Abstract
Depressed calcium handling by the sarcoplasmic reticulum (SR) Ca-ATPase and its regulator phospholamban (PLN) is a key characteristic of human and experimental heart failure. Accumulating evidence indicates that increases in the relative levels of PLN to Ca-ATPase in failing hearts and resulting inhibition of Ca sequestration during diastole, impairs contractility. Here, we identified a genetic variant in the PLN promoter region, which increases its expression and may serve as a genetic modifier in dilated cardiomyopathy (DCM). The variant AF177763.1:g.203A>C (at position −36bp relative to the PLN transcriptional start site) was found only in the heterozygous form in 1 out of 296 normal subjects and in 22 out of 381 cardiomyopathy patients (heart failure at age of 18–44 years, ejection fraction=22±9%). In vitro analysis, using luciferase as a reporter gene in rat neonatal cardiomyocytes, indicated that the PLN-variant increased activity by 24% compared to the wild type. Furthermore, the g.203A>C substitution altered the specific sequence of the steroid receptor for the glucocorticoid nuclear receptor (GR)/transcription factor in the PLN promoter, resulting in enhanced binding to the mutated DNA site. These findings suggest that the g.203A>C genetic variant in the human PLN promoter may contribute to depressed contractility and accelerate functional deterioration in heart failure.
Keywords: promoter, polymorphism, transcriptional factor, GR, GRE, cardiomyopathy, PLN, SR Ca-ATPase
INTRODUCTION
Heart failure is a multifactorial syndrome in which intrinsic myocardial dysfunction contributes to cardiac dilation and diminished ejection performance, leading to progressive cardiac deterioration or sudden death [Richardson et al., 1996; Seidman and Seidman, 2001]. Genes causally associated with cardiomyopathy have been identified through nonbiased genetic analysis or by candidate gene studies in experimental system [Geisterfer-Lowrance et al., 1996; Franz et al., 2001]. Thus, molecular modifiers of heart failure include mutations of genes that encode cytoskeletal, sarcomeric, nuclear membrane, and calcium handling sarcoplasmic reticulum (SR) proteins. These findings have implicated pathogenic mechanisms whereby perturbation of structural integrity, contractile force dynamics, and calcium regulation within the cardiac myocyte intrinsically contribute to myocardial disease.
Abnormal calcium homeostasis is a prototypical mechanism for contractile dysfunction in failing cardiomyocytes. Depressed calcium cycling in experimental and human heart failure reflects, at least in part, impaired calcium sequestration by the SR [Chien, 2000; MacLennan and Kranias, 2003]. Calcium sequestration is mediated by a Ca-transport ATPase (SERCA2a), whose activity is modulated by alteration in the expression and phosphorylation of phospholamban (PLN; MIM# 172405) [Luo et al., 1996; Simmerman and Jones, 1998]. In experimental models, expression levels of PLN closely correlate with basal contractile parameters and their responses to β-agonists [Luo et al., 1994; Kadambi et al., 1996; Brittsan et al., 2000; Dash et al., 2001]. In human heart failure, the levels of PLN are increased relative to SERCA2a, resulting in higher inhibition of the Ca-pump’s Ca-affinity, which impairs relaxation [Beuckelmann et al., 1992; Meyer et al., 1995; Hasenfuss, 1998]. As a double insult, the phosphorylation status of PLN is decreased, leading to increased inhibitory function and further depression of SR Ca-cycling. Thus, PLN is a major Ca-regulatory protein and efforts have concentrated on identifying naturally occurring mutations in the human PLN gene, which may perturb its activity and contribute to dilated cardiomyopathy (DCM). Indeed, three mutations in the coding region of the human PLN gene have been identified that are associated with familial cardiomyopathy [Haghighi et al., 2003, 2006; Schmitt et al., 2003]. However, parallel studies on genetic variants in the PLN promoter region, which may alter its expression levels, are limiting.
The PLN gene is located on human chromosome 6 [Fujii et al., 1991] and the 200 bp of its 5′ flanking region exhibits significant sequence homology between human, rabbit, rat, and mouse [Fujii et al., 1991; Haghighi et al., 1997; McTiernan et al., 1999a, 1999b]. Importantly, this segment of the 5′ upstream region of the human PLN gene contains conserved consensus motifs for GATA, CP1/NF-y, M-CAT, and E-box elements, which are also found in other mammalian species [Haghighi et al., 1997; McTiernan et al., 1999a]. However, the importance of these elements in regulation of PLN gene expression under physiological and pathophysiological conditions remains uninvestigated. Indeed, most studies indicate that cardiac PLN expression levels are maintained under stress and remodeling conditions, including pressure overload, hypertrophy, and failure [Ito et al., 2001; Kogler et al., 2003; Mills et al., 2006]. A recent study reported the presence of a rare human mutation in this highly conserved PLN promoter region (A>G at −77 bp), which was associated with increased PLN (1.5-fold) expression [Minamisawa et al., 2003]. This variant was found in 1 out of 87 hypertrophic cardiomyopathy patients, suggesting a role of the PLN promoter mutant in depressed Ca cycling, leading to hypertrophy.
In this study, we sought to identify naturally occurring PLN promoter mutations in nonfamilial heart failure patients. A novel point genetic variant (A>C) at position AF177763.1:g.203A>C (at −36bp relative to the PLN transcriptional start site: −36A>C) in the 5′ UTR region of the PLN gene was detected only in the heterozygous state in 22 heart failure patients and one normal subject. In vitro studies on the functional significance of this genetic variant revealed that it increases PLN expression levels by altering glucocorticoid nuclear receptor (GR)-mediated regulation of transcription.
