Abstract
Targeted subunit vaccines for cancer immunotherapy do not capture tumor antigenic complexity, while approaches employing tumor lysate are often limited by inefficient antigen uptake and presentation, and low immunogenicity. Here, whole cancer cells were processed to generate antigen-rich, membrane-enclosed subcellular particles, termed “reduced cancer cells”, that reflect the diversity and breadth of the parent cancer cell antigen repertoire, and could be loaded with disparate adjuvant payloads. These vesicular particles enhanced uptake of the adjuvant payload, and potentiated the activation of primary dendritic cells in vitro. Similarly, reduced cancer cell-associated antigens were more efficiently presented by primary dendritic cells in vitro than were soluble counterparts or lysate control. In mice, vaccination using adjuvant-loaded reduced cancer cells facilitated the induction of antigen-specific cellular and humoral immune responses. Taken together, these observations demonstrate that adjuvant-loaded reduced cancer cells could have utility in cancer vaccines as an alternative to lysate.
Keywords: cancer vaccine, immunotherapy, adjuvant, cancer cell vesicles, lysate
ToC summary
Whole cancer cells were processed to generate immunogenic subcellular vesicular particles that can be used to stimulate antigen-specific immune responses in vitro and in vivo. The particles retain a broad antigen repertoire representative of the parent cells, and can be loaded with disparate adjuvant payloads. These particles may represent a superior alternative to lysate, the standard for primary antigen preparations, in various cancer vaccine applications.
1. Introduction
Therapeutic cancer vaccines immunize the patient against cancer antigens in order to generate immune effector cells that can recognize and eliminate the cancer cells. Cancer vaccination is challenging for several reasons. First, tumors are heterogeneous populations with high degrees of both inter- and intra-tumoral antigen variability.[1] Second, tumors possess tolerogenic mechanisms that enable them to avoid immune destruction.[2] Third, tumors are dynamic populations that can evolve in response to weak selective pressures induced by suboptimal vaccination.[3] In order to overcome these challenges, it is important for a cancer vaccine to elicit a response that is personalized to the patient’s unique cancer antigen repertoire, and broad enough to capture the antigenic diversity of the tumor. The vaccine must also facilitate efficient antigen presentation and promote robust cytotoxic effector cell function in order to overcome the immunosuppressive mechanisms imposed by the tumor.
In general, there exist two approaches to cancer vaccination: 1) targeted approaches, which immunize against defined tumor antigens and 2) broad vaccination against a breadth of undefined tumor antigens. Targeted approaches, such as subunit vaccines, are attractive because they generally target antigenic determinants that are known to be uniquely overexpressed and immunogenic. However, because targeted vaccines do not reflect the antigenic complexity of the tumor, suboptimal vaccinations that fail to induce efficient antigen spread can potentially facilitate the selection of lowly immunogenic subpopulations, leading to immune escape.[3] Additionally, targeted approaches are commonly non-personalized in that they immunize against prototypic cancer antigens, which precludes the use of such approaches for a large patient population that either has a cancer for which a characteristic target has not been identified, or is refractory for the particular known target. Alternatively, strategies that utilize material derived from the tumor as an antigen source vaccinate against a breadth of prospective tumor antigens in a patient-specific manner. The current standard for such primary antigen preparations is tumor lysate, the soluble fraction obtained after freeze-thawing cancer cells. Lysate-based approaches have the advantage of capturing both the uniqueness and breadth of the tumor antigen repertoire, without the need for pre-knowledge of specific antigen targets. This is particularly advantageous in cancer types with high mutational loads (e.g. melanoma), in which patients frequently have unique and extensive mutanomes encoding neoantigens not subject to central tolerance, that could potentially facilitate efficient vaccination.[4, 5] Despite these advantages, tumor lysate possesses limited immunogenicity, and the soluble lysate constituents are not efficiently taken up by antigen-presenting cells, limiting the immune response generated.[6]
Previous work has demonstrated that the physical association of antigen with a molecular danger signal, such as a Toll-like receptor (TLR) agonist, for example through chemical conjugation[7, 8] or co-loading onto a nano- or microscale colloidal scaffold,[9–12] significantly increases its presentation efficiency, even when compared to the same amount of soluble antigen and danger cue admixed but not physically associated. Indeed, the presence of danger signal and antigen in the same phagosomal compartment has been shown to be important for the efficient presentation of that antigen.[13, 14] It has also been shown that compared to a soluble counterpart, antigens associated with a “phagocytic substrate” such as an engineered colloid,[9, 10, 15] or cell debris,[16] or that is packaged into apoptotic blebs,[17] or tumor exosomes,[18] are more efficiently presented by antigen presenting cells. The application of nano- or microscale materials as delivery vehicles in cancer vaccines has allowed for the development of many promising targeted vaccines, but non-targeted approaches based on such platforms are much more limited, largely owing to the difficulty of loading complex undefined mixtures of proteins into such systems while maintaining a reasonable degree of colloidal stability. In addition, the use of exogenous materials as colloidal delivery vehicles can potentially lead to challenges associated with scale-up and reproducibility, as well as regulatory hurdles to translation.
In light of these observations, the goal of this work was to develop an alternative method for processing cancer cells in which the antigen diversity of the parent cells is maintained but the antigen content is reformulated to facilitate efficient antigen presentation. To this end, we hypothesized that whole cancer cells could be broken down into membrane-enclosed vesicular compartments and loaded with adjuvant. The benefits of such a system are that (1) the unique antigen repertoire of the parent cells is reflected, (2) the breadth of antigenic diversity of the parent cell population is captured, (3) associated cancer antigens are presented in a highly immunogenic format, in contrast to lysate, the current standard, and (4) endogenous cell material functions as the delivery vehicle, obviating the input of exogenous carrier material. In this work, we investigated two approaches to mechanically disrupt whole cancer cells, extrusion and sonication, in order to generate vesicular compartments we termed “reduced cancer cells” (RCCs). We demonstrate that in contrast to extruded RCCs, sonicated RCCs retain protein from diverse intracellular compartments with a protein distribution that is highly representative of the parent whole cells, and leverage this property for vaccine design. We show that sonicated RCCs can be loaded with diverse payloads, and that these adjuvant-loaded RCCs are highly immunogenic and facilitate more efficient antigen presentation in vitro than cell lysate or purified soluble antigen. When administered as a vaccine in vivo, RCCs can also be used to stimulate antigen-specific cellular and humoral immune responses. Overall, this work describes an approach for reformulating whole cancer cells into immunogenic antigen-rich particles that may represent a superior alternative to the derivation of lysate.
2. Results
2.1. Derivation and Characterization of RCCs
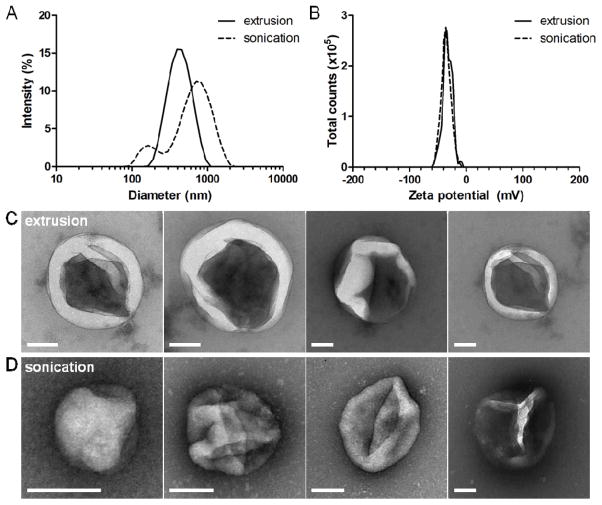
Whole B16-F10 melanoma cells were disrupted via either mechanical extrusion (eRCCs) or sonication (sRCCs) to generate RCCs which were subsequently purified using density centrifugation. Both methods generated RCCs with nano-microscale size distributions (Figure 1A). Dynamic light scattering (DLS) measurements indicated that eRCCs had an average hydrodynamic diameter of ~ 400 nm whereas sRCCs had an average hydrodynamic diameter of ~ 500 nm associated with a broader, less monodisperse distribution and a higher PDI (Figure 1A, S1A, S1B). Both eRCCs and sRCCs had comparable zeta potentials of ~ −30 mV (Figure 1B, S1C), consistent with what has been reported previously for purified mammalian cell membranes.[19, 20] Examination of RCCs via TEM revealed structures within the size range predicted by DLS. Consistent with DLS, TEM showed that eRCC structures appeared generally more monodisperse whereas sRCC structures were more variable in size (Figure 1C, 1D, S1D, S1E). Enhancement of contrast upon negative staining with uranyl formate suggests that both eRCCs and sRCCs represent enclosed structures as would be expected of membrane-bound vesicles. Although partial membrane collapse of the RCCs was observed following staining and dehydrating during TEM sample preparation, it is likely that the RCCs are spherical structures in the hydrated state. These observations suggest that by disrupting whole cancer cells via either mechanical extrusion or sonication, membrane-enclosed vesicular particles are obtained. Importantly, these particles may retain endogenous cell membrane-associated proteins in the membranous “shell” of the particles, as well as soluble intracellular proteins in the aqueous “core”.
