Abstract
Although data are available on the change of expression/activity of drug-metabolizing enzymes in liver cirrhosis patients, corresponding data on transporter protein expression are not available. Therefore, using quantitative targeted proteomics, we compared our previous data on noncirrhotic control livers (n = 36) with the protein expression of major hepatobiliary transporters, breast cancer resistance protein (BCRP), bile salt export pump (BSEP), multidrug and toxin extrusion protein 1 (MATE1), multidrug resistance–associated protein (MRP)2, MRP3, MRP4, sodium taurocholate–cotransporting polypeptide (NTCP), organic anion–transporting polypeptides (OATP)1B1, 1B3, 2B1, organic cation transporter 1 (OCT1), and P-glycoprotein (P-gp) in alcoholic (n = 27) and hepatitis C cirrhosis (n = 30) livers. Compared with control livers, the yield of membrane protein from alcoholic and hepatitis C cirrhosis livers was significantly reduced by 56 and 67%, respectively. The impact of liver cirrhosis on transporter protein expression was transporter-dependent. Generally, reduced protein expression (per gram of liver) was found in alcoholic cirrhosis livers versus control livers, with the exception that the expression of MRP3 was increased, whereas no change was observed for MATE1, MRP2, OATP2B1, and P-gp. In contrast, the impact of hepatitis C cirrhosis on protein expression of transporters (per gram of liver) was diverse, showing an increase (MATE1), decrease (BSEP, MRP2, NTCP, OATP1B3, OCT1, and P-gp), or no change (BCRP, MRP3, OATP1B1, and 2B1). The expression of hepatobiliary transporter protein differed in different diseases (alcoholic versus hepatitis C cirrhosis). Finally, incorporation of protein expression of OATP1B1 in alcoholic cirrhosis into the Simcyp physiologically based pharmacokinetics cirrhosis module improved prediction of the disposition of repaglinide in liver cirrhosis patients. These transporter expression data will be useful in the future to predict transporter-mediated drug disposition in liver cirrhosis patients.
Introduction
Liver cirrhosis resulting from chronic liver injury (e.g., chronic infection with hepatitis virus or excess alcohol use), characterized by loss of functional hepatocytes and formation of scar tissue (fibrosis), can lead to significant morphologic changes in the architecture of the liver (Schuppan and Afdhal, 2008). At the end-stage of chronic liver disease, liver cirrhosis may impair hepatic and renal function, reduce plasma protein concentrations, elevate intrahepatic resistance (resulting in portal hypertension), and increase cardiac output. These outcomes can have a profound impact on the pharmacokinetics of drugs administered to patients with liver cirrhosis, which may result in unexpected adverse effects (Bosch and Garcia-Pagan, 2000; Dincer et al., 2005; Schuppan and Afdhal, 2008) and/or potential change of drug efficacy. For example, compared with healthy subjects, the plasma area under the curve (AUC) of clomethiazole, used in the treatment of alcoholism, is increased by 17-fold in liver cirrhosis patients (Pentikainen et al., 1980). Indeed, according to the FDA Guidance on pharmacokinetics in patients with impaired hepatic function, dose adjustment is recommended when at least a 2-fold change in plasma AUC is observed in patients with hepatic impairment (http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm072123).
A considerable amount of data shows that drug-metabolizing enzyme (e.g., CYP1A2, 2C9, 2E1, and 3A4) expression is decreased in cirrhotic human livers (George et al., 1995). Clinical studies confirm that the urinary metabolic ratio of caffeine, mephenytoin, debrisoquine, and chlorzoxazone, which are in vivo probes of CYP1A2, 2C9, 2D6, and 2E1, respectively, is reduced (Ohnishi et al., 2005; Frye et al., 2006). However, there is a paucity of data on the effect of cirrhosis on expression and activity of hepatic drug transporters (Ogasawara et al., 2010; More et al., 2013). Therefore, using quantitative proteomics, we quantified the protein expression of the major hepatobiliary transporters in liver tissue obtained from patients with alcoholic and hepatitis C cirrhosis. The results obtained were then compared with our previous data on the expression of these transporters in noncirrhotic human livers (controls) determined using the same methodology (Prasad et al., 2016). Finally, we used these data to predict changes in the disposition of a drug, repaglinide, in patients with liver cirrhosis.
Materials and Methods
Chemicals and Reagents.
Synthetic signature peptides for quantified transporters (Table 1) were obtained from New England Peptides (Boston, MA). The corresponding stable isotope-labeled internal standards (AQUA QuantPro, ± 25% precision) were purchased from Thermo Fisher Scientific (Rockford, IL). These peptides were identical to those used previously to quantify hepatobiliary transporters in control livers (Prasad et al., 2013, 2014; Wang et al., 2015). The ProteoExtract native membrane protein extraction kit was obtained from Calbiochem (Temecula, CA). Sodium deoxycholate (98% purity) was purchased from MP Biomedicals (Santa Ana, CA). Iodoacetamide, dithiothreitol, ammonium bicarbonate (98% purity), BCA protein assay kit, and in-solution trypsin digestion kit were obtained from Pierce Biotechnology (Rockford, IL). High-performance liquid chromatography–grade acetonitrile (99.9% purity), methanol (99.9% purity), and formic acid (≥99.5% purity) were purchased from Fischer Scientific (Fair Lawn, NJ). Deionized water was obtained from a Q-Gard 2 Purification Cartridge water purifying system (Millipore, Bedford, MA).
TABLE 1 .