MATERIALSANDMETHODS
Mutation Identification
Informed consent was obtained from participating subjects. All protocols were approved by the institutional review board of the Onassis Cardiac Surgery Center (Athens, Greece) or the University of Cincinnati College of Medicine, (Cincinnati, OH). Genomic DNA was isolated either from whole blood or from paraffin blocks containing heart tissue. The genomic reference with GenBank accession number AF177763.1 was used to retrieve the PLN sequence corresponding to proximal promoter and exon 1. A 600-bp fragment of the PLN gene, containing the PLN promoter region was amplified by PCR, using 60 ng of genomic DNA and a high-fidelity Taq polymerase. The primers were: sense, 5′CTAAGCCTGAAGATGC3′ and antisense, 5′CCAGTAACCA GGATC3′, tagged with M13 forward and reverse primer sequences, respectively. The conditions were: one cycle at 94°C for 3 min, linked to 30 cycles at 94°C for 1 min, 47°C for 1 min, and 72°C for 1 min, followed by one cycle at 94°C for 1 min, 53°C for 1 min, and 72°C for 10 min. The gel purified PLN DNA fragment was sequenced using automated dye-primer chemistry. The generated sequences were compared with the reported human PLN sequences by a computational method and the electropherograms were inspected individually for confirmation. The GenBank accession number AF177763.1 was used as a reference for numbering the PLN promoter polymorphism.
Echocardiography
Comprehensive 2D and Doppler echocardiography was performed according to the recommendations of the American Society of Echocardiography [Levy et al., 1990]. Left ventricular dimensions (interventricular septum end-diastolic thickness [IVEDT], left ventricular posterior wall end-diastolic thickness [PWEDT], left ventricular end-systolic and end-diastolic diameter [LVESD and LVEDD]) were measured with M-mode echocardiography, using the left parasternal window. Left ventricular volumes and ejection fraction (LVEF) were determined by apical two-and four-chamber views using the modified Simpson rule [Levy et al., 1990].
Cloning of the Human PLN Gene Promoter-Reporter Constructs
A PCR-based strategy was employed using high-fidelity DNA polymerase to amplify the mutant region from human PLN genomic DNA, comprising the upstream PLN promoter. A 510-bp DNA fragment was PCR-amplified from normal and DCM genomic DNA utilizing the primers 5′-TACCTGTGTTTATTTTTCTC-3′ and 5′-AAGAAGAATTACCAAAGTCAGC3′. To facilitate cloning, Kpn I and Xho I sites were added to the beginning of the primers. The 510-bp fragment containing the engineered Kpn I and Xho I sites was subcloned into the pBlueScript vector (Stratagene, La Jolla, CA). The upstream PLN promoter region was verified by DNA sequence analysis. Then, the PLN promoter fragment containing either the nucleotide transition, −36A>C, or the wild type sequences of the PLN gene was digested with Sac I and Pst I, and cloned into pGL3-Basic (Promega, Madison, WI) to create the PLN promoter-luciferase reporter constructs.
About 600 bp of 5′ upstream of the PLN gene sequences were scanned for putative transcription factor binding sites, using public domain software (Transcription Element Search Software; www.cbil.upenn.edu/tess; TFBLAST of TRANSFAC 6.0; Biobase Corporation, Beverly, MA; www.gene-regulation.com/cgi-bin/pub/programs/tfblast/tfblast.cgi).
Cardiomyocyte Culture, Transient Transfection, and Luciferase Assays
Ventricular myocytes were isolated from 1-day-old Sprague-Dawley rats and cultured as described [Minamisawa et al., 2003]. For promoter-reporter studies, after 24 hr incubation with serum-free medium, the myocytes were transiently cotransfected with 300 ng of each PLN luciferase test plasmid and 75 ng of phRL-TK control plasmid (Promega). The cells were harvested in Passive Lysis Buffer (Promega) 48 hr after transfection, and were stored at −80°C until processed for the luciferase assay. The cells were allowed to grow in the absence or presence of 3 μM dexamethasone for the last 45 hr of the 48-hr incubation period. Luciferase assays were performed according to the protocol of the Dual Luciferase Assay System (Promega). Each data point represents the mean and the standard error of the mean (SEM) of seven experiments.
Electrophoretic Mobility Shift Assays
Nuclear extracts from ventricular tissue samples were prepared as described previously [Brown et al., 2005] with modifications. Briefly, ventricular tissue was pulverized at liquid N2 temperatures, homogenized at low speed in buffer containing 10mM HEPES (pH 9), 1.5mM MgCl2, 10mM KCl, 0.5mM dithiothreitol (DTT), 25 μg/ml leupeptin, 0.2mM sodium orthovanadate, and 0.1% (vol/vol) Triton X, then vortexed and incubated on ice for 10 min. After centrifugation (5,000 g for 10 min), the pellet was suspended in solution containing 20mM HEPES (pH 7.9), 25% (vol/vol) glycerol, 0.6M KCl, 1.5mM MgCl2, 0.2mM EDTA, 0.5mM phenylmethanesulphonylfluoride (PMSF), 0.5mM DTT, 25 μg/ml leupeptin, and 0.2mM sodium orthovanadate, and then vortexed. This suspension was incubated on ice for 40 min with rigorous vortexing every 10 min. After centrifugation (10,000 g for 15 min), the supernatant was retained as a crude nuclear extract. Protein concentrations were determined using a Bio-Rad (Hercules, CA) protein assay with bovine serum albumin as a standard.
A double-stranded 20-bp oligodeoxynucleotide, containing the PLN promoter wild type (5′-CCTCCCTAG}{A} {ACACTTTTTC-3′; underlined, glucocorticoid binding element) or mutant form (5′-CCTCCCTAG}{C}{ACACTTTTTC-3′; bold, mutated nucleotide) was end-labeled using [γ-32P]ATP and T4 polynucleotide kinase (Promega), and was purified using a G-50 Sephadex column (Amersham Pharmacia Biotech, Piscataway, NJ). The binding reactions were performed in a final volume of 10 μl that contained 20 μg of nuclear protein, 10mM Tris · HCl (pH 7.5), 50mM NaCl, 1mM MgCl2, 0.5mM EDTA, 0.5mM DTT, 4% glycerol (vol/vol), and 1 μg of poly(dI-dC). After a 10-min preincubation at room temperature, the labeled probe (1 × 105 cpm/reaction) was added to each reaction and the reactions were incubated for an additional 20 min at room temperature. The DNA-protein complexes were separated on 6% nondenaturing polyacrylamide gels in 1× Tris borate-EDTA buffer. Gels were vacuum-dried and exposed to X-ray film at −20°C, using intensifying screens. Competition assays with 100-fold molar excess of unlabeled consensus oligodeoxynucleotide or control nonspecific oligodeoxynucleotide were performed to ensure that the signal was specific. The commercially available oligonucleotide containing the common glucocorticoid consensus, 5′-GACGGTACAAAATGTTCTAGG-3′ (Active Motif, Carlsbad, CA) and antiglucocorticoid antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) were used for specific binding activity confirmation. A double-stranded 22-bp oligodeoxynucleotide (5′-AGTTGAGGGGACTTTCCCAGGC-3′) containing a consensus nuclear factor-κB (NF-κB) binding site (underlined) was used as positive control.