Figure 1.
Derivation of RCCs. (A) Representative size intensity distributions of eRCCs and sRCCs as measured by DLS. (B) Representative zeta potential distributions of eRCCs and sRCCs as measured by DLS. Representative TEM images of eRCCs (C) and sRCCs (D). All scale bars = 100 nm.
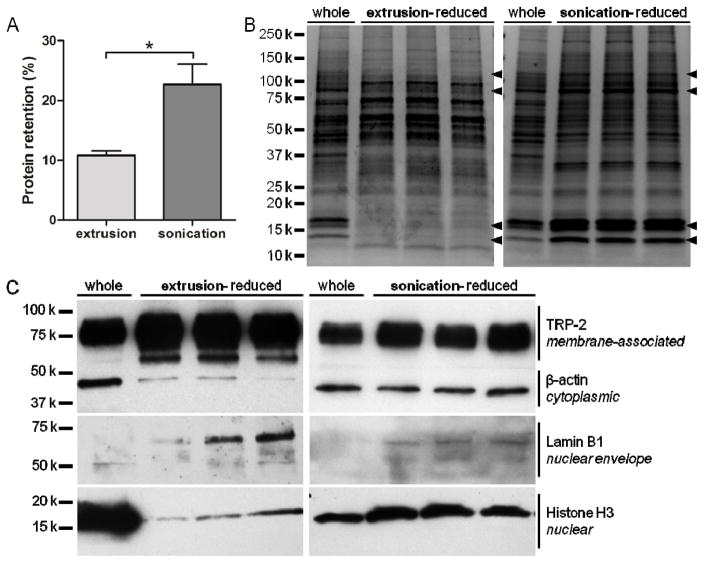
In order to investigate the prospective antigen content of eRCCs and sRCCs, total and specific RCC protein retention was characterized. eRCCs were found to retain 10.8 ± 0.7% of total parent cell protein content whereas sRCCs were found to retain 22.7 ± 3.4% (Figure 2A). Qualitative analysis of total protein content via coomassie staining showed that eRCCs exhibited the depletion of a number of prominent bands observed in the parent whole cell lysate whereas sRCCs maintained a protein distribution qualitatively more similar to the parent whole cell lysate (Figure 2B). When the same amount of total protein was loaded, western blotting for proteins localized to various cellular compartments indicated that while eRCCs showed a marked enrichment of membrane-associated proteins with a concomitant loss of proteins in other compartments, notably cytoplasmic and nuclear, protein retained by sRCCs was highly representative of all compartments of the parent cells based on the markers evaluated (Figure 2C). Given that many tumor antigens are localized to the cytoplasm and the nucleus,[21] the ability to retain these fractions is likely to be functionally advantageous for a cancer vaccine. Because of this, as well as superior gross protein retention, sRCCs were used for all subsequent analyses. For all functional assays, sRCCs were typically prepared and used fresh. Although sRCCs were observed to settle over time, particles suspended in PBS could be stored for several days at 4°C and redispersed with brief bath sonication without a significant change in average particle size (Figure S2A). Because the RCCs are comprised entirely of cell material, it is expected that they can be completely degraded when taken up by phagocytic cells such as antigen-presenting cells (APCs). In contrast, we observed that in the absence of cells, RCCs were not significantly degraded in the presence of serum for several days, although serum-mediated aggregation was evident over this time (Figure S2B).
Figure 2.
Protein retention of RCCs. (A) Percent protein retention of eRCCs and sRCCs relative to protein content of intact cells, analyzed via BCA assay. (B) Coomassie stain of three separate eRCC and sRCC samples as compared to the parent cells (whole) from which they were derived. Arrowheads indicate bands that were present in whole parent cells and largely depleted by eRCCs but maintained by sRCCs. (C) Western blot of three separate eRCC and sRCC samples as compared to the parent cells (whole) from which they were derived, probing for proteins localized to various cellular compartments: TRP-2 (membrane-associated), β-actin (cytoplasmic), lamin B1 (nuclear envelope), and histone H3 (nuclear). Data represent the mean ± SD, *p<0.05.
2.2. RCCs as Adjuvant Delivery Vehicles
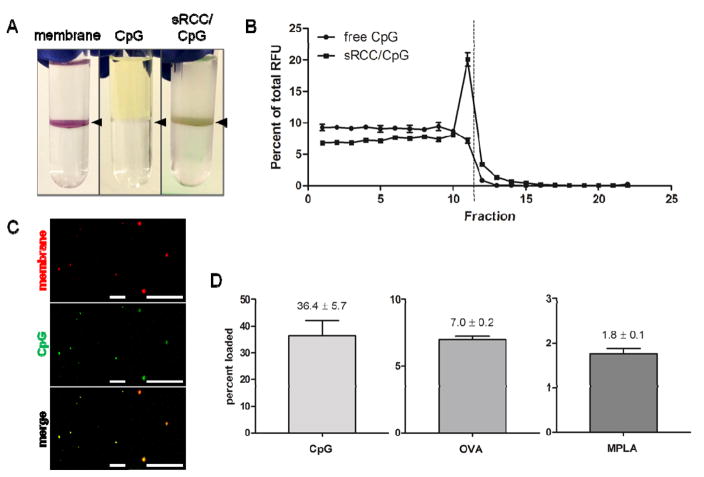
To evaluate whether sRCCs could be loaded with adjuvant, a 5′ fluorophore-conjugated CpG DNA was used as a model adjuvant, and loaded into sRCCs by mixing the parent cells with CpG and sonicating the CpG/cell suspension. CpG is a hydrophilic oligodeoxynucleotide-based adjuvant and a ligand for the endosomally-localized TLR9, an immune-activating pattern recognition receptor (PRR). CpG has been shown to be an effective adjuvant in cancer vaccine formulations.[22–26] When sRCCs were stained with a membrane dye and subjected to density centrifugation in a two-step (10%/50%) iodixanol gradient, significant enrichment of the dye was observed at the interface of the steps, representing the membrane-enclosed sRCC-rich fraction (Figure 3A). While free fluorescent CpG did not localize to a specific fraction upon density centrifugation, when CpG was mixed with the cell suspension and sRCCs were derived from the CpG/cell mixture, a fraction of the CpG was found to colocalize with the interfacial RCC band following density centrifugation (Figure 3A, B), suggesting CpG loading of the resultant sRCCs. Confocal microscopy of sRCCs also revealed colocalization of a membrane-specific dye and the fluorescent CpG (Figure 3C). To preclude the possibility that the fluorescent tag was facilitating loading, sRCCs were also loaded with untagged CpG and loading was quantified using HPLC and measuring absorbance at 260 nm. Loading of fluorescent CpG was similarly evaluated using HPLC and measuring absorbance at 260 nm as well as fluorescence at 488 nm. Comparison of tagged and untagged CpG did not reveal any differences in loading (Figure S3). Furthermore, no change in elution time was observed when HPLC was used to compare the fluorescent CpG stock and fluorescent CpG that had been sRCC-loaded and recovered following RCC lysing, suggesting that intact CpG is loaded into the RCCs (Figure S4). CpG loading of sRCCs was evaluated as a function of cell input, and 5e6 cells/ml was found to be the optimal cell concentration among those tested (Figure S5). This cell concentration was used for subsequent experiments involving CpG-loaded sRCCs. Based on such analyses, it was found CpG could be loaded into sRCCs at 36.4% efficiency, the lipophilic adjuvant monophosphoryl lipid A (MPLA) could be loaded at 1.8% efficiency, and the model antigen ovalbumin (OVA) could be loaded at 7.0% efficiency (Figure 3D).
