Abstract
Fibronectin is a modular extracellular matrix protein that is essential for vertebrate development. The 3rd type III domain (3FN3) in fibronectin interacts with other parts of fibronectin and with anastellin, a protein fragment that causes fibronectin aggregation. 3FN3 opens readily both as an isolated domain in solution and when part of fibronectin in stretched fibrils, and it was proposed that this opening is important for anastellin binding. We determined the structure of 3FN3 using nuclear magnetic resonance spectroscopy and we investigated its stability, folding and unfolding. Similar to most other FN3 domains, 3FN3 contains two antiparallel β-sheets that are composed of three (A, B and E) and four (C, D, F and G) β-strands, respectively, and are held together by a conserved hydrophobic interface. Cis-trans isomerization of P847 at the end of β-strand C leads to observable conformational heterogeneity in 3FN3, with a cis peptide bond present in almost one quarter of the molecules. The chemical stability of 3FN3 is relatively low, but the folding rate constant in the absence of denaturant is in the same range as those of other, more stable FN3 domains. Interestingly, the unfolding rate constant in the absence of denaturant is several orders of magnitude higher than the unfolding rate constants of other FN3 domains investigated to date. This unusually fast rate is comparable to the rate of 3FN3 binding to anastellin at saturating anastellin concentrations, consistent with the model that 3FN3 has to unfold in order to interact with anastellin.
Fibronectin is an essential extracellular matrix protein (reviewed in 1-3) that plays an important role in cell adhesion and migration and affects cell proliferation, survival and differentiation. It is required for vertebrate development, and mice that lack fibronectin die during embryogenesis with shortened anterior-posterior axis, absent notochord and somites, and abnormal heart and vasculature 4. Aside from development, fibronectin serves an important function in tissue repair 5-7.
Fibronectin is composed of 12 fibronectin type I (FN1), two fibronectin type II (FN2) and 15-17 fibronectin type III (FN3) homologous repeats, and is secreted as a ~500 kDa disulfide-linked dimer. Formation of fibrils from soluble fibronectin dimers is not spontaneous and generally occurs only in the presence of cells, which assemble fibronectin fibrils around themselves. The molecular basis of the conversion of soluble fibronectin into insoluble fibrillar aggregates is poorly understood, and the atomic resolution structure of the fibrils still remains to be elucidated 3, 8.
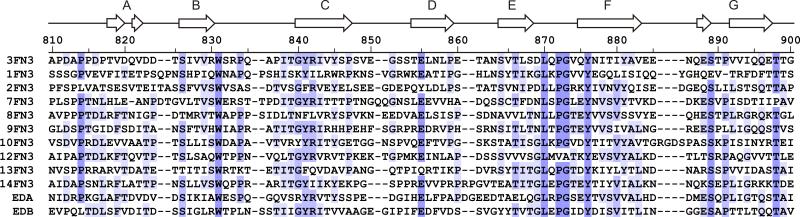
Some parts of fibronectin have been studied extensively, but relatively little information is available about the 3rd FN3 domain (3FN3) (Fig. 1). Together with the 2nd FN3 domain, 3FN3 was proposed to maintain soluble fibronectin dimers in a compact conformation by ionic interactions with the 12th-14th FN3 domains 9 and the 2nd-5th FN1 domains 10. 3FN3 was also reported to mediate cell adhesion and migration through binding to a so far unidentified β1 integrin 11. Finally, 3FN3 and several other FN3 domains were shown to interact with a small fragment of fibronectin called anastellin 12,13.
Figure 1.
Structure-based sequence alignment of FN3 domains from human fibronectin. The sequences are colored by percent identity 57. The sequence of 3FN3 and a diagram of its secondary structure are shown at the top. EDA and EDB stand for the alternatively spliced extra domain A and B, respectively. The 4th-6th, 11th and 15th FN3 domains are not included because their structures are not available.
Anastellin exhibits anti-tumor, anti-metastatic and anti-angiogenic properties 14, 15 and requires endogenous fibronectin for these in vivo activities 16, 17. The binding of anastellin to fibronectin leads to conversion of the soluble fibronectin dimers to insoluble aggregates that are reminiscent of fibronectin fibrils deposited by cells 18, 19. It is not known how anastellin interacts with its target FN3 domains and why this interaction causes aggregation of fibronectin. It was proposed that transient opening of the target FN3 domains is important for binding to anastellin 12, 13, 20.
We employed nuclear magnetic resonance (NMR) spectroscopy, equilibrium denaturation and stopped-flow methods to characterize 3FN3 and its opening behavior. We determined the solution structure of 3FN3, and we identified a minor 3FN3 conformer that is populated under native conditions. We also measured the stability of 3FN3 and investigated its folding and unfolding kinetics, and we show that the unfolding rate constant in the absence of denaturant is significantly higher than the unfolding rate constants reported to date for other isolated FN3 domains. This unusually fast unfolding rate is in the same range as the rate of 3FN3 binding to anastellin at saturating anastellin concentrations, consistent with the model that 3FN3 has to unfold in order to interact with anastellin.
Experimental Procedures
Expression and purification of 3FN3
The pET15b expression vector encoding 3FN3 was provided to us by Dr. Tomoo Ohashi and Dr. Harold Erickson (Duke University) 12, 13. The His-tagged fusion protein was expressed in Escherichia coli BL21-CodonPlus(DE3)-RIPL cells (Agilent) and purified by affinity chromatography on a HisTrap FF column (GE Healthcare). The N-terminal His-tag was cleaved off with thrombin, and 3FN3 was separated from thrombin and the His-tag by affinity chromatography on HiTrap Benzamidine FF (high sub) (GE Healthcare) and HisTrap FF columns respectively. 3FN3 was then further purified by size-exclusion chromatography on Superdex 75 resin (GE Healthcare). The recombinant 3FN3 protein consisted of six extraneous amino acids at the N-terminus, residues 808-905 of human fibronectin (P02751), and two extraneous amino acids at the C-terminus (gshmgtTTAP...PRSDgt).
NMR spectroscopy
Samples for NMR spectroscopy typically contained 0.6 mM unlabeled, uniformly 15N-labeled or uniformly 13C/15N-labeled 3FN3 in phosphate buffered saline (PBS) (pH 7.5) supplemented with 10% 2H2O. Most NMR data were acquired at 25°C on a Varian 600 MHz NMR System that was equipped with a triple resonance probe or a 13C-enhanced salt-tolerant cold probe. NMR experiments that were used to assign 1H, 13C and 15N chemical shifts and to generate restraints for structure calculations included 2-dimensional (2D) 1H-15N heteronuclear single-quantum correlation (HSQC), 1H-13C aliphatic and aromatic HSQC, double-quantum-filtered correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY), nuclear Overhauser effect spectroscopy (NOESY), HBCB(CGCD)HD, 1H-15N heteronuclear multiple-quantum correlation (HMQC) optimized for histidine imidazole rings, 3-dimensional (3D) HNCACB, CBCA(CO)NH, C(CO)NH, H(CCO)NH, HNCO, HN(CA)CO, CCH-TOCSY, HCCH-TOCSY, 15N-edited TOCSY, 15N-edited NOESY, 13C-edited aliphatic NOESY (with and without sensitivity enhancement) and 13C-edited aromatic NOESY. In addition, a 2D 13C/15N-filtered, 13C/15N-edited NOESY spectrum 21 was acquired on a Varian 800 MHz NMR System equipped with a salt-tolerant 13C-enhanced cold probe for a sample containing 0.9 mM 13C/15N-labeled 3FN3 and 0.9 mM unlabeled 3FN3. To evaluate backbone conformational flexibility, steady-state 1H-15N nuclear Overhauser effect (NOE) was measured using 2D heteronuclear correlation experiments. All data were processed with NMRPipe 22 and analyzed with CcpNmr Analysis version 2.2.2 23. Φ and Ψ dihedral angle restraints were generated with TALOS-N 24.
