Abstract
Srpr is a gene encoding α subunit of the signal recognition particle receptor which is involved in the targeting and translocation of nascent secretory and membrane proteins to the endoplasmic reticulum. Previous studies showed aberrant expression of Srpr in several cell types with abnormal growth rate. Although Srpr is expressed in various tissues including skin, the role of Srpr in keratinocytes and regulation of its expression by miRNAs have not been studied. In this study, we investigated the role of SRPR and regulation of its expression by miRNA in skin keratinocytes. We found that SRPR was highly expressed in epidermal keratinocytes and regulated keratinocyte proliferation by affecting cell cycle progression. We also demonstrated that miR-330-5p directly inhibits Srpr expression. These data suggest that miR-330-5p-mediated regulation of the SRPR level is needed for the regulation of proliferation of epidermal keratinocytes.
Introduction
Srpr is a gene that encodes signal recognition particle receptor alpha (SRPRα, also called docking protein); it forms a heterodimer with SRPRβ, which becomes the signal recognition particle receptor (SR). SR is involved in the targeting and translocation of nascent secretory and membrane proteins to the endoplasmic reticulum membrane, which aids the spatial control of protein synthesis [1,2]. SR function is mediated in conjunction with the signal recognition particle (SRP) and is well conserved from bacteria to eukaryotes. SRP binds to the signal sequence of a newly synthesized peptide and slows protein synthesis (elongation arrest). This ribosome-nascent chain complex is targeted by SR to the protein-conducting channel, known as the translocon, in the endoplasmic reticulum membrane [3]. Aberrant Srpr expression has been detected in several cell types that have abnormal growth rate, suggesting that Srpr is essential for cell survival [4,5].
MicroRNAs (miRNAs) are non-coding RNA molecules (17–25 nucleotides) that regulate gene expression at the post-transcriptional level. This regulation is mediated through the recognition and annealing of complementary sequences (referred to as ‘seed’ regions) of the miRNAs to target sites, which comprise 6–8 nucleotides in the 3′ untranslated regions (UTRs) of the target mRNAs [6]. miRNAs play important roles in various biological processes including cell proliferation, apoptosis, and metastasis, and also affect sensitivity to chemotherapy and radiotherapy in multiple cancers [7,8]. In skin biology, the specific regulation of miRNAs is associated with keratinocyte proliferation, migration, and differentiation, and also with the development of skin disease. For example, up-regulation of miR-330-5p inhibits keratinocyte proliferation and migration by targeting Pdia3 expression [9]. Overexpression of miR-199a-5p inhibits keratinocyte migration [10]. miR-378b promotes keratinocyte differentiation by targeting NKX3.1 [11]. Down-regulation of miR-31-5p accelerates hair follicle growth and alters hair shaft formation [12]. Therefore, it is considered that miRNAs have critical roles in skin biology and hair cycle regulation.
Although Srpr is expressed in various tissues including skin, the role of Srpr in keratinocytes and regulation of its expression by miRNAs have not been studied. In the present study, we investigated the role of Srpr in skin keratinocytes and miRNAs that regulate its expression in keratinocytes. We found that Srpr is abundantly expressed in epidermal keratinocytes and regulates their proliferation by promoting cell cycle progression. We also showed that miR-330-5p directly suppresses Srpr expression by targeting its 3′ UTR. These data suggest that miR-330-5p-mediated regulation of SRPR controls proliferation of epidermal keratinocytes.
Materials and Methods
Mice
The BALB/C mice (total 8 of male or female, P10-P49 of age, 15–25 g) were bred in the barrier system under specific-pathogen-free conditions with regulated light (07:00–19:00 h), temperature (23°C), humidity (50%), and ventilation (10–12 times per hour). In each cage, less than 5 mice were housed. Food and water were received ad libitum. To obtain the dorsal skin, each animal were sacrificed with CO2 inhalation. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Catholic University of Korea. All experiments were carried out in accordance with the guidelines for animal experimentation and all efforts were made to minimize suffering.
Cell culture and transfection experiment
PAM212 (mouse keratinocyte) and 3T3-L1 (mouse adipocyte) cells were maintained in Dulbecco's Modified Eagle Medium (Invitrogen) containing 10% fetal bovine serum with 5% CO2 in a 37°C incubator. For the inhibition of Srpr expression, total of 5×105 cells were plated in a 60 mm dish. After 24 hrs, Srpr siRNA (Ambion) was transfected into these cells. MiR-330-5p mimic (Dharmacon) or miR-330-5p inhibitor (Dharmacon) was used for miR-330-5p over-expression or suppression, respectively. These experiments were carried out using DharmaFECT 1 transfection reagent (Dharmacon) following the manufacturer’s instruction. The negative mimic (Dharmacon) was used as control at the same concentration. At 72hr post-transfection, cells were harvested and used to extract total RNA or protein. For the over-expression of Srpr, the full length Srpr-cDNA construct was purchased (Sino biological) and transfected into cells using the Lipofectamine 2000 reagent. All experiments were repeated in triplicate. Cell images were captured at three different fields which were located at the relatively similar position in each plate using inverted microscope (Leica).
