ZmMADS1 is activated in leaves during shoot apical meristem transition and acts as a positive flowering time regulator in day-neutral maize.
Abstract
Flowering time (FTi) control is well examined in the long-day plant Arabidopsis (Arabidopsis thaliana), and increasing knowledge is available for the short-day plant rice (Oryza sativa). In contrast, little is known in the day-neutral and agronomically important crop plant maize (Zea mays). To learn more about FTi and to identify novel regulators in this species, we first compared the time points of floral transition of almost 30 maize inbred lines and show that tropical lines exhibit a delay in flowering transition of more than 3 weeks under long-day conditions compared with European flint lines adapted to temperate climate zones. We further analyzed the leaf transcriptomes of four lines that exhibit strong differences in flowering transition to identify new key players of the flowering control network in maize. We found strong differences among regulated genes between these lines and thus assume that the regulation of FTi is very complex in maize. Especially genes encoding MADS box transcriptional regulators are up-regulated in leaves during the meristem transition. ZmMADS1 was selected for functional studies. We demonstrate that it represents a functional ortholog of the central FTi integrator SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) of Arabidopsis. RNA interference-mediated down-regulation of ZmMADS1 resulted in a delay of FTi in maize, while strong overexpression caused an early-flowering phenotype, indicating its role as a flowering activator. Taken together, we report that ZmMADS1 represents a positive FTi regulator that shares an evolutionarily conserved function with SOC1 and may now serve as an ideal stating point to study the integration and variation of FTi pathways also in maize.
Flowering time (FTi) in crops is an important agronomical trait critical for harvesting date, biomass yield, crop rotation schemes, and terminal drought avoidance (Jung and Müller, 2009; Bendix et al., 2015). For flowering, the shoot apical meristem (SAM) has to change from vegetative to generative growth, a process called the floral transition. This process is controlled by various key players representing components of different signaling pathways triggered by both external and endogenous factors. The main environmental influence on FTi is derived from photoperiods or daylength as well as temperature. Unlike Arabidopsis (Arabidopsis thaliana) and some winter-annual crops like wheat (Triticum aestivum) and barley (Hordeum vulgare) that require vernalization by cold temperature periods for floral induction, this does not seem to play an essential role for flowering in maize (Zea mays). Furthermore, during thousands of years of breeding history, cultivated temperate maize is thought to have lost its photoperiod sensitivity and is well adapted to growth under long-day (LD) conditions. Tropical lines flower under short-day (SD) conditions and depend on photoperiods to flower (Colasanti and Coneva, 2009). Especially in temperate maize lines, endogenous signals related to the developmental stage of the plant appear to play the main role in floral induction, like the autonomous, the aging, and the GA3 signaling pathways (for review, see Khan et al., 2014).
Most information about factors controlling the transition from the vegetative to the floral phase was gained from research with the LD-adapted model plant Arabidopsis. It was demonstrated that the different signaling pathways mediating both environmental and endogenous cues converge at a few floral integrators (for an overview of this complex network in Arabidopsis, see Blümel et al., 2015). One of these integrators is FLOWERING LOCUS T (FT), a phosphatidylethanolamine-binding protein, which represents the mobile signal moving from the leaves through the phloem toward the SAM (Kardailsky et al., 1999; Jaeger and Wigge, 2007). Expression of FT at high levels is mainly induced by LD conditions mediated by the zinc finger transcriptional regulator CONSTANS (CO; Kardailsky et al., 1999; Kobayashi et al., 1999). CO itself is positively regulated by the large circadian clock-associated protein GIGANTEA (GI), and in accordance with this, it is stabilized in the afternoon in a circadian rhythm (Song et al., 2012). By linking the circadian clock with flowering-inducing pathways, CO represents another important key integrator for the floral transition (Suárez-López et al., 2001). In the SAM, FT forms a complex with the bZIP transcription factor (TF) FLOWERING LOCUS D (FD), triggering the floral transition through the activation of meristem identity genes such as APETALA1 (AP1; Abe et al., 2005; Wigge et al., 2005). The major direct target of the FT-FD complex in the SAM is the MIKCC-type II MADS box TF SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), which is expressed mainly in developing leaves and meristems (Borner et al., 2000). SOC1 is supposed to represent the main key integrator of all five flowering-inducing pathways in Arabidopsis: the photoperiodic, the vernalization, the aging, the GA3, and the autonomous pathways. Expression of SOC1 is negatively regulated by the MADS box TF FLOWERING LOCUS C (FLC; Searle et al., 2006). By forming a complex with the MADS box TF SHORT VEGETATIVE PHASE (SVP), FLC represses not only the expression of SOC1 but also that of FT and FD, thus representing a negative regulator of the floral transition (Li et al., 2008). FLC mediates signaling from the autonomous and the vernalization pathways, while SVP is regulated by the autonomous and the GA3 pathways and regulates GA3 biosynthesis at the shoot apex (Amasino, 2004; Li et al., 2008; Andrés et al., 2014). Furthermore, SOC1 is positively regulated in an age-dependent manner by SQUAMOSA BINDING FACTOR-LIKE9, thereby integrating all five flowering-inducing pathways (Wang et al., 2009).