Statistics
Data are presented as mean±SEM. Statistical analysis was performed using two-way analysis of variance (ANOVA) followed by Student-Newman-Keuls test. A P value of <0.05 was considered statistically significant. The agreement with the Hardy-Weinberg expectations (HWE) of genotype frequencies was determined using the chi-squared test based on the number of observed and expected heterozygotes and the exact test based on the number of observed and expected genotypes [Guo and Thompson, 1992].
RESULTS
Clinical History
A total of 381 DCM patients and 296 normal subjects without any known cardiomyopathy history were recruited from the University Hospital, Cincinnati Heart Failure/Transplant Program (Cincinnati, OH) and the Onassis Cardiac Surgery Center (Athens, Greece). The clinical characteristics and the demographic data for the DCM populations are summarized in Table 1. Comorbid conditions in the cohorts included: hypertension (8%), diabetes (6%), hypercholesterolemia (12%), and atrial fibrillation (12%). The medications used by the DCM patients were angiotensin-converting enzyme (ACE) inhibitors (97%), diuretics (94%), digoxin (98%), beta blockers (75%), Ca-channel blockers (12%), and antiarrhythmics (45%).
TABLE 1.
Clinical Characteristics of the United States and Greek DCM Patients With Heart Failure
| Ethnicity
|
||
|---|---|---|
| United States | Greek | |
| n | 163 | 218 |
| Age (years) | 44.95±3.3 | 40±6.2 |
| Gender (%) | ||
| Male | 72 | 82 |
| Female | 28 | 18 |
| Etiology (%) | ||
| Dilated cardiomyopathy | 94.27 | 100 |
| Ischemic cardiomyopathy | 5.76 | — |
| Functional class (% NYHA III/IV) | 67.2 | 70.6 |
| LVEF (%) | 23.6±8 | 26.4±6 |
DCM, dilated cardiomyopathy; LVEF, left ventricular ejection fraction; NYHA, New York Heart Association Classification.
Identification of a Genetic Variant in the Human PLN Promoter Region
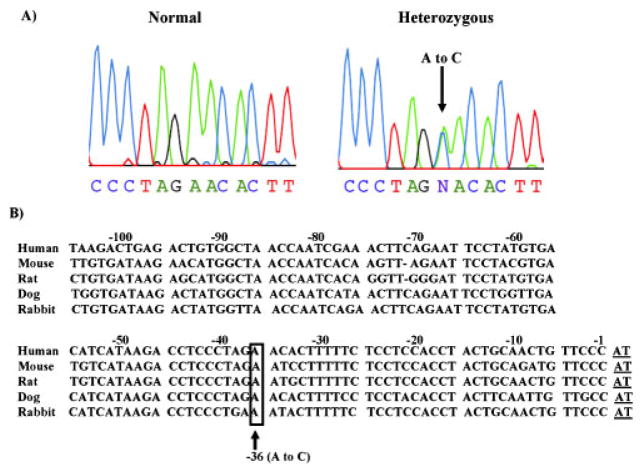
In the initial discovery study, the PLN gene promoter region 600 bp upstream from the transcriptional start site [McTiernan et al., 1999a] was sequenced in 40 unrelated Greek DCM patients. The sequencing of this region identified a single nucleotide transition from A>C at position AF177763.1:g.203A>C (at −36bp relative to the PLN transcriptional start site: −36A>C) (Fig. 1A). We subsequently screened an additional 178 Greek DCM patients to determine the frequency of this PLN genetic variant. The −36A>C substitution was found in another 15 patients (16/218 total) and it was only present in the heterozygous form, reflecting an allelic frequency of 3.66% in the Greek DCM population. To confirm the presence of this novel PLN promoter variant in a different heart failure population, 163 Caucasian DCM patients (University Hospital, Cincinnati Heart Failure/Transplant Program, University of Cincinnati, OH) were also screened. The −36A>C variant was found in the heterozygous form in six patients, reflecting an allelic frequency of 1.84%. The characteristics of the patients with the identified transition in the PLN gene in the two cohorts were similar (Table 2). The PLN −36A>C variant carriers presented with heart failure symptoms and were diagnosed with cardiomyopathy at ages ranging from 18 to 44 years. Echocardiography studies indicated severe left ventricular dilatation and systolic dysfunction (e.g., ejection fraction of 22±9%). Their symptoms remained under control with drug treatments. However, some patients’ symptoms progressively deteriorated (New York Heart Association [NYHA] Classification, NYHA class III), leading to the death of one patient at the age of 48 years and heart transplantation in another patient at the age of 46 years. The promoter variant −36A>C was found in only 1 normal control subject out of 296 screened. There were no departures from Hardy-Weinberg equilibrium for allelic frequencies in either DCM or control populations.
FIGURE 1.
Genomic DNA sequence analysis of the PLN promoter region. A: Partial nucleotide sequences of the PLN promoter region in normal subjects and DCM patients heterozygous for the AF177763.1:g.203A>C (at −36bp from transcriptional start site: −36A>C) substitution. B: Sequence comparison of the proximal mammalian PLN promoter sequences was performed by the FASTA program (http://fasta.bioch.virginia.edu/fasta_www2/fasta_list2.shtml) (GenBank reference sequence numbers AF177763.1, AF037348.1, L03381.1, and M63600.1). The numbers correspond to human nucleotides upstream of exon 1 (transcription start site, underlined). The position of the A>C transition (boxed) is indicated. Gaps are shown by dash. Polymorphism numbering is based on using the GenBank accession numberAF177763.1for human PLN sequence corresponding to proximal promoter and exon1and the transcription start site as a reference. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
TABLE 2.