Cell proliferation assay
PAM212 cells (2×104) were seeded in a 96-well plate and incubated for 24 hrs at 37°C. Transfection experiments were performed with Srpr siRNA or the miR-330-5p mimic using DharmaFECT 1 transfection reagent. After 72 hrs, the relative viability of the cells was measured using an EZ-Cytox Cell viability assay kit which measures mitochondrial reduction of WST-8 (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt) (DoGen) according to the manufacturer’s protocol.
RT-PCR and Real-time PCR
Total RNAs were prepared from the cells or dorsal skins of BALB/C mice of various ages using the QIAzol reagent (Qiagen). Then cDNA was synthesized using the PrimeScript 1st strand cDNA Synthesis kit (Takara). Realtime PCR was carried out with the SYBR Premix Ex Taq (Takara) in an Mx3000P (Stratagene) and the annealing temperatures given in the Table 1. All the expression levels were normalized against glyceraldehyde-3-phosphatedehydrogenase gene (Gapdh) expression using the comparative ΔΔCt method [13]. Results are the average of three independent experiments performed in duplicate.
Table 1. List of gene specific primers for Realtime PCR.
| Genes | Accession Number | Sequences | size (bp) | Tm (°C) |
|---|---|---|---|---|
| Srpr | NM_026130.1 | F: atgatgaaggggccactcaa | 171 | 60 |
| R cagcagccacattcttagca | ||||
| Srpr (3’UTR) | NM_026130.1 | F: atgtggctcttgcctaatacca | 908 | 62 |
| R: atttctctgggcagacagcc | ||||
| SRPR | NM_003139.3 | F: cagaagcatgggaggggtat | 200 | 60 |
| R: ccagtccctcgaatcaggtt | ||||
| Gapdh | NM_008084 | F: aactttggcattgtggaagg | 223 | 62 |
| R: acacattgggggtaggaaca | ||||
| GAPDH | NM_002046 | F: gagtcaacggatttggtcgtt | 238 | 60 |
| R: ttgattttggagggatctcg |
Western blot analysis
Total lysated PAM212 cells were prepared using radioimmunoprecipitation assay buffer (150 mM sodium chloride, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris-HCl [pH 8.0]) following the standard method at 72 hrs post-transfection. Total eighty micrograms of protein were loaded to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. This membrane was incubated with a rabbit polyclonal SRPR antibody (1:1000, Abcam) or a mouse polyclonal GAPDH antibody (1:5000, Applied Biological Materials) following the standard protocol. The signal was detected using an enhanced chemiluminescence system (Amersham Bioscience). The relative quantities of proteins were determined using ImageJ software (http://imagej.nih.gov/ij/index.html).
Plasmid Construction
The cDNAs of the full length 3’ UTR of Srpr was amplified from the dorsal skin cDNA of wild-type mice by PCR using PrimeSTAR DNA Polymerase (Takara). The gene-specific primers are listed in the Table 1. The product was cloned into pGEMT-easy vector and subsequently subcloned into the psiCHECK-2 vector (Promega) using Not I site.
Dual luciferase reporter assay
For luciferase assay, PAM212 cells (5 × 105/dish) were plated in 60 mm dishes and incubated for 24 hrs at 37°C. Then the construct containing the full length 3’ UTR of Srpr (+1987 bp -+2904 bp, Table 1) was transfected into cells with miR-330-5p mimic or control mimic using the Lipofectamine 2000 reagent. Luciferase activity was measured at 48 hrs post-transfection using the Dual-Luciferase Reporter Assay reagent (Promega).
Cell cycle assay
Srpr siRNA treated PAM212 cells were collected at 72hr post-transfection and washed with PBS twice. Then cells were fixed in 70% ethanol at 4°C overnight and incubated in 1 mg/ml RNase A at 37°C for 30min. These cells were re-suspended in a propidium iodide staining solution (50 μg/ml). The distribution of cells in each phase of the cell cycle was measured using FACSCanto II (BD Biosciences).
Immunohistochemistry
Mouse dorsal skins of BALB/C mice at postnatal days 7 (P7), 17 (P17), 21 (P21), 28 (P28), 35 (P35), 42 (P42) and 49 (P49) were prepared and each tissue section was deparaffinized, rehydrated, and washed with PBS, sequentially. Then, antigen retrieval was carried out by treating slides with 10 mM sodium citrate buffer for 15 min. Subsequently, the slides were incubated with an antibody against SRPR (1:100, Thermofisher) for overnight at room temperature. After washing three times with PBS, each slide was incubated with a horseradish peroxidase-conjugated antibody (Dako) for 1 hr at room temperature. The signals were detected with a fluorescent microscope (Olympus) after a color reaction using diaminobenzidine as a chromogen (Dako).
