While L-amino acids are the building blocks for proteins synthesized in ribosomes in all kingdoms of life, D-amino acids (D-aa) have important non-ribosome based functions1. Mammals synthesize D-Ser and D-Asp, primarily in the central nervous system, where D-Ser is critical for neurotransmission2. Bacteria synthesize a largely distinct set of D-aa, which become integral components of the cell wall and are also released as free D-aa3,4. However, the impact of free microbial D-aa on host physiology at the host-microbial interface has not been explored. Here, we show that the mouse intestine is rich in free D-aa that are derived from the microbiota. Furthermore, the microbiota induces production of D-amino acid oxidase (DAO) by intestinal epithelial cells, including goblet cells, which secrete the enzyme into the lumen. Oxidative deamination of intestinal D-aa by DAO, which yields the antimicrobial product H2O2, protects the mucosal surface in the small intestine from the cholera pathogen. DAO also modifies the composition of the microbiota and is associated with microbial induction of intestinal sIgA. Collectively, these results identify D-aa and DAO as previously unrecognized mediators of microbe-host interplay and homeostasis on the epithelial surface of the small intestine.
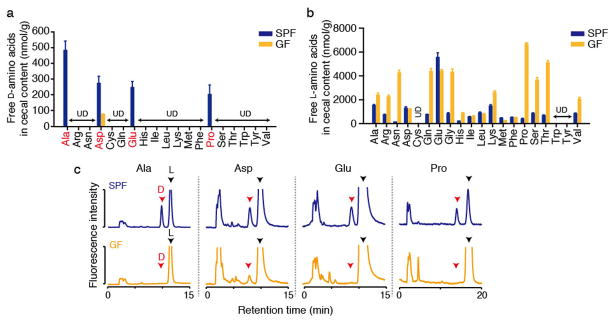
Mammals recognize and respond to diverse microbial products, including fragments of the cell wall5,6. However, the prevalence and role of free microbial D-aa on host physiology in the intestinal milieu has not been explored. Given the abundance and diversity of bacteria present in the mammalian intestinal tract7, we first assessed if microbe-generated free D-aa are present within the intestine. D- and L-forms of all proteinogenic amino acids were quantified by two-dimensional HPLC8 in the cecal contents of specific pathogen-free (SPF) mice and germ-free (GF) mice. D-Ala, D-Asp, D-Glu, and D-Pro were detected in SPF mice (~200–500 nmol/g), whereas ceca of GF mice contained only low levels of D-Asp (Fig. 1ac). In contrast, abundant L-amino acids were detected in both SPF and GF animals, but cecal L-amino acid levels were generally lower in SPF mice compared with GF mice (Fig. 1b), likely due to their utilization by the gut microbiota. Chow for SPF and GF mice contained comparable amounts of D/L-aa, and is thus not responsible for the disparity between GF and SPF animals in intestinal amino acid content (Supplementary Fig. 1ab). These observations, coupled with knowledge regarding host D-aa synthesis9, strongly suggest that free D-aa in the murine intestinal tract are primarily microbial products.
Figure 1. Free D-aa in the intestinal tract are produced by the gut microbiota.
Free D- and L-amino acids (a and b, respectively) were quantified in cecal contents from adult mice with resident microbiota (specific pathogen free; SPF) and from germ free (GF) mice using 2D-HPLC. (n = 3 mice each). ‘UD’, undetectable. Error bars, mean ± s.e.m. c, Representative chromatograms for D/L- Ala, Asp, Glu, and Pro.
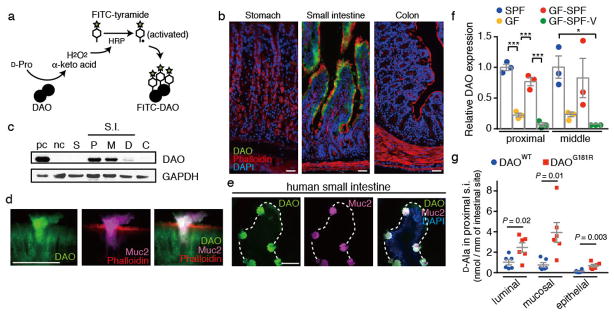
In mammals, endogenous D-aa levels are regulated by the action of D-amino acid oxidase (DAO), which converts neutral D-aa, such as D-Ser, into H2O2 and α-keto acids10. Given the abundance of microbiota-derived D-aa in the gut, we asked whether DAO, which is known to be primarily expressed in the CNS and kidney10, is also present in the intestine. Using an activity-based assay11, we detected DAO exclusively associated with the villous epithelium of the small intestine (SI) of SPF mice (Fig. 2ab and Supplementary Fig. 2ab). Immunoblotting confirmed the presence of DAO in the proximal and middle SI and its absence from other regions of the GI tract (Fig. 2c and Supplementary Fig. 2cd). In the proximal SI, DAO activity was detected primarily near the apical border of enterocytes and in goblet cells, and in the middle SI DAO activity was observed in both secretory vesicles of goblet cells and in mucus (Fig. 2d and Supplementary Fig. 2bfg). DAO was also detected within human goblet cells, and its presence in human small intestinal lysates was confirmed using immunoprecipitation and immunoblotting (Fig. 2e and Supplementary Fig. 3). These data demonstrate the expression of active DAO in the mouse and human intestine and that, compared to its localization in peroxisomes of CNS astrocytes10, some portion of intestinal DAO appears to be secreted into the lumen by goblet cells (Fig. 2d and Supplementary Figs. 2e–g). Bioinformatic analyses of the murine and human DAO amino acid sequences revealed the presence of a signal peptide and a predicted cleavage site near their respective N termini, consistent with their being secreted proteins (Supplementary Fig. 4).