Clinical Characteristics of Dilated Cardiomyopathy Patients With the −36A>C Substitution in the PLN Gene*
| Wild-type allele | Mutant allele | |
|---|---|---|
| n | 359 | 22 |
| Age (years) | 42±3.1 | 40±6 |
| Etiology (%) | ||
| Dilated cardiomyopathy | 94.27 | 100 |
| Functional class (% NYHA III/IV) | 67.2 | 66.66 |
| LVEF (%) | 24.5±8 | 22±9 |
Polymorphism numbering is based on using the GenBank accession number AF177763.1 for human PLN sequence, corresponding to proximal promoter and exon 1 as a reference.
DCM, dilated cardiomyopathy; LVEF, left ventricular ejection fraction; NYHA, New York Heart Association Classification.
The promoter region of the human PLN gene, containing the genetic variant is a highly conserved region among species (Fig. 1B) [McTiernan et al., 1999a]. Therefore, it was hypothesized that this change in nucleotide sequence might alter PLN promoter activity and consequently its regulation of SERCA2a, and thus contribute to the pathophysiology of heart failure.
In Vitro Assays of PLN Promoter Activity
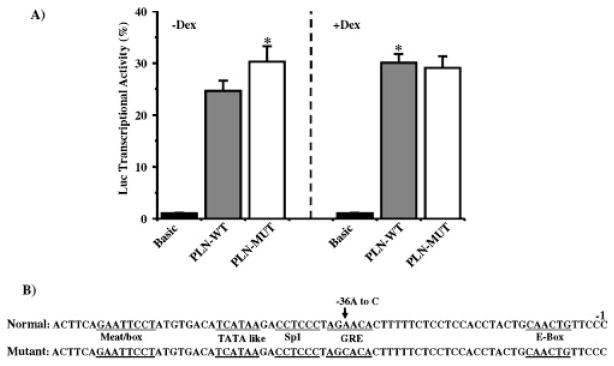
To determine the potential functional importance of the identified genetic variation on PLN transcriptional regulation, we generated reporter constructs that expressed luciferase under the control of the putative promoter sequences from the human PLN gene. When neonatal rat cardiomyocytes were transiently transfected with luciferase reporters under the control of wild-type (PLN-WT) or “mutant” PLN (PLN-MT) promoters, the −36A>C transition resulted in a significant increase of 24% in transcriptional activity, compared to the wild-type promoter (Fig. 2A). To examine whether the −36A>C point transition in the PLN gene may alter regulation by any of the sequence-specific DNA-binding proteins, such as transcription factors, we performed a computer sequence search for putative regulatory binding sites. We identified a potential sequence for the glucocorticoid response element (GRE) within the mouse PLN promoter (Fig. 2B). Our DNA scanning revealed that the −36A>C substitution was within the putative glucocorticoid receptor binding site of the PLN promoter gene. To further investigate the regulation of PLN gene expression by the glucocorticoid response element, the luciferase reporter constructs of PLN-WT and PLN-MT were transiently transfected into rat neonatal cardiac cells in the absence or presence of dexamethasone. The induced luciferase activity of PLN-WT was significantly increased when dexamethasone was present, while there was no effect of dexamethasone on the PLN-MT, compared to basal levels (Fig. 2A). The lack of luciferase activity induction in PLN-MT following stimulation of transfected cells by dexamethasone may indicate that the genetic variant abolished the direct or indirect mediation of the dexamethasone-mediated enhancement of the reporter gene activity.
FIGURE 2.
Effect of the −36A>C genetic variant on human PLN promoter activity. A: Rat neonatal cardiomyocytes were transiently transfected with a luciferase expression vector driven by PLN-WT or PLN-MT (−36A>C) promoters, and were cultured in the absence (left) or presence (right) of 3 μM dexamethasone (Dex) for 45 hr. Transcriptional activity of the promoters was defined as a ratio of firefly luciferase activity to Renilla luciferase activity in the same cells, and normalized to the mean basal transcriptional activity of the promoter-less pGL3-basic vector. B: Sequence alignment of the normal and mutant human PLN upstream promoter regions. The relative positions of the promoter starting site (-1) and of the potential regulatory sequences (underlined) are indicated. The values are expressed as means±SEM (n=7). *P<0.05 vs. PLN-WT without Dex (two-way ANOVA and Student-Neuman-Keuls test). Polymorphism numbering is based on using the GenBank accession number AF177763.1for human PLN-sequence corresponding to proximal promoter and exon1as a reference.
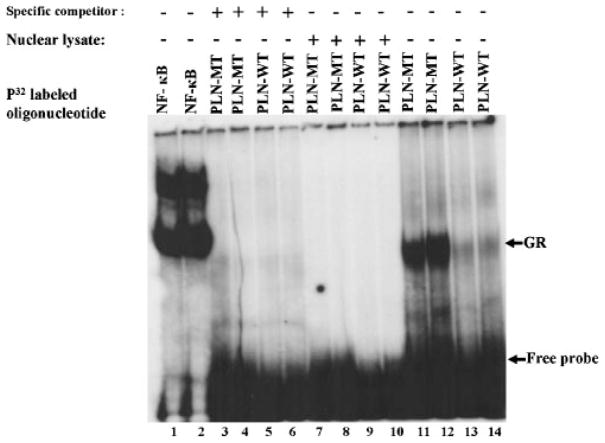
To further examine the functional significance of the −36A>C PLN promoter variant, gel mobility shift assays were employed. Using nuclear extracts from mouse heart, the binding assays showed that both synthetic WT (Fig. 3; lanes 13 and 14) and MT (Fig. 3; lanes 11 and 12) oligonucleotides were able to form a DNA-protein complex, indicating transcription factor binding. However, stronger binding was observed with the PLN-MT oligonucleotide, demonstrating that this sequence has a higher affinity for transcription factor binding. Binding was completely blocked in the presence of 100-fold excess of the cold-labeled WT (Fig. 3; lanes 3 and 4) or MT oligonucleotide (Fig. 3; lanes 5 and 6), used as specific competitors. Nuclear lysate was used as a negative control and it did not form any complexes in the presence of either synthetic WT or MT oligonucleotide (Fig. 3; lanes 7–10). However, an oligonucleotide containing a consensus NF-kB binding site, used as a positive control for nuclear lysate activity, yielded DNA-protein complexes in the lysates. These findings suggest that the quality of the nuclear lysates and the binding conditions were appropriate (Fig. 3; lanes 1 and 2).