Multiple reaction monitoring parameters of peptides (calibrator and internal standard) selected for targeted analysis of human hepatobiliary transporters
The labeled amino acid residue of the internal standard is shown in bold and italic.
| Transporter | Signature Peptides | Parent Ion | Product Ions |
Cone (V) | CE (V) | RT | On-Column Calibration Rangea | |
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | |||||||
| z = 2 | z = 1 | z = 1 | min | femtomole | ||||
| BCRP | SSLLDVLAAR | 523.0 | 757.4 | 644.3 | 40 | 18 | 21.0 | 2.76–88 |
| SSLLDVLAAR | 527.8 | 654.5 | 767.5 | |||||
| BSEP | STALQLIQR | 515.2 | 529.1 | 657.1 | 32 | 18 | 15.4 | 2.92–93 |
| STALQLIQR | 520.3 | 539.4 | 667.4 | |||||
| MATE1 | GGPEATLEVR | 514.7 | 816.9 | 688.0 | 30 | 14 | 12.2 | 3.46–111 |
| GGPEATLEVR | 519.8 | 627.4 | 698.4 | |||||
| MRP2 | LTIIPQDPILFSGSLR | 885.4 | 989.1 | 1329.7 | 30 | 26 | 22.2 | 2.29–73 |
| LTIIPQDPILFSGSLR | 890.5 | 999.6 | 1339.7 | |||||
| MRP3 | ADGALTQEEK | 531.1 | 634.3 | 747.3 | 30 | 16 | 8.2 | 1.96–125 |
| ADGALTQEEK | 535.3 | 642.3 | 755.4 | |||||
| MRP4 | AEAAALTETAK | 538.2 | 733.0 | 875.4 | 30 | 17 | 9.8 | 6.41–103 |
| AEAAALTETAK | 542.3 | 741.4 | 883. 5 | |||||
| NTCP | GIYDGDLK | 440.7 | 547.1 | 710.1 | 30 | 13 | 12.5 | 3.42–219 |
| GIYDGDLK | 444.7 | 555.3 | 718.3 | |||||
| OATP1B1 | NVTGFFQSFK | 587.9 | 860.5 | 961.4 | 35 | 17 | 20.1 | 1.33–85 |
| NVTGFFQSFK | 591.9 | 868.5 | 969.5 | |||||
| OATP1B3 | IYNSVFFGR | 551.8 | 826.5 | 526.6 | 30 | 15 | 18.0 | 1.31–84 |
| IYNSVFFGR | 556.8 | 635.2 | 836.6 | |||||
| OATP2B1 | VLAVTDSPAR | 514.8 | 646.3 | 816.4 | 30 | 16 | 11.2 | 1.56–100 |
| VLAVTDSPAR | 519.9 | 656.3 | 826.4 | |||||
| OCT1 | LSPSFADLFR | 576.7 | 476.6 | 768.0 | 25 | 23 | 21.9 | 1.38–88 |
| LSPSFADLFR | 581.8 | 778.4 | 865.4 | |||||
| P-gp | NTTGALTTR | 467.6 | 618.3 | 719.4 | 30 | 16 | 8.4 | 1.50–96 |
| NTTGALTTR | 472.6 | 628.3 | 729.5 | |||||
CE, Collision energy; RT, retention time.
The low end of this range reflects the lower limit of quantification of the assay.
Human Liver Tissue.
Twenty-seven frozen alcoholic cirrhosis human liver tissues were obtained from the Liver Tissue Cell Distribution System, University of Minnesota. In addition, 30 frozen hepatitis C cirrhosis liver tissues were obtained from the Liver Center Tissue Bank of University of Kansas Medical Center. These tissues were obtained from patients undergoing liver transplant. The donor demographics and clinical characteristics of the liver donors are shown in Table 2. Most clinical pharmacokinetic studies for liver cirrhosis patients use the Child-Pugh score to indicate the level of liver impairment (Rodighiero, 1999; Hatorp et al., 2000; Stangier et al., 2000; Hui et al., 2005). However, the Child-Pugh scores of the donors of liver tissues were not available. As these tissues were obtained from donors who received a liver transplant, it is highly probable that they belong to the Child-Pugh score C category, which represents end-stage liver disease. These livers were obtained under protocols approved by the appropriate committees for the conduct of human research. All liver tissues were stored at –80°C until analysis. Demographics, procurement, and storage of the control (noncirrhotic) livers have been published before (Paine et al., 1997). In brief, most of these livers were obtained from vehicular accident victims who were otherwise healthy.
TABLE 2.
Demographics of liver tissue donors
| Alcoholic Cirrhosis | Hepatitis C Cirrhosis | Control | |
|---|---|---|---|
| Sample size | 27 | 30 | 36 |
| Age (years) | 55 ± 8 | 53 ± 8 | 47 ± 14 |
| Sex | Male: 25; Female: 2 | Male: 18; Female: 12 | Male: 18; Female: 18 |
| Race | C: 25; AI: 1; NR: 1 | C: 25; AA: 4; H: 1 | C: 33; AA: 2; A: 1 |
| Albumin (g/dl) | 2.9 ± 0.6 | 2.6 ± 0.6 | NR |
| Alkaline phosphatase (units per liter) | 171 ± 96 | 112 ± 54 | NR |
| Alanine transaminase (units per liter) | NR | 146 ± 243 | NR |
| Aspartate aminotransferase (units per liter) | 219 ± 550 | 223 ± 388 | NR |
| Creatine (mg/dl) | 1.6 ± 0.7 | NR | NR |
| INR | 2.6 ± 1.1 | 1.7 ± 0.5 | NR |
| Total bilirubin (mg/dl) | 9.5 ± 8.9 | 3.6 ± 2.9 | NR |
A, Asian; AA, African American; AI, American Indian; C, Caucasian; H, Hispanic; NR, not reported; INR, International normalized ratio.
Peptide Selection, Membrane Protein Extraction, and Trypsin Digestion.
Signature peptides were selected for quantification of each transporter (Table 1) on the basis of previously reported criteria (Kamiie et al., 2008; Prasad and Unadkat, 2014). Isolation of total membrane protein (in triplicate), methanol-chloroform precipitation, and trypsin digestion were performed as described previously (Wang et al., 2015). After total membrane protein isolation, 100 μl of 2.0 mg/ml (or lower concentration) membrane protein was used for further treatment. Finally, 5 μl of the processed sample was injected onto the liquid chromatography–tandem mass spectrometry system.