Figure 2. Intestinal epithelial cells produce DAO in response to the microbiota.
a, Schematic depicting activity-based labeling of DAO. b, d, and e, Active DAO (green), f-actin (red), Muc2 (magenta) and nuclei (blue) were labeled in adult murine stomach, small intestine, and colon (b, d) and in human small intestine (e). Scale bars, 50 μm. Images are based on n = 5 (b), 5 (c), 10 (d), and 3 (e) biological replicates, respectively. c, Immunoblotting of DAO in epithelial lysates from mouse gut. pc/nc, positive (cerebellum)/negative (cerebral cortex) control; S, stomach; P, proximal; M, middle; D, distal; S.I., small intestine; C, colon. f, Relative DAO levels in epithelial lysates of murine proximal and middle small intestine (Supplementary Fig. 6ab). Specific pathogen free (SPF), germ free (GF), GF with SPF bedding (GF-SPF), and vancomycin-treated GF-SPF (GF-SPF-V) mice were assessed (n = 3 mice each). g, Abundance of D-Ala in epithelial, mucosal, and luminal samples from proximal small intestine of DAOWT and DAOG181R mice (n = 6 mice each). Error bars, mean ± s.e.m. *P < 0.05, **P < 0.01, and ***P < 0.001, analyzed with one-way ANOVA (f) or Wilcoxon Rank Sum test (g).
We investigated whether the microbiota, which generates DAO’s substrates, also influences production of intestinal DAO and found that enterocytes and goblet cells of SPF mice contained far more DAO than did those of GF mice (Fig. 2f and Supplementary Fig. 5a–c). Conventionalization of GF mice induced DAO to nearly the levels observed in SPF animals (Fig. 2f and Supplementary Fig. 6ab), providing additional support for the idea that the microbiota modulate production of DAO in the intestine. In particular, a vancomycin-sensitive subset of the microbiota appears linked to DAO production, both in conventionalized GF mice and in SPF mice, as DAO levels were markedly lower in animals treated with vancomycin than in untreated controls (Supplementary Fig. 6a–c). DAO levels were not reduced in the CNS or kidneys of GF mice relative to wt mice, suggesting that microbiota control of DAO expression is limited to the intestine (Supplementary Fig. 5de). The precise pathways by which the microbiota regulates DAO production in the intestine are unknown; however, induction appears to be largely independent of MyD88 and RIPK2, which mediate two major innate immune recognition pathways12 (Supplementary Fig. 6d).
Using mice harboring a natural point mutation (DAOG181R) that abolishes DAO activity13, we assessed whether DAO modulates intestinal D-aa content. In the proximal SI, where DAO activity is high, wild-type (WT) mice had lower luminal, mucosal and intra-epithelial concentrations of D-Ala compared to DAOG181R animals (Fig. 2g and Supplementary Fig. 1ceg). Differences were particularly marked for the mucosal and epithelial samples, compared to within the lumen, consistent with the fact that DAO activity requires oxygen, which is more readily available near the epithelial surface14. In contrast, SI samples from WT and mutant mice contained similar levels of D-Asp and D-Glu, which are not oxidized by DAO, as well as L-aa (Supplementary Fig. 1c–h). Consistent with the absence of DAO expression in the colon, we did not observe differences between fecal concentrations of D-aa in WT and mutant mice (Supplementary Fig. 1ij). Together, these data demonstrate that intestinal DAO can regulate the abundance of its substrate D-aa in a region-specific manner.
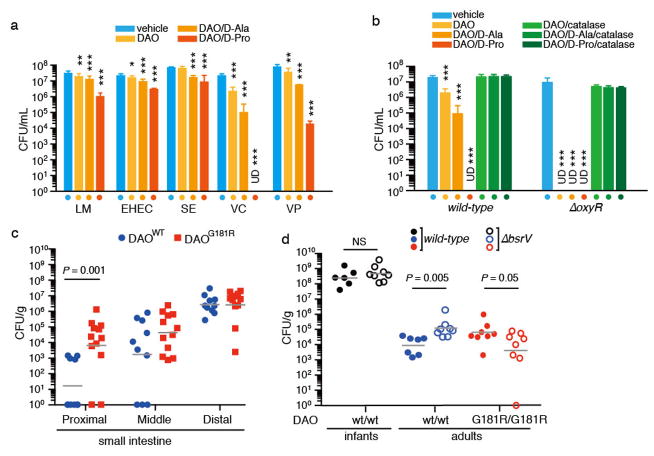
Since oxidative deamination of D-aa by DAO yields H2O2, which is known to have bactericidal activity and to be an important factor in host defense15, we tested the effect of DAO +/- D-aa on the growth of several common enteric pathogens. In vitro, purified porcine DAO supplemented with D-Pro or D-Ala had modest cytotoxicity for several organisms that colonize the GI tract, resulting in up to 30-fold reduction in colony forming units (CFU) (Fig. 3a). However, Vibrio cholerae and V. parahaemolyticus, diarrheal pathogens that colonize the SI and cause cholera and enteritis, respectively, were particularly sensitive to DAO and D-Pro. V. cholerae, a non-invasive pathogen that proliferates adjacent to but does not disrupt the epithelial barrier, was also susceptible to DAO alone, even without addition of D-aa, likely because the substantial amounts of D-aa it releases3 enable greater H2O2 production (Supplementary Fig. 7b–c). Consistent with this idea, we found that an amino acid racemase-deficient V. cholerae mutant (bsrV), which releases less D-aa than the wt strain3,16 and therefore generates less H2O2 in the presence of exogenous DAO (Supplementary Fig. 7d), shows reduced susceptibility to DAO toxicity in the absence of exogenous D-aa (Supplementary Fig. 7f). V. cholerae’s sensitivity to DAO (+/- D-aa) was blocked by catalase, an enzyme that destroys H2O2 (Fig. 3b), providing further support for the idea that DAO kills V. cholerae via production of H2O2. In addition, V. cholerae mutants lacking oxyR, which encodes a positive regulator of H2O2-inducible genes17, showed enhanced sensitivity to DAO (Fig. 3b). Collectively, these findings demonstrate that in vitro, DAO can exert bactericidal effects on several enteric pathogens through production of H2O2 from oxidation of D-aa.