FIGURE 3.
Electrophoretic mobility gel shift assay of wild-type and genetically-altered glucocorticoid elements in the PLN promoter sequences. Electrophoretic mobility gel shift assays were used to determine DNA–protein complex formation using nuclear extracts from mouse hearts. NF-k B was used as a positive technical control (lanes1and 2); nonlabeled wild-type (PLN-WT, lanes 3 and 4) and altered (PLN-MT, lanes 5 and 6) were used as specific competitors; and nuclear lysate as a negative control (lanes 7–10); PLN-MT oligonucleotide (lanes11and12) and PLN-WT oligonucleotide (lanes13 and14). Duplicate samples were assayed for each treatment.
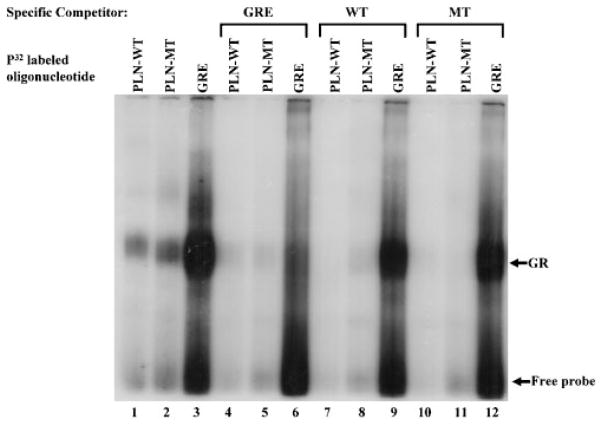
To verify whether the binding activity from heart nuclear extracts reflects a specific interaction between the GR with the PLN-WT and PLN-MT probes, as predicted based upon computer searches, we employed a commercially available oligonucleotide, containing a known consensus GRE sequence. This oligonucleotide was used in DNA binding and competition studies, designed to assess specificity of our DNA-protein complexes. The GRE consensus oligonucleotide displayed a strong DNA-protein binding complex in nuclear extracts (Fig. 4; lane 3). Furthermore, this commercially available oligonucleotide could completely block DNA-protein complex formation with PLN-WT, PLN-MT, and the GRE oligonucleotide in the nuclear extracts (100-fold excess; Fig. 4; lanes 4–6). The consensus GRE containing oligonucleotide and the PLN promoter-derived sequences (PLN-WT and PLN-MT) demonstrated identical migration of the DNA-protein complexes (Figs. 3 and 4). It was interesting to note that the PLN-WT or PLN-MT oligonucleotides could not completely compete with the common glucocorticoid consensus (Fig. 4; lanes 9 and 12). Taken together, these results indicate that the GR binds specifically to the PLN-WT and PLN-MT promoter sequences, albeit at lower affinity than the consensus GRE employed.
FIGURE 4.
Electrophoretic mobility gel shift assay of the PLN wild-type and genetically-altered glucocorticoid element specificity in the presence of common glucocorticoid consensus. Consensus glucocorticoid receptor element (GRE), PLN-WT, and PLN-MT motifs were used to examine binding specificity in cardiac nuclear extracts from wild-type mice. Lanes1–3: cardiac nuclear extracts reacted with radiolabeled oligonucleotides of PLN-WT, PLN-MT, and consensus GRE sequences. Lanes 4–6:consensus GRE oligonucleotides were used as specific competitor (100 × unlabeled GRE oligonucleotides). Lanes 7–9 and lanes 10–12: PLN-WT and PLN-MT oligonucleotides were used as competitors (100×-fold unlabeled oligonucleotide), respectively.
DISCUSSION
In this study, we identified a novel variant (−36A>C) in the human PLN promoter region in 22 heart failure patients and one normal subject, which appears to enhance promoter activity and alter the GR receptor binding element. Importantly, this PLN promoter variant was identified in two heart failure populations. The allelic frequencies in two ethnic populations and in controls were in Hardy-Weinberg equilibrium, indicating that this genetic variant is heritable and a combination of the −36A>C PLN variant with other genetic and environmental modifies may contribute to the time course of the disease in the patients. The identified nucleotide substitution is in close proximity to the putative TATA (5′-TCATAA-3′) boxes at position −48 to −53 in an evolutionarily conserved PLN gene region between species, and may play a significant role in regulating PLN gene expression. Indeed, in vitro studies of this genetic variant indicated that it may increase PLN expression levels and consequently, depress SR Ca cycling associated with cardiomyopathy. The functional significance of increased PLN levels in cardiac muscle has previously been demonstrated through the generation and characterization of transgenic mouse models [Kadambi et al., 1996; Dash et al., 2001]. Consistent with findings in transgenic mice, an increase in the apparent stoichiometry of PLN to SERCA2a, as a result of the PLN promoter genetic variant, may contribute to the depressed Ca cycling and deterioration of cardiac function.