To date, little is known about FTi regulation in maize. Some genes encoding proteins homologous to FTi components of Arabidopsis could be identified (Dong et al., 2012), including the photoperiodically regulated genes GIGZ1a and GIGZ1b encoding homologs of Arabidopsis GI (Miller et al., 2008). Interestingly, loss of GIGZ1a function results in an early-flowering phenotype under LD conditions, whereas Arabidopsis gi mutants show late flowering. As GIGZ1a activity can rescue gi loss-of-function mutants in Arabidopsis, it is likely that GI acts as a molecular switch, with its function depending on a species-specific gene regulatory network (Bendix et al., 2013). Furthermore, there are genes involved in FTi regulation that do not have any homologs in Arabidopsis, such as the C2H2 zinc finger protein INDETERMINATE1 (ID1), which seems to represent a master FTi regulator in maize (Colasanti et al., 1998). ID1 acts upstream of ZEA MAYS CENTRORADIALES8 (ZCN8), which is considered the most likely maize homolog of the FTi regulator FT, as it is able to rescue the late-flowering phenotype of Arabidopsis ft mutants (Danilevskaya et al., 2008a; Lazakis et al., 2011; Meng et al., 2011). Overexpression of ZCN8 leads to an early-flowering phenotype, whereas down-regulation results in late flowering (Meng et al., 2011). Similar to FT, ZCN8 is expected to act as a mobile cue, and it was demonstrated to interact with the maize FD homolog DELAYED FLOWERING1 (DLF1; Muszynski et al., 2006; Meng et al., 2011). Recent research indicated that the ZCN8 paralog ZCN7 might represent a second maize florigen (Mascheretti et al., 2015). A floral meristem identity gene acting downstream of the floral activators DLF1 and ID1 is ZEA MAYS MADS4 (ZMM4), which leads to an early-flowering phenotype if overexpressed in maize (Danilevskaya et al., 2008b). However, knowledge about FTi regulation in maize is still very limited, and more information about key players is necessary to begin establishing the complex gene regulatory network responsible for the floral initiation in this species. In a screen for FTi regulators in 29 maize inbreds, including temperate and tropical lines, we determined the time point of the floral transition and identified the MADS box TF ZmMADS1, also known as ZmM5, as the functional maize homolog of SOC1. ZmMADS1 is able to fully rescue the late-flowering phenotype of Arabidopsis soc1-2 mutants. By modifying ZmMADS1 expression levels, the time point of the floral transition and the number of leaves are altered significantly.