Figure 3.
Loading of RCCs with diverse payloads. (A) Visualization of RCCs stained with membrane dye (left), free fluorescent CpG (middle), and RCCs loaded with fluorescent CpG (right) following density centrifugation. Black arrowheads point to location of interface. (B) Quantification of fluorescence as a function of fraction following density centrifugation of free fluorescent CpG or fluorescent CpG loaded into sRCCs. Dashed line represents location of interface. (C) Co-localization of a membrane-associated dye and fluorescent CpG following loading into sRCCs at low magnification (left) and high magnification (right). All scale bars = 10 μm. (D) Loading efficiency of CpG, OVA, and MPLA into sRCCs. Numbers represent mean loading efficiency ± SD.
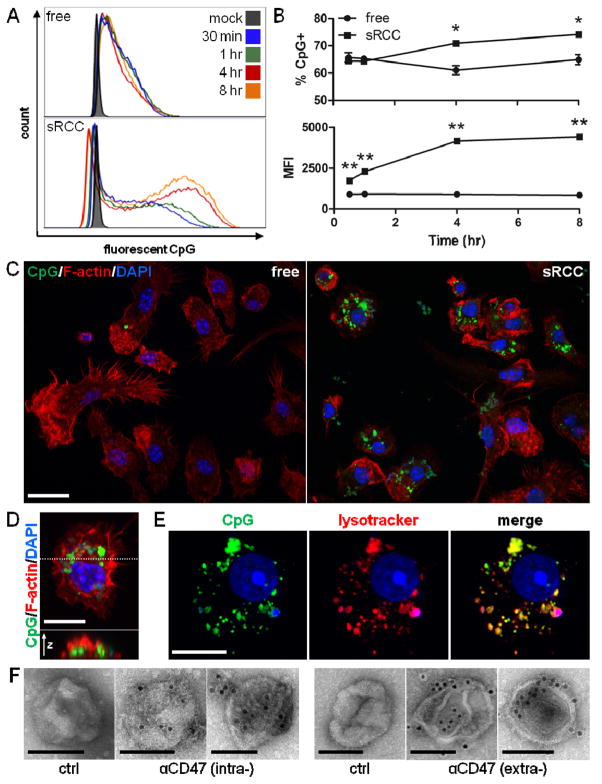
The ability of RCCs to facilitate the uptake of associated adjuvant by APCs was next evaluated using fluorescent CpG as a model adjuvant and primary bone marrow-derived dendritic cells (DCs) as a model APC. CpG associated with RCCs was taken up by DCs with faster kinetics and at a higher magnitude than a comparable amount of free CpG, with ~10% more DCs being CpG+ by 4 hours and beyond, and a greater than 4 and 5-fold higher MFI being observed among CpG+ cells at 4 and 8 hours, respectively (Figure 4A, B). These observations were confirmed with confocal microscopy wherein significantly more CpG signal was visible in DCs treated with RCC-associated versus free CpG at all timepoints evaluated (Figure 4C, S6A). Taking an orthogonal slice through a CpG+ DC, CpG fluorescence was apparent in the interior of the cell (Figure 4D) demonstrating true internalization of the CpG. This was further confirmed by visualizing slices through the z-axis of CpG+ DCs as CpG-associated fluorescence was observed throughout the cell volume (Figure S6B). CpG was found to be localized within the endolysosomal compartment of DCs after treatment for 4 hours (Figure 4E), which is likely to have functional implications as TLR9 is localized in the endosomal compartment. Taken together, these data show faster uptake kinetics and a higher amount of CpG internalization with RCC-associated CpG compared to free CpG, demonstrating a particle-mediated enhancement of payload uptake by DCs.
Figure 4.
BMDC uptake of sRCC-associated CpG. (A) Flow cytometry analysis of BMDC uptake of free or sRCC-associated fluorescent CpG at various timepoints. (B) Quantification of data shown in (A) presented as percent of cells CpG+ (top), and MFI (geometric mean) of CpG+ population (bottom). Data represent the mean ± SD, *p<0.05, **p<0.001. (C) Confocal microscopy of BMDC uptake of free or sRCC-associated fluorescent CpG after 4 hour incubation. Scale bar = 20 μm. (D) High magnification image of a CpG+ BMDC (top), and an orthogonal slice through the cell (bottom) taken at the position indicated by the dashed line. Scale bar = 10 μm. (E) Confocal image demonstrating colocalization of fluorescent CpG and lysotracker signals after 4 hour incubation. Scale bar = 10 μm. (F) Immunoelectron microscopy of sRCC stained with a primary antibody specific for either the intracellular (left) or extracellular (right) domains of CD47 followed by an appropriate 10 nm gold colloid-conjugated secondary antibody. Primary antibody was omitted for controls. Two representative sRCC images are shown for each antibody stain. Scale bars = 100 nm.
RCC sidedness, the orientation of the RCC membranes, could potentially have functional implications, and so was also analyzed. In particular, CD47 is a cell surface transmembrane protein that inhibits phagocytosis, and its upregulation is a common mechanism of immune evasion by tumor cells.[27] Immunoelectron microscopy of sRCCs was performed using antibodies that target either the extracellular or intracellular portion of CD47 (Figure 4F). sRCCs were stained by both antibodies suggesting that plasma membrane-derived structures among sRCCs were likely present in a mixture of right-side out and inside-out orientations. This contrasts with previous reports that plasma membrane vesicles derived via mechanical extrusion are primarily maintained in a right-side out orientation,[28, 29] which could potentially hinder uptake by APCs if functional CD47 is retained.
2.3. Immunologic Functionality of Adjuvant-Loaded RCCs in vitro
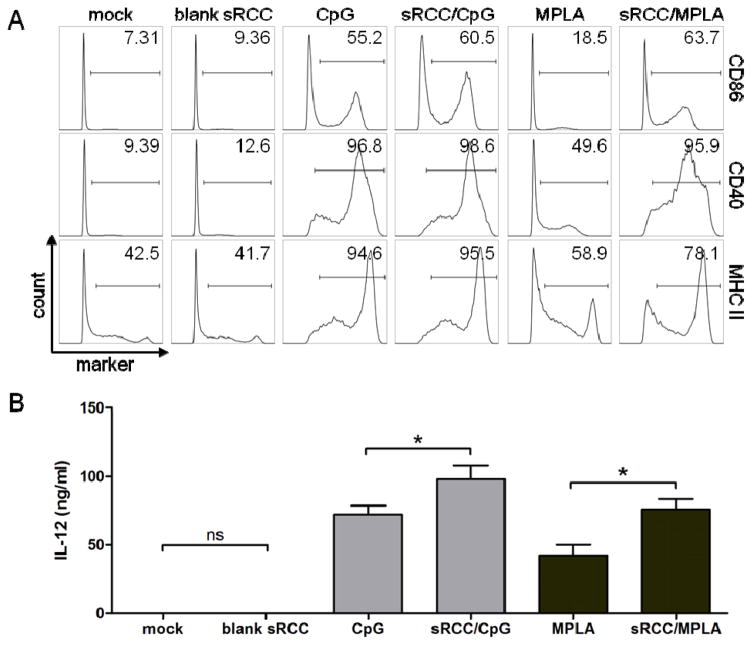
To evaluate the functionality of RCC-loaded adjuvants, DCs were treated with CpG and MPLA in either free or RCC-associated forms, and assayed for activation based on upregulation of surface activation markers and production of the proinflammatory cytokine IL-12. Treatment of DCs with blank sRCCs did not result in any increase in surface activation marker expression or IL-12 production compared to mock treatment. In contrast, treatment with sRCCs loaded with either CpG or MPLA significantly upregulated surface expression of MHC II and the costimulatory molecules CD86 and CD40 on DCs, to a similar or greater degree than the respective free adjuvant (Figure 5A). Notably, treatment of DCs with sRCC-associated adjuvants promoted significantly greater production of IL-12 than did the same amount of the respective free adjuvants, with a ~36% and ~80% increase observed with CpG and MPLA, respectively (Figure 5B). Treatment of DCs with CpG-loaded sRCCs also promoted a change in cell morphology characterized by increased spreading and aspect ratio relative to unactivated DCs (Figure S7). These morphology changes are consistent with what has been previously reported to take place upon in vitro DC activation.[30]
Figure 5.