Calculation of 3FN3 structure
Structure calculations were performed using Aria 2.3 25 in conjunction with CNS 1.21 26, with peaks from the NOESY spectra and dihedral angle restraints from TALOS-N as input (Table 1). Manual assignments of the NOESY peaks were retained. 1H-15N NOE values and TALOS-N predictions indicated that the N- and C-terminal tails were dynamic, and no medium or long-range connectivities were identified for residues preceding A810 or following T901 during manual inspection of the NOESY spectra. Aria calculations nevertheless tended to create restraints between the tails and the rest of the protein even when other, more likely assignments were available. To prevent the generation of such artefactual restraints for the tails, only chemical shifts for residues T809-P902 were included in the calculations. In addition, manually identified intraresidual and sequential NOESY peaks stemming from the dynamic residues outside of the T809-P902 region were omitted from the input.
Table 1.
Experimental restraints and structural statistics for 3FN3
| Number of peaks in NOESY spectra | |
| 3D 15N-edited NOESY | 1057 |
| 3D 13C-edited aliphatic NOESY (without sensitivity enhancement) | 1722 |
| 3D 13C-edited aliphatic NOESY (sensitivity-enhanced) | 906 |
| 3D 13C-edited aromatic NOESY | 64 |
| 2D NOESY in H2O | 1912 |
| 2D NOESY in D2O | 1343 |
| 2D 13C/15N-filtered, 13C/15N-edited NOESY | 25 |
| Number of experimental restraints | |
| NOE distance restraints in the last ARIA iteration | |
| Intraresidual | 873 |
| Sequential | 454 |
| Medium range (2 ≤ |i-j| ≤5) | 171 |
| Long range (6 ≤ |i-j|) | 758 |
| Ambiguous | 1179 |
| Dihedral angle restraints (Φ and Ψ) | 168 |
| Hydrogen bond distance restraints | 60 |
| Number of experimental restraint violations | |
| Distance (NOE and hydrogen bond) violations > 0.5 Å | 0.2 ± 0.4 |
| Dihedral angle violations > 5° | 1.1 ± 0.9 |
| Root mean square deviation (RMSD) from experimental restraints | |
| Distance (NOE and hydrogen bond) restraints (Å) | 0.040 ± 0.002 |
| Dihedral angle restraints (°) | 0.9 ± 0.1 |
| RMSD from idealized geometry | |
| Bonds (Å) | 0.0053 ± 0.0001 |
| Angles (°) | 0.63 ± 0.01 |
| Impropers (°) | 1.75 ± 0.06 |
| RMSD of residues 811-901 from mean coordinates | |
| Backbone atoms (N, Ca, C’) (Å) | 0.33 ± 0.08 |
| Heavy atoms (Å) | 0.73 ± 0.06 |
| Distribution of Φ and Ψ dihedral angles of residues 811-901 in Ramachandran plot 56 | |
| Most favored regions (%) | 83.6 |
| Additional allowed regions (%) | 16.1 |
| Generously allowed regions (%) | 0.1 |
| Disallowed regions (%) | 0.3 |
The calculation protocol employed spin-diffusion correction 27, quadrupled number of cooling steps during simulated annealing 28 and otherwise mostly default Aria parameters and options. Hydrogen bond connectivity in the β-sheets was established based on NOE patterns, delayed 1H/2H exchange and the presence of the hydrogen bonds in preliminary structures. Distance restraints were then introduced in subsequent Aria runs to reinforce the expected hydrogen bonds. In the final run, a total of 80 structures were calculated and 25 of them were refined in explicit water. The statistics for the 25 refined structures are summarized in Table 1. Figures depicting the structures were prepared with the molecular graphics program Molmol 2K.1 29. The coordinates and chemical shifts were deposited in the Protein Data Bank (PDB ID 2N1K) and the Biomagnetic Resonance Bank (BMRB ID 25564).
Equilibrium denaturation studies of 3FN3
Samples for equilibrium denaturation studies contained 1 μM 3FN3 and 0-6 M guanidine hydrochloride (GuHCl) in PBS (pH 7.4). After mixing, the samples were allowed to equilibrate at room temperature for at least 2 hours, and their fluorescence at 350 nm was then measured in a JASCO FP-8200 spectrofluorometer at 24°C, using excitation at 280 nm. For each GuHCl concentration tested, fluorescence intensities from triplicate samples were averaged. The concentration of GuHCl at which half of the protein is unfolded, [GuHCl]50%, and the proportionality constant between the free energy of unfolding and the GuHCl concentration, mD-N, were obtained by fitting equation 11 from 30 to the denaturation data. The fitting was done in Matlab version 2012a (MathWorks Inc., Natick, MA). The free energy of unfolding in the absence of denaturant was calculated as 30.
Folding and unfolding kinetics
The changes in fluorescence that accompany folding and unfolding of 3FN3 were monitored in an Applied Photophysics SX20 stopped-flow spectrometer at 23°C, using 280 nm for excitation and a 320 nm cutoff filter to detect emission. Unfolding was initiated by rapidly mixing one volume of 10.6 μM 3FN3 in PBS (pH 7.4) with 10 volumes of PBS containing 1.40-5.00 M GuHCl. To initiate refolding, one volume of 10.6 μM 3FN3 and 3.00 M GuHCl in PBS was mixed with 10 volumes of PBS containing 0-1.14 M GuHCl. Five 5 s fluorescence traces were collected and averaged for each final GuHCl concentration. A double exponential function was then fitted to the unfolding traces and a triple exponential to the refolding traces. A 3 minute trace that was recorded for folding in 0.60 M GuHCl was fitted with a quadruple exponential function. The fitting was performed with the Pro-Data Viewer 4.1.18 software provided with the stopped-flow instrument. The unfolding and refolding rate constants in the absence of denaturant, and , the proportionality constants between ln(ku) or ln(kf) and GuHCl concentration, mku and mkf, and a constant describing the curvature of the unfolding rate, c, were obtained by fitting equations 1 and 2 from 31 to the rate constants for the major unfolding and refolding phases, respectively. The fitting was carried out in Matlab.