Recently, there has been a considerable upsurge of interest in the influence of cis-acting genetic variations on gene transcription. Furthermore, these mutations and polymorphisms, found in various gene promoter regions, have been reported to affect gene expression and impact function [Collins et al., 2003; Hudson, 2003; Buckland et al., 2004; Guy et al., 2004; Schulz et al., 2006]. Importantly, the PLN promoter variant (A>C, underlined below), identified herein, was within a novel consensus sequence segment that matched a glucocorticoid receptor-binding site (5′-AGAAGA-3′). Previous studies have shown that thyroid hormone and glucocorticoids regulate the expression of several genes, including calcium cycling proteins [Kiss et al., 1994, 1998; Smith and Smith, 1994; Brittsan et al., 1999; Muangmingsuk et al., 2000]. Thyroid hormone was reported to mediate changes in PLN protein levels [Kiss et al., 1994, 1998; Brittsan et al., 1999] possibly through interaction with thyroid hormone elements residing in the PLN promoter region. Glucocorticoids downregulate Na-Ca exchanger mRNA levels and activity in aortic myocytes [Smith and Smith, 1994], while they increase expression of alpha-myosin heavy chain (MHC) and decreased expression of beta-MHC in neonatal rat cardiomyocytes [Muangmingsuk et al., 2000]. These changes suggest that, similar to thyroid hormone-mediated transcriptional activation, the glucocorticoid effects may also be mediated in part through transcriptional mechanisms. Indeed, the level of PLN transcripts was significantly decreased, when rat neonatal cardiomyocytes were treated with cytokines (interleukin [IL]-1β, tumor necrosis factor [TNF]) [McTiernan et al., 1997], while dexamethasone significantly elevated the levels of PLN transcripts [McTiernan et al., 1997], indicating the direct effects of dexamethasone on PLN gene regulation. In this report, similar results were obtained with dexamethasone induction of PLN-WT promoter expression. In contrast, dexamethasone did not increase the luciferase transcriptional activity of the PLN-MT promoter, suggesting that the −36A>C substitution may have abolished the interaction site for glucocorticoid receptor elements in the PLN gene.
The role of transacting elements in the transcriptional activity of the PLN gene remains poorly understood and the nuclear proteins involved in the regulation of the gene through binding to these elements are unknown. Our previous studies on characterization of the mouse PLN promoter indicated that 200 bp proximal to the transcriptional initiation site is sufficient for moderate (40%) expression of PLN levels [Haghighi et al., 1997]. The dexamethasone-responsive PLN gene sequences are located within the 200-bp proximal promoter region of the mouse and human PLN gene, which are highly conserved between species [Haghighi et al., 1997; McTiernan et al., 1999a]. Increased luciferase activity in the promoter-reporter studies suggest that GREs within this region may contribute to the modulation of transcriptional regulation via DNA–protein interactions of the PLN gene as further supported by electrophoretic mobility gel shift assay studies. Obviously, the limitation of this study is that the upregulation of the PLN promoter activity presented here is primarily from in vitro studies; in vivo relevance of these finding could not be performed due to lack of cardiac biopsies from affected individuals.
The glucocorticoid receptor is a ligand-dependent transcription factor with both hormone and DNA binding domains, affecting the transcription of specific genes [Schoneveld et al., 2004]. Specifically, glucocorticoid hormones are the major mediators of systemic stress responses [Brent et al., 1991] and it has been suggested that they may regulate SR function and cellular calcium homeostasis in the myocardium [Rao et al., 2001; Aoyama et al., 2005]. The possible mechanisms may involve modulation of PLN phosphorylation through Ca/calmodulin-dependent protein kinase II (CaM kinase II) [Rao et al., 2001]. Interestingly, the CaM kinase II dependent phosphorylation site of PLN, Thr17, has been implicated in stress responses of the cardiomyocytes [Hagemann et al., 2000; Zhao et al., 2004]. Therefore, under stress conditions, GR modulation of PLN activity and/or expression levels may influence SR Ca cycling and myocardial function, which may be beneficial during early cardiac remodeling but deleterious under pathophysiological conditions. However, the abolished PLN GRE site by the −36A>C genetic variant eliminates the GR-mediated regulation, resulting in chronic increases in PLN expression levels and inhibition of SERCA activity, which may accelerate deterioration of function in DCM.
Acknowledgments
Grant sponsors: Leducq Foundation; National Institutes of Health (NIH); Grant numbers: HL-77101; HL-026057; HL-64018; HL-77101; and HL-52318.
We thank all the patients for their participation and we thank the physicians in the Department of Medicine at the University of Cincinnati, Cincinnati, OH, and the Onassis Cardiac Surgery Center in Athens, Greece, for their collaboration in this study. This research was supported by grants from the NIH (HL-77101 [to E.G.K. and G.W.D.]; HL-026057 [to E.G.K.]; HL-64018 [to E.G.K.]; and HL-52318 [to G.W.D.]) and from the Leducq Foundation (to E.G.K. and G.W.D.).
References
- Aoyama T, Matsui T, Novikov M, Park J, Hemmings B, Rosenzweig A. Serum and glucocorticoid-responsive kinase-1 regulates cardiomyocyte survival and hypertrophic response. Circulation. 2005;111:1652–1659. doi: 10.1161/01.CIR.0000160352.58142.06. [DOI] [PubMed] [Google Scholar]