DC activation in response to adjuvant-loaded sRCCs. (A) Flow cytometry analysis of DC surface activation marker expression, and (B) quantification of IL-12 production via ELISA, following mock treatment or treatment with blank sRCCs, treatment with 0.2 μM CpG in free or sRCC-associated form, or treatment with 0.02 μM MPLA in free or sRCC-associated form, after 12–15 hour incubation. Numbers in (A) represent average percent positive cells based on gate shown. Values in (B) represent the mean ± SD, *p<0.05.
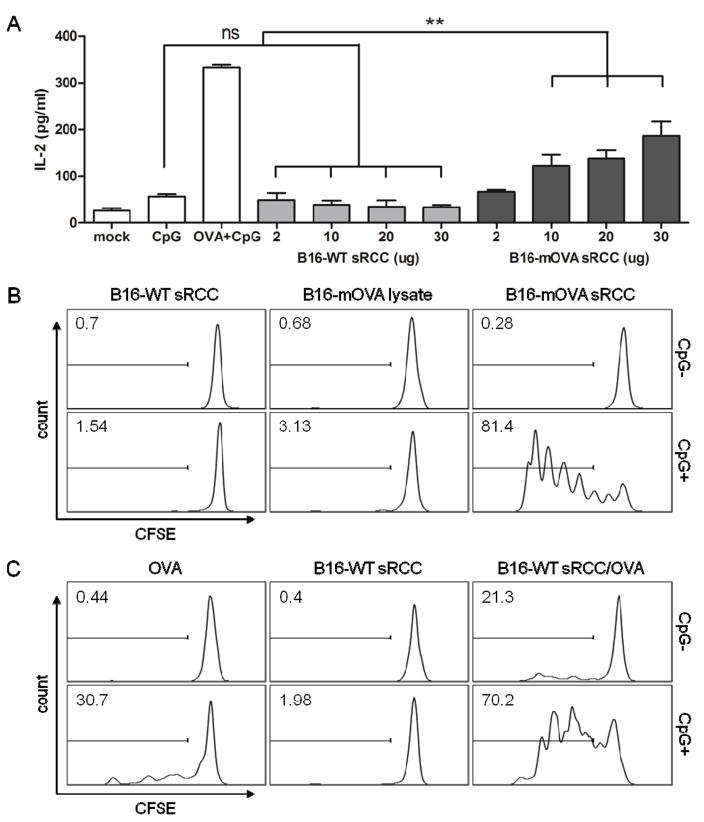
Next, the ability of RCCs to facilitate antigen presentation by APCs was evaluated using the model antigen OVA. To evaluate presentation of RCC-associated antigens on MHC II, DCs were loaded with sRCCs derived from either wild-type B16-F10 cells (B16-WT), or B16-F10 cells transduced to express a membrane-bound form of OVA (B16-mOVA) (Figure S8), and co-cultured with MF2.2D hybridoma T cells. MF2.2D cells recognize an OVA-derived peptide presented on MHC II, and produce IL-2 in response to T cell receptor (TCR) stimulation. Whereas no increase in IL-2 production was observed over control conditions when DCs were treated with B16-WT sRCCs, a dose-dependent increase in IL-2 production was observed with B16-mOVA sRCC treatment over the concentration range tested (Figure 6A).
Figure 6.
T cell response to sRCC-treated DCs. (A) Quantification of IL-2 production by MHC II-restricted MF2.2D hybridoma T cells in response to co-culture with DCs treated with B16-WT- or B16-mOVA-derived sRCCs. CpG samples were treated with 0.8 μM CpG, OVA samples were treated with 2.8 μM OVA, and OVA + CpG samples were treated with 2.8 μM OVA and 0.8 μM CpG. sRCC samples were treated with between 2–30 μg of sRCCs containing between 0.1–1.5 μg (78.6 nM–1.2 μM) CpG based on a total protein/CpG mass ratio of 1/0.05. (B) Representative histograms in which sRCC conditions represent T cell response to BMDCs treated with 13.9 μg sRCCs (+/− 0.3 μg CpG; 0.2 μM). Lysate conditions represent T cell response to BMDCs treated with lysate derived from an equivalent number of whole cells, and admixed with an equivalent amount of CpG. (C) Representative histograms in which OVA conditions represent T cell response to BMDCs treated with 44.4 nM free OVA with or without 78.6 nM free CpG, an equivalent amount of OVA and CpG loaded into B16-WT sRCCs, or an equivalent amount of blank B16-WT sRCCs. All co-culture experiments were carried out in 0.2 ml volume. Gates and values in (B–C) indicate the mean percentage positive cells in respective sample. Data in (A) represent the mean ± SD, analyzed using one-way ANOVA, followed by Tukey post-test, *p<0.05.
To evaluate MHC I-mediated cross-presentation of RCC-associated antigens, a process critical for robust anti-tumor immunity, DCs treated with various stimuli were co-cultured with CFSE-stained primary CD8+ T cells derived from OT-I mice, which express a transgenic T cell receptor specific for an OVA peptide presented on MHC I. T cell activation was evaluated by measuring CFSE dilution as a proxy for T cell proliferation. First, DCs were treated with either B16-WT sRCCs, B16-mOVA sRCCs, or cancer cell lysate from the same number of starting B16-mOVA parent cells. sRCC samples were either unloaded, or loaded with CpG, and lysate samples were either used as-prepared, or admixed with the same amount of soluble CpG. Whereas DCs treated with B16-WT sRCCs or B16-mOVA lysate, with or without CpG, facilitated minimal OT-I CD8+ T cell proliferation, DCs treated with B16-mOVA sRCCs loaded with CpG facilitated extremely robust OT-I CD8+ T cell proliferation (Figure 6B).
Because it is possible that the difference in antigen presentation efficiency between lysate and RCC could be due to differences in the level of the relevant protein retained by the two derivation procedures, the antigen presentation efficiency of soluble OVA protein was next compared to the same amount of OVA protein loaded into B16-WT sRCCs. Consistent with observations made with endogenously expressed OVA, it was observed that whereas DCs treated with soluble OVA admixed with CpG could facilitate moderate levels of OT-I CD8+ T cell proliferation, DCs treated with OVA-loaded and OVA/CpG-co-loaded B16-WT sRCCs facilitated significantly greater levels of OT-I CD8+ T cell proliferation relative to the respective soluble OVA conditions (Figure 6C). Taken together, these data demonstrate that antigens associated with adjuvant-loaded sRCCs are efficiently presented by DCs in vitro in a functional manner, and that the presentation of these sRCC-associated antigens is significantly more efficient than antigens delivered in a soluble format.