Figure 3. DAO has antimicrobial activity in vitro and in vivo.
a, The indicated bacterial species were treated with vehicle, DAO, or DAO and D-aa, and surviving CFU were enumerated. LM, Listeria monocytogenes; EHEC, enterohemorrhagic Escherichia coli; SE, Salmonella enterica; VC, Vibrio cholerae; VP, Vibrio parahaemolyticus. b, Wild-type and ΔoxyR V. cholerae were treated as in (a) or treatment was supplemented with catalase (a, b; n = 3 biological replicates each). Addition of D-aa alone had no effect on bacterial viability (Supplementary Fig. 7e). c Intestinal colonization of streptomycin-treated adult DAOWT or DAOG181R mice by WT V. cholerae (n = 10, 12 mice); d, Intestinal colonization of infant or streptomycin-treated adult DAOWT or DAOG181R mice by WT or ΔbsrV V. cholerae (n = 6, 8, 7, 8, 8, 8 mice, from left to right group, respectively). Geometric means (a–d) with 95% confidence interval (a, b) are shown. ‘UD’, undetectable. *P < 0.05, **P < 0.01, and ***P < 0.001, analyzed with one-way ANOVA (a, b) or Wilcoxon Rank Sum test (c, d).
Previous studies have suggested that DAO may contribute to systemic defense against pathogens via the activity of neutrophil DAO18,19. To test whether DAO activity is important to mucosal defense in vivo, we explored the effect of DAO and D-aa on V. cholerae intestinal colonization in streptomycin-treated adult mice. Adult animals were used rather than the infants commonly used to study V. cholerae colonization because infant mice produce markedly less DAO and no detectable secreted DAO (Supplementary Fig. 8). Strikingly, comparison of V. cholerae intestinal colonization in wt and DAOG181R littermate mice from heterozygote breeders revealed that CFU were ~1000x more abundant in the proximal SI of DAOG181R animals (Fig. 3c). In contrast, colonization of the distal SI, which does not express DAO, did not differ for the wt and mutant animals. These data strongly suggest that the presence of DAO has a profound effect on V. cholerae’s ability to survive in the proximal SI. To further investigate the effects of D-aa oxidation on mucosal defense, we compared colonization of wt and DAOG181R animals by WT and bsrV V. cholerae. In adult wt mice, the bsrV mutant colonized the proximal/middle SI to a greater extent (~15x) than did WT V. cholerae. No such increased colonization by the bsrV mutant was detected in infant wt or adult DAOG181R mice (Fig. 3d). Collectively, these results indicate that enhanced colonization by ΔbsrV V. cholerae is not an intrinsic feature of this strain, and are consistent with the idea that V. cholerae survival in vivo is impaired by the presence of active DAO and its microbially generated D-aa substrates. Infection of adult mice with V. cholerae does not induce intestinal pathology, so the effect of DAO on disease can’t be assessed, but DAO is the first intestinal epithelial-derived factor found to restrict V. cholerae intestinal colonization. Our data raises the possibility that DAO plays a general role in protection at the mucosal surface.
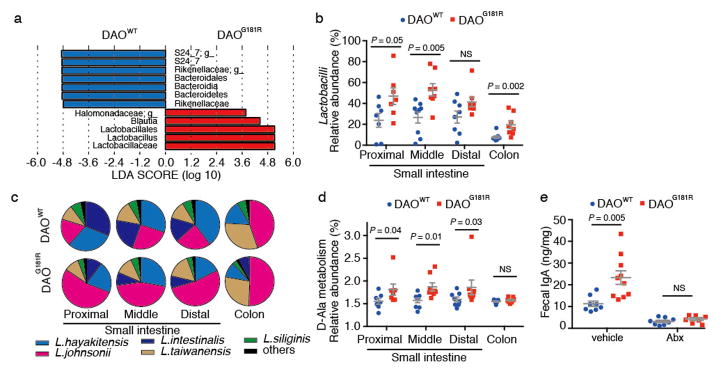
To further investigate the effect of DAO on intestinal microbes, metagenomic analysis of 16s rRNA gene sequences was used to characterize the composition of the intestinal microbiota in DAOWT and DAOG181R littermate mice from heterozygote breeders. While the overall beta-diversity and total numbers of intestinal bacteria did not differ between groups (Supplementary Fig. 9ab), DAO null animals had an increased abundance of Lactobacillales and decreased abundance of Bacteroidales in both the proximal and middle SI (Fig. 4ab and Supplementary Fig. 9c–e). The elevated abundance of Lactobacilli in the SI was also detected using fluorescent in situ hybridization (Supplementary Fig. 9f). Thus, DAO activity in the intestinal tract modulates the composition of the microbiota as well as the abundance of the cholera pathogen. The increased abundance of commensal Lactobacillales in DAOG181R mice could in principle reflect decreased H2O2-mediated killing of these commensal organisms. However, Lactobacillales are relatively resistant to H2O220 and we found that L. johnsonii, which was particularly prominent in DAOG181R mice (Fig. 4c and Supplementary Fig. 9g) was not killed when aerobically mixed DAO and D-Ala was added to an anaerobic culture (Supplementary Fig. 10a). Thus, additional mechanisms besides direct killing mediated by H2O2 likely contribute to the altered microbiota in the absence of DAO. Such mechanisms may include the capacity of the elevated D-Ala in DAO null animals to support growth of particular bacterial species such as L. johnsonii, an Ala auxotroph21,22 (Supplementary Fig. 10b). Indeed, PICRUSt23/HUMAnN24-based comparisons of the potential metabolic capacities of the microbiota found in the small intestine of DAOWT and DAOG181R mice suggest that there is greater potential for D-Ala metabolism in the DAO null animals (Fig. 4d and Supplementary Fig. 10d). However, utilization of D-Ala or other D-aa as nutrients is unlikely to account for the enhanced growth of V. cholerae in the DAOG181R mice, since supplementation of in vitro cultures with D-Ala did not alter the pathogen’s growth (Supplementary Fig. 10c).