Results and Discussion
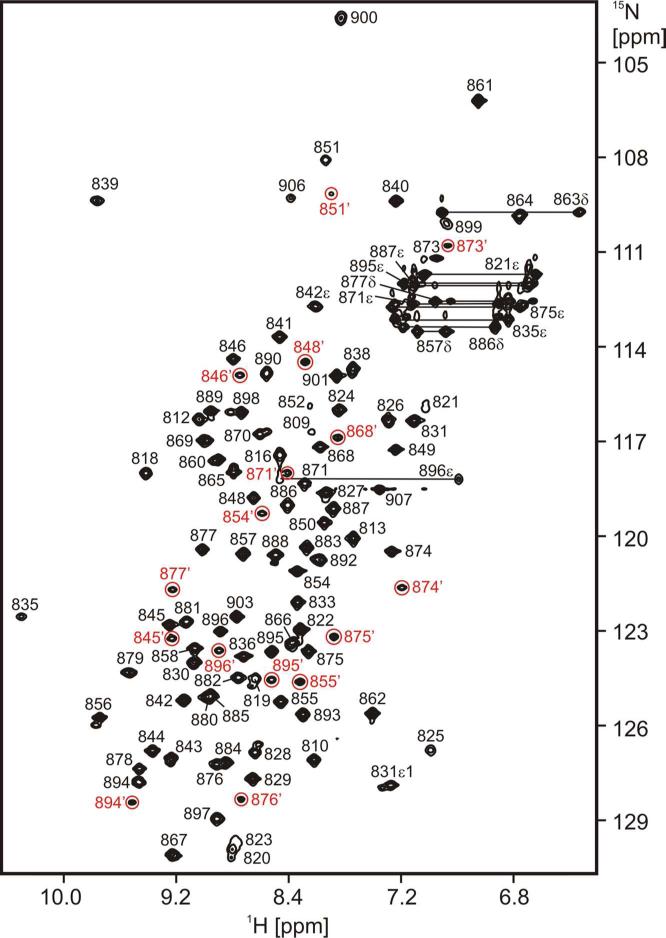
3FN3 is very soluble and stable and its NMR spectra are of high quality (Fig. 2). We were therefore able to assign the majority of its 1H, 13C and 15N chemical shifts and we determined its solution structure on the basis of 2D and 3D homo- and heteronuclear NMR experiments.
Figure 2.
2D 1H-15N HSQC spectrum of 3FN3. The assignments are indicated. Crosspeaks from the minor species that do not adjoin their respective major peaks are highlighted in red and denoted by an apostrophe (’). Signals from T825 and R842ε are aliased from 97.76 ppm and 83.67 ppm, respectively.
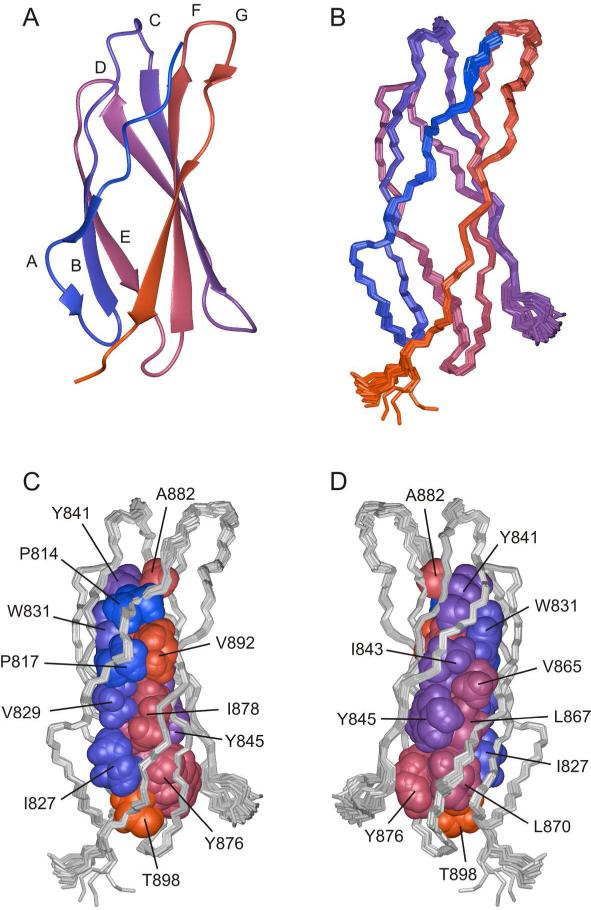
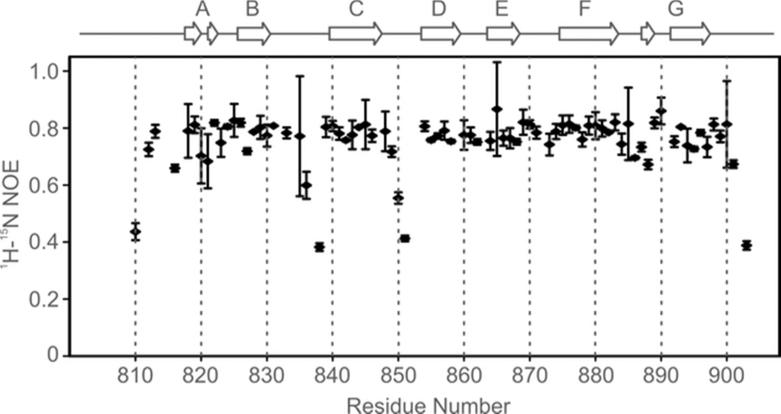
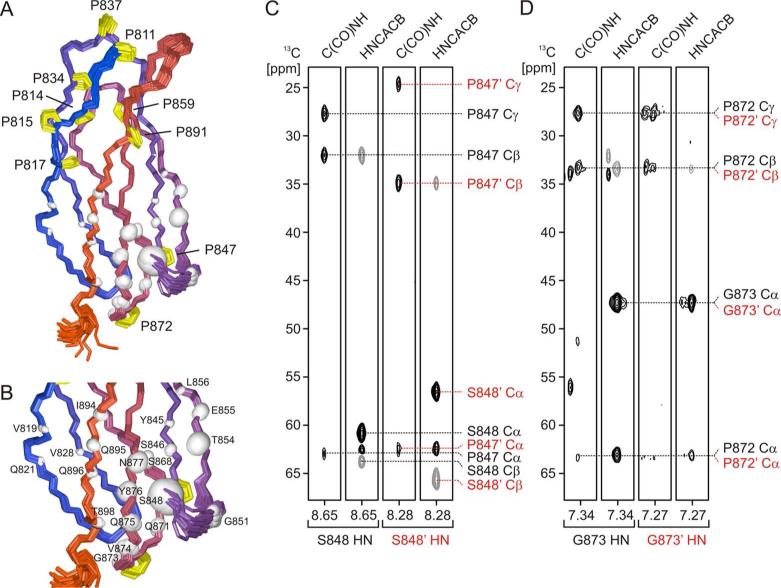
The structure of 3FN3 is shown in Fig. 3. Similar to most other FN3 domains 32-43, 3FN3 contains 7 β-strands (labeled A-G) that form a three-stranded antiparallel β-sheet (β-strands A, B and E) and a four-stranded antiparallel β-sheet (β-strands C, D, F and G); β-strands A and G each contain a bulge (Fig. 3A,B). The two β-sheets are held together by an extensive hydrophobic interface that includes several residues highly conserved among FN3 domains, in particular P814, W831, Y841, L870, Y876 and T898 (Fig. 3C,D). The hydroxyl groups of Y841 and T898 are buried, and the signals from the hydroxyl hydrogens are observable in several NMR spectra (Fig. 4). The loops between β-strands A and B, D and E, E and F, and F and G are well-ordered, but the inter-sheet loop connecting β-strands B and C and the intra-sheet loop connecting β-strands C and D exhibit increased mobility, as indicated by the 1H-15N NOE data (Fig. 5).