2.4. Immunologic Functionality of Adjuvant-Loaded RCCs in vivo
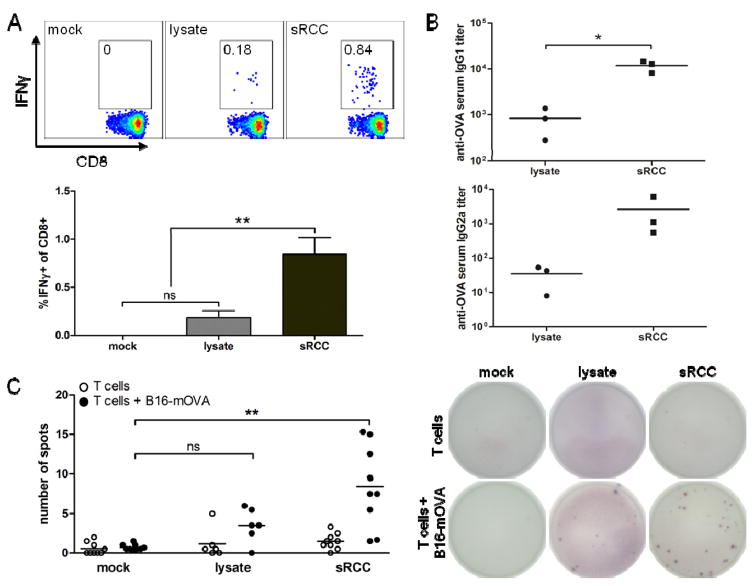
The in vivo functionality of the RCCs was evaluated by performing vaccinations of naive mice and measuring antigen-specific cellular and humoral immune responses. First, vaccinations were performed either with B16-WT-derived lysate admixed with OVA protein and MPLA, or with B16-WT sRCCs co-loaded with the same amount of OVA and MPLA. Analysis of T cells in the peripheral blood 8 days post-vaccination showed significantly greater frequencies of functional IFNγ-producing CD8+ T cells in samples from mice treated with sRCCs as compared to mock-treated or lysate-treated mice, following ex vivo stimulation of peripheral blood mononuclear cells (PBMCs) with a relevant MHC I-restricted OVA peptide (Figure 7A). Consistent with this, sRCC vaccination also facilitated the induction of OVA-specific IgG1 and IgG2a antibody titers comparable to, or greater than lysate (Figure 7B).
Figure 7.
Cellular and humoral response to sRCC vaccinations in naive mice. (A)–(B) Cellular and humoral response to vaccination with sRCCs or cell lysate derived from B16-WT cells, and loaded or admixed, respectively, with exogenous OVA protein. (A) Representative flow cytometry plots with mean frequency of IFNγ-positive cells among live CD8+ cells in peripheral blood (top), and quantification of data (bottom). Cells were stimulated in vitro with peptide. (B) anti-OVA IgG1 and IgG2a serum titers, on day 8 post-vaccination. Naive mice were vaccinated subcutaneously with either 370 μg B16-WT sRCCs loaded with 70 μg OVA and 90 ng MPLA, or cell lysate derived from the same number of B16-WT cells (5e6) and admixed with the same amount of OVA and MPLA. (C) Cellular response to vaccination with sRCCs or cell lysate derived from B16-mOVA cells. ELISPOT analysis of IL-2-producing T cells from spleens of mice following overnight co-culture with parent B16-mOVA cells on day 4 post-boost (day 22 post-vaccination). Naive mice were vaccinated subcutaneously on days 0 and 2 (prime), and days 16 and 18 (boost), with either 370 μg B16-mOVA sRCCs loaded with 18 μg CpG and 180 ng MPLA, or cell lysate derived from the same number of B16-mOVA cells (5e6) and admixed with the same amount of CpG and MPLA. Quantification of spots (left) and representative images of ELISPOT membranes (right). Values represent the mean ± SD, *p<0.05, **p<0.001. Data in (C) analyzed using one-way ANOVA, followed by Tukey post-test.
Lastly, to evaluate the immune response to endogenous parent cell antigens elicited by sRCCs, naive mice were vaccinated with either B16-mOVA-derived lysate admixed with CpG and MPLA, or with B16-mOVA sRCCs co-loaded with the same amount of CpG and MPLA. Because immune responses can be induced against a diversity of different parent cell antigens, the cellular immune response was evaluated by co-culturing T cells isolated from spleens of treated mice with irradiated B16-mOVA parent cells, and T cell stimulation evaluated via IL-2 production was measured using ELISPOT. sRCC vaccination facilitated the induction of functional B16-mOVA-specific T cells at frequencies comparable to, or greater than lysate (Figure 7C). Taken together, these data demonstrate that RCCs can facilitate the induction of antigen-specific cellular and humoral responses in vivo.
3. Discussion
In this work, we show that whole cancer cells can be processed to produce antigen-rich particles termed RCCs. Although these structures can be generated through various methods, a derivation method based on sonication led to the retention of greater than 20% of total protein content. Proteins from different intracellular compartments were retained in a distribution similar to the parent cells, providing a broad antigen pool for anti-cancer immunization. In contrast, RCCs generated via extrusion using the method employed in this work retained significantly less total protein, with significant loss of non-membrane-associated protein. The difference in protein retention between the eRCCs and the sRCCs likely stems from the different mechanisms of cell disruption that take place during extrusion versus sonication. Cell disruption during extrusion is expected to take place via mild mechanical shear, likely resulting in brief disruption of membrane-enclosed compartments as the cells are forced through the pores. In this work, cells were extruded through membranes with a minimal pore size of 1 μm, which is small enough to disrupt membrane-bound organelles, potentially resulting in the release of their contents. Indeed, while the majority of the lipid content is recovered following density centrifugation, the significant loss of cystolic and nuclear proteins, but not membrane-associated (including nuclear envelope-associated) proteins suggests that this is the case. An additional factor contributing to protein loss could be fouling of the filter membrane during extrusion. In contrast, cell disruption during sonication takes place primarily via cavitation, involving the continuous mass nucleation, expansion, and implosion of microbubbles throughout the medium, which causes complete disintegration of membranes and the release of soluble protein content. A possible mechanism for the observed retention of soluble protein by the sonicated RCCs could involve the recapture of a fraction of the freed protein during the thermodynamically favored reformation of membrane-enclosed structures following membrane disintegration.
This work also demonstrated that sRCCs can be loaded with both hydrophilic (CpG) and lipophilic (MPLA) adjuvants, illustrating the versatility of this approach. This allows the flexibility to load adjuvants that differentially regulate the generated immune response, as well as diverse combinations of different adjuvants that activate nonredundant signaling pathways. The latter is particularly interesting as it could allow, for example, for the generation of RCCs loaded with multiple TLR or other PRR agonists in combinations that mimic natural pathogens. Indeed, it has been shown that the delivery of multiple PRR agonists enhances the resultant cytokine response by immune cells and that certain combinations show greater synergy than others.[12, 31] In the vaccination studies evaluating the immune response to endogenous parent cell antigens, combinatorial delivery of CpG and MPLA was employed, allowing for activation of both the MyD88- and TRIF-dependent pathways. Further studies exploring the synergy of various adjuvant combinations could allow for further optimization of this system.
It was observed that adjuvant was taken up more efficiently by DCs when it was associated with the sRCCs than when it was administered in free form. Treatment of DCs with adjuvant-loaded sRCCs enhanced the expression of surface activation markers and the production of the proinflammatory cytokine IL-12 compared to soluble adjuvant, and enhanced the efficiency of antigen presentation compared to soluble antigen sources such as lysate. The enhancement in payload uptake is likely due to the concentration of the adjuvant and antigen in a colloidal vehicle, which is consistent with previous findings.[32] Enhancement of DC activation by CpG, which signals via the endosomal TLR9, is likely also related to the enhanced uptake, and is also consistent with previous findings that association of CpG with a colloidal vehicle enhances its adjuvant activity.[33] Enhancement of DC activation by MPLA, which signals via the cell surface TLR4, is likely related to an enhancement of MPLA avidity on the surface of the sRCCs, which can facilitate TLR4 clustering and subsequent activation.[34, 35] Notably, it was observed that nonadjuvanted sRCCs did not activate DCs compared to mock treatment. This demonstrates that nonadjuvanted sRCCs are not inherently immunostimulatory and should not promote adverse inflammatory reactions, for example, due to the retention of endogenous danger associated molecular patterns (DAMPs). Rather, loading the sRCCs with exogenous adjuvants is necessary to impart them with immunostimulatory capacity, and the ability to load these structures with adjuvant using this derivation method is critical to the utility of the generated structures for vaccination.