Liquid Chromatography–Tandem Mass Spectroscopy Analyses.
Waters Xevo TQS tandem mass spectrometer coupled to Waters Acquity UPLC System (Waters, Hertfordshire, UK) operated in electrospray positive ionization mode was used for analyses of the signature peptides. The mass spectrometry conditions were as follows: capillary 3.5 kV, source offset 30 V, source temperature 500°C. Analyte-specific parameters are listed in Table 1. The transitions from doubly charged parent ion to singly charged product ions for the analyte peptides and their respective stable isotope-labeled peptides were monitored (Table 1). These peptides were separated on a ultra-high-performance liquid chromatography (UPLC) column (Acquity UPLC HSS T3 1.8 μm, 2.1 × 100 mm, Waters) with a Security Guard column (C18, 4 mm × 2.0 mm) from Phenomenex (Torrance, CA). Mobile phases (0.3 ml/min) consisted of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). The linear gradient was: 0–3 minutes: 3% B; 3–10 minutes: 3–13% B; 10–20 minutes: 13–25% B; 20–24 minutes: 25–50% B; 24–24.1 minutes: 50–80% B; 24.1–25 minutes: 80% B; 25–25.1 minutes: 80–3% B; and 25.1–29 minutes: 3% B. Intraday accuracy and precision was determined using quality control (QC) samples. Coefficients of variation of ≤15% and ≤25% for these QC samples, prepared in extraction buffer II and pooled liver membrane, respectively, was considered acceptable. The signal from the endogenous expression of the transporter were subtracted when the signal in the QC samples prepared in pooled liver membrane was calculated. The stability (freeze and thaw, bench-top, and autosampler conditions) of the selected peptides has been reported in previous publications (Prasad et al., 2013, 2014; Wang et al., 2015).
Genotyping Methods and Genotype-Dependent Changes in Organic Anion–Transporting Polypeptide 1B1 Protein Expression.
The SLCO1B1 single-nucleotide polymorphisms (SNPs) rs4149015, rs2306283, rs4149056, rs4149057, and rs2291075 define the variants –11187G>A, 388A>G, 521T>C, 571T>C, and 597C>T, respectively. These SNPs were genotyped using validated TaqMan assays from Applied Biosystems (Foster City, CA). The cycling conditions for polymerase chain reaction (PCR) amplification were one cycle at 95°C for 10 minutes followed by 50 cycles of 95°C for 15 seconds and 60°C for 90 seconds in a reaction volume of 10 μl containing 1 μl of genomic DNA (∼25–100 ng) and 1× final concentrations of TaqMan universal PCR master mix and SNP assay primers and probes. The allelic discrimination was determined in a post-PCR analysis on a Bio-Rad (Hercules, CA) CFX Connect Real-Time System running software CFM Manager V 3.1 with allele-specific probes for the common and variant SNPs (FAM and VIC). The SLCO1B1 SNP rs11045819 (463C>A) was determined by PCR amplification and DNA sequencing using forward and reverse oligonucleotides flanking the variant. In brief, DNA (∼25–100 ng) was subjected to an amplification profile: 95°C for 3 minutes, 40 cycles of PCR cycling (95°C for 30 seconds, 57.6°C for 30 seconds, and 72°C for 1 minutes), 72°C for 10 minutes and a final 4°C hold. Each PCR reaction was composed of: 1.0 μl genomic DNA (∼25–100 ng), 2.0 μl Pfu 10× buffer (Agilent Technologies., La Jolla, CA), 200 μM dNTPs, 300 nM forward (TTTCTAGGAAAAGTGAAAATATTCAGTAGATAAGC) and reverse (TTGCTAATGAATATCACAACAATTTTTAGAGATGT) primers, and 0.2 μl (0.5 IU) Pfu HotStart Turbo DNA polymerase (Agilent Technologies) in a total volume of 20 μl. The 274-bp amplicon was purified using QIAquick spin column (Qiagen Inc., Valencia, CA) and DNA sequencing determined using an internal forward primer (CAGTGATGTTCTTACAGTTACAG) and BigDye V3.1 chemistry (Applied Biosystems, Foster City, CA); allele calls were made on the basis of visual inspection of resultant electropherograms.
Statistical Analyses.
Data are reported as mean ± S.D. Statistical difference (P < 0.05) in the expression of transporters among different sample types (noncirrhotic control livers, alcoholic cirrhosis livers, and hepatitis C cirrhosis livers) or organic anion–transporting polypeptide (OATP)1B1 genotype/haplotype was assessed using the Kruskal-Wallis test followed by Dunn’s multiple comparison correction.
Physiologically Based Pharmacokinetics Simulations.
Using the Simcyp cirrhosis population module (version 14 release 1; Simcyp Limited, Sheffield, UK), the effect of alcoholic liver cirrhosis on the pharmacokinetics of OATP1B1 substrate repaglinide was predicted. This module incorporates the physiologic changes in liver cirrhosis (Child-Pugh score: B and C), including the impact on the expression/activity of cytochrome P450 enzymes (Johnson et al., 2010). Liver weight was assumed not to be altered in cirrhotic patients. To conduct these simulations, repaglinide parameters, shown in Supplemental Table 1, were obtained from a previous publication (Varma et al., 2013) and used without any modifications. The mean expression of OATP1B1 in alcoholic cirrhotic livers versus control livers, obtained from the present study, was used and assumed to be the same for Child-Pugh score B and C. Data for 120 subjects (ratio of Child-Pugh B versus C, 3:1; age, 42–62 years; proportion of females, 0), given a single dose of repaglinide (4 mg, PO), were simulated (10 trials × 12 subjects). The simulated pharmacokinetic profiles and parameters were compared with the observed data from a previous clinical study of repaglinide in 12 healthy subjects and 12 liver cirrhosis patients (n = 9 and 3 for Child-Pugh score B and C, respectively; the authors did not specify the cause of cirrhosis) (Hatorp et al., 2000).