RESULTS
Transition of the SAM to Reproductive Growth Strongly Depends on the Genetic Background in Maize
In order to identify leaf-expressed genes involved in activation or repression of the floral transition, 29 maize inbred lines were chosen based on a genetic diversity study (Liu et al., 2003). Plants grown under LD conditions in the greenhouse were analyzed by stereomicroscopy to identify the time point of SAM transition from vegetative to reproductive growth. The expression of genes coding for regulators of this floral transition is expected to change shortly before the SAM switch. Therefore, our aim was to identify clusters of related inbred lines for transcriptome analysis with a strong temporal difference regarding their floral transition. Based on the closely related genetic background, other genes than those coding for floral regulators were not expected to change their expression levels in photosynthetically active leaves. Table I indicates the days after sowing (DAS) as well as the developmental stage, according to the nomenclature of Abendroth et al. (2011), at which the floral transition was observed. Most of the selected maize lines grow under temperate climate conditions, like U.S. dent, S.A. (for South American) popcorn, European flint, and lines originally derived from Argentina (Yan et al., 2009). The other lines are adapted to tropical conditions, like U.S. tropical, Nigeria dent, and Tropical (dent). As expected, most plants of a similar genetic background, such as NC320 and NC332 of the same pedigree (according to www.maizegdb.org), displayed the SAM transition (switch) at similar times. The SAM transition in B73 plants that were propagated for many generations either exclusively in the field or in the greenhouse occurred on the same day, 26 DAS. The switch occurred in flint, dent, and popcorn maize lines adapted to a temperate climate earliest at 18 DAS (V3/V4) and latest at 29 DAS (V7). In contrast, all tropical lines were observed to be late flowering under LD greenhouse conditions, exhibiting transition of the SAM to reproductive growth earliest at 31 DAS (V7) and latest at 40 DAS (V11). These observations indicate that FTi is genetically stable but highly variable depending on the genetic background, which changed during the domestication process generating numerous inbred lines adapted to various environments.
Table I. Transition of the SAM from vegetative to reproductive growth in maize.
Developmental stage (according to Abendroth et al., 2011) and time points (indicated in DAS) of the SAM transition were determined in 29 inbred lines (www.maizegdb.org). Two inbred lines from Argentina and two moderate U.S. dent lines (in boldface) showing strong differences in the SAM transition were selected for transcriptome analyses. Seeds were obtained from the stock center except for B73 (Rgbg.) and A188 (Rgbg.), originating from plants that were grown for 10 years exclusively in the greenhouse in Regensburg.
| Genotype | Stage | Transition Time Point | Classificationa |
|---|---|---|---|
| EP1 | V3/V4 | 18 | European flint |
| F7 | V3/V4 | 18 | European flint |
| A188 (Rgbg.) | V5 | 19 | U.S. dent |
| VA26 | V4 | 20 | U.S. dent |
| PA762 | V4/V5 | 20 | U.S. dent |
| OH43 | V4/V5 | 20 | U.S. dent |
| Wf9 | V5/V6 | 23 | U.S. dent |
| 4F-240 BX 16 | V5 | 24 | Argentina |
| B77 | V6 | 24 | U.S. dent |
| R229 | V5/V6 | 25 | U.S. dent |
| Mo1W | V5/V6 | 26 | U.S. dent |
| B73 | V5/V6 | 26 | U.S. dent |
| B73 (Rgbg) | V6 | 26 | U.S. dent |
| VA102 | V5 | 26 | U.S. dent |
| IDS69 | V6 | 28 | S.A. popcorn |
| SA24 | V7 | 29 | S.A. popcorn |
| NC332 | V5/V6 | 29 | U.S. dent |
| NC320 | V7 | 29 | U.S. dent |
| NC350 | V7 | 31 | Tropicalb |
| Ki11 | V7 | 33 | Tropical flintc |
| E2558W | V8 | 33 | U.S. dent |
| Tzi25 | V8 | 33 | Nigeria dent |
| 4F-350 CN 2 | V8 | 35 | Argentina |
| NC304 | V9 | 35 | Tropicalb |
| Tzi16 | V10 | 36 | Nigeria dent |
| Ki3 | V10 | 36 | Tropical flintc |
| A6 | V10 | 40 | U.S. tropical |
| Tzi10 | V11 | 40 | Nigeria dent |
According to the MaizeGDB stock database and the Germplasm Resources Information Network. bFrom El Salvador. cFrom Thailand
Based on this observation, it was difficult to select genetically closely related inbred lines that strongly varied in SAM transition time points. Therefore, the two Argentinian lines 4F-240 BX 16 and 4F-350 CN 2 that were collected in the same region, showing a switch at V5/V8 at 24 and 35 DAS, respectively, were selected for further analysis. Additionally, we have chosen the temperate U.S. dent lines B77 and E2558W, where the switch at V6/V8 occurred at 24 and 33 DAS, respectively (Table I). According to Liu et al. (2003), these two lines display a moderate genetic similarity. These four lines were chosen for further transcriptome analyses. To compare whether growth conditions show an impact on FTi behavior of these lines, time points of floral transitions in LD greenhouse conditions were compared with those in LD and SD illumination in growth chambers (Supplemental Table S1). Compared with greenhouse-grown plants, the differences in floral transition within each pair were reduced slightly, with SD illumination generally resulting in an earlier floral transition than LD illumination. Nevertheless, these results confirmed the strong differences in SAM transition observed using LD greenhouse conditions.