Collectively, the observations presented herein suggest that adjuvant-loaded sRCCs potentially represent an efficient and versatile cancer vaccine platform. Indeed, previous work employing material isolated from deconstructed cancer cells supports the idea that such material, when appropriately adjuvanted, can be employed as a cancer antigen source for vaccination both in vitro and in vivo.[20, 36] The generation of adjuvant-loaded sRCCs is a method for processing cancer cells in which the antigen diversity of the parent cells is maintained, but the antigen content is reformulated into cell material-derived particles that facilitate efficient antigen presentation. This is in contrast to standard bottom-up design approaches for targeted vaccines, which do not recapitulate this degree of antigen diversity. Notably, sRCC derivation does not involve the input of any exogenous material, and it is therefore expected that the RCCs can be fully degraded by APCs without any harmful degradation products. Further, as simply a method for processing cancer cells, sRCC derivation is analogous to the preparation of tumor lysate, which is also a form of processed cancer cell material, and is the current standard for primary cancer antigen mixtures. Importantly, lysate is commonly employed in diverse cancer vaccine systems in both the preclinical and clinical settings,[22–24, 37–39] demonstrating that such tumor-derived material is generally considered a safe antigen source for vaccines. Tumor lysate as an antigen source is most commonly used in DC vaccines, vaccines that activate and load DCs with cancer antigen ex vivo for subsuquent reinfusion. The in vitro observations reported in this study are compelling evidence that adjuvant-loaded RCCs may represent a superior alternative to tumor lysate in such vaccine approaches by facilitating enhanced uptake and cross-presentation of cancer antigens. Notably, the in vivo data also suggests that RCCs may be more effective than lysate in this context, although the relatively low frequency of antigen-specific T cells generated against endogenous cell antigens indicates that the RCCs will likely need to be optimized for direct in vivo applications. One suboptimal property of the RCCs for in vivo applications is their broad size distribution, which extends far beyond a feasible size range for passive drainage to the lymph nodes via the lymphatics.[40, 41] In order to optimize the RCCs for standalone vaccination, it may be necessary to improve the size distribution of the particles via further processing, for example, through post-sonication extrusion. Alternatively, improving the active trafficking of the RCCs to the lymph node by APCs could be achieved via surface modification with cues that enhance APC uptake,[36] or by incorporating the RCCs into material scaffolds that bring large numbers of relevant APCs to the vaccination site.[22–24, 26, 37]
4. Conclusions
In summary, whole cancer cells can be processed to produce subcellular vesicular particles within the nano- to microscale size range, that retain a broad distribution of proteins representative of the parent cells from which they are derived. When these structures, termed RCCs, are loaded with adjuvant, they facilitate enhanced DC activation and antigen presentation when compared to soluble adjuvant and antigen, respectively. Vaccination of mice with these structures also facilitates the induction of antigen specific cellular and humoral immune responses. Based on these observations, we propose that RCCs represent an attractive platform for cancer vaccination, in particular, as an alternative to tumor lysate in vaccine approaches that currently use lysate as an antigen source. RCCs could also have utility in various materials-based approaches that function to bring large numbers of APCs to the vaccination site, and may be potentially useful as a standalone particulate cancer vaccine system with optimization.
5. Experimental Section
Animals
All work with C57BL/6J and C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-I) mice (The Jackson Laboratory) was performed in compliance with National Institutes of Health and institutional guidelines.
Cells and Reagents
The B16-F10 murine melanoma cell line (ATCC) was cultured in DMEM supplemented with 10% heat inactivated FBS (HI-FBS) and 1% penicillin-streptomycin. The MF2.2D murine T cell hybridoma cell line, kindly provided by Dr. Kenneth Rock, was cultured in RPMI 1640 supplemented with 10% HI-FBS, 2 mM L-glutamine, 55 μM beta-mercaptoethanol, 1x non-essential amino acids, 10mM HEPES, and 1% penicillin-streptomycin. The B3Z murine T cell hybridoma cell line, kindly provided by Dr. Nilabh Shastri, was cultured in RPMI 1640 supplemented 10% HI-FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 50 μM beta-mercaptoethanol, and 1% penicillin-streptomycin.
Primary CD8+ T cells were isolated from the spleens of OT-I mice and enriched using an anti-CD8 MACS isolation kit (Miltenyi Biotec). T cells were used immediately for co-culture experiments and were cultured in RPMI 1640 supplemented with 10% HI-FBS, 1% penicillin-streptomycin, 1 mM sodium pyruvate, 5 mM HEPES, and 50 μM beta-mercaptoethanol.
CpG and CpG-FAM (fluorescent CpG) were synthesized by Integrated DNA Technologies based on the described B class 1826 sequence (tccatgacgttcctgacgtt). MPLA derived from Salmonella minnesota R595 was purchased from Invivogen. Unless otherwise stated, antibodies were purchased from eBioscience.
Derivation of RCCs
B16-F10 or B16-mOVA cells were grown in T150 tissue culture flasks until near-confluency. Cells were non-enzymatically removed by incubating cells with Ca-/Mg- PBS at 4°C for 5 minutes and washed three times with PBS. After the final wash, cells were resuspended at 5e6 cells/ml in PBS supplemented with a protease inhibitor tablet (Roche Life Science) and CpG or OVA, if applicable. Cell suspensions were cooled on ice until disruption. Cell suspensions were maintained at 5e6 cells/ml for eRCC derivation as this concentration has previously been shown to be efficient for whole cell extrusion,[28, 42] and higher initial cell concentrations were found to result in significant clogging of the extruder membrane (data not shown). To generate eRCCs, 1 ml aliquots of pre-cooled cell suspensions were serially extruded using a mini extruder (Avanti Polar Lipids) through 10 μm, 5 μm, and 1 μm polycarbonate filters. To generate sRCCs, 1 ml aliquots of pre-cooled cell suspensions were transferred to round-bottom tubes and sonicated (Qsonica Q700 equipped with 3.2 mm stepped microtip probe) for a total process time of 45 seconds in alternating 15 second burst and rest cycles of 60% amplitude and no sonication, respectively. Raw RCC suspensions were subsequently fractionated in a discontinuous iodixanol (Sigma-Aldrich) gradient containing 10% and 50% iodixanol layers. Following centrifugation at 100,000 rcf for at least 2 hours at 4°C, RCCs were collected from the interface of the layers. Where applicable, RCCs were incubated overnight at room temperature with MPLA. The collected fraction was subsequently washed by diluting at least 20-fold in PBS and pelleted by centrifuging at 95,000 rcf for 20 minutes at 4°C. The supernatant was completely decanted, and the RCC pellets were resuspended by slowly running the collected material through a 29 gauge syringe. For western blot and CpG loading studies, RCCs were lysed in RIPA buffer (Amresco) supplemented with protease inhibitor (Roche). For all other studies, RCCs were resuspended in PBS at approximately 0.9 mg protein/ml, unless otherwise stated. RCCs were stored at 4°C and used within a few days.
Characterization of RCCs
RCC hydrodynamic size and zeta potential were evaluated via dynamic light scattering (DLS) using a Malvern Nano ZS. Size and zeta potential measurements were taken with RCCs suspended at ~0.005 mg/ml in PBS or H2O, respectively. For stability studies in which DLS measurements were taken over time, sRCCs were suspended at ~0.05 mg/ml in either PBS, or DMEM supplemented with 10% HI-FBS, and stored at 4°C, or 37°C, respectively. At the indicated timepoints, samples were bath sonicated for 5 minutes, and size measurements were taken.