Results
Analytical.
The calibration curves for the signature peptide was linear (r2 > 0.995) over two orders of magnitude, and the lower limit of quantification for all signature peptides was in the low femtomole range (Table 1). The accuracy and precision (% CV) of the assay, on the basis of QC samples, was 87–115% and <20%, respectively.
Comparison of Membrane Protein Yield between Control and Cirrhotic Human Livers.
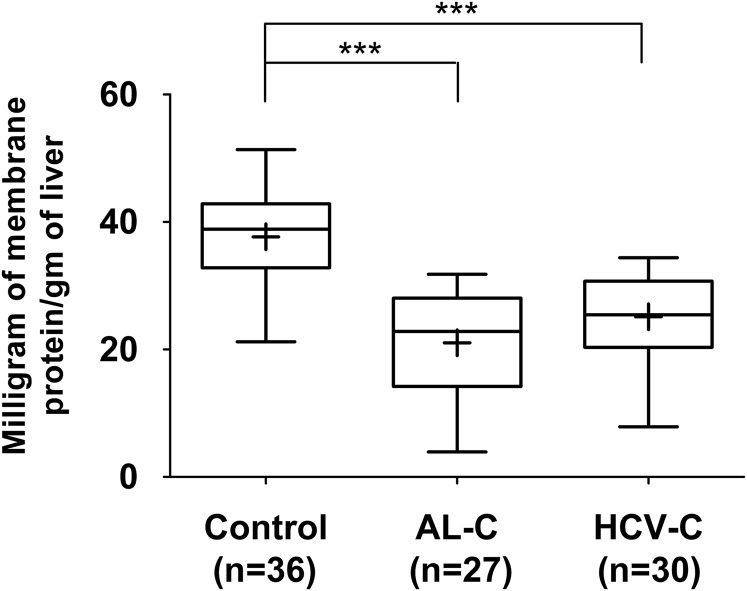
We previously quantified the expression of hepatobiliary transporters in 36 control (noncirrhotic) human livers from the human liver bank of the School of Pharmacy, University of Washington (Prasad et al., 2016). The yield of membrane protein from these control livers was 37.7 ± 7.0 mg/gm of liver (Fig. 1). The yield of membrane protein from alcoholic and hepatitis C cirrhosis human livers was significantly lower, i.e., 21.1 ± 7.9 and 25.1 ± 8.3 mg/gm of liver, respectively (Fig. 1).
Fig. 1.
The yield of membrane protein from alcoholic cirrhosis (AL-C) and hepatitis C cirrhosis (HCV-C) livers was significantly lower than that from control livers. Horizontal line, median; +, mean; boxes, 25th to 75th percentiles; whiskers, non-outlier range. ***p < 0.001.
Comparison of Expression of Hepatobiliary Transporters between Control and Cirrhotic Human Livers.
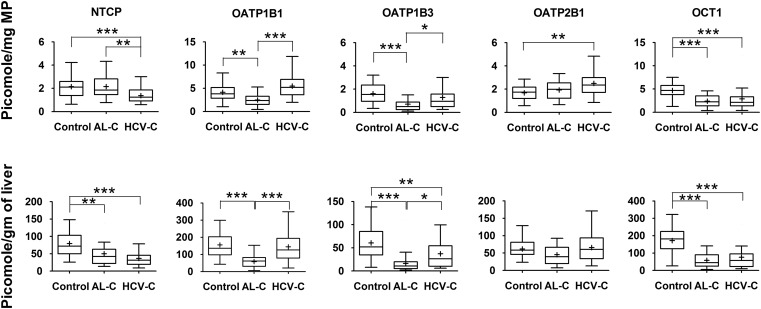
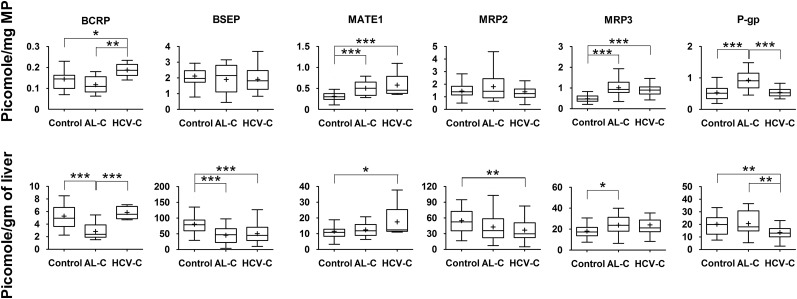
All the investigated transporters could be quantified with the exception of multidrug resistance–associated protein (MRP)4, which was below the lower limit of quantification (on column LLOQ = 6.41 femtomole) for all livers. When the expression of the transporters was normalized to milligrams of membrane protein, a number of transporters demonstrated altered expression in cirrhotic livers compared with control livers (Figs. 2–3). Specifically, in alcoholic cirrhosis livers, the expression of OATP1B1, OATP1B3, and organic cation transporter 1 (OCT1) was significantly reduced, by 40, 56, and 49%, respectively, whereas the expression of multidrug and toxin extrusion protein 1 (MATE1), MRP3, and P-glycoprotein (P-gp) was significantly increased by 67, 110, and 75%, respectively. In contrast, in hepatitis C cirrhosis livers, the expression of only sodium taurocholate–cotransporting polypeptide (NTCP) and OCT1 was decreased by 36 and 38%, respectively, whereas the expression of breast cancer resistance protein (BCRP), MATE1, MRP3, and OATP2B1 was increased by 36, 87, 88, and 50%, respectively. However, to use these findings in physiologically based pharmacokinetics (PBPK) predictions of in vivo transporter-mediated drug disposition, these data need to be expressed per gram of tissue, which can then be scaled up to the whole liver (assuming that the weight of the liver does not change owing to cirrhosis). When this was done for each individual, the pattern of differences between cirrhotic and control livers changed. All investigated transporters showed reduced protein expression (37–73%) in alcoholic cirrhosis livers versus control livers, with the exception that the expression of MATE1, MRP2, OATP2B1, and P-gp was not affected, whereas MRP3 (32%) was increased (Figs. 2–3). In contrast, the expression of six transporters, including bile salt export pump (BSEP), NTCP, OATP1B3, OCT1, MRP2, and P-gp exhibited significant decrease (32–56%) in hepatitis C cirrhosis livers versus control livers, whereas the expression of BCRP, OATP 1B1, and 2B1 was not affected, and the expression of MATE1 (46%) was increased (Figs. 2–3) . The expression of none of the transporters was significantly (P < 0.05) correlated with clinical markers of liver function listed in Table 2.