Leaf-Expressed MADS Box TF Genes Are Up-Regulated before the SAM Transition
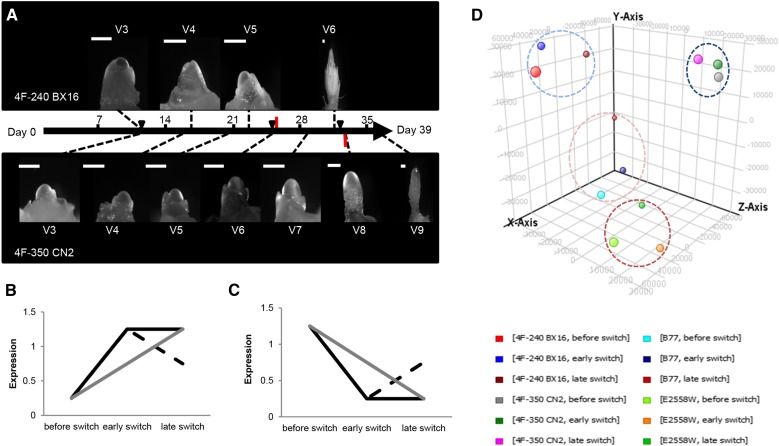
For transcriptome analyses, leaf samples of the uppermost leaf were taken for RNA extraction from the following three developmental stages after Abendroth et al. (2011): I, at stage V3 (none of the lines has switched from vegetative to generative growth); II, 1 d before the floral transition occurred in the early-switcher plants (4F-240 BX 16 and B77); and III, 1 d before the transition occurred in the late-switcher plants (4F-350 CN 2 and E2558W). Figure 1A shows as an example the microscopic comparison of the developing SAM of inbred lines 4F-240 BX 16 and 4F-350 CN 2 until 39 DAS. The scale indicates the time points of floral transition as well as sample collection. Three biological replicates of each collection time point were hybridized to a maize gene chip containing more than 42,000 genes developed by the OPTIMAS consortium (Colmsee et al., 2012). Expected expression profiles of putative FTi activators and repressors are shown in Figure 1, B and C. According to the model, FTi regulators were expected to be expressed at the same level in all lines in sample I (before the switch in all lines) and their expression was expected to increase/decrease in sample II (early switch: still before the switch in late-flowering lines and after the transition in early-flowering lines), with the early switchers showing a stronger regulation. In sample III (late switch: after the floral transition in late-flowering lines), late-switching lines display their strongest expression, while the expression of early switchers either keeps the level reached in sample II or becomes less regulated. Excluding FTi regulators, the transcriptomic profiles in photosynthetically active leaves were expected to be comparable within each pair of genetically closely related plants. To verify this assumption, a principal component analysis was performed (Fig. 1D). The transcriptome data of the three collection time points of each maize line were clustered together, whereas the different genotypes were clearly separated from each other. This indicated that the general genotype-dependent differences in gene expression pattern have a stronger influence on the transcriptomic profile of leaves than the different developmental stages used to study the floral transition in maize. Therefore, a more stringent filter method was applied. For each line, the transcriptomic profile of sample I was compared with that of sample II for early switchers and sample III for late switchers to identify differentially expressed genes. These sets of genes were then compared between pairs 4F-350 CN2/4F-240 BX 16 and E2558W/B77.
Figure 1.