Figure 3.
Solution structure of 3FN3. A. A ribbon model of 3FN3 (residues 811-901). The color changes smoothly from blue at the N terminus to red at the C terminus, and the individual β-strands are labeled. B. 25 superimposed backbone traces of 3FN3. The coloring scheme and orientation are the same as in panel A. C and D. The hydrophobic core in 3FN3. To illustrate packing at the interface between the two β-sheets, the side chains of buried residues are shown in a space filling representation and are colored as in panels A and B. The backbone traces are grey. The orientation in C is the same as in panels A and B, and the view in D is related by a 180° rotation around the vertical axis.
Figure 4.
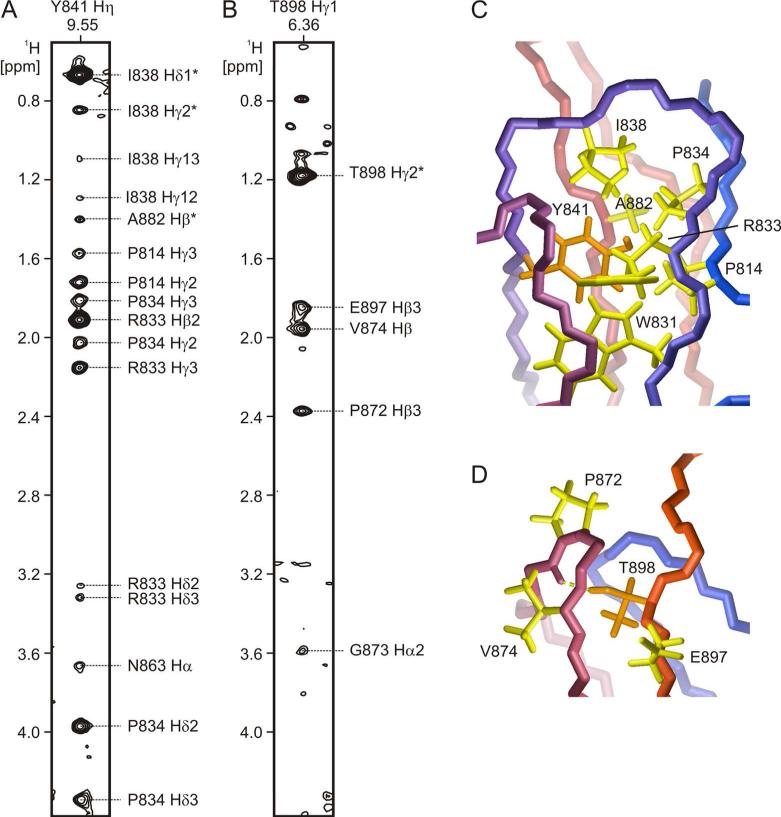
NMR signals from buried hydroxyl groups in 3FN3. A and B. Strips from a 2D 13C/15N-filtered, 13C/15N-edited NOESY spectrum of 3FN3 (mixing time 200 ms) with crosspeaks between Y841 Hη (panel A) or T898 Hγ1 (panel B) and nearby aliphatic hydrogens. The assignments for all observed signals are indicated. The NOESY spectrum also contains crosspeaks between Y841 Hη and W831 Hε1, and between T898 Hγ1 and V874 HN (not shown). C and D. A close-up view of Y841 (panel C) and T898 (panel D) in the 3FN3 structure. The backbone is colored as in Fig. 3A-B, and the side chains of Y841 and T898 are orange. The side chains of residues that are near the hydroxyl hydrogens according to the NOESY data are shown in yellow color. T898 Hγ1 (panel D) forms a hydrogen bond (yellow dashed line) to the backbone carbonyl oxygen of Q871.
Figure 5.
Backbone amide dynamics in 3FN3: 1H-15N NOE data. The secondary structure of 3FN3 is outlined on top. The 1H-15N NOE values for D905, g906 and t907 are negative and are not included in the figure.
The 2D 1H-15N HSQC spectrum contains a set of secondary, weaker crosspeaks that stem from a minor 3FN3 species (Fig. 2). The population of this species is 23±3% as judged from peak volumes. The residues whose backbone amide chemical shifts differ the most between the minor and major forms are clustered in the C-terminal half of the four-stranded β-sheet (Fig. 6A). There are a total of 11 prolines in 3FN3, and we therefore investigated whether proline cis-trans isomerization might be responsible for the heterogeneity. The majority of the prolines are located at the N-terminal end of the domain, and only two, P847 and P872, are in the part of 3FN3 for which the heterogeneity in NMR spectra is observed (Fig. 6A). The Cα, Cβ and Cγ chemical shifts of P872 in the minor species are virtually identical to those in the major form (Fig. 6D). In contrast, the chemical shifts of P847 in the minor species (Cα 62.48 ppm, Cβ 34.93 ppm, Cγ 24.59 ppm) are significantly different from those in the major form (Cα 62.70 ppm, Cβ 32.08 ppm, Cγ 27.75 ppm) (Fig. 6C), and they are indicative of the S846-P847 peptide bond in the minor species being in a cis conformation 44. The largest backbone amide chemical shift differences between the two forms are observed for S848, which immediately follows P847, for T854 in β-strand D, which flanks P847 on one side, and for Y876 and N877 in β-strand F, which flank P847 on the other side (Fig. 6B). Hence, all our data point to the cis-trans isomerization of the S846-P847 peptide bond as the primary source of the conformational heterogeneity in 3FN3.
Figure 6.
Conformational heterogeneity in 3FN3. A. Backbone amide chemical shift heterogeneity mapped onto the 3D structure of 3FN3. The backbone amide nitrogen of each residue with a minor crosspeak in the 2D 1H-15N HSQC spectrum (Fig. 2) is displayed as a white sphere. The volume of the sphere is proportional to the chemical shift difference between the crosspeaks of the major and minor form. The backbone traces are colored as in Fig. 3A-B. Proline side chains are highlighted in yellow and labeled to help visualize their positions relative to the residues whose chemical shifts differ between the two forms. P902, which is located in the dynamic C-terminal tail, is not shown. The view is related to the orientation in Fig. 3A-C by a 40° rotation around the vertical axis. B. A close-up view with the residues labeled. C and D. 13C chemical shifts of P847 (panel C) or P872 (panel D) as observed in the 3D C(CO)NH and HNCACB spectra. The left two strips in each panel are for the major conformer, and the right two strips are for the minor conformer. Positive contour levels are black and negative levels are grey. The chemical shift assignments for the minor conformer are denoted with an apostrophe (’) and are colored red, and the assignments for the major conformer are shown in black.