Figure 4. Genetic inactivation of DAO alters microbiota composition and secretory IgA levels.
a, LEfSe-based29 comparisons of the relative abundance of bacterial taxa in proximal/middle small intestine from DAOWT and DAOG181R mice. Taxa significantly enriched in DAOWT and DAOG181R mice are depicted in blue and red, respectively. b, Abundance of Lactobacilli relative to total intestinal microbiota in DAOWT and DAOG181R mice. c, Average Lactobacilli species composition in intestines of DAOWT and DAOG181R mice. d, Relative abundance of KEGG pathways involved in D-Ala metabolism in DAOWT and DAOG181R mice, predicted with PICRUSt23 followed by HUMAnN24. (b–d; n = 8, 7 mice, respectively) e, Fecal IgA levels in vehicle- or antibiotic-treated DAOWT and DAOG181R mice (vehicle, n = 9, 10 mice; Abx, n = 8, 7 mice, respectively). Error bars, mean ± s.e.m. P values analyzed with Wilcoxon Rank Sum test (b, d, e).
Secretory immunoglobulin A (sIgA) has been shown to be an important host factor in determining the relative proportions of commensal bacteria in the gut25. To determine if differences in commensal-specific IgA contributed to the observed differences in microbiota composition between WT and DAOG181R mice, we performed IgA-SEQ26. We did not observe differences between the composition of fecal IgA-positive bacteria in the DAOWT and DAOG181R mice (Supplementary Fig. 11a–c). Notably, however, the lack of DAO activity was associated with ~2-fold higher fecal IgA and increased numbers of small intestinal IgA-positive cells than in DAOWT mice (Fig. 4e and Supplementary Fig. 11d). Antibiotic treatment significantly reduced the fecal IgA levels, abolishing the difference between DAOWT and DAOG181R mice (Fig. 4e), excluding the possibility of an intrinsic IgA-increase in DAO-null mice. Increased sIgA levels have also been observed in mice with reduced innate host defense27,28, and thus may reflect impaired mucosal defense in DAOG181R animals.
Collectively, our findings reveal that DAO is a microbiota-regulated host innate immune factor that modulates growth of both pathogens and commensals, primarily in the small intestine. Given the protein’s distribution and dependence on oxygen, DAO likely limits bacterial growth close to the absorptive epithelial surface. The enzyme DAO likely regulates the intestinal microbial compartment by several means, including via production of H2O2, and modulation of its activity may provide a means for controlling microbial homeostasis (Supplementary Fig. 12).
Methods
Antibodies
Polyclonal antiserum against a C-terminal epitope (CLEEKKLSRLPPSHL) of mouse DAO was generated in rabbits and affinity-purified (GenScript, Piscataway, NJ, USA) (validated in Supplementary Fig. 2c). Goat polyclonal antiserum to human DAO (EB11100) was obtained from Everest Biotech (Oxfordshire, UK). A rabbit monoclonal antibody to GAPDH (14C10) was purchased from Cell Signaling Tech (Danvers, MA, USA). Goat polyclonal antiserum to Mucin 2 was from Santa Cruz (H-300, Santa Cruz Biotech, Dallas, TX, USA). A goat polyclonal anti-mouse IgA antibody conjugated with FITC was obtained from Sigma-Aldrich (St Louis, MO, USA).
Animals
All animal experiments were performed in accordance with a protocol approved by the Harvard IACUC.
DAOG181R mice on the ddY background13 were a kind gift from R. Konno. The ddY/DAOG181R mice were backcrossed with C57BL6 mice (Jackson Laboratory, Bar Harbor, ME, USA) more than 10 times. All DAOG181R mice and DAOWT mice used in this study were the offspring of heterozygote breeders (mixed sex).
Germ-free (GF) C57BL6 male mice were obtained from the Harvard Digestive Disease Center Gnotobiotic and Microbiology Core. C57BL6 male mice from Jackson Lab were used as SPF mice. Six week old GF mice were conventionalized (GF-SPF mice) by placing the mice in cages with bedding from SPF mice. The GF-SPF vancomycin-treated mice (GF-SPF-V) were conventionalized similarly to GF-SPF mice but with vancomycin (0.5 g/L, Sigma-Aldrich) in their drinking water for two weeks.
To test the effect of vancomycin on DAO expression in adult SPF, C57BL6 mice (6 weeks old) were treated with 0.5 g/L vancomycin in their drinking water for two weeks.
MyD88-knockout (B6.129P2(SJL)-Myd88tm1.1Defr/J) (8 weeks old, male) and RIPK2-knockout (B6.129S2-Ripk2tm1Flv/J) (8 weeks old, male) mice were purchased from Jackson Lab, and C57BL6 mice (8 weeks old, male) from Jackson Lab were used as their control. Genotypes of the knockout mice were determined according to the Jackson Lab’s genotyping protocols.
Isolation of luminal, mucosal, and epithelial layers in the small intestine
Slightly modified versions of protocols described by Vaishnava et al.27 were used to isolate luminal and mucosal layers in the small intestine; the protocol of Roche30 was followed for isolation of the epithelial layer.
Freshly dissected small intestines were divided equally into three parts (proximal, middle, and distal), and each part was further subdivided into three pieces. ‘Luminal contents’ of each piece were washed out with 2 mL of ice-cold sterile PBS, which resulted in 6 mL of luminal washout for each region. The tissue pieces were opened longitudinally, and placed in 15 mL conical tubes with 2 mL of ice-cold sterilized PBS. Tubes were inverted twenty times and vortexed for 10 sec and then the liquid was collected in a fresh tube and defined as ‘mucosal content’. Then, the remaining tissue pieces were further cut into approximately 3–5 mm square patches, gathered in a 15 mL conical tube with 10 mL of an isolation buffer (0.5 mM DTT/5 mM EDTA in PBS), and incubated with constant shaking at 37°C for 20 min. The epithelial cells were collected by pipetting ten times with a 10 mL pipet and the sample was filtered through a 70-μm cell strainer (Corning; Corning, NY, USA) to remove tissue fragments. The cells in suspension were pelleted by centrifugation at 6,000 × g at 4°C for 10 min and defined as the ‘epithelial layer’.