Statistical analysis
Statistical significance was determined by Student's t-tests and values < 0.05 were regarded as statistically significant.
Results
SRPR was expressed in epidermal keratinocyte in mouse skin
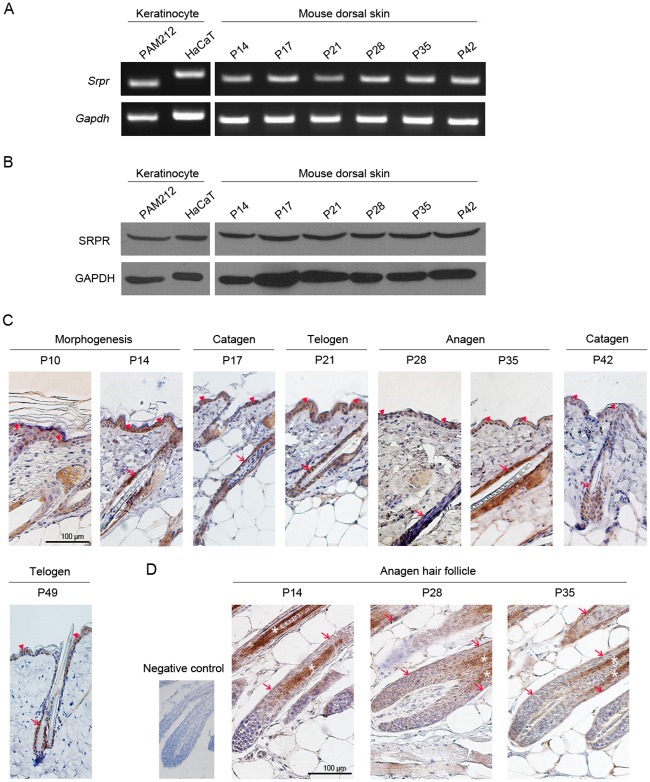
To determine whether SRPR has a function in keratinocytes, we first examined the Srpr expression in keratinocytes and mouse dorsal skin. We used mouse (PAM212) and human (HaCaT) keratinocyte cell lines and mouse dorsal skin at various hair cycle stages (P10, P14, P17, P21, P28, P35, and P42). RT-PCR and western blot analysis revealed that both Srpr mRNA and the SRPR protein were abundantly expressed in both keratinocytes and mouse dorsal skin (Fig 1A and 1B). In addition, we performed real-time PCR to compare expression level of Srpr with those of several genes whose expressions were regulated by miR-330-5p in keratinocyte [9]. As shown in the S1 Fig, Srpr was abundantly expressed to the level similar to that of Integrin5A. Next, we performed immunohistochemistry to further analyze the expression pattern of SRPR in wild-type mouse dorsal skin and found the strong SRPR expression in the interfollicular epidermis, hair cortex, and outer root sheath (ORS) at all hair cycle stages (Fig 1C and 1D). In contrast, we did not find SRPR expression in the hair matrix, or dermis including dermal papillae. These results indicate that SRPR is mainly expressed in epidermal keratinocytes in mouse skin.
Fig 1. Abundant expression of SRPR in mouse epidermal keratinocyte.
(A-B) Srpr was abundantly expressed in keratinocyte (PAM212 and HaCaT cell) and mouse dorsal skin at both mRNA by RT-PCR (A) and protein level by western blot analysis (B). (C-D) SRPR expression in dorsal skin (C) and HF (D) of BALB/C mice at postnatal days P10, P14, P17, P21, P28, P35, P42 and P49 by immunohistochemistry. Brown signals indicated the SRPR-positive epidermis cells (arrowhead), HF (arrow) and hair cortex (star). Scale bar = 100 μm.