Cis-trans isomerization of peptidyl-prolyl bonds in the context of a native state was observed previously in other folded proteins and, in some cases, was shown to be functionally important and act as a molecular switch 45-49. The functional significance of the cis-trans isomerization of P847 in 3FN3, however, is not clear at this point, and the heterogeneity may just be a consequence of the relatively low stability of 3FN3 (see below) and the inability to stabilize one isomer strongly over the other 48, 49. Interestingly, a number of the FN3 domains in fibronectin contain a proline residue at the position equivalent to P847 (Fig. 1), but a population of molecules with a cis peptide bond has so far not been identified in any of the structures determined by NMR spectroscopy 40 or X-ray crystallography 35, 37, 42, 43.
3FN3 interacts with anastellin, and it was proposed that the rate of binding at saturating anastellin concentrations (~ 0.02 s−1) is determined by the rate at which 3FN3 spontaneously opens and exposes an otherwise cryptic binding site 13. However, the rates of unfolding that have been reported so far for other FN3 domains under comparable conditions are significantly slower than 0.02 s−1 (7.21 ± 0.26 × 10−5 s−1 for the 3rd FN3 domain from tenascin 50 and 2.3 ± 1.2 × 10−4 s−1 for the 10th FN3 domain from fibronectin, 10FN3 51). Experiments probing the accessibility of engineered buried cysteines in FN3 domains suggested that 3FN3 opens readily, but the opening and closing frequency was not measured 52. This lack of information about the opening rates and opening equilibrium prompted us to undertake a rigorous investigation of the unfolding and refolding behavior of 3FN3.
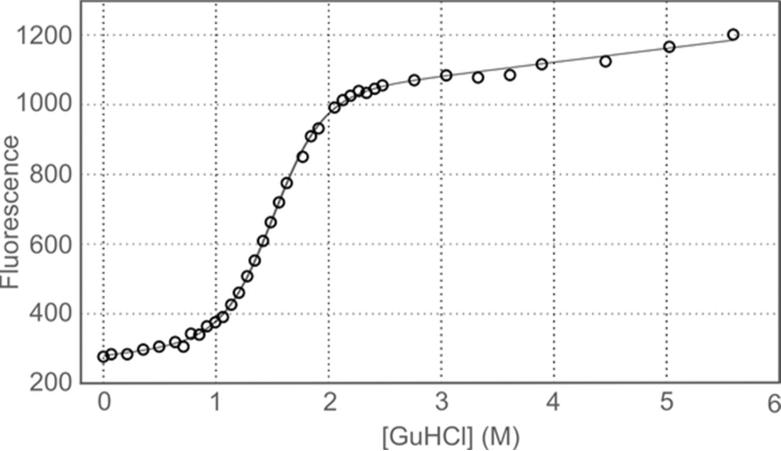
To evaluate the stability of 3FN3, we measured the intrinsic fluorescence of the single tryptophan, W831, as a function of GuHCl concentration. The aromatic ring of W831 is buried in the hydrophobic interface between the two β-sheets (Fig. 3) and becomes exposed to solvent when 3FN3 unfolds; this is accompanied by a large increase in fluorescence emission at 350 nm (Fig. 7). Fitting of the fluorescence denaturation curve yielded the concentration of GuHCl at which half of 3FN3 is unfolded, [GuHCl]50%, of 1.51 ± 0.02 M, and the proportionality constant between the free energy of unfolding and GuHCl concentration, mD-N, of 2.65 ± 0.17 kcal mol−1 M−1. The free energy of unfolding in the absence of denaturant, , was then calculated from these values to be 4.00 ± 0.26 kcal mol−1. This corresponds to about 1 out of every 880 molecules being unfolded under native conditions. No deviation from a two-state transition was apparent, suggesting that the stabilities of the major and minor conformer are comparable.
Figure 7.
Equilibrium denaturation of 3FN3 in GuHCl. The black curve represents the equation fitted to the data.
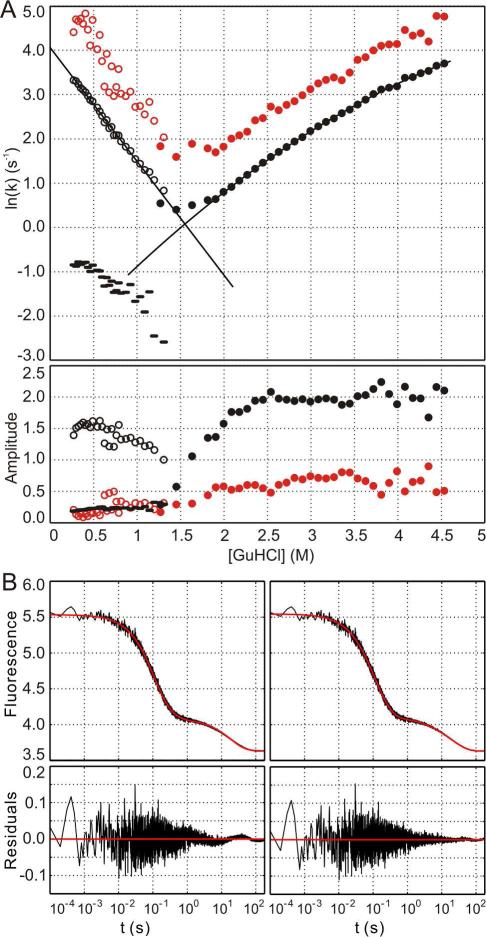
The kinetics of 3FN3 unfolding and refolding were also monitored using W831 fluorescence. Two unfolding and three refolding phases were resolved in 5 s fluorescence traces at all GuHCl concentrations investigated, and an additional slow refolding phase was detected in a 3 minute experiment (Fig. 8). Outside of the transition region, the amplitudes of the individual phases are relatively independent of GuHCl concentration. The natural logarithm of the rate constant for the major refolding phase (empty black circles in Fig. 8A) varies linearly with GuHCl concentration. A linear equation, , was therefore fitted to the data, yielding the refolding rate constant in the absence of denaturant, , of 58 ± 3 s−1, and the proportionality constant, mkf, of 2.57 ± 0.08 M−1. In contrast, the natural logarithm of the rate constant for the major unfolding phase (filled black circles in Fig. 8A) displays non-linear dependence on GuHCl concentration, and a second degree polynomial, , was used to fit the data. The fitting yielded the unfolding rate constant in the absence of denaturant, , of 0.057 ± 0.010 s−1, the proportionality constant, mku, of 2.1 ± 0.1 M−1, and the curvature, c, of 0.15 ± 0.02 M−2. The rate of 3FN3 folding is within the range observed for other FN3 domains but the rate of unfolding is several orders of magnitude higher than those reported previously 50, 51 and is comparable to the rate of anastellin binding to 3FN3 at saturating anastellin concentrations 13.
Figure 8.