Transcriptome profiling of leaf tissue during the SAM transition from vegetative to reproductive growth in maize. A, Transition of the SAM of the inbred lines 4F-240 BX 16 (top) and 4F-350 CN 2 (bottom) on a time scale of 39 DAS. The transition of the SAM is marked by red bars, and black triangles indicate times of tissue sampling of the upper most fully expanded leaf. Bars = 200 µm. B and C, Expected expression profiles for putative FTi activators (B) and putative FTi repressors (C). Black lines represent expected expression profiles in genotypes with early SAM transition, and gray lines represent those for genotypes with late transition. D, Principal component analysis of transcriptome data from four selected inbred lines (see Table I). Principal component analysis depicts the mean of transcriptome triplicates per time point and line. The x axis shows component 1 (27.72%), the y axis shows component 2 (18.85%), and the z axis shows component 3 (17.2%). Data sets for 4F-350 CN 2 are circled in dark blue, those for 4F-240 BX 16 are circled in light blue, those for E2558W are circled in dark red, and those for B77 are circled in light red.
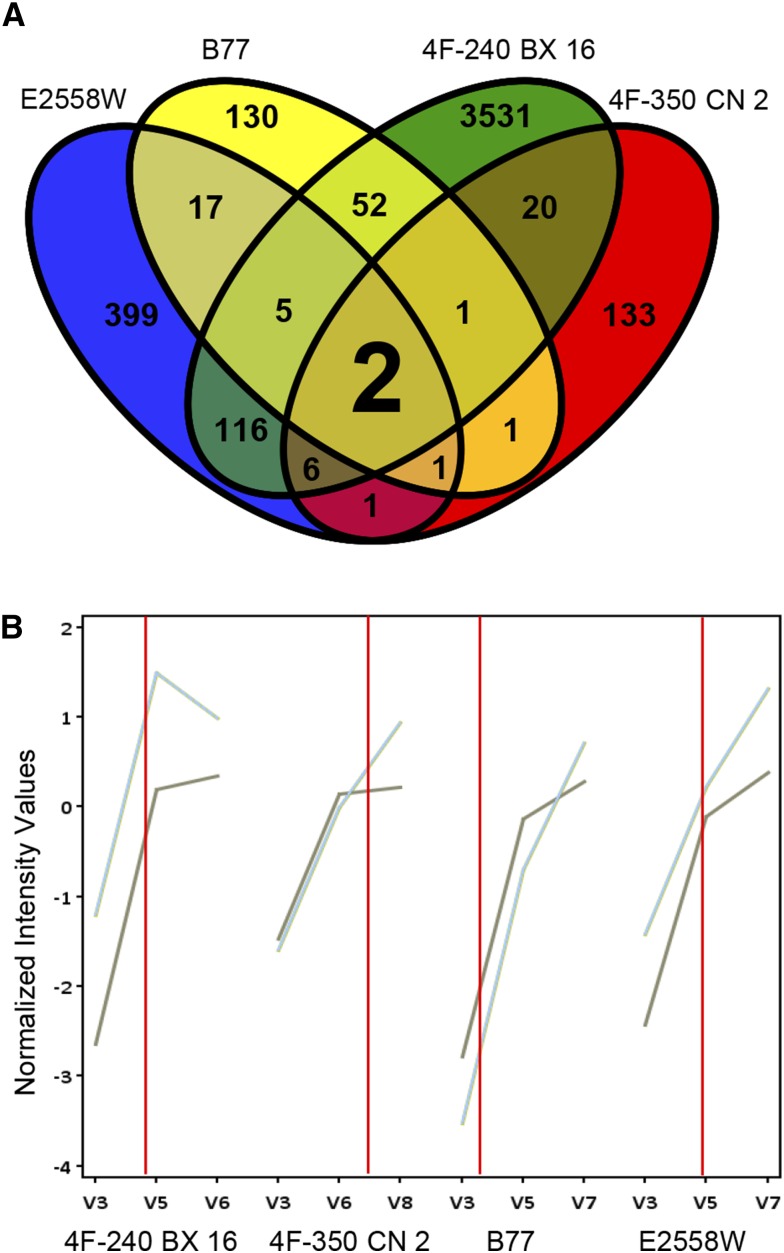
This strategy resulted in a list of 28 genes with significant fold change (greater than 2) of transcription shortly before the SAM transition in the pair 4F-350 CN 2/4F-240 BX 16 (Table II) and 25 genes in the pair E2558W/B77 (Table III). Interestingly, several MADS box TF genes (four and five, respectively) could be identified in both gene sets, all being up-regulated. Both lists were compared with each other to reveal genes that are differentially regulated in all four maize lines. Only two genes changed their expression level in all four maize lines shortly before the flowering switch occurred (Fig. 2A), which both encode for MADS box TFs. The first candidate is ZmMADS1, also known as ZMM5, which was cloned more than a decade ago as a MADS domain-containing gene with a broad expression pattern (Heuer et al., 2001). The second candidate gene encodes for ZMM26, also described as ZmMADS22. Both genes show a strong induction prior to the SAM transition resembling the expression pattern described in Figures 1B and 2B. This finding indicates that they might act as activators in FTi regulation. Their expression profiles and those of randomly selected additional genes from Tables II and III were verified by RT-qPCR analysis using novel biological samples (Supplemental Fig. S1).