Determination of D- and L-amino acid levels by two-dimensional HPLC
Cecal luminal contents were scooped out and mixed with 20-fold volume/weight of ice-cold sterile PBS, incubated on ice for 30 min, vortexed for 10 sec, and centrifuged at 4°C at 8,000 × g for 5 min. For amino acids in chow, pellets were mixed with 20-fold volume/weight of ice-cold sterile PBS, kept on ice for 30 min, homogenized by bead-beating, and centrifuged at 4°C at 13,000 × g for 5 min. Supernatants were stored at -80°C until use. For amino acids in small intestinal layers, luminal/mucosal contents in PBS were centrifuged at 4°C at 6,000 × g for 5 min, and epithelial pellets were homogenized in 300 μl PBS and centrifuged at 4°C at 12,000 × g for 5 min; their supernatants were stored at -80°C until used.
D- and L-amino acid concentrations in the PBS solutions were determined using a two-dimensional HPLC-based method as described8,31 with some minor modifications. Briefly, PBS homogenates (20 μL) were mixed with 20 μL of H2O and 360μL of methanol. The mixture was vortexed vigorously and centrifuged at 4°C to obtain supernatant. Aliquots of the supernatant (10 μL for cecal contents and 100 μL for SI samples) were dried under reduced pressure, and the residues were re-dissolved in 20 μL of 200 mM Na-borate buffer (pH 8.0). The amino acids were then derivatized with 4-fluoro-7-nitro-2,1,3-benzoxadiazole (NBD-F) by adding 5 μL of 40 mM NBD-F (in acetonitrile) and heated at 60°C for 2 min. After adding 75 μL of 0.1% trifluoroacetic acid in H2O, 2 μL of the reaction mixture was subjected to the two-dimensional HPLC system (NANOSPACE SI-2 series, Shiseido, Tokyo, Japan) combining a capillary-monolithic ODS column (0.53 mm ID × 1000 mm, prepared in a fused silica capillary, provided from Shiseido) and a narrowbore-enantioselective column (KSAACSP-001S, 1.5 mm ID × 250 mm, self-packed, material was provided from Shiseido, or Sumichiral OA-3200S, 1.5 mm ID × 250 mm, self-packed, material was provided from Sumika Chemical Analysis Service, Osaka, Japan). The ODS column yielded fractions of NBD-derivatized amino acids as D plus L mixtures, and the enantiomers were further separated and determined by narrowbore-enantioselective columns. Fluorescence detection of the NBD-amino acids was carried out at 530 nm with excitation at 470 nm.
Histology
DAO enzyme-histochemistry and multiple fluorescence staining
Activity-based DAO labeling was performed similarly to a previously reported method11. Deeply anesthetized mice were perfused transcardially with ice-cold PBS and subsequently with 2% paraformaldehyde in PBS. Tissues were dissected and cryoprotected in 20% sucrose in PBS at 4°C until they sank. They were frozen in a mixture (2:5) of 20% sucrose in PBS and Tissue-Tek O.C.T. Compound (Sakura Finetek, Tokyo, Japan). Sections (10 μm) were sliced on a cryostat (Leica, Wetzlar, Germany) at -24°C and stored at -80°C until they were used. Sections were rinsed in PBS and immersed in DAO-activity staining solution [7 mM sodium pyrophosphate pH 8.3, 0.1% horseradish peroxidase, 0.065% sodium azide, 0.6% nickel ammonium sulfate, 22 mM D-Pro (or L-Pro as a negative control), 20 μM FAD, and FITC-tyramide (1:400; Perkin-Elmer, Waltham, MA, USA)] for 10 min.
For multiple staining, the sections were rinsed in PBS twice, and then incubated in PBS containing 5 unit/ml Alexa fluor 568 Phalloidin (Life Technologies, Carlsbad, CA, USA), and/or 0.1 mg/ml Alexa fluor 633 Wheat Germ Agglutinin (Life Technologies) for 20 min. Then, the sections were rinsed in PBS twice and cover-slipped with ProLong Gold Antifade Reagent with DAPI (Life Technologies). Fluorescence signals were visualized using a confocal microscope (Nikon Eclipse Ti, Nikon, Tokyo, Japan). Each section being compared was imaged under identical conditions.
Immunofluorescence staining of human samples
Human small intestine samples were obtained from Biospecimen Bank at the Brigham and Women’s Hospital approved by The Partners Human Research Committee (Institutional Review Board). Informed consent was obtained from all individuals. Tissues were fixed in 4% paraformaldehyde in PBS at room temperature for 2 hours. After fixation, cryosections were made as for DAO enzyme-histochemistry. Tissue sections were rinsed in PBS twice, permeabilized in 0.3% TritonX PBS for 15 min, and then blocked in serum diluted in PBS for 3 hours at room temperature. The sections were immersed in primary antibodies (goat anti-human DAO, 1:100; rabbit anti-Mucin-2, 1:20) at 4°C overnight. They were rinsed in PBS twice, and incubated in secondary antibodies [anti-goat (Fab′)-FITC, 1:250, Sigma; anti-rabbit (Fab′)-Alexa594, 1:250, Life Technologies] for 40 min at room temperature. The sections were rinsed in PBS and cover-slipped with ProLong Gold Antifade Reagent with DAPI and imaged as described above.
Image analysis
The intensity of cellular DAO fluorescence was quantified with imageJ. The signal intensity in enterocytes and goblet cells were measured, divided by cellular dimensions, and standardized with the averaged value of intensity/area in the control region for enterocytes.