Kinetics of 3FN3 folding and unfolding. A. The natural logarithm of the rate constants (upper panel) and the amplitudes (lower panel) are shown as a function of GuHCl concentration for two unfolding (black and red filled circles) and three refolding phases (black and red empty circles and black dashes). The slowest refolding phase that was only detected in a 3 minute experiment is not included in the plot. The solid black lines represent the equations fitted to the data for the major unfolding and refolding phases. B. A triple exponential function (left panel) or a quadruple exponential function (right panel) was fitted to a 3 minute refolding trace in 0.60 M GuHCl. The residuals are shown in the lower panels. The rate constants and amplitudes for the quadruple exponential function are k1 = 17.5 ± 2.2 s−1, a1 = 0.38 ± 0.09 (20%), k2 = 6.7 ± 0.3 s−1, a2 = 1.06 ± 0.09 (55%), k3 = 0.28 ± 0.06 s−1, a3 = 0.07 ± 0.01 (4%), k4 = 0.047 ± 0.002 s−1, a4 = 0.40 ± 0.01 (21%).
Interestingly, we also observed a minor unfolding and a minor refolding phase whose rate constants were slightly higher than those for the major phase (filled and empty red circles in Fig. 8A). The slopes and the positions of the minima in the ln(k) versus [GuHCl] plot for the minor and the major phases are very similar, and, consequently, the plot for these phases has a “nested chevron” appearance (Fig. 8A). We attribute the minor unfolding and refolding phase to unfolding and refolding of the minor 3FN3 conformer that contains a cis S846-P847 peptide bond. This interpretation is consistent with the stabilities of the minor and major conformer being indistinguishable in equilibrium denaturation experiments, and with the structure of the minor conformer overall closely resembling that of the major species (~70% of backbone amide chemical shifts are identical in the two forms). The minor kinetic phase accounts for 25 ± 5% of the unfolding amplitude, in excellent agreement with the population of the minor conformer that was estimated from the NMR data.
In addition to the major phase and the fast minor phase described above, we identified two slow minor refolding phases (Fig. 8B and black dashes in Fig. 8A). In 0.6 M GuHCl, these slow phases represent ~25% of the total refolding amplitude (Fig. 8B). We attribute the slow phases to processes in which proline cis-trans isomerization is the rate-limiting step, i.e. to refolding of denatured 3FN3 in which some of the prolines are in the non-native cis conformation 53.
The kinetic data are in general consistent with a two-state folding transition, as demonstrated using several well established tests 31, 50, 54. We do not observe a significant burst phase or deviation from linearity in the ln(kf) versus [GuHCl] plot for the major refolding phase. In addition, the equilibrium unfolding constant derived from the kinetic data and adjusted for proline isomerization in the denatured and native state corresponds to free energy of unfolding, , of 3.89 ± 0.12 kcal mol−1. This value compares well with the free energy of unfolding from equilibrium experiments, 4.00 ± 0.26 kcal mol−1. Finally, the m-value calculated from the kinetic data, mk = RT(mkf + mku), is 2.76 ± 0.08 kcal mol−1 M−1, in good agreement with the m-value obtained from equilibrium experiments, 2.65 ± 0.17 kcal mol−1 M−1.
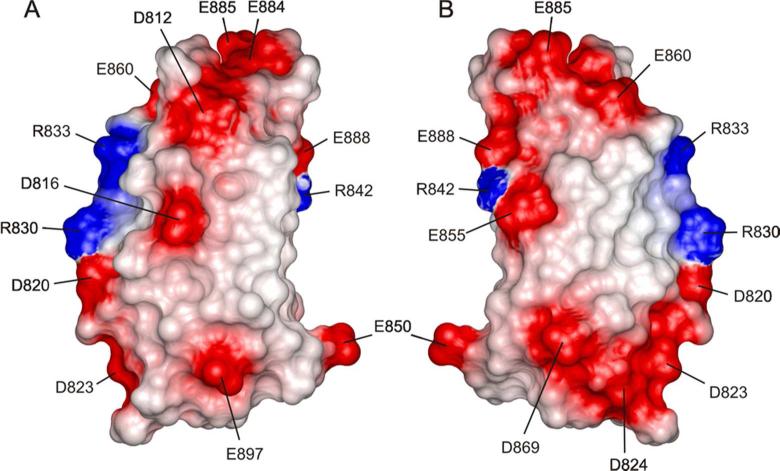
We have inspected the sequence and structure of 3FN3 for features that might explain the relatively low stability and high unfolding rate. The structure of 3FN3 is very similar to that of other FN3 domains, and most residues in its hydrophobic core are identical to those in the markedly more stable 10FN3 or are conservatively substituted (Fig. 1 and 3). The only residue in the hydrophobic core that significantly differs from 10FN3 and from the FN3 consensus is P817. In 10FN3 and most other FN3 domains, this position is occupied by leucine (Fig. 1). In addition to hydrophobic core packing, electrostatic interactions are also known to play a role in protein stability 55. 3FN3 is highly acidic, with 6 aspartates, 7 glutamates, 3 arginines and no lysines or histidines within the structured region (Fig. 9). It is therefore likely that unfavorable electrostatic interactions contribute to the lower stability and faster unfolding rate. The only residue that has so far been shown to destabilize 3FN3, D869, is in fact acidic 13. Most other FN3 domains contain a glycine or an asparagine at this position (Fig. 1), and a D869G mutation in 3FN3 increases the chemical stability of the domain and decreases the rate of binding to anastellin 13.
Figure 9.
Distribution of charged residues in 3FN3. The solvent accessible surface of 3FN3 is colored according to electrostatic potential, with areas of negative, positive, or neutral electrostatic potential depicted in red, blue, or white, respectively. In panel A, the orientation is the same as in Fig. 3A-C, and the view in panel B is related by a 180° rotation around the vertical axis.
Overall our results demonstrate that while the structure and folding behavior of 3FN3 are similar to those of other FN3 domains, 3FN3 differs from previously characterized FN3 domains in several aspects. Firstly, 3FN3 is conformationally heterogeneous due to a cis-trans peptide bond isomerization at the semi-conserved P847. Secondly, 3FN3 displays relatively low stability and significantly higher unfolding rate than other isolated FN3 domains investigated to date. Ohashi et al. proposed that when 3FN3 interacts with anastellin, the rate of binding at saturating anastellin concentrations (~ 0.02 s−1) is determined by the rate at which 3FN3 spontaneously opens 13. We have now shown that the experimentally determined rate of 3FN3 unfolding (~ 0.06 s−1) is comparable to the rate observed by Ohashi, consistent with the model that 3FN3 has to open in order to bind anastellin.
Acknowledgements
We are grateful to Dr. Tomoo Ohashi and Dr. Harold Erickson (Duke University) for providing the vector for expression of 3FN3.
Funding
Research reported in this publication was supported by the National Science Foundation Grant MCB-0846132 and the National Institutes of Health Grant P20GM103546. The Magnetic Resonance Core Facility at the University of Montana, at which some of the work was performed, also received support from the National Institutes of Health Grant P20GM103546. The 600 MHz NMR spectrometer at the University of Montana was purchased with funds from the National Science Foundation Grant CHE-0321002 and the Murdock Charitable Trust. The W. M. Keck High Field 800 MHz NMR Facility at the University of Colorado at Boulder was established using funds from the W. M. Keck Foundation.