Fig. 2.
Hepatic uptake transporter protein expression in alcoholic cirrhosis (AL-C), and hepatitis C cirrhosis (HCV-C) livers differed from that in control livers. This difference was transporter- and disease-dependent and also depended on whether the data were normalized to milligram of membrane protein (MP; upper panel) or gram of liver (lower panel). Horizontal line, median; +, mean; boxes, 25th to 75th percentiles; whiskers, non-outlier range. *p < 0.05; **p < 0.01; ***p < 0.001. MP, Membrane protein.
Fig. 3.
Hepatic efflux transporter protein expression in alcoholic cirrhosis (AL-C) and hepatitis C cirrhosis (HCV-C) livers differed from that in control livers. This difference was transporter- and disease-dependent and also depended on whether the data were normalized to milligram of membrane protein (upper panel) or gram of liver (lower panel). Horizontal line, median; +, mean; boxes, 25th to 75th percentiles; whiskers, non-outlier range. *p < 0.05; **p < 0.01; ***p < 0.001. MP, Membrane protein.
Protein-Protein Expression Correlation of Transporters in Cirrhotic Livers.
The protein expression (normalized to gram of liver) of the following transporters was correlated with r2 > 0.5 in both alcoholic and hepatitis C cirrhosis livers: BSEP versus NTCP, BSEP versus OATP1B1, BSEP versus OATP2B1, and NTCP versus OATP1B1. In addition, the expression of MRP2 versus NTCP and MRP2 versus OATP2B1 in hepatitis C cirrhosis livers, as well as BCRP versus MRP2 and BSEP versus OCT1 in alcoholic cirrhosis livers showed a correlation of r2 > 0.5. Protein-protein expression correlation of the remaining transporters in cirrhotic human livers was poor or nonexistent (r2 < 0.5).
Effect of OATP1B1 Genotype on Hepatic Transporter Protein Expression in Cirrhotic Livers.
The protein expression of OATP1B1 in cirrhotic livers was not dependent (data not shown) on the observed SNPs (Table 3). More importantly, the protein expression of OATP1B1 in these diseased human livers was not dependent on SLCO1B1 haplotypes of the three previously described key variants (c.388A>G, c.463C>A, and c.521T>C) (data not shown). The sample size of 1a/*1a, *1b/*1a, *14/*1a, and *15/*1a haplotypes was 11, 4, 2, and 4 for alcoholic cirrhosis livers and 6, 4, 6, and 4 for hepatitis C cirrhosis livers.
TABLE 3 .
Frequency of OATP1B1SNPs detected in the alcoholic and hepatitis C cirrhosis human liver tissue
| Marker ID | Variant | Change for Variant | Frequency |
||
|---|---|---|---|---|---|
| Wild-Type | Heterozygous Variant | Homozygous Variant | |||
| n | n | n | |||
| Alcoholic cirrhosis | |||||
| rs4149015 | −11187G>A | Promoter | 23 | 4 | 0 |
| rs2306283 | 388A>G | N130D | 12 | 12 | 3 |
| rs4149056 | 521T>C | V174A | 19 | 7 | 1 |
| rs4149057 | 571T>C | L191L | 6 | 10 | 10 |
| rs2291075 | 597C>T | F199F | 10 | 13 | 3 |
| rs11045819 | 463C>A | P155T | 23 | 4 | 0 |
| Hepatitis C cirrhosis | |||||
| rs4149015 | −11187G>A | Promoter | 24 | 6 | 0 |
| rs2306283 | 388A>G | N130D | 7 | 14 | 9 |
| rs4149056 | 521T>C | V174A | 25 | 5 | 0 |
| rs4149057 | 571T>C | L191L | 10 | 12 | 8 |
| rs2291075 | 597C>T | F199F | 8 | 14 | 8 |
| rs11045819 | 463C>A | P155T | 22 | 7 | 1 |
Quantitative Impact of Liver Cirrhosis on the Pharmacokinetics of OATP1B1 Substrate Repaglinide.