Western blotting
COS7 cells transfected with plasmids (pFLAG-CMV5) encoding cDNA of mouse or human DAO, or epithelial cells isolated from the small intestine were homogenized in a lysis buffer [150 mM sodium chloride, 1.0% NP-40, 50 mM Tris (pH 8.0), and a protease inhibitor cocktail, Complete EDTA-free (Roche, Basel, Switzerland)] and spun down at 15,000 × g at 4°C for 5 min. A cerebellar lysate was used as a positive control and cerebral cortical lysate as negative control. Supernatants were subjected to SDS-PAGE and protein was transferred to nitrocellulose membranes. Blots were blocked in 5% skim milk in Tris-buffered saline with 0.1% Tween-20 (TBST) with constant shaking for 1 hour. The membranes were rinsed in TBST and immersed in TBST with an appropriate primary antibody at 4°C overnight. Membranes were subsequently rinsed in TBST, then incubated with an appropriate secondary antibody conjugated with horseradish peroxidase for 1 hour. The membranes were rinsed in TBST and bound antibodies detected with SuperSignal West Pico (Thermo Scientific, Waltham, MA, USA). For the quantification of protein levels, the density of each band was measured using ImageJ 1.49c and DAO expression was standardized with that of GAPDH.
Bacterial strains, growth media and conditions
All Vibrio cholerae strains are derivatives of El Tor clinical isolate N1696132. The oxyR deletion mutant was generated using standard allele replacement protocol33 with a derivative of the suicide plasmid pCVD442. The ΔbsrV strain has been described previously3. Listeria monocytogenes (mutant strain, Lm 10403S InlAm)34 was grown in Brain Heart Infusion broth (BHIB). Lactobacillus johnsonii (wild-type strain, VPI7960)35 was cultured in MRS broth (MRSB) unless otherwise described. The remaining bacterial strains, including Enterohaemorrhagic E. coli (wild-type strain, EDL933)36, Salmonella enterica serovar Typhimurium (wild-type strain, ATCC 14028), Vibrio parahaemolyticus (wild-type strain, VP47)37, Staphylococcus aureus (wild-type strain, MT8)38, Pseudomonas aeruginosa (wild-type strain, PA01)39 were grown in Luria broth (LB) at 37°C.
DAO bactericidal activity
Overnight cultures of the tested strains were diluted 500-fold in LB or BHIB and incubated with constant shaking at 37°C with or without 500 μM D-Ala or D-Pro in the presence or absence of 100 μg/ml porcine DAO (Sigma-Aldrich). For rescue from DAO toxicity, 50μg/ml bovine catalase (Sigma-Aldrich) was added to the diluted overnight culture. The strains receiving various treatments were aerobically grown until O.D.600 of the control (vehicle treated) sample reached 0.10 (~ 2 hours). Then, cells were serial diluted, plated, and incubated at 37°C.
V. cholerae intestinal colonization
Streptomycin resistant wild-type and ΔbsrV V. cholerae were cultured in LB media overnight at 26°C. The culture were pelleted and then resuspended in 2.6% (w/v) NaHCO3 to O.D.600 = 1.5. Either 4 month old DAOWT or DAOG181R, 7 week old C57BL6 mice (male), or 5 day old suckling C57BL6 mice (mixed sex) were orogastrically inoculated with ~ 2 x108 (for adults) or 1 x106 (for infants) CFU of WT or ΔbsrV V. cholerae as described40,41. Prior to inoculation of the adult mice, they were treated for 24 hours with streptomycin (5 g/L, Life Technologies) in their drinking water. The inoculated adult mice were fed ad libitum and provided with water containing 0.2 g/L streptomycin. Suckling mice were not treated with streptomycin. After 24 hr, the animals were euthanized, and the proximal two thirds of small intestines were dissected, homogenized in 5 mL, serial-diluted, plated and incubated at 37°C on LB agar plates with 200 μg/ml streptomycin. CFU were counted and normalized to the weight of the tissue.
16S rRNA gene sequencing and analysis
Intestinal luminal or mucosal contents were homogenized in 10-fold volume/weight PBS with stainless steel beads (3.2 mm diameter, Biospec) using the bead-beater (Biospec) for 20 sec. A mixture of 300 μl of the homogenate, 400 μl of an extraction buffer (200 mM Tris pH 8.0, 200 mM NaCl, 20 mM EDTA), 50 μl of 10% SDS, a 300 μl slurry of glass beads (0.1 mm diameter, Biospec), and 500 μl of PCI solution (phenol:chloroform:isoamyl-alcohol, 25:24:1; Invitrogen) was further homogenized by the bead-beater (Biospec) for 3 min and spun down at 14,000 × g for 5 min. Subsequently, 500 μl of the supernatant was mixed with the same volume of chloroform, vortexed for 10 sec, and centrifuged at 14,000 × g for 5 min. The DNA was precipitated by adding 300 μl of isopropanol to 300 μl of the aqueous phase, pelleted by centrifugation at 14,000 × g for 5 min, and resuspended with 100 μl of 10 mM Tris-HCl (pH 8.5). Then, the DNA extract was mixed with 5 volumes of PB solution (QIAGEN, Venlo, Netherlands) and purified with a Spin Smart column (CM 400-50, Denville, South Plainfield, NJ, USA). After washing the column with PE solution (QIAGEN), bacterial genomic DNA was eluted with 10 mM Tris-HCl (pH 8.5). Then, the V3–V4 region of 16S ribosomal RNA was PCR amplified (12.5 ng purified DNA per reaction; Phusion polymerase, New England Biolab, Ipswich, MA, USA) (25 cycles: 95°C for 30 sec, 50°C for 30 sec, and 72°C for 30 sec) (primer pair: 341F/805R42 with overhang adapters) (adapter-341F: 5′-tcg tcg gca gcg tca gat gtg tat aag aga cag CCT ACG GGN GGC WGC AG-3′; adapter-805R: 5′-gtc tcg tgg gct cgg aga tgt gta taa gag aca gGA CTA CHV GGG TAT CTA ATC C-3′). PCR products were purified (MinElute, QIAGEN) and resuspended in 25 μl of 10 mM Tris-HCl pH 8.5. The V3–V4 PCR products were indexed with the Nextera XT Index kit (Illumina, San Diego, CA, USA) by PCR (2.5 μl PCR product; Nextera XT Index primers; Phusion polymerase) (8 cycles: 95°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec). The 16S rRNA amplicon with indices were purified (MinElute, QIAGEN), resuspended in 25 μl of 10 mM Tris-HCl pH 8.5, quantified with a Qubit 2.0 Fluorometer (Life Technologies), pooled at a concentration of 4 nM, denatured, diluted to a final concentration of 4 pM, and sequenced using the MiSeq Reagent Kit v3 (600-cycle, paired-end, Illumina) on a MiSeq sequencer (Illumina).