Abbreviations
- 2D
two-dimensional
- 3D
three-dimensional
- 10FN3
the tenth fibronectin type III domain from human fibronectin
- 3FN3
the third fibronectin type III domain from human fibronectin
- COSY
correlation spectroscopy
- EDA
extra domain A
- EDB
extra domain B
- FN1
fibronectin type I
- FN2
fibronectin type II
- FN3
fibronectin type III
- GuHCl
guanidine hydrochloride
- HMQC
heteronuclear multiple-quantum correlation
- HSQC
heteronuclear single-quantum correlation
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- NOESY
nuclear Overhauser effect spectroscopy
- PBS
phosphate-buffered saline
- TOCSY
total correlation spectroscopy
References
- 1.Mosher DF. Fibronectin. Academic Press; New York: 1989. [Google Scholar]
- 2.Hynes RO. Fibronectins. Springer-Verlag Inc.; New York: 1990. [Google Scholar]
- 3.Singh P, Carraher C, Schwarzbauer JE. Assembly of Fibronectin Extracellular Matrix. Annu. Rev. Cell Dev. Biol. 2010;26:397–419. doi: 10.1146/annurev-cellbio-100109-104020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.George EL, Georges-Labouesse EN, Patel-King RS, Rayburn H, Hynes RO. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993;119:1079–1091. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- 5.Sakai T, Johnson KJ, Murozono M, Sakai K, Magnuson MA, Wieloch T, Cronberg T, Isshiki A, Erickson HP, Fässler R. Plasma fibronectin supports neuronal survival and reduces brain injury following transient focal cerebral ischemia but is not essential for skin-wound healing and hemostasis. Nat. Med. 2001;7:324–330. doi: 10.1038/85471. [DOI] [PubMed] [Google Scholar]
- 6.Ni H, Yuen PS, Papalia JM, Trevithick JE, Sakai T, Fässler R, Hynes RO, Wagner DD. Plasma fibronectin promotes thrombus growth and stability in injured arterioles. Proc. Natl. Acad. Sci. USA. 2003;100:2415–2419. doi: 10.1073/pnas.2628067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konstandin MH, Toko H, Gastelum GM, Quijada P, De La Torre A, Quintana M, Collins B, Din S, Avitabile D, Völkers M, Gude N, Fässler R, Sussman MA. Fibronectin is essential for reparative cardiac progenitor cell response after myocardial infarction. Circ. Res. 2013;113:115–125. doi: 10.1161/CIRCRESAHA.113.301152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Früh SM, Schoen I, Ries J, Vogel V. Molecular architecture of native fibronectin fibrils. Nat. Commun. 2015;6:7275. doi: 10.1038/ncomms8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson KJ, Sage H, Briscoe G, Erickson HP. The compact conformation of fibronectin is determined by intramolecular ionic interactions. J. Biol. Chem. 1999;274:15473–15479. doi: 10.1074/jbc.274.22.15473. [DOI] [PubMed] [Google Scholar]
- 10.Vakonakis I, Staunton D, Ellis IR, Sarkies P, Flanagan A, Schor AM, Schor SL, Campbell ID. Motogenic sites in human fibronectin are masked by long range interactions. J. Biol. Chem. 2009;284:15668–15675. doi: 10.1074/jbc.M109.003673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Obara M, Sakuma T, Fujikawa K. The third type III module of human fibronectin mediates cell adhesion and migration. J. Biochem. 2010;147:327–335. doi: 10.1093/jb/mvp168. [DOI] [PubMed] [Google Scholar]
- 12.Ohashi T, Erickson HP. Domain unfolding plays a role in superfibronectin formation. J. Biol. Chem. 2005;280:39143–39151. doi: 10.1074/jbc.M509082200. [DOI] [PubMed] [Google Scholar]
- 13.Ohashi T, Augustus AM, Erickson HP. Transient opening of fibronectin type III (FNIII) domains: the interaction of the third FNIII domains of FN with anastellin. Biochemistry. 2009;48:4189–4197. doi: 10.1021/bi900001g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pasqualini R, Bourdoulous S, Koivunen E, Woods VL, Jr., Ruoslahti E. A polymeric form of fibronectin has antimetastatic effects against multiple tumor types. Nat. Med. 1996;2:1197–1203. doi: 10.1038/nm1196-1197. [DOI] [PubMed] [Google Scholar]
- 15.Yi M, Ruoslahti E. A fibronectin fragment inhibits tumor growth, angiogenesis, and metastasis. Proc. Natl. Acad. Sci. USA. 2001;98:620–624. doi: 10.1073/pnas.98.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yi M, Sakai T, Fässler R, Ruoslahti E. Antiangiogenic proteins require plasma fibronectin or vitronectin for in vivo activity. Proc. Natl. Acad. Sci. USA. 2003;100:11435–11438. doi: 10.1073/pnas.1635112100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Åkerman ME, Pilch J, Peters D, Ruoslahti E. Angiostatic peptides use plasma fibronectin to home to angiogenic vasculature. Proc. Natl. Acad. Sci. USA. 2005;102:2040–2045. doi: 10.1073/pnas.0409844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morla A, Zhang Z, Ruoslahti E. Superfibronectin is a functionally distinct form of fibronectin. Nature. 1994;367:193–196. doi: 10.1038/367193a0. [DOI] [PubMed] [Google Scholar]
- 19.Briknarová K, Åkerman ME, Hoyt DW, Ruoslahti E, Ely KR. Anastellin, an FN3 fragment with fibronectin polymerization activity, resembles amyloid fibril precursors. J. Mol. Biol. 2003;332:205–215. doi: 10.1016/s0022-2836(03)00890-8. [DOI] [PubMed] [Google Scholar]
- 20.Ohashi T, Erickson HP. Fibronectin aggregation and assembly. The unfolding of the second fibronectin type III domain. J. Biol. Chem. 2011;286:39188–39199. doi: 10.1074/jbc.M111.262337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zwahlen C, Legault P, Vincent SJF, Greenblatt J, Konrat R, Kay LE. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: application to a bacteriophage λ N-peptide/boxB RNA complex. J. Am. Chem. Soc. 1997;119:6711–6721. [Google Scholar]
- 22.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 23.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinás M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 24.Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin TE, Nilges M. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- 26.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system (CNS): A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Linge JP, Habeck M, Rieping W, Nilges M. Correction of spin diffusion during iterative automated NOE assignment. J. Magn. Reson. 2004;167:334–342. doi: 10.1016/j.jmr.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 28.Fossi M, Oschkinat H, Nilges M, Ball LJ. Quantitative study of the effects of chemical shift tolerances and rates of SA cooling on structure calculation from automatically assigned NOE data. J. Magn. Reson. 2005;175:92–102. doi: 10.1016/j.jmr.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 29.Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 30.Clarke J, Fersht AR. Engineered disulfide bonds as probes of the folding pathway of barnase: Increasing the stability of proteins against the rate of denaturation. Biochemistry. 1993;32:4322–4329. doi: 10.1021/bi00067a022. [DOI] [PubMed] [Google Scholar]