For quantification of payload loading, payloads were loaded into sRCCs and purified sRCCs were subsequently lysed in RIPA buffer. CpG loading was quantified through a combination of fluorescent measurements of a fluorescently tagged CpG conjugate using a standard plate reader, and quantification with HPLC. HPLC analysis was conducted on an Agilent 1100 HPLC equipped with an XTerra MS C18 5 micron, 4.6x250 mm Column (Waters Cat# 186000494). Samples were analyzed using 0.1M triethylammonium acetate, pH 7 (Mobile Phase A) and 90% acetonitrile in 0.1M triethylammonium acetate, pH 7 (Mobile Phase B) with the following gradient: 0min-2%B, 4min-2%B, 45min-55%B, 50min-90%B, 51min-2%B, 60min-2%B. The flow rate was 0.5 mL/min, column temperature was 30°C, samples were injected without dilution at 50 μl, and detection was conducted at 260 and 488 nm. MPLA loading was quantified through a combination of fluorescent measurements of a fluorescent MPLA conjugate using a standard plate reader, and measurements of untagged MPLA using an Endosafe-MCS system (Charles River). Ovalbumin loading was quantified through fluorescent measurements of a fluorescent ovalbumin conjugate (invitrogen) using a standard plate reader. Data shown are means of loading studies conducted with fluorescent conjugates. Alternative approches were performed to ensure consistency of loading measurements.
RCC Uptake Studies
Uptake of RCC-loaded CpG was evaluated using a 5′ fluorophore conjugated CpG (Integrated DNA Technologies). Primary DCs were treated with either free or RCC-associated fluorescent CpG for various lengths of time and then analyzed via either flow cytometry or confocal microscopy for uptake. For flow cytometry analysis of uptake, treated DCs were removed by scraping and stained for viability (Biolegend) and CD11c. Cells were analyzed on a BD LSRFortessa and the percentage of CpG+ cells among live CD11c+ cells was quantified. To evaluate uptake via confocal microscopy, cells were analyzed either live or fixed. For live cell imaging, DCs were seeded on glass bottom tissue culture plates (MatTek). Adjuvant-loaded cells were treated with Hoescht 33342 (Life Technologies) for 15 minutes followed by imaging on a Leica SP5 X MP Inverted Confocal Microscope equipped with a Ludin incubation chamber. For fixed cell imaging, DCs were seeded on glass coverslips. At the time of analysis, adjuvant-loaded DCs were fixed in 4% paraformaldehyde/PBS for 10 minutes at room temperature followed by permeabilization in 0.1% Triton X/PBS for 15 minutes. Cells were then stained with Alexa Fluor 594-phalloidin (Life Technologies) and Hoescht 33342 (Life Technologies) and mounted on glass slides using ProLong Gold antifade reagent (Life Technologies). Cells were subsequently imaged on a Zeiss LSM 710 confocal microscope.
DC Activation and Antigen Presentation Studies
For DC activation studies, adjuvant loaded sRCCs were prepared at total protein/adjuvant mass ratio of 1/0.05 for CpG, and 1/0.002 for MPLA. To evaluate the stimulatory functionality of RCC-loaded adjuvants, primary DCs were plated in non-tissue culture-treated plates, and CpG (0.2 μM) or MPLA (0.02 μM) was added in free or RCC-associated form. The CpG stock used for these studies was tested for endotoxin content and found to have <0.05 EU/mg endotoxin contamination. After a 12–15 hour stimulation, media samples were collected for IL-12 quantification via ELISA (Peprotech), and cells were removed by scraping and stained for viability, CD11c, MHC II, and the costimulatory molecules CD40 and CD86. DC activation was evaluated via flow cytometry based on the proportion of live CD11c+ cells that were positive for the activation markers.
Adjuvant loaded RCCs were prepared at total protein/CpG mass ratios of 1/0.05 for hybridoma T cell and OT-I CD8+ T cell co-culture studies. To evaluate the capacity of RCC-delivered antigen to be presented in MHC I and II, primary DCs were plated in non-tissue culture treated plates and treated with either free or RCC-associated antigens for 3 hours. Loaded DCs were subsequently co-cultured with either MF2.2D cells to investigate MHC II presentation, or primary CD8+ OT-I T cells to investigate MHC I cross-presentation, in a total culture volume of 0.2 ml. For co-culture experiments involving the use of the MF2.2D hybridoma line, co-culture was carried out for 12–15 hours after which media was collected for IL-2 quantification via ELISA (Peprotech). For CD8+ OT-I T cell co-culture, primary CD8+ T cells were isolated from OT-I mice as described above and stained with CFSE according to the manufacturer’s instructions. CFSE-stained T cells were then co-cultured with treated DCs for 3 days. After the co-culture period, T cells were collected and stained for viability and CD8α and analyzed via flow cytometry. CFSE dilution of live CD8+ cells was evaluated.
Vaccination Studies
6–10 week old C57BL/6J mice were used for all vaccination studies. Mice were vaccinated subcutaneously in the flank, typically with 370 μg of RCCs per dose, or lysate derived from the same starting number of parent cells.
Ex Vivo Peptide Stimulation and Intracellular Cytokine Staining
For detection of functional peripheral blood OVA-specific CD8+ T cells, peripheral blood was collected at indicated time points, and erythrocytes were lysed with ACK buffer (Lonza). PBMCs were stimulated with 1 μM SIINFEKL peptide for 4 hours in the presence of GolgiPlug (BD), stained with a live/dead stain (eBioscience) and for surface CD8, fixed/permeabilized, and stained for IFNγ (gating scheme shown in Figure S9).
Detection of Serum Anti-OVA Antibodies
Sera was collected by centrifuging peripheral blood at 550 rcf for 5 minutes. Anti-OVA antibodies were detected by incubating diluted sera with OVA-coated plates overnight, and subsequently detecting relevant IgG subclasses using appropriate anti-mouse secondary antibodies.
IL-2 ELISPOT
Spleens were processed by mashing through a 70 μm nylon cell strainer and red blood cell lysing with ACK buffer. T cells were subsequently purified using a pan T cell negative selection MACS kit (Miltenyi Biotec). T cells were seeded at 500,000 cells per well in 96 well plates coated with anti-IL2 antibody (BD) and co-cultured for 18 hours with 50,000 irradiated (10,000 rad) B16-mOVA cells that had been treated overnight with 10 ng/ml IFNγ to increase MHC I presentation. Captured IL-2 was subsequently detected using an HRP-conjugated IL-2 antibody (BD). Automated spot quantification was performed using a CTL ImmunoSpot S4 analyzer.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism. Unless otherwise stated, data were compared using the unpaired two-tailed t test and p-values less than 0.05 were considered to be statistically significant. Where applicable, data are reported as the mean ± SD.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant R01 EB015498 to D. Mooney. S. Koshy was supported by an HHMI ISRF. This work was performed in part at the Center for Nanoscale Systems (CNS) at Harvard University, a member of the National Nanotechnology Infrastructure Network (NNIN), which is supported by the National Science Foundation under NSF award no. ECS-0335765. The authors thank Dr. Steven Perrault, Dr. Alex Watters, Dr. Deniz Yuksel, Dr. Luo Gu, Dr. Omar Ali, Dr. Martin Thelin, Aileen Li, and Chris Johnson for scientific discussions, and Dr. Dennis Kasper for use of the CTL ImmunoSpot S4 analyzer. The authors especially thank Dr. Kenneth Rock and the Dana-Farber Cancer Institute for providing the MF2.2D cell line, and Dr. Nilabh Shastri for providing the B3Z cell line.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Alexander S. Cheung, John A. Paulson School of Engineering and Applied Sciences, Harvard University, Cambridge, MA 02138, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA
Sandeep T. Koshy, John A. Paulson School of Engineering and Applied Sciences, Harvard University, Cambridge, MA 02138, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA. Harvard-MIT Division of Health Sciences and Technology, Cambridge, MA 02139, USA
Alexander G. Stafford, Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA
Dr. Maartje M.C. Bastings, Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA. Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA. Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA 02115, USA
Prof. David J. Mooney, Email: mooneyd@seas.harvard.edu, John A. Paulson School of Engineering and Applied Sciences, Harvard University, Cambridge, MA 02138, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA
References
- 1.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P. New Engl J Med. 2012;366:883. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabinovich GA, Gabrilovich D, Sotomayor EM. Annu Rev Immunol. 2007;25:267. doi: 10.1146/annurev.immunol.25.022106.141609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mittal D, Gubin MM, Schreiber RD, Smyth MJ. Curr Opin Immunol. 2014;27:16. doi: 10.1016/j.coi.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schumacher TN, Schreiber RD. Science. 2015;348:69. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 5.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Cell. 2015;160:48. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.González FE, Gleisner A, Falcón-Beas F, Osorio F, López MN, Salazar-Onfray F. Hum Vaccin Immunother. 2014;10:3261. doi: 10.4161/21645515.2014.982996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tighe H, Takabayashi K, Schwartz D, Marsden R, Beck L, Corbeil J, Richman DD, Eiden JJ, Jr, Spiegelberg HL, Raz E. Eur J Immunol. 2000;30:1939. doi: 10.1002/1521-4141(200007)30:7<1939::AID-IMMU1939>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 8.Heit A, Schmitz F, O’Keeffe M, Staib C, Busch DH, Wagner H, Huster KM. J Immunol. 2005;174:4373. doi: 10.4049/jimmunol.174.7.4373. [DOI] [PubMed] [Google Scholar]
- 9.Moon JJ, Suh H, Bershteyn A, Stephan MT, Liu H, Huang B, Sohail M, Luo S, Um SH, Khant H. Nat Mater. 2011;10:243. doi: 10.1038/nmat2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott EA, Stano A, Gillard M, Maio-Liu AC, Swartz MA, Hubbell JA. Biomaterials. 2012;33:6211. doi: 10.1016/j.biomaterials.2012.04.060. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, Tan MT, Keegan BP, Barry MA, Heffernan MJ. Biomaterials. 2014;35:8385. doi: 10.1016/j.biomaterials.2014.05.067. [DOI] [PubMed] [Google Scholar]
- 12.Black M, Trent A, Tirrell M, Olive C. Expert Rev Vaccines. 2010;9:157. doi: 10.1586/erv.09.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blander JM, Medzhitov R. Nature. 2006;440:808. doi: 10.1038/nature04596. [DOI] [PubMed] [Google Scholar]
- 14.Burgdorf S, Schölz C, Kautz A, Tampé R, Kurts C. Nat Immunol. 2008;9:558. [Google Scholar]
- 15.Rock KL. Eur J Immunol. 1994;24:2421. doi: 10.1002/eji.1830241024. [DOI] [PubMed] [Google Scholar]
- 16.Li M, Davey GM, Sutherland RM, Kurts C, Lew AM, Hirst C, Carbone FR, Heath WR. J Immunol. 2001;166:6099. doi: 10.4049/jimmunol.166.10.6099. [DOI] [PubMed] [Google Scholar]
- 17.Ruben JM, van den Ancker W, Bontkes HJ, Westers TM, Hooijberg E, Ossenkoppele GJ, de Gruijl TD, van de Loosdrecht AA. Cancer Immunol, Immunother. 2014;63:335. doi: 10.1007/s00262-013-1515-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andre F, Schartz N, Chaput N, Flament C, Raposo G, Amigorena S, Angevin E, Zitvogel L. Vaccine. 2002;20:A28. doi: 10.1016/s0264-410x(02)00384-5. [DOI] [PubMed] [Google Scholar]
- 19.Parodi A, Quattrocchi N, van de Ven AL, Chiappini C, Evangelopoulos M, Martinez JO, Brown BS, Khaled SZ, Yazdi IK, Enzo MV. Nat Nanotechnol. 2013;8:61. doi: 10.1038/nnano.2012.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang RH, Hu CMJ, Luk BT, Gao W, Copp JA, Tai Y, O’Connor DE, Zhang L. Nano Lett. 2014;14:2181. doi: 10.1021/nl500618u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vigneron N, Stroobant V, Van den Eynde BJ, van der Bruggen P. Cancer Immun. 2013;13:15. [PMC free article] [PubMed] [Google Scholar]
- 22.Ali OA, Emerich D, Dranoff G, Mooney DJ. Sci Transl Med. 2009;1:8ra19. doi: 10.1126/scitranslmed.3000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ali OA, Huebsch N, Cao L, Dranoff G, Mooney DJ. Nat Mater. 2009;8:151. doi: 10.1038/nmat2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ali OA, Tayalia P, Shvartsman D, Lewin S, Mooney DJ. Adv Funct Mater. 2013;23:4621. doi: 10.1002/adfm.201203859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gungor B, Yagci FC, Tincer G, Bayyurt B, Alpdundar E, Yildiz S, Ozcan M, Gursel I, Gursel M. Sci Transl Med. 2014;6:235ra61. doi: 10.1126/scitranslmed.3007909. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Li WA, Choi Y, Lewin SA, Verbeke CS, Dranoff G, Mooney DJ. Nat Biotechnol. 2015;33:64. doi: 10.1038/nbt.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiskopf K, Ring AM, Ho CCM, Volkmer JP, Levin AM, Volkmer AK, Özkan E, Fernhoff NB, van de Rijn M, Weissman IL. Science. 2013;341:88. doi: 10.1126/science.1238856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang SC, Kim OY, Yoon CM, Choi DS, Roh TY, Park J, Nilsson J, Lötvall J, Kim YK, Gho YS. ACS Nano. 2013;7:7698. doi: 10.1021/nn402232g. [DOI] [PubMed] [Google Scholar]
- 29.Hu CMJ, Fang RH, Luk BT, Chen KNH, Carpenter C, Gao W, Zhang K, Zhang L. Nanoscale. 2013;5:2664. doi: 10.1039/c3nr00015j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xing F, Wang J, Hu M, Yu Y, Chen G, Liu J. Nanoscale Res Lett. 2011;6:1. doi: 10.1186/1556-276X-6-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Q, Egelston C, Vivekanandhan A, Uematsu S, Akira S, Klinman DM, Belyakov IM, Berzofsky JA. Proc Natl Acad Sci USA. 2008;105:16260. doi: 10.1073/pnas.0805325105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brewer JM, Pollock KG, Tetley L, Russell DG. J Immunol. 2004;173:6143. doi: 10.4049/jimmunol.173.10.6143. [DOI] [PubMed] [Google Scholar]
- 33.De Titta A, Ballester M, Julier Z, Nembrini C, Jeanbart L, van der Vlies AJ, Swartz MA, Hubbell JA. Proc Natl Acad Sci USA. 2013;110:19902. doi: 10.1073/pnas.1313152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Triantafilou M, Miyake K, Golenbock DT, Triantafilou K. J Cell Sci. 2002;115:2603. doi: 10.1242/jcs.115.12.2603. [DOI] [PubMed] [Google Scholar]
- 35.Meraz IM, Hearnden CH, Liu X, Yang M, Williams L, Savage DJ, Gu J, Rhudy JR, Yokoi K, Lavelle EC. PloS one. 2014;9:e94703. doi: 10.1371/journal.pone.0094703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Broekhoven CL, Parish CR, Demangel C, Britton WJ, Altin JG. Cancer Res. 2004;64:4357. doi: 10.1158/0008-5472.CAN-04-0138. [DOI] [PubMed] [Google Scholar]
- 37.Ali OA, Verbeke C, Johnson C, Sands RW, Lewin SA, White D, Doherty E, Dranoff G, Mooney DJ. Cancer Res. 2014;74:1670. doi: 10.1158/0008-5472.CAN-13-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prins RM, Wang X, Soto H, Young E, Lisiero DN, Fong B, Everson R, Yong WH, Lai A, Li G. J Immunother. 2013;36:152. doi: 10.1097/CJI.0b013e3182811ae4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poschke I, Lövgren T, Adamson L, Nyström M, Andersson E, Hansson J, Tell R, Masucci GV, Kiessling R. Cancer Immunol, Immunother. 2014;63:1061. doi: 10.1007/s00262-014-1575-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reddy ST, van der Vlies AJ, Simeoni E, Angeli V, Randolph GJ, O’Neil CP, Lee LK, Swartz MA, Hubbell JA. Nat Biotechnol. 2007;25:1159. doi: 10.1038/nbt1332. [DOI] [PubMed] [Google Scholar]
- 41.Mueller SN, Tian S, DeSimone JM. Mol Pharm. 2015;12:1356. doi: 10.1021/mp500589c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu H, Oliver AE, Ngassam VN, Yee CK, Parikh AN, Yeh Y. Integr Biol. 2012;4:685. doi: 10.1039/c2ib20022h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.