Srpr knockdown induced inhibition of proliferation of mouse keratinocyte
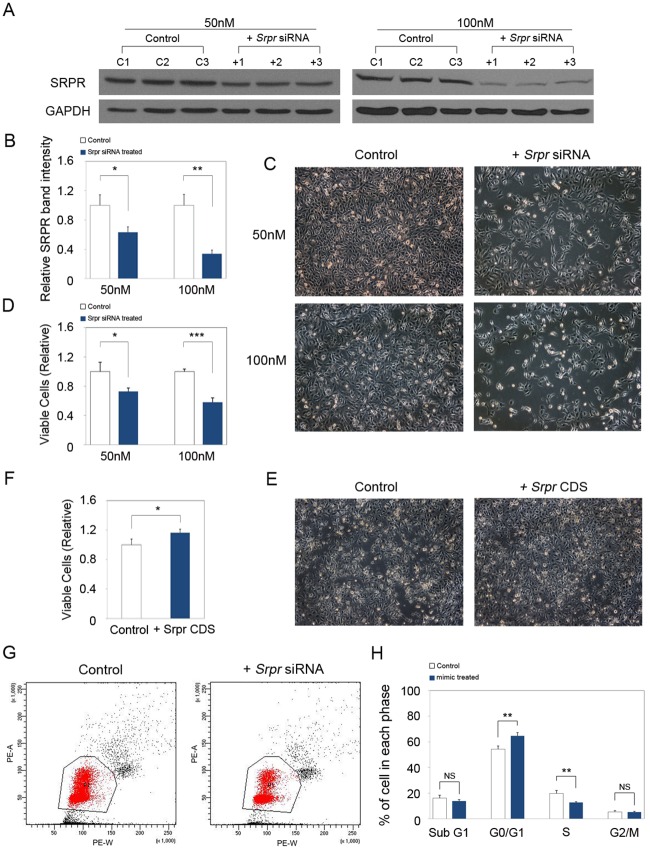
SRPR was previously shown to be important for cell growth in Saccharomyces cerevisiae [4]. As cells in the basal layer of the epidermis and in the ORS continuously proliferate throughout the lifetime, we hypothesized that SRPR affects keratinocyte proliferation in the mouse skin. To test this hypothesis, we utilized a mouse keratinocyte cell line, PAM212 as an experimental system. We transfected PAM212 cells with Srpr siRNA (50 or 100 nM). Western blot analysis confirmed a decreased expression of SRPR in Srpr siRNA-transfected cells in comparison with the control cells at both siRNA concentrations (Fig 2A and 2B). Proliferation assays showed that SRPR knockdown inhibited the proliferation of PAM212 cells transfected with either 50 nM or 100 nM siRNA in a concentration-dependent manner (Fig 2C and 2D). To further investigate whether keratinocyte proliferation is affected by the SRPR expression level, we induced SRPR expression by transfecting PAM212 cells with a Srpr cDNA expression construct. The total number of PAM212 cells increased in comparison with that in the mock transfection control (Fig 2E and 2F).
Fig 2. SRPR regulates the proliferation of keratinocyte.
(A) Western blot analysis showed that SRPR protein expression was significantly decreased at 50nM and 100nM Srpr siRNA transfected PAM212 cells. GAPDH was used as a loading control. (B) Quantitative analysis of western blot using ImageJ software. The data was normalized against GAPDH expression. (C) Proliferation assay revealed that Srpr siRNA transfection inhibited proliferation of PAM212 cells in comparison with control at both 50 nM and 100 nM in a dose-dependent manner. (D) Relative viable cells were measured at 48 h after transfection. (E, F) In contrast, relative viable PAM212 cells were increased by transfection of Srpr CDS construct. (G, H) Cell cycle assay was performed and revealed that Srpr knockdown induced cell cycle arrest at G0/G1 and number of cell at S phase were decreased. (A-H) Results are the average of three independent experiments *P<0.05; **P<0.01; ***P<0.001. NS = no significant.
Next, we examined the cell cycle distribution of the cells to determine whether inhibition of proliferation by decreased SRPR expression was caused by cell cycle arrest. The number of cells in G0/G1 phase was significantly higher and that in S phase was markedly lower among cells transfected with Srpr siRNA than among the control cells (Fig 2G and 2H). This data suggests that SRPR plays a role in cell proliferation by promoting the progression from G1 to S phase of the cell cycle.
