Bastos de Oliveira FM, Harris MR, de Bruin RAM. The checkpoint transcriptional response: Make sure to turn it off once you are satisfied. Cell Cycle. 2012;11:3166–3174. doi: 10.4161/cc.21197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dutta C, Patel PK, Rosebrock A, Oliva A, Leatherwood J, Rhind N. The DNA Replication Checkpoint Directly Regulates MBF-Dependent. Molecular and Cellular Biology. 2008;28:5977–5985. doi: 10.1128/MCB.00596-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–7. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 23.Kaufmann WK, Cleaver JE. Mechanisms of inhibition of DNA replication by ultraviolet light in normal human and xeroderma pigmentosum fibroblasts. Journal of Molecular Biology. 1981;149:171–87. doi: 10.1016/0022-2836(81)90297-7. [DOI] [PubMed] [Google Scholar]
- 24.Kaufmann WK, Cleaver JE, Painter RB. Ultraviolet Radiation Inhibits Replicon Initiation In S Phase Human Cells. Biochimica et Biophysica Acta. 1980;608:191–195. doi: 10.1016/0005-2787(80)90147-1. [DOI] [PubMed] [Google Scholar]
- 25.Merrick CJ, Jackson D, Diffley JFX. Visualization of altered replication dynamics after DNA damage in human cells. The Journal of Biological Chemistry. 2004;279:20067–75. doi: 10.1074/jbc.M400022200. [DOI] [PubMed] [Google Scholar]
- 26.Falck J, Petrini JHJ, Williams BR, Lukas J, Bartek J. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nature Genetics. 2002;30:290–4. doi: 10.1038/ng845. [DOI] [PubMed] [Google Scholar]
- 27.Santocanale C, Diffley JF. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–8. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 28.Chastain PD, Nevis KR, Lin L, Kaufmann WK, Kaufman DG. Checkpoint Regulation of Replication Dynamics in UV-Irradiated Human Cells. Cell Cycle. 2006;5:2160–2167. doi: 10.4161/cc.5.18.3236. [DOI] [PubMed] [Google Scholar]
- 29.Seiler JA, Conti C, Syed A, Aladjem MI, Pommier Y. The Intra-S-Phase Checkpoint Affects both DNA Replication Initiation and Elongation_: Single-Cell and - DNA Fiber Analyses. Molecular and Cellular Biology. 2007;27:5806–5818. doi: 10.1128/MCB.02278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao HUI, Piwnica-Worms H. ATR-Mediated Checkpoint Pathways Regulate Phosphorylation and Activation of Human Chk1. Molecular and Cellular Biology. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falck J, Mailand N, Syljuåsen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410:842–7. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 32.Sørensen CS, Syljuåsen RG, Falck J, Schroeder T, Rönnstrand L, Khanna KK, Zhou BB, Bartek J, Lukas J. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell. 2003;3:247–58. doi: 10.1016/s1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 33.Heffernan TP, Ünsal-Kaçmaz K, Heinloth AN, Dennis A, Paules RS, Sancar A, Cordeiro-Stone M, William K. Cdc7/Dbf4 and the Human S Checkpoint Response to UVC. Journal of Biological Chemistry. 2007;282:9458–9468. doi: 10.1074/jbc.M611292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zegerman P, Diffley JFX. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature. 2010;467:474–478. doi: 10.1038/nature09373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez-Mosqueda J, Maas NL, Jonsson ZO, Defazio-Eli LG, Wohlschlegel J, Toczyski DP. Damage-induced phosphorylation of Sld3 is important to block late origin firing. Nature. 2010;467:479–483. doi: 10.1038/nature09377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–21. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 37.Tercero JA, Longhese MP, Diffley JFX. A Central Role for DNA Replication Forks in Checkpoint Activation and Response. Molecular Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 38.Szyjka SJ, Aparicio JG, Viggiani CJ, Knott S, Xu W, Tavaré S, Aparicio OM. Rad53 regulates replication fork restart after DNA damage in Saccharomyces cerevisiae. Genes & Development. 2008;22:1906–20. doi: 10.1101/gad.1660408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willis N, Rhind N. Regulation of DNA replication by the S-phase DNA damage checkpoint. Cell Division. 2009;4:1–10. doi: 10.1186/1747-1028-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unsal-Kaçmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Molecular and Cellular Biology. 2007;27:3131–42. doi: 10.1128/MCB.02190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutation Research. 