- 31.Clarke J, Hamill SJ, Johnson CM. Folding and stability of a fibronectin type III domain of human tenascin. J. Mol. Biol. 1997;270:771–778. doi: 10.1006/jmbi.1997.1147. [DOI] [PubMed] [Google Scholar]
- 32.Leahy DJ, Hendrickson WA, Aukhil I, Erickson HP. Structure of a fibronectin type III domain from tenascin phased by MAD analysis of the selenomethionyl protein. Science. 1992;258:987–991. doi: 10.1126/science.1279805. [DOI] [PubMed] [Google Scholar]
- 33.Main AL, Harvey TS, Baron M, Boyd J, Campbell ID. The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions. Cell. 1992;71:671–678. doi: 10.1016/0092-8674(92)90600-h. [DOI] [PubMed] [Google Scholar]
- 34.Dickinson CD, Veerapandian B, Dai XP, Hamlin RC, Xuong NH, Ruoslahti E, Ely KR. Crystal structure of the tenth type III cell adhesion module of human fibronectin. J. Mol. Biol. 1994;236:1079–1092. doi: 10.1016/0022-2836(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 35.Leahy DJ, Aukhil I, Erickson HP. 2.0 Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synenergy region. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- 36.Copié V, Tomita Y, Akiyama SK, Aota S, Yamada KM, Venable RM, Pastor RW, Krueger S, Torchia DA. Solution structure and dynamics of linked cell attachment modules of mouse fibronectin containing the RGD and synergy regions: comparison with the human fibronectin crystal structure. J. Mol. Biol. 1998;277:663–682. doi: 10.1006/jmbi.1998.1616. [DOI] [PubMed] [Google Scholar]
- 37.Sharma A, Askari JA, Humphries MJ, Jones EY, Stuart DI. Crystal structure of a heparin- and integrin-binding segment of human fibronectin. EMBO J. 1999;18:1468–1479. doi: 10.1093/emboj/18.6.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fattorusso R, Pellecchia M, Viti F, Neri P, Neri D, Wüthrich K. NMR structure of the human oncofoetal fibronectin ED-B domain, a specific marker for angiogenesis. Structure. 1999;7:381–390. doi: 10.1016/s0969-2126(99)80051-3. [DOI] [PubMed] [Google Scholar]
- 39.Niimi T, Osawa M, Yamaji N, Yasunaga K, Sakashita H, Mase T, Tanaka A, Fujita S. NMR structure of human fibronectin EDA. J. Biomol. NMR. 2001;21:281–284. doi: 10.1023/a:1012947209393. [DOI] [PubMed] [Google Scholar]
- 40.Gao M, Craig D, Lequin O, Campbell ID, Vogel V, Schulten K. Structure and functional significance of mechanically unfolded fibronectin type III1 intermediates. Proc. Natl. Acad. Sci. USA. 2003;100:14784–14789. doi: 10.1073/pnas.2334390100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vakonakis I, Staunton D, Rooney LM, Campbell ID. Interdomain association in fibronectin: insight into cryptic sites and fibrillogenesis. EMBO J. 2007;26:2575–2583. doi: 10.1038/sj.emboj.7601694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bencharit S, Cui CB, Siddiqui A, Howard-Williams EL, Sondek J, Zuobi-Hasona K, Aukhil I. Structural insights into fibronectin type III domain-mediated signaling. J. Mol. Biol. 2007;367:303–309. doi: 10.1016/j.jmb.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schiefner A, Gebauer M, Skerra A. Extra-domain B in oncofetal fibronectin structurally promotes fibrillar head-to-tail dimerization of extracellular matrix protein. J. Biol. Chem. 2012;287:17578–17588. doi: 10.1074/jbc.M111.303131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schubert M, Labudde D, Oschkinat H, Schmieder P. A software tool for the prediction of Xaa-Pro peptide bond conformations in proteins based on 13C chemical shift statistics. J. Biomol. NMR. 2002;24:149–154. doi: 10.1023/a:1020997118364. [DOI] [PubMed] [Google Scholar]
- 45.Evans PA, Dobson CM, Kautz RA, Hatfull G, Fox RO. Proline isomerism in staphylococcal nuclease characterized by NMR and site-directed mutagenesis. Nature. 1987;329:266–268. doi: 10.1038/329266a0. [DOI] [PubMed] [Google Scholar]
- 46.Andreotti AH. Native state proline isomerization: An intrinsic molecular switch. Biochemistry. 2003;42:9515–9524. doi: 10.1021/bi0350710. [DOI] [PubMed] [Google Scholar]
- 47.Schmid F. Prolyl isomerization in protein folding. In: Buchner J, Kiefhaber T, editors. Protein folding handbook. Part I. Wiley-VCH Verlag GmbH; Weinheim, Germany: 2005. pp. 916–945. [Google Scholar]
- 48.Schmidpeter PAM, Schmid FX. Prolyl isomerization and its catalysis in protein folding and protein function. J. Mol. Biol. 2015;427:1609–1631. doi: 10.1016/j.jmb.2015.01.023. [DOI] [PubMed] [Google Scholar]
- 49.Schmidpeter PAM, Koch JR, Schmid FX. Control of protein function by prolyl isomerization. Biochim. Biophys. Acta. 2015;1850:1973–1982. doi: 10.1016/j.bbagen.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 50.Hamill SJ, Meekhof AE, Clarke J. The effect of boundary selection on the stability and folding of the third fibronectin type III domain from human tenascin. Biochemistry. 1998;37:8071–8079. doi: 10.1021/bi9801659. [DOI] [PubMed] [Google Scholar]
- 51.Cota E, Clarke J. Folding of beta-sandwich proteins: Three-state transition of a fibronectin type III module. Protein Sci. 2000;9:112–120. doi: 10.1110/ps.9.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lemmon CA, Ohashi T, Erickson HP. Probing the folded state of fibronectin type III domains in stretched fibrils by measuring buried cysteine accessibility. J. Biol. Chem. 2011;286:26375–26382. doi: 10.1074/jbc.M111.240028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jackson SE, Fersht AR. Folding of chymotrypsin inhibitor 2. 2. Influence of proline isomerization on the folding kinetics and thermodynamic characterization of the transition state of folding. Biochemistry. 1991;30:10436–10443. doi: 10.1021/bi00107a011. [DOI] [PubMed] [Google Scholar]
- 54.Jackson SE, Fersht AR. Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry. 1991;30:10428–10435. doi: 10.1021/bi00107a010. [DOI] [PubMed] [Google Scholar]
- 55.Strickler SS, Gribenko AV, Gribenko AV, Keiffer TR, Tomlinson J, Reihle T, Loladze VV, Makhatadze GI. Protein stability and surface electrostatics: a charged relationship. Biochemistry. 2006;45:2761–2766. doi: 10.1021/bi0600143. [DOI] [PubMed] [Google Scholar]
- 56.Laskowski RA, Rullmann JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality or protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 57.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]