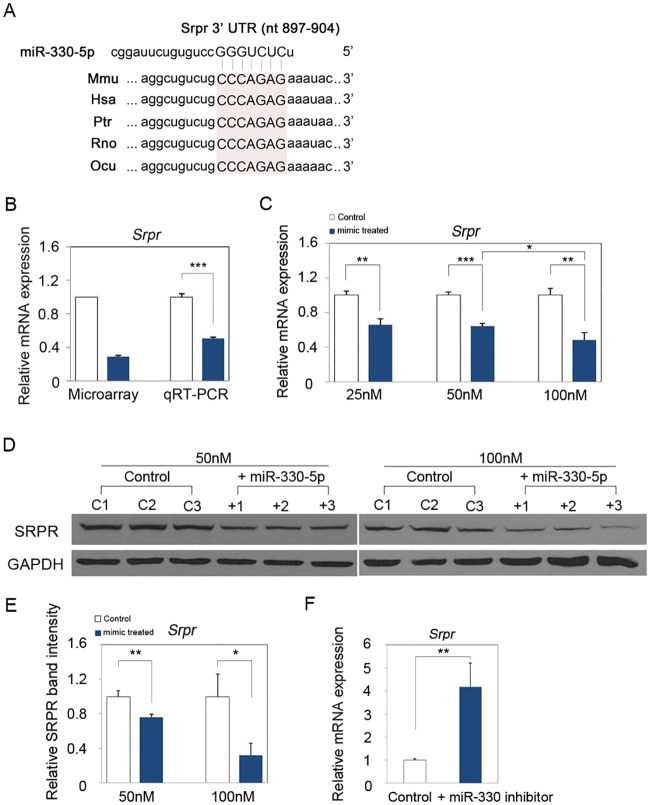
MiR-330-5p regulates endogenous Srpr expression in mouse keratinocyte
Using microarray analysis, we recently reported that expression of many genes was significantly changed in miR-330-5p-overexpressing PAM212 cells compared to the control [9]. In particular, we found that Srpr expression was down-regulated by miR-330-5p. In line with this finding, the 3′ UTR of Srpr mRNA contains a seed sequence for miR-330-5p, which is conserved in various species (Fig 3A). We previously showed that miR-330-5p inhibits proliferation of mouse keratinocytes [9] and have confirmed this effect in the present study (S2 Fig). On the basis of these observations, we hypothesized that the regulation of cell proliferation by SRPR might be mediated by miR-330-5p. To test this hypothesis, we first validated Srpr down-regulation by miR-330-5p using real-time quantitative PCR on total RNAs that were used for microarray analysis in our previous study [9]. We found that the expression of Srpr mRNA was significantly decreased in PAM212 cells transfected with a miR- 330-5p mimic than in the control-transfected cells (Fig 3B). Next, we analyzed the effect of miR-330-5p on Srpr expression in keratinocytes at the mRNA and protein levels using real-time quantitative PCR and western blotting, respectively. Expression of Srpr mRNA was consistently decreased in PAM212 cells transfected with the miR-330-5p mimic in comparison with control-transfected cells (Fig 3C). Similarly, SRPR expression was reduced by miR-330-5p mimic treatment (Fig 3D and 3E). In order to confirm these findings, we also investigated the expression of Srpr using a miR-330-5p inhibitor. We found that transfection of miR-330-5p inhibitor led to a significant induction of the endogenous Srpr expression (Fig 3F). These data indicate that SRPR expression is regulated by miR-330-5p at both mRNA and protein levels.
Fig 3. MiR-330-5p regulates Srpr expression in mouse epidermal keratinocyte.
(A) Targetscan algorithm predicted that a conserved binding sequence of miR-330-5p was present in the 3’ UTR of Srpr mRNA. (B) Real-time quantitative PCR was performed to validate the result of previous study using the same total RNA used for microarray analysis. (C) MiR-330-5p down-regulated the Srpr expression at 25nM, 50nM and 100nM mimic transfected PAM212 cells. The data was normalized against Gapdh expression. (D) Western blot analysis also revealed that SRPR protein level was decreased at 50nM and 100nM miR-330-5p mimic transfected PAM212 cells in comparison to the control. β-actin was used as a loading control. (E) Quantitative analysis of western blot using ImageJ software. The SRPR band intensity was normalized against GAPDH expression. (F) Srpr expression was significantly increased by inhibition of miR-330-5p. (B-F) Results are the average of three independent experiments. *P<0.05; **P<0.01; ***P<0.001.
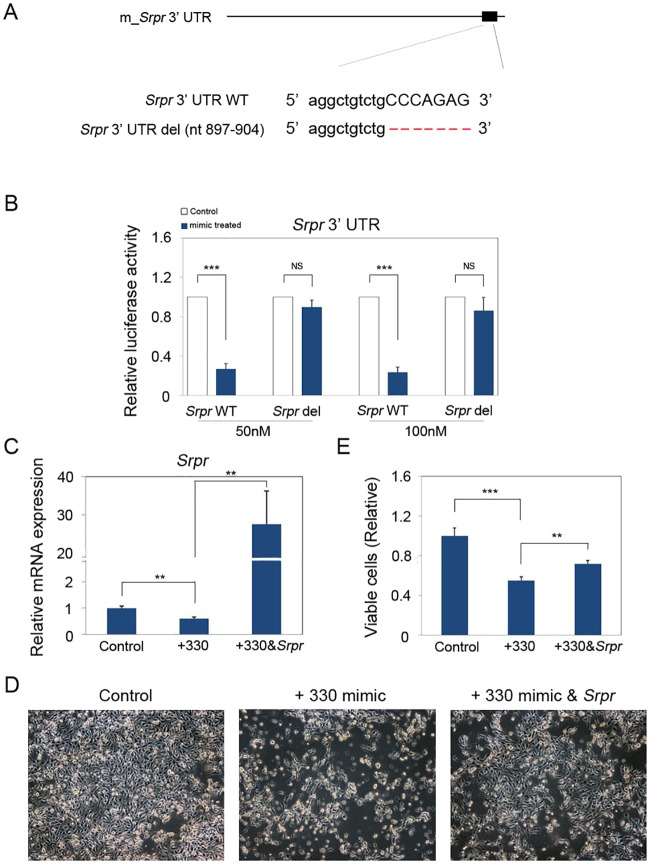
3’ UTR of Srpr is a direct target of miR-330-5p in mouse keratinocyte
The web-based target prediction software programs (TargetScan and microRNA.org) suggested that a binding site for miR-330-5p is present in the 897–904 bp region of the Srpr 3′ UTR (Fig 4A). To examine whether SRPR expression is directly regulated by miR-330-5p, we performed a luciferase reporter assay using constructs generated in the psiCHECK-2 dual luciferase vector. As shown in Fig 4B, the wild-type construct with full-length 3′ UTR showed significantly decreased luciferase activity when cells were co-transfected with the miR-330-5p mimic at both 50 nM and 100 nM concentrations. However, luciferase activity did not change significantly when cells were co-transfected with the deletion mutant lacking the miR-330-5p binding site and the miR-330-5p (Fig 4B). These data suggest that Srpr is a direct target of miR-330-5p.