Paired end reads were initially demultiplexed using MiSeq Reporter v2.0, merged with FastqJoin, and quality filtered with a Q-score cutoff of 20. The closed-reference OTU picking workflow in QIIME (versions 1.8 and 1.9) and the Greengenes reference database were used to cluster the reads into 97% identity Operational Taxonomic Units (OTUs). The May 2013 Greengenes taxonomy was used to assign taxonomy to representative OTUs43,44. For analysis including samples from wt and DAOG181R mice, samples were subsampled to 70,000 reads/sample. OTUs with less than 0.005% frequency were filtered out from OTU tables45. Diversity analysis was performed with QIIME. The Linear Discriminant Analysis Effect Size (LEfSe) Galaxy module (http://huttenhower.sph.harvard.edu/galaxy/) was used for additional statistical analysis29. Significance levels for LEfSe were p < 0.05 and Linear Discriminant Analysis (LDA) > 2.0. Relative abundance of taxa found significant in LEfSe was further analyzed with a Wilcoxon rank-sum tests using Prism (Graphpad Software). Species level analysis of Lactobacillus was performed with 16S Metagenomics v1.0.1, an application in the BaseSpace (Illumina).
For inference of metagenomic information from 16S amplicon sequencing data, the algorithm (phylogenetic investigation of communities by reconstruction of unobserved states; PICRUSt)23 (Galaxy module) was performed using the default settings. The resulting metagenomic data were submitted for further analysis to the HMP unified metabolic analysis network (HUMAnN)24 pipeline (version 0.99) to determine the presence/absence and abundance of microbial pathways by sorting individual genes into Kyoto encyclopedia of genes and genomes (KEGG) pathways. The LEfSe was used for statistical analysis of outputs from HUMAnN (significance levels: p < 0.05 and LDA score > 2.0).
ELISA for fecal IgA
Six week old DAOwt and DAOG181R mice were treated with a mixture of antibiotics (1 g/L ampicillin, 1 g/L neomycin, 1 g/L metronidazole, and 0.5 g/L vancomycin) for 1 week and then with the same antibiotics at one half the initial concentrations for 2 additional weeks in their drinking water. Antibioics were diluted in 5% sucrose, which was used as vehicle control. Feces were homogenized in 10x volume (v/w) of PBS containing a protease inhibitor cocktail, Complete EDTA-free (Roche), using a bead-beater (Biospec, Bartlesville, OK, USA) for 20 sec, and centrifuged at 3,000 rpm for 15 min at 4°C. Supernatants were further spun down at 10,000 rpm for 5 min at 4°C, and the IgA level in the final supernatant was determined using the Mouse IgA ELISA Ready-SET-Go! kit (eBioscience, San Diego, CA, USA), according to the manufacturer’s protocol. Fecal IgA levels were standardized per weight of feces.
Statistics
No statistical methods were used to predetermine sample size for animal experiments. The sample size for experiments of V. cholerae colonization in mice was based on previous studies40, 41. Animal experiments (Fig. 2g, 3c, 4a–d) were performed blind to group allocations. Blinding was not possible for other animal experiments. No randomization was used. Prism (Graphpad software) was used for data plotting and statistical analyses. Statistical significance was determined by one-way ANOVA with Tukey post-test when more than 2 groups with normal distribution were compared (Fig. 2f, 3ab), or Wilcoxon Rank Sum test to compare two groups when data were not normally distributed (Fig. 2g, 3cd, 4bde).
Supplementary Material
Acknowledgments
We thank M. Nakane (Shiseido Co. Ltd.) for technical support with 2D-HPLC; L. Comstock for help with anaerobic cultures; R. Konno for providing DAOG181R mice; Waldor lab colleagues and M. Silverman for their helpful comments on the manuscript; Q. Wang for help creating ΔoxyR strain; L. Bry for germ-free mice and G. Abu-Ali/Huttenhower Group for assistance with LEfSe and 16S metagenomic analysis and S. Aiso for constant support. This work was supported by Howard Hughes Medical Institute (M.K.W.), NIH grant R37 AI-042347 (M.K.W.), and the Moritani Scholarship Foundation (J.S.).
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
J.S. and M.K.W. conceived and designed the study. J.S. carried out histological experiments, biochemical analyses, microbiological studies, sequence analyses, animal experiments with T.Z., and figure preparation. Y.M. and K.H. performed HPLC quantifications of chiral amino acids with technical support by M.M.. S.R. and B.M.D. provided scientific advice. M.K.W. supervised the experiments and directed the analysis. J.S., S.R., B.M.D., and M.K.W. wrote the manuscript.
Additional information
16S rRNA sequences, generated from intestinal samples in raw format prior to post-processing and data analysis, have been deposited at the European Nucleotide Archive (accession number, PRJEB12079).
Supplementary information is available in the online version of the paper.