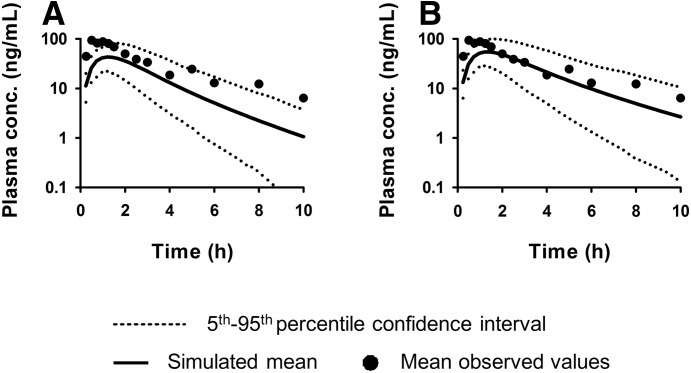
Utilizing the protein expression data (Fig. 3 and Supplemental Table 1), we predicted the impact of liver cirrhosis on the human pharmacokinetics of an OATP1B1 substrate repaglinide. To do so, we used the Simcyp cirrhosis population module which incorporates changes in physiology (e.g., hepatic blood flow) and cytochrome P450 expression/activity attributable to cirrhosis, with the assumption that liver weight was not altered in cirrhotic liver. When our transporter data were not incorporated in this PBPK model, the simulated mean plasma concentrations of repaglinide underestimated the observed plasma concentrations, with ratio of observed-versus-simulated AUC of 2.5 (Fig. 4A). Assuming that the subjects in the reported study were alcoholic cirrhosis patients and that repaglinide was predominately transported by OATP1B1, we next incorporated into the PBPK model the relative mean expression of OATP1B1 in alcoholic cirrhotic livers versus control livers. Clearly, incorporation of change in OATP1B1 expression improved the ability to predict the observed repaglinide plasma concentrations-versus-time profiles, with the ratio of the observed-versus-simulated AUC of repaglinide improved from 2.5 to 1.5 (Fig. 4B).
Fig. 4.
The Simcyp liver cirrhosis population PBPK module, which incorporates changes in physiology including cytochrome P450 expression/activity owing to cirrhosis but not changes in transporter expression, was used to predict repaglinide disposition in liver cirrhosis patients (A). Incorporation in the model of changes in OATP1B1 expression owing to cirrhosis improved the prediction of repaglinide disposition in liver cirrhosis patients (B). The observed data are from Hatorp et al. (2000).
Discussion
We present here the protein expression of major hepatobiliary transporters in alcoholic and hepatitis C cirrhosis livers. The impact of liver cirrhosis on transporter protein expression exhibited transporter-dependent pattern irrespective of whether it was normalized to milligrams of membrane protein or grams of liver. For example, when normalized to the former, the expression of OCT1 was significantly reduced and the expression of MATE1 and MRP3 was significantly increased in both alcoholic and hepatitis C cirrhosis livers. In contrast, the expression of OATP1B1and OATP1B3 was significantly decreased, whereas that of P-gp was significantly increased in only the alcoholic cirrhosis livers. Likewise, the expression of BCRP and OATP2B1 was significantly increased in only the hepatitis C cirrhosis livers. The expression of other transporters was not affected by cirrhosis. These results differ from previous studies in which the mRNA and protein expression of some of these transporters was determined (Ogasawara et al., 2010; More et al., 2013). Ogasawara et al. reported that the mRNA expression of BCRP, MATE1, MRP2, MRP3, OATP1B1, 1B3, 2B1, and OCT1 was significantly reduced in hepatitis C cirrhosis livers versus control livers. Another study, in a limited number of alcoholic cirrhosis livers (n = 10), found that BCRP, MRP1, and MRP3–5 protein expression (but not that of MRP2 or MRP6) was elevated (More et al., 2013). The reasons for the discrepancies between these data and our data are not known but could be differences in ethnicity (Japanese versus Caucasian liver tissues), endpoints (mRNA versus protein expression), and severity of the disease or sample size.
To predict transporter-mediated drug disposition in vivo through PBPK simulation, protein expression in vitro (in liver tissue) needs to be scaled to that in vivo. To do so, the yield of the membrane protein needs to be taken into consideration. In cirrhotic livers, scar tissue is diffused throughout the liver and therefore could lead to reduced yield of membrane protein. Indeed, in the current study, the yield of membrane protein from cirrhotic livers was 56–67% lower than from the control livers. This finding is consistent with other studies in which the functional hepatocyte volume (measured by ultrasound or binding of a highly selective radioligand to a specific hepatocyte surface protein) in cirrhotic livers with Child-Pugh score B and C was found to be 56–67% lower than the functional hepatocyte volume of control livers (Johnson et al., 2010).
To take into consideration the above lower membrane protein yield in cirrhotic livers, we expressed our transporter expression data per gram of liver tissue. In doing so, the pattern of liver cirrhosis on transporter expression was altered. For example, the expression of BCRP and NTCP in alcoholic cirrhosis was not significantly different from that in control livers when normalized to milligrams of membrane protein, but it was when normalized to gram of liver. These differences highlight the importance of normalizing transporter expression data to grams of tissue for the purpose of predicting in vivo transporter-mediated drug disposition. In doing so, the effect of cirrhosis on transporter protein expression was also transporter-dependent. For example, although OATP1B1 and 1B3 expression was reduced in alcoholic cirrhosis versus control livers, the same was not true for OATP2B1. These data suggest differential regulation of transporters in liver cirrhosis. We also found that the expression of hepatic transporters was cirrhosis type–dependent. For example, the expression of BCRP and OATP1B1 (normalized to gram of liver tissue) was reduced only in alcoholic cirrhosis livers and not in hepatitis C cirrhosis livers, whereas MATE1 and MRP2 expression was significantly altered only in hepatitis C cirrhosis livers. This difference could be attributable to different mechanisms of regulation of transporters in alcoholic versus hepatitis C virus cirrhosis. These findings suggest that the pharmacokinetics of drugs cleared by hepatobiliary transporters will differ between alcoholic and hepatitis C cirrhosis patients. Therefore, we recommend that patients be classified into these two diseases when pharmacokinetic studies are conducted in the clinic. Unfortunately, the current classification of Child-Pugh score does not distinguish between these two diseases.
The above interpretation of OATP1B1 protein expression data in the cirrhotic livers was not confounded by genotype. We found that neither the six OATP1B1 SNPs (rs4149015, rs2306283, rs4149056, rs4149057, rs2291075, and rs11045819) nor SLCO1B1 haplotypes (1a/*1a, *1b/*1a, *14/*1a, and *15/*1a) significantly affected OATP1B1 protein expression in the cirrhotic livers. These findings differ from our previously published data in a larger number of control livers (Prasad et al., 2014), probably owing to the smaller sample size of the cirrhotic livers.