Fig 4. Srpr is a direct target of miR-330-5p in mouse keratinocyte.
(A) Location of the binding site for miR-330-5p in 3’UTR of Srpr mRNA. Deletion is depicted in red hyphen. (B) Dual Luciferase assay showed that miR-330-5p directly inhibited the luciferase activity by targeting the binding site in Srpr 3’UTR. However, luciferase activity did not change significantly when cells were co-transfected with the deletion mutant lacking the miR-330-5p-binding site and the miR-330-5p. (C-E) Mir-330-5p suppressed the proliferation of PAM212 cells through inhibition of SRPR expression. (C) Real-time quantitative PCR showed that cotransfection of miR-330-5p and the full length SRPR-cDNA (Srpr CDS) was able to abolish the decreased expression of Srpr by miR-330-5p alone. (D) Proliferation of PAM212 cells was decreased by miR-330-5p. Co-transfection with the Srpr CDS construct rescued inhibition of cell proliferation by miR-330-5p. (E) Relative viable cells were measured at 72 h after transfection. (B-E) Results are the average of three independent experiments. **P<0.01; ***P<0.001. NS = no significance.
Finally, we used proliferation assay to investigate whether miR-330-5p inhibits keratinocyte proliferation by targeting SRPR expression. We compared proliferation of cells transfected with the miR-330-5p mimic alone or co-transfected with the full length SRPR-cDNA (Srpr CDS) construct and the miR-330-5p mimic. Co-transfection with the Srpr CDS construct rescued inhibition of cell proliferation by miR-330-5p (Fig 4C–4E). Therefore, we conclude that SRPR mediates the inhibitory effect of miR-330-5p on mouse keratinocyte proliferation.
Discussion
For sustained cell growth, cellular machinery for targeting nascent secretory proteins and membrane proteins to their proper ER locations is essential. SRP and SRPR are part of this cellular machinery and play a critical role in this process [14]. In the present study, we found that SRPR is abundantly expressed in epidermal keratinocytes, including those in the interfollicular epidermis and ORS. The epidermis is a continuously regenerating tissue derived from transient amplifying progenitors located in its basal layer. ORS is similar to the epidermis in that it is formed by proliferation of progenitor cells located in the bulge; ORS plays an indirect paracrine role in the regulation of hair growth [15,16]. These notions suggest that SRPR is expressed in proliferating keratinocytes and may modulate their proliferation. In addition, SRPR expression does not seem to be limited to basal cells. SRPR expression is also present in the suprabasal layer cells (Fig 1C). Thus, SRPR may play another role in epidermis such as differentiation, cell-cell communication and so on.
Together with SRP, SRPR as a docking protein involves in recognition and translocation of proteins to ER membrane. Thus SRPR plays a basic function for the cell. In the current study, we found that Srpr knockdown inhibited proliferation of mouse keratinocytes in vitro by arresting cell cycle at G0/G1 phase, thus reducing the number of cells in S phase. Moreover, this cell cycle arrest did not increased the apoptotic cell death (Fig 2H). Although G0/G1 arrest is a common response in cells that are perturbed by a variety of means, our results suggest that SRPR directly or indirectly controls cell proliferation by promoting the progression of cells from G1 to S phase. Our finding is in line with the previous report on the relationship between SRPR and retinoblastoma protein (pRb), an important regulator of cell proliferation and differentiation. In G0/G1 phase, pRb was shown to bind the Srpr promoter together with E2F1, an S phase-promoting transcription factor; binding of these proteins showed inhibition of Srpr transcription in human leukemia cells [17]. pRb restricts DNA replication by preventing progression from G1 to S phase [18]. Thus, collectively these findings suggest that SRPR plays a role in the regulation of cell proliferation by affecting the cell cycle, specifically transition from G1 to S phase. Further mechanistic studies will be needed in order to elucidate the mechanisms how SRPR affects these processes in keratinocytes.