References
- 1.Fujii N, Saito T. Homochirality and life. Chemical record. 2004;4:267–278. doi: 10.1002/tcr.20020. [DOI] [PubMed] [Google Scholar]
- 2.Wolosker H, Dumin E, Balan L, Foltyn VN. D-amino acids in the brain: D-serine in neurotransmission and neurodegeneration. The FEBS journal. 2008;275:3514–3526. doi: 10.1111/j.1742-4658.2008.06515.x. [DOI] [PubMed] [Google Scholar]
- 3.Lam H, et al. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cava F, de Pedro MA, Lam H, Davis BM, Waldor MK. Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. The EMBO journal. 2011;30:3442–3453. doi: 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 6.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annual review of immunology. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 7.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyoshi Y, et al. Chiral amino acid analysis of Japanese traditional Kurozu and the developmental changes during earthenware jar fermentation processes. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences. 2014;966:187–192. doi: 10.1016/j.jchromb.2014.01.034. [DOI] [PubMed] [Google Scholar]
- 9.Ohide H, Miyoshi Y, Maruyama R, Hamase K, Konno R. D-Amino acid metabolism in mammals: biosynthesis, degradation and analytical aspects of the metabolic study. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences. 2011;879:3162–3168. doi: 10.1016/j.jchromb.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 10.Pollegioni L, Piubelli L, Sacchi S, Pilone MS, Molla G. Physiological functions of D-amino acid oxidases: from yeast to humans. Cellular and molecular life sciences: CMLS. 2007;64:1373–1394. doi: 10.1007/s00018-007-6558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasabe J, et al. D-amino acid oxidase controls motoneuron degeneration through D-serine. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:627–632. doi: 10.1073/pnas.1114639109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Konno R, Yasumura Y. Mouse mutant deficient in D-amino acid oxidase activity. Genetics. 1983;103:277–285. doi: 10.1093/genetics/103.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espey MG. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free radical biology & medicine. 2013;55:130–140. doi: 10.1016/j.freeradbiomed.2012.10.554. [DOI] [PubMed] [Google Scholar]
- 15.Nathan C, Cunningham-Bussel A. Beyond oxidative stress: an immunologist’s guide to reactive oxygen species. Nature reviews. Immunology. 2013;13:349–361. doi: 10.1038/nri3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espaillat A, et al. Structural basis for the broad specificity of a new family of amino-acid racemases. Acta crystallographica. Section D, Biological crystallography. 2014;70:79–90. doi: 10.1107/S1399004713024838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, et al. Catalases promote resistance of oxidative stress in Vibrio cholerae. PloS one. 2012;7:e53383. doi: 10.1371/journal.pone.0053383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuinema BR, Reid-Yu SA, Coombes BK. Salmonella evades D-amino acid oxidase to promote infection in neutrophils. mBio. 2014;5:e01886. doi: 10.1128/mBio.01886-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura H, Fang J, Maeda H. Protective role of D-amino acid oxidase against Staphylococcus aureus infection. Infection and immunity. 2012;80:1546–1553. doi: 10.1128/IAI.06214-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serata M, Iino T, Yasuda E, Sako T. Roles of thioredoxin and thioredoxin reductase in the resistance to oxidative stress in Lactobacillus casei. Microbiology. 2012;158:953–962. doi: 10.1099/mic.0.053942-0. [DOI] [PubMed] [Google Scholar]
- 21.Pridmore RD, et al. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2512–2517. doi: 10.1073/pnas.0307327101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Kaaij H, Desiere F, Mollet B, Germond JE. L-alanine auxotrophy of Lactobacillus johnsonii as demonstrated by physiological, genomic, and gene complementation approaches. Applied and environmental microbiology. 2004;70:1869–1873. doi: 10.1128/AEM.70.3.1869-1873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langille MG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature biotechnology. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abubucker S, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS computational biology. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fagarasan S. Evolution, development, mechanism and function of IgA in the gut. Current opinion in immunology. 2008;20:170–177. doi: 10.1016/j.coi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Palm NW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zelante T, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome biology. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roche JK. Isolation of a purified epithelial cell population from human colon. Methods in molecular medicine. 2001;50:15–20. doi: 10.1385/1-59259-084-5:15. [DOI] [PubMed] [Google Scholar]
- 31.Hamase K, et al. Simultaneous determination of hydrophilic amino acid enantiomers in mammalian tissues and physiological fluids applying a fully automated micro-two-dimensional high-performance liquid chromatographic concept. Journal of chromatography. A. 2010;1217:1056–1062. doi: 10.1016/j.chroma.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Heidelberg JF, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infection and immunity. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wollert T, et al. Extending the host range of Listeria monocytogenes by rational protein design. Cell. 2007;129:891–902. doi: 10.1016/j.cell.2007.03.049. [DOI] [PubMed] [Google Scholar]
- 35.Fujisawa T, Benno Y, Yaeshima T, Mitsuoka T. Taxonomic study of the Lactobacillus acidophilus group, with recognition of Lactobacillus gallinarum sp. nov. and Lactobacillus johnsonii sp. nov. and synonymy of Lactobacillus acidophilus group A3 (Johnson et al. 1980) with the type strain of Lactobacillus amylovorus (Nakamura 1981) International journal of systematic bacteriology. 1992;42:487–491. doi: 10.1099/00207713-42-3-487. [DOI] [PubMed] [Google Scholar]
- 36.Perna NT, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 37.Makino K, et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet. 2003;361:743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 38.Schlievert PM, Blomster DA. Production of staphylococcal pyrogenic exotoxin type C: influence of physical and chemical factors. J Infect Dis. 1983;147:236–242. doi: 10.1093/infdis/147.2.236. [DOI] [PubMed] [Google Scholar]
- 39.Holloway BW, Krishnapillai V, Morgan AF. Chromosomal genetics of Pseudomonas. Microbiological reviews. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nygren E, Li BL, Holmgren J, Attridge SR. Establishment of an adult mouse model for direct evaluation of the efficacy of vaccines against Vibrio cholerae. Infection and immunity. 2009;77:3475–3484. doi: 10.1128/IAI.01197-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Angelichio MJ, Spector J, Waldor MK, Camilli A. Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infection and immunity. 1999;67:3733–3739. doi: 10.1128/iai.67.8.3733-3739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herlemann DP, et al. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. The ISME journal. 2011;5:1571–1579. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME journal. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bokulich NA, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.