Repaglinide is used for the treatment of type 2 diabetes. After oral administration, repaglinide is absorbed rapidly and eliminated predominantly by CYP3A4- and CYP2C8-mediated hepatic metabolism (Hatorp, 2002). Clinical studies have shown that OATP1B1 polymorphism significantly impacts the AUC of repaglinide (60–190% increase or 30% decrease, depending on OATP1B1 genotype), indicating that OATP1B1-mediated transport is the rate-determining step for the hepatic uptake of repaglinide (Niemi et al., 2005; Zhang et al., 2006; Kalliokoski et al., 2008, 2010). The plasma AUC of repaglinide is increased by 4-fold in patients with cirrhosis (Hatorp et al., 2000). This increase may be attributable to a decrease in the expression of CYP3A4, 2C8, OATP1B1, or all three (Johnson et al., 2010). Therefore, in addition to changes in cytochrome P450 expression as a result of cirrhosis, we investigated whether incorporation of cirrhosis-induced changes in OATP1B1 expression in the PBPK model improved the prediction of plasma concentration of repaglinide. Indeed it did (Fig. 4B). This demonstrates the potential of incorporating transporter expression data in PBPK models to improve prediction of drug disposition in patients with cirrhosis.
We found significant correlation between protein expression of some of the hepatobiliary transporters (e.g., NTCP versus OATP1B1; r2 > 0.5) in both alcoholic and hepatitis C cirrhosis livers. This is in contrast to our findings in noncirrhotic livers, where this correlation was poor (r2 < 0.3) or nonexistent (Prasad et al., 2014; Wang et al., 2015). These data suggest that cirrhosis may downregulate the expression of multiple transporters, resulting in a correlation when none existed in noncirrhotic livers. Such correlation data are important when simulating via PBPK modeling and simulation the interindividual variability in disposition of drugs within a population.
In summary, this is the first report to quantify the abundance of major hepatobiliary transporters in liver cirrhosis patients using quantitative proteomics. These transporter expression data should facilitate prediction of transporter-mediated drug disposition in liver cirrhosis patients through PBPK modeling and simulation.
Acknowledgments
The authors thank Dr. Bhagwat Prasad and Tob Bui for their technical assistance and Dr. Sarah Billington for her help in manuscript preparation. The authors also thank Benjamin Roberts and Marion Namenwirth for handling and shipping liver cirrhosis samples.
Abbreviations
- AUC
area under the curve
- BCRP
breast cancer–resistance protein
- BSEP
bile salt export pump
- MATE1
multidrug and toxin extrusion protein 1
- MRP
multidrug resistance–associated protein
- NTCP
sodium taurocholate–cotransporting polypeptide
- OATP
organic anion–transporting polypeptide
- OCT1
organic cation transporter 1
- P-gp
P-glycoprotein
- PBPK
physiologically based pharmacokinetics
- PCR
polymerase chain reaction
- QC
quality control
- SNPs
single-nucleotide polymorphisms
- UPLC
ultra-high-performance liquid chromatography
Authorship Contributions
Participated in research design: Wang, Collins, Kelly, Chu, Ray, Salphati, Xiao, Lee, Lai, Liao, Mathias, Evers, Humphreys, Hop, Unadkat.
Conducted experiments: Wang, Kelly.
Contributed new reagents or analytic tools: Kumer.
Performed data analysis: Wang.
Wrote or contributed to the writing of the manuscript: Wang, Collins, Kelly, Chu, Ray, Salphati, Xiao, Lee, Lai, Liao, Mathias, Evers, Humphreys, Hop, Unadkat.
Footnotes
This work was supported by UWRAPT (University of Washington Research Affiliate Program on Transporters sponsored by Ardea Biosciences, Biogen, Bristol-Myers Squibb, Genentech, Gilead Sciences, Merck & Co., and Takeda (https://sop.washington.edu/department-of-pharmaceutics/research-affiliate-program-on-transporters-uwrapt). Cirrhotic human liver tissues were obtained through the Liver Tissue Cell Distribution System, Minneapolis, Minnesota, which was funded by a National Institutes of Health contract [Grant HHSN276201200017C].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Bosch J, García-Pagán JC. (2000) Complications of cirrhosis. I. Portal hypertension. J Hepatol 32(1, Suppl)141–156. [DOI] [PubMed] [Google Scholar]
- Dincer D, Besisk F, Demirkol O, Demir K, Kaymakoglu S, Cakaloglu Y, Okten A. (2005) Relationships between hemodynamic alterations and Child-Pugh Score in patients with cirrhosis. Hepatogastroenterology 52:1521–1525. [PubMed] [Google Scholar]
- Frye RF, Zgheib NK, Matzke GR, Chaves-Gnecco D, Rabinovitz M, Shaikh OS, Branch RA. (2006) Liver disease selectively modulates cytochrome P450--mediated metabolism. Clin Pharmacol Ther 80:235–245. [DOI] [PubMed] [Google Scholar]
- George J, Murray M, Byth K, Farrell GC. (1995) Differential alterations of cytochrome P450 proteins in livers from patients with severe chronic liver disease. Hepatology 21:120–128. [PubMed] [Google Scholar]
- Hatorp V. (2002) Clinical pharmacokinetics and pharmacodynamics of repaglinide. Clin Pharmacokinet 41:471–483. [DOI] [PubMed] [Google Scholar]
- Hatorp V, Walther KH, Christensen MS, Haug-Pihale G. (2000) Single-dose pharmacokinetics of repaglinide in subjects with chronic liver disease. J Clin Pharmacol 40:142–152. [DOI] [PubMed] [Google Scholar]