A similar observation has been reported in S. cerevisiae, in which Srpr disruption decreased growth rate [4], suggesting that SRPR is required for cell proliferation. On the contrary, down-regulation of Srpr was detected in a rat model of cyclosporine A-induced gingival overgrowth [5], suggesting that SRPR may not be the only one ER targeting regulator affecting cell proliferation. While there are no other loci known for Srpr in mouse, rat and human, there are 2 isoforms reported in both rat and human. Thus, it raises a possibility that an isoform may play a redundant function in rat and human. Obviously, a more detailed examination of the function of SRPR in cell proliferation is needed to elucidate this complex mechanism.
Previous reports indicated that deregulated miR-330-5p expression controls cell proliferation in colorectal and prostate cancers [19,20]. Recently, we also showed that miR-330-5p inhibits keratinocyte proliferation by arresting cells in G0/G1 phase [9]. Moreover, in this study, we demonstrated that miR-330-5p directly down-regulates the expression of SRPR by targeting its 3′ UTR, suggesting that the inhibitory effect of SRPR down-regulation on cell proliferation is mediated by miR-330-5p. Similar to SRPR, knockdown of Pdia3 expression by miR-330-5p inhibits proliferation of mouse keratinocytes [9], which explains why Srpr overexpression did not fully rescue the inhibitory effect of miR-330-5p on keratinocyte proliferation. Therefore, miR-330-5p is a key regulator of cell cycle because it targets genes associated with this process.
We also showed that decreased expression of Srpr inhibits proliferation of 3T3-L1 adipocytes (S3 Fig). In this cell line, Srpr expression was also down-regulated by miR-330-5p through direct binding to its 3′ UTR (S4 Fig). These results raise a possibility that regulation of Srpr by miR-330-5p occurs in various cell types and plays important roles in regulation of cell proliferation. Further studies are required to elucidate the detailed mechanisms underlying these regulatory effects.
In summary, our current study suggests that SRPR plays a critical role in keratinocyte proliferation by affecting the cell cycle. This effect of SRPR is controlled by miR-330-5p. These findings provide evidence for a new role of SRPR and miR-330-5p in keratinocytes and, more generally, in skin biology.
Supporting Information
Real-time PCR was performed to compare expression level of Srpr with several genes whose expressions were regulated by miR-330-5p in keratinocyte. Results are the average of three independent experiments.
(DOC)
(A) Over-expression of miR-330-5p inhibited proliferation of PAM212 cells in a dose dependant manner. (B) Relative cell viability was determined at 72 h post transfection with 50 nM and 100 nM RNAs. Results are the average of three independent experiments. *P<0.05; ***P<0.001.
(DOC)
(A) Srpr siRNA transfection induced the inhibition of proliferation. (B) Relative viable cells were measured after 48 h transfection. Results are the average of three independent experiments. **P<0.01; ***P<0.001.
(DOC)
(A) MiR-330-5p over-expression induced Proliferation inhibition of the 3T3-L1 cells. (B) Relative viable cells were counted after 72 h transfection. (C) MiR-330-5p down-regulated the Srpr expression in 3T3-L1 cells. The data was normalized against Gapdh expression. (D) Dual Luciferase assay revealed that miR-330-5p significantly inhibited the luciferase activity in the 3T3-L1 cells containing full length of Srpr 3’UTR. (B-D) Results are the average of three independent experiments. **P<0.01; ***P<0.001.
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF- 2013R1A1A2011821).