- Beuckelmann DJ, Nabauer M, Erdmann E. Intracellular calcium handling in isolated ventricular myocytes from patients with terminal heart failure. Circulation. 1992;85:1046–1055. doi: 10.1161/01.cir.85.3.1046. [DOI] [PubMed] [Google Scholar]
- Brent GA, Moore DD, Larsen PR. Thyroid hormone regulation of gene expression. Annu Rev Physiol. 1991;53:17–35. doi: 10.1146/annurev.ph.53.030191.000313. [DOI] [PubMed] [Google Scholar]
- Brittsan AG, Kiss E, Edes I, Grupp IL, Grupp G, Kranias EG. The effect of isoproterenol on phospholamban-deficient mouse hearts with altered thyroid conditions. J Mol Cell Cardiol. 1999;31:1725–1737. doi: 10.1006/jmcc.1999.1010. [DOI] [PubMed] [Google Scholar]
- Brittsan AG, Carr AN, Schmidt AG, Kranias EG. Maximal inhibition of SERCA2 Ca affinity by phospholamban in transgenic hearts overexpressing a non-phosphorylatable form of phospholamban. J Biol Chem. 2000;275:12129–12135. doi: 10.1074/jbc.275.16.12129. [DOI] [PubMed] [Google Scholar]
- Brown M, McGuinness M, Wright T, Ren X, Wang Y, Boivin GP, Hahn H, Feldman AM, Jones WK. Cardiac-specific blockade of NF-kappaB in cardiac pathophysiology: differences between acute and chronic stimuli in vivo. Am J Physiol Heart Circ Physiol. 2005;289:H466–H476. doi: 10.1152/ajpheart.00170.2004. [DOI] [PubMed] [Google Scholar]
- Buckland PR, Coleman SL, Hoogendoorn B, Guy C, Smith SK, O’Donovan MCA. High proportion of chromosome 21 promoter polymorphisms influences transcriptional activity. Gene Expr. 2004;11:233–239. doi: 10.3727/000000003783992225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien KR. Genomic circuits and the integrative biology of cardiac diseases. Nature. 2000;407:227–232. doi: 10.1038/35025196. [DOI] [PubMed] [Google Scholar]
- Collins FS, Green ED, Guttmacher AE, Guyer MS. A vision for the future of genomics research. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- Dash R, Kadambi V, Schmidt AG, Tepe NM, Biniakiewicz D, Gerst MJ, Canning AM, Abraham WT, Hoit BD, Liggett SB, Lorenz JN, Dorn GW, II, Kranias EG. Interactions between phospholamban and beta-adrenergic drive may lead to cardiomyopathy and early mortality. Circulation. 2001;103:889–896. doi: 10.1161/01.cir.103.6.889. [DOI] [PubMed] [Google Scholar]
- Franz WM, Muller OJ, Katus HA. Cardiomyopathies: from genetics to the prospect of treatment. Lancet. 2001;358:1627–1637. doi: 10.1016/S0140-6736(01)06657-0. [DOI] [PubMed] [Google Scholar]
- Fujii J, Zarain-Herzberg A, Willard HF, Tada M, MacLennan DH. Structure of the rabbit phospholamban gene, cloning of the human cDNA, and assignment of the gene to human chromosome 6. J Biol Chem. 1991;266:11669–11675. [PubMed] [Google Scholar]
- Geisterfer-Lowrance AA, Christe M, Conner DA, Ingwall JS, Schoen FJ, Seidman CE, Seidman JG. A mouse model of familial hypertrophic cardiomyopathy. Science. 1996;272:731–734. doi: 10.1126/science.272.5262.731. [DOI] [PubMed] [Google Scholar]
- Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportions for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- Guy CA, Hoogendoorn B, Smith SK, Coleman S, O’Donovan MC, Buckland PR. Promoter polymorphisms in glutathione-S-transferase genes affect transcription. Pharmacogenetics. 2004;14:45–51. doi: 10.1097/00008571-200401000-00005. [DOI] [PubMed] [Google Scholar]
- Hagemann D, Kuschel M, Kuramochi T, Zhu W, Cheng H, Xiao RP. Frequency-encoding Thr17 phospholamban phosphorylation is independent of Ser16 phosphorylation in cardiac myocytes. J Biol Chem. 2000;275:22532–22536. doi: 10.1074/jbc.C000253200. [DOI] [PubMed] [Google Scholar]
- Haghighi K, Kadambi VJ, Koss KL, Luo W, Harrer JM, Ponniah S, Zhou Z, Kranias EG. In vitro and in vivo promoter analyses of the mouse phospholamban gene. Gene. 1997;203:199–207. doi: 10.1016/s0378-1119(97)00514-3. [DOI] [PubMed] [Google Scholar]
- Haghighi K, Kolokathis F, Pater L, Lynch RA, Asahi M, Gramolini AO, Fan GC, Tsiapras D, Hahn HS, Adamopoulos S, Liggett SB, Dorn GW, II, MacLennan DH, Kremastinos DT, Kranias EG. Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J Clin Invest. 2003;111:869–876. doi: 10.1172/JCI17892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighi K, Kolokathis F, Gramolini AO, Waggoner JR, Pater L, Lynch RA, Fan GC, Tsiapras D, Parekh RR, Dorn GW, II, MacLennan DH, Kremastinos DT, Kranias EG. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc Natl Acad Sci USA. 2006;103:1388–1393. doi: 10.1073/pnas.0510519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenfuss G. Alterations of calcium-regulatory proteins in heart failure. Cardiovasc Res. 1998;37:279–289. doi: 10.1016/s0008-6363(97)00277-0. [DOI] [PubMed] [Google Scholar]
- Hudson TJ. Wanted: regulatory SNPs. Nat Genet. 2003;33:439–440. doi: 10.1038/ng0403-439. [DOI] [PubMed] [Google Scholar]
- Ito K, Yan X, Feng X, Manning WJ, Dillmann WH, Lorell BH. Transgenic expression of sarcoplasmic reticulum Ca-ATPase modifies the transition from hypertrophy to early heart failure. Circ Res. 2001;89:422–429. doi: 10.1161/hh1701.095522. [DOI] [PubMed] [Google Scholar]
- Kadambi VJ, Ponniah S, Harrer JM, Hoit BD, Dorn GW, II, Walsh RA, Kranias EG. Cardiac-specific overexpression of phospholamban alters calcium kinetics and resultant cardiomyocyte mechanics in transgenic mice. J Clin Invest. 1996;97:533–539. doi: 10.1172/JCI118446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss E, Jakab G, Kranias EG, Edes I. Thyroid hormone-induced alterations in phospholamban protein expression. Regulatory effects on sarcoplasmic reticulum Ca transport and myocardial relaxation. Circ Res. 1994;75:245–251. doi: 10.1161/01.res.75.2.245. [DOI] [PubMed] [Google Scholar]