- Hui CK, Cheung BM, Lau GK. (2005) Pharmacokinetics of pitavastatin in subjects with Child-Pugh A and B cirrhosis. Br J Clin Pharmacol 59:291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TN, Boussery K, Rowland-Yeo K, Tucker GT, Rostami-Hodjegan A. (2010) A semi-mechanistic model to predict the effects of liver cirrhosis on drug clearance. Clin Pharmacokinet 49:189–206. [DOI] [PubMed] [Google Scholar]
- Kalliokoski A, Neuvonen M, Neuvonen PJ, Niemi M. (2008) The effect of SLCO1B1 polymorphism on repaglinide pharmacokinetics persists over a wide dose range. Br J Clin Pharmacol 66:818–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliokoski A, Neuvonen PJ, Niemi M. (2010) SLCO1B1 polymorphism and oral antidiabetic drugs. Basic Clin Pharmacol Toxicol 107:775–781. [DOI] [PubMed] [Google Scholar]
- Kamiie J, Ohtsuki S, Iwase R, Ohmine K, Katsukura Y, Yanai K, Sekine Y, Uchida Y, Ito S, Terasaki T. (2008) Quantitative atlas of membrane transporter proteins: development and application of a highly sensitive simultaneous LC/MS/MS method combined with novel in-silico peptide selection criteria. Pharm Res 25:1469–1483. [DOI] [PubMed] [Google Scholar]
- More VR, Cheng Q, Donepudi AC, Buckley DB, Lu ZJ, Cherrington NJ, Slitt AL. (2013) Alcohol cirrhosis alters nuclear receptor and drug transporter expression in human liver. Drug Metab Dispos 41:1148–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi M, Backman JT, Kajosaari LI, Leathart JB, Neuvonen M, Daly AK, Eichelbaum M, Kivistö KT, Neuvonen PJ. (2005) Polymorphic organic anion transporting polypeptide 1B1 is a major determinant of repaglinide pharmacokinetics. Clin Pharmacol Ther 77:468–478. [DOI] [PubMed] [Google Scholar]
- Ogasawara K, Terada T, Katsura T, Hatano E, Ikai I, Yamaoka Y, Inui K. (2010) Hepatitis C virus-related cirrhosis is a major determinant of the expression levels of hepatic drug transporters. Drug Metab Pharmacokinet 25:190–199. [DOI] [PubMed] [Google Scholar]
- Ohnishi A, Murakami S, Akizuki S, Mochizuki J, Echizen H, Takagi I. (2005) In vivo metabolic activity of CYP2C19 and CYP3A in relation to CYP2C19 genetic polymorphism in chronic liver disease. J Clin Pharmacol 45:1221–1229. [DOI] [PubMed] [Google Scholar]
- Paine MF, Khalighi M, Fisher JM, Shen DD, Kunze KL, Marsh CL, Perkins JD, Thummel KE. (1997) Characterization of interintestinal and intraintestinal variations in human CYP3A-dependent metabolism. J Pharmacol Exp Ther 283:1552–1562. [PubMed] [Google Scholar]
- Pentikäinen PJ, Neuvonen PJ, Jostell KG. (1980) Pharmacokinetics of chlormethiazole in healthy volunteers and patients with cirrhosis of the liver. Eur J Clin Pharmacol 17:275–284. [DOI] [PubMed] [Google Scholar]
- Prasad B, Evers R, Gupta A, Hop CE, Salphati L, Shukla S, Ambudkar SV, Unadkat JD. (2014) Interindividual variability in hepatic organic anion-transporting polypeptides and P-glycoprotein (ABCB1) protein expression: quantification by liquid chromatography tandem mass spectroscopy and influence of genotype, age, and sex. Drug Metab Dispos 42:78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B, Gaedigk A, Vrana M, Gaedigk R, Leeder JS, Salphati L, Chu X, Xiao G, Hop C, Evers R, et al. (2016) Ontogeny of hepatic drug transporters as quantified by LC-MS/MS proteomics. Clin Pharmacol Ther DOI: (published ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B, Lai Y, Lin Y, Unadkat JD. (2013) Interindividual variability in the hepatic expression of the human breast cancer resistance protein (BCRP/ABCG2): effect of age, sex, and genotype. J Pharm Sci 102:787–793. [DOI] [PubMed] [Google Scholar]
- Prasad B, Unadkat JD. (2014) Optimized approaches for quantification of drug transporters in tissues and cells by MRM proteomics. AAPS J 16:634–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodighiero V. (1999) Effects of liver disease on pharmacokinetics. An update. Clin Pharmacokinet 37:399–431. [DOI] [PubMed] [Google Scholar]
- Schuppan D, Afdhal NH. (2008) Liver cirrhosis. Lancet 371:838–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangier J, Su CA, Schöndorfer G, Roth W. (2000) Pharmacokinetics and safety of intravenous and oral telmisartan 20 mg and 120 mg in subjects with hepatic impairment compared with healthy volunteers. J Clin Pharmacol 40:1355–1364. [PubMed] [Google Scholar]
- Varma MV, Lai Y, Kimoto E, Goosen TC, El-Kattan AF, Kumar V. (2013) Mechanistic modeling to predict the transporter- and enzyme-mediated drug-drug interactions of repaglinide. Pharm Res 30:1188–1199. [DOI] [PubMed] [Google Scholar]
- Wang L, Prasad B, Salphati L, Chu X, Gupta A, Hop CE, Evers R, Unadkat JD. (2015) Interspecies variability in expression of hepatobiliary transporters across human, dog, monkey, and rat as determined by quantitative proteomics. Drug Metab Dispos 43:367–374. [DOI] [PubMed] [Google Scholar]
- Zhang W, He YJ, Han CT, Liu ZQ, Li Q, Fan L, Tan ZR, Zhang WX, Yu BN, Wang D, et al. (2006) Effect of SLCO1B1 genetic polymorphism on the pharmacokinetics of nateglinide. Br J Clin Pharmacol 62:567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]