References
- 1.Schwartz T, Blobel G (2003) Structural basis for the function of the beta subunit of the eukaryotic signal recognition particle receptor. Cell 112: 793–803. [DOI] [PubMed] [Google Scholar]
- 2.Nagai K, Oubridge C, Kuglstatter A, Menichelli E, Isel C, et al. (2003) Structure, function and evolution of the signal recognition particle. EMBO J 22: 3479–3485. 10.1093/emboj/cdg337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egea PF, Stroud RM, Walter P (2005) Targeting proteins to membranes: structure of the signal recognition particle. Curr Opin Struct Biol 15: 213–220. 10.1016/j.sbi.2005.03.007 [DOI] [PubMed] [Google Scholar]
- 4.Ogg SC, Poritz MA, Walter P (1992) Signal recognition particle receptor is important for cell growth and protein secretion in Saccharomyces cerevisiae. Mol Biol Cell 3: 895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marley JJ, Phenix KV, Irwin CR, Thompson J, Robinson PA, et al. (1999) Signal recognition particle receptor (SRPR) is downregulated in a rat model of cyclosporin A-induced gingival overgrowth. J Periodontal Res 34: 188–196. [DOI] [PubMed] [Google Scholar]
- 6.Fabian MR, Sonenberg N, Filipowicz W (2010) Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79: 351–379. 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- 7.Shenouda SK, Alahari SK (2009) MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev 28: 369–378. 10.1007/s10555-009-9188-5 [DOI] [PubMed] [Google Scholar]
- 8.Ferracin M, Veronese A, Negrini M (2010) Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn 10: 297–308. 10.1586/erm.10.11 [DOI] [PubMed] [Google Scholar]
- 9.Kim BK, Yoo HI, Choi K, Yoon SK (2015) miR-330-5p inhibits proliferation and migration of keratinocytes by targeting Pdia3 expression. FEBS J 282: 4692–4702. 10.1111/febs.13523 [DOI] [PubMed] [Google Scholar]
- 10.Kim BK, Kim I, Yoon SK (2015) Identification of miR-199a-5p target genes in the skin keratinocyte and their expression in cutaneous squamous cell carcinoma. J Dermatol Sci 79: 137–147. 10.1016/j.jdermsci.2015.05.005 [DOI] [PubMed] [Google Scholar]
- 11.Wang XL, Zhang T, Wang J, Zhang DB, Zhao F, et al. (2015) MiR-378b Promotes Differentiation of Keratinocytes through NKX3.1. PLoS One 10: e0136049 10.1371/journal.pone.0136049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mardaryev AN, Ahmed MI, Vlahov NV, Fessing MY, Gill JH, et al. (2010) Micro-RNA-31 controls hair cycle-associated changes in gene expression programs of the skin and hair follicle. FASEB J 24: 3869–3881. 10.1096/fj.10-160663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 14.Akopian D, Shen K, Zhang X, Shan SO (2013) Signal recognition particle: an essential protein-targeting machine. Annu Rev Biochem 82: 693–721. 10.1146/annurev-biochem-072711-164732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Legue E, Nicolas JF (2005) Hair follicle renewal: organization of stem cells in the matrix and the role of stereotyped lineages and behaviors. Development 132: 4143–4154. 10.1242/dev.01975 [DOI] [PubMed] [Google Scholar]
- 16.Yu BD, Mukhopadhyay A, Wong C (2008) Skin and hair: models for exploring organ regeneration. Hum Mol Genet 17: R54–59. 10.1093/hmg/ddn086 [DOI] [PubMed] [Google Scholar]
- 17.Wells J, Yan PS, Cechvala M, Huang T, Farnham PJ (2003) Identification of novel pRb binding sites using CpG microarrays suggests that E2F recruits pRb to specific genomic sites during S phase. Oncogene 22: 1445–1460. 10.1038/sj.onc.1206264 [DOI] [PubMed] [Google Scholar]
- 18.Goodrich DW, Wang NP, Qian YW, Lee EY, Lee WH (1991) The retinoblastoma gene product regulates progression through the G1 phase of the cell cycle. Cell 67: 293–302. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Zhu X, Xu W, Wang D, Yan J (2013) miR-330 regulates the proliferation of colorectal cancer cells by targeting Cdc42. Biochem Biophys Res Commun 431: 560–565. 10.1016/j.bbrc.2013.01.016 [DOI] [PubMed] [Google Scholar]
- 20.Mao Y, Chen H, Lin Y, Xu X, Hu Z, et al. (2013) microRNA-330 inhibits cell motility by downregulating Sp1 in prostate cancer cells. Oncol Rep 30: 327–333. 10.3892/or.2013.2452 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Real-time PCR was performed to compare expression level of Srpr with several genes whose expressions were regulated by miR-330-5p in keratinocyte. Results are the average of three independent experiments.
(DOC)
(A) Over-expression of miR-330-5p inhibited proliferation of PAM212 cells in a dose dependant manner. (B) Relative cell viability was determined at 72 h post transfection with 50 nM and 100 nM RNAs. Results are the average of three independent experiments. *P<0.05; ***P<0.001.
(DOC)
(A) Srpr siRNA transfection induced the inhibition of proliferation. (B) Relative viable cells were measured after 48 h transfection. Results are the average of three independent experiments. **P<0.01; ***P<0.001.
(DOC)
(A) MiR-330-5p over-expression induced Proliferation inhibition of the 3T3-L1 cells. (B) Relative viable cells were counted after 72 h transfection. (C) MiR-330-5p down-regulated the Srpr expression in 3T3-L1 cells. The data was normalized against Gapdh expression. (D) Dual Luciferase assay revealed that miR-330-5p significantly inhibited the luciferase activity in the 3T3-L1 cells containing full length of Srpr 3’UTR. (B-D) Results are the average of three independent experiments. **P<0.01; ***P<0.001.
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.