1985;150:77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- 42.Johnson RE, Yu SL, Prakash S, Prakash L. A role for yeast and human translesion synthesis DNA polymerases in promoting replication through 3-methyl adenine. Molecular and Cellular Biology. 2007;27:7198–205. doi: 10.1128/MCB.01079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore PD, Bose KK, Rabkin SD, Strauss BS. Sites of termination of in vitro DNA synthesis on ultraviolet- and N-acetylaminofluorene-treated ΦX174 templates by prokaryotic and eukaryotic DNA polymerases. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:110–114. doi: 10.1073/pnas.78.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friedberg EC, Lehmann AR, Fuchs RPP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Molecular Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 45.Lambert S, Froget B, Carr A. Arrested replication fork processing: Interplay between checkpoints and recombination. DNA Repair. 2007;6:1042–1061. doi: 10.1016/j.dnarep.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 46.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Molecular Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 47.Lindsay HD, Griffiths DJF, Edwards RJ, Christensen PU, Murray JM, Osman F, Walworth N, Carr AM. S-phase specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes & Development. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willis N, Rhind N. Mus81,Rhp51(Rad51), and Rqh1 form an Epistatic Pathway Required for the S-Phase DNA Damage Checkpoint. Molecular Biology of the Cell. 2009;20:819–833. doi: 10.1091/mbc.E08-08-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhind N, Russell P. The Schizosaccharomyces pombe S-Phase Checkpoint Differentiates Between Different Types of DNA Damage. Genetics. 1998;149:1729–1737. doi: 10.1093/genetics/149.4.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshizawa-Sugata N, Masai H. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. The Journal of Biological Chemistry. 2007;282:2729–40. doi: 10.1074/jbc.M605596200. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Guan J, Hu B, Weiss RS, Iliakis G, Wang Y. Involvement of Hus1 in the chain elongation step of DNA replication after exposure to camptothecin or ionizing radiation. Nucleic Acids Research. 2004;32:767–75. doi: 10.1093/nar/gkh243. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Patro BS, Frøhlich R, Bohr VA, Stevnsner T. WRN helicase regulates the ATR-CHK1-induced S-phase checkpoint pathway in response to topoisomerase-I-DNA covalent complexes. Journal of Cell Science. 2011;124:3967–79. doi: 10.1242/jcs.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16090–16095. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–7. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 55.Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes & Development. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–61. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 57.Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. The EMBO journal. 2003;22:4325–36. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, Gasser SM. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes & Development. 2005;19:3055–69. doi: 10.1101/gad.361805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Piccoli G, Katou Y, Itoh T, Nakato R, Shirahige K, Labib K. Replisome Stability at Defective DNA Replication Forks Is Independent of S Phase Checkpoint Kinases. Molecular Cell. 2012;45:696–704. doi: 10.1016/j.molcel.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 60.Rhind N, Russell P. Checkpoints_: It takes more than time to heal some wounds. Current Biology. 2000:10. doi: 10.1016/s0960-9822(00)00849-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sedgwick SG. Genetic and kinetic evidence for different types of postreplication repair in Escherichia coli B. Journal of Bacteriology. 1975;123:154–61. doi: 10.1128/jb.123.1.154-161.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Courcelle CT, Chow KH, Casey A, Courcelle J. Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9154–9. doi: 10.1073/pnas.0600785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jossen R, Bermejo R. The DNA damage checkpoint response to replication stress: A Game of Forks. Frontiers in Genetics. 2013;4:26. doi: 10.3389/fgene.2013.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 65.Blasius M, Forment JV, Thakkar N, Wagner SA, Choudhary C, Jackson SP. A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biology. 2011;12:R78. doi: 10.1186/gb-2011-12-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]