- Kiss E, Brittsan AG, Edes I, Grupp IL, Grupp G, Kranias EG. Thyroid hormone-induced alterations in phospholamban-deficient mouse hearts. Circ Res. 1998;83:608–613. doi: 10.1161/01.res.83.6.608. [DOI] [PubMed] [Google Scholar]
- Kogler H, Hartmann O, Leineweber K, Nguyen P, Schott P, Brodde OE, Hasenfuss G. Mechanical load-dependent regulation of gene expression in monocrotaline-induced right ventricular hypertrophy in the rat. Circ Res. 2003;93:230–237. doi: 10.1161/01.RES.0000085042.89656.C7. [DOI] [PubMed] [Google Scholar]
- Levy D, Garrison RJ, Savage DD, Kannel WB, Castell WP. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N Engl J Med. 1990;322:1561–1566. doi: 10.1056/NEJM199005313222203. [DOI] [PubMed] [Google Scholar]
- Luo W, Grupp IL, Harrer J, Ponniah S, Grupp G, Duffy JJ, Doetschman T, Kranias EG. Targeted ablation of the phospholamban gene is associated with markedly enhanced myocardial contractility and loss of beta-agonist stimulation. Circ Res. 1994;75:401–409. doi: 10.1161/01.res.75.3.401. [DOI] [PubMed] [Google Scholar]
- Luo W, Wolska BM, Grupp IL, Harrer JM, Haghighi K, Ferguson DG, Slack JP, Grupp G, Doetschman T, Solaro RJ, Kranias EG. Phospholamban gene dosage effects in the mammalian heart. Circ Res. 1996;78:839–847. doi: 10.1161/01.res.78.5.839. [DOI] [PubMed] [Google Scholar]
- MacLennan DH, Kranias EG. Phospholamban: a crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol. 2003;4:566–577. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- McTiernan CF, Bonnie H, Lemster Frye C, Brooks S, Combes A, Feldman AM. Interleukin-1β inhibits phospholamban gene expression in cultured cardiomyocytes. Circ Res. 1997;81:493–503. doi: 10.1161/01.res.81.4.493. [DOI] [PubMed] [Google Scholar]
- McTiernan CF, Frye CS, Lemster BH, Kinder EA, Ogletree-Hughes ML, Moravec CS, Feldman AM. The human phospholamban gene: structure and expression. Mol Cell Cardiol. 1999a;31:679–692. doi: 10.1006/jmcc.1998.0904. [DOI] [PubMed] [Google Scholar]
- McTiernan CF, Lemster BH, Frye CS, Johns DC, Feldman AM. Characterization of proximal transcription regulatory elements in the rat phospholamban promoter. J Mol Cell Cardiol. 1999b;31:2137–2153. doi: 10.1006/jmcc.1999.1042. [DOI] [PubMed] [Google Scholar]
- Meyer M, Schillinger W, Pieske B, Holubarsch C, Heilmann C, Posival H, Kuwajima G, Mikoshiba K, Just H, Hasenfuss G. Alterations of sarcoplasmic reticulum proteins in failing human dilated cardiomyopathy. Circulation. 1995;92:778–784. doi: 10.1161/01.cir.92.4.778. [DOI] [PubMed] [Google Scholar]
- Mills GD, Kubo H, Harris DM, Berretta RM, Piacentino V, III, Houser SR. Phosphorylation of phospholamban at threonine-17 reduces cardiac adrenergic contractile responsiveness in chronic pressure overload-induced hypertrophy. Am J Physiol Heart Circ Physiol. 2006;291:H61–H70. doi: 10.1152/ajpheart.01353.2005. [DOI] [PubMed] [Google Scholar]
- Minamisawa S, Sato Y, Tatsuguchi Y, Fujino T, Imamura S, Uetsuka Y, Nakazawa M, Matsuoka R. Mutation of the phospholamban promoter associated with hypertrophic cardiomyopathy. Biochem Biophys Res Commun. 2003;304:1–4. doi: 10.1016/s0006-291x(03)00526-6. [DOI] [PubMed] [Google Scholar]
- Muangmingsuk S, Ingram P, Gupta MP, Arcilla RA, Gupta M. Dexamethasone induced cardiac hypertrophy in newborn rats is accompanied by changes in myosin heavy chain phenotype and gene transcription. Mol Cell Biochem. 2000;209:165–173. doi: 10.1023/a:1007128300430. [DOI] [PubMed] [Google Scholar]
- Rao MK, Xu A, Narayanan N. Glucocorticoid modulation of protein phosphorylation and sarcoplasmic reticulum function in rat myocardium. Am J Physiol Heart Circ Physiol. 2001;281:H325–H333. doi: 10.1152/ajpheart.2001.281.1.H325. [DOI] [PubMed] [Google Scholar]
- Richardson P, McKenna W, Bristow M, Maisch B, Mautner B, O’Connell J, Olsen E, Thiene G, Goodwin J, Gyarfas I, Martin I, Nordet P. Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the definition and classification of cardiomyopathies. Circulation. 1996;93:841–842. doi: 10.1161/01.cir.93.5.841. [DOI] [PubMed] [Google Scholar]
- Schmitt JP, Kamisago M, Asahi M, Li GH, Ahmad F, Mende U, Kranias EG, MacLennan DH, Seidman JG, Seidman CE. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299:1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- Schoneveld OJ, Gaemers IC, Lamers WH. Mechanisms of glucocorticoid signalling. Biochim Biophys Acta. 2004;1680:114–128. doi: 10.1016/j.bbaexp.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Schulz V, Hendig D, Henjakovic M, Szliska C, Kleesiek K, Gotting C. Mutational analysis of the ABCC6 gene and the proximal ABCC6 gene promoter in German patients with pseudoxanthoma elasticum (PXE) Hum Mutat. 2006;27:831–844. doi: 10.1002/humu.9444. [DOI] [PubMed] [Google Scholar]
- Seidman JG, Seidman C. Genetic basis for cardiomyopathy from mutation identification to mechanistic paradigms. Cell. 2001;104:557–567. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- Simmerman HK, Jones LR. Phospholamban: protein structure, mechanism of action, and role in cardiac function. Physiol Rev. 1998;78:921–947. doi: 10.1152/physrev.1998.78.4.921. [DOI] [PubMed] [Google Scholar]
- Smith L, Smith JB. Regulation of sodium-calcium exchanger by glucocorticoid and growth factor in vascular smooth muscle. J Biol Chem. 1994;269:27527–27531. [PubMed] [Google Scholar]
- Zhao W, Uehara Y, Chu G, Song Q, Qian J, Young K, Kranias EG. Threonine-17 phosphorylation of phospholamban: a key determinant of frequency-dependent increase of cardiac contractility. J Mol Cell Cardiol. 2004;37:607–612. doi: 10.1016/j.yjmcc.2004.05.013. [DOI] [PubMed] [Google Scholar]