Table II. Candidate FTi regulatory genes differentially regulated during the floral transition in leaves of the maize lines 4F-240 BX 16 and 4F-350 CN 2.
The table shows the identifier of the oligonucleotide on the microarray chip, transcript identifier (MaizeSequence version 4a.53) of the coding sequence from which the oligonucleotide was derived, and, for both lines, fold change (FC), P value, regulation, and annotation. Values were calculated between transcriptome data obtained from leaf samples at V3 and at 1 d before the transition of the SAM from vegetative to flowering meristem. MADS box TFs (MADS-TF) are indicated in boldface. POUF, Protein of unknown function.
| OPTIMAS Identifier | 4a.53 Coding Sequence | 4F 240 BX 16 |
4F-350 CN 2 |
Annotation | ||||
|---|---|---|---|---|---|---|---|---|
| FC | P | Regulation | FC | P | Regulation | |||
| OptiV1N35942 | GRMZM2G125853_T06 | 45.72 | 0.02 | Down | 72.40 | 0.04 | Down | POUF |
| OptiV1C09461 | GRMZM2G053977_T01 | 40.18 | 0.02 | Up | 13.11 | 0.05 | Up | Short-chain dehydrogenase/reductase family protein |
| OptiV1C01178 | GRMZM2G152801_T02 | 26.68 | 0.02 | Down | 11.75 | 0.04 | Down | FLS (FLAVONOL SYNTHASE) |
| OptiV1S17892 | GRMZM2G060866_T01 | 24.65 | 0.02 | Down | 30.02 | 0.04 | Down | Anther-specific Pro-rich protein |
| OptiV1S18863 | GRMZM2G700665_T02 | 19.07 | 0.02 | Down | 4.16 | 0.03 | Down | RAP2.7 |
| OptiV1S26761 | GRMZM2G152801_T01 | 16.36 | 0.03 | Down | 17.64 | 0.04 | Down | FLS (FLAVONOL SYNTHASE) |
| OptiV1C11740 | GRMZM2G036203_T01 | 13.25 | 0.05 | Up | 4.29 | 0.04 | Up | Cys proteinase, putative |
| OptiV1C06259 | GRMZM2G113480_T01 | 10.27 | 0.03 | Down | 6.50 | 0.04 | Down | POUF |
| OptiV1S29412 | GRMZM2G088964_T01 | 9.02 | 0.04 | Up | 4.05 | 0.05 | Up | ATKT1 (POTASSIUM TRANSPORTER1) |
| OptiV1C17237 | GRMZM2G171365_T01 | 8.73 | 0.02 | Up | 5.78 | 0.02 | Up | MADS-TF (MADS1) |
| OptiV1S20394 | GRMZM2G363229_T01 | 7.87 | 0.02 | Down | 3.07 | 0.04 | Down | MATE efflux family protein |
| OptiV1C05995 | GRMZM2G070034_T01 | 7.77 | 0.02 | Up | 43.86 | 0.01 | Up | MADS-TF (AtAGL20) |
| OptiV1S21916 | GRMZM2G046885_T02 | 7.59 | 0.02 | Up | 3.24 | 0.04 | Up | MADS-TF (M26) |
| OptiV1C10423 | GRMZM2G029912_T02 | 6.81 | 0.03 | Up | 11.23 | 0.04 | Up | CER3 (ECERIFERUM3) |
| OptiV1S18200 | GRMZM2G055446_T01 | 5.99 | 0.02 | Up | 12.07 | 0.01 | Up | UDP-glycosyltransferase/sinapate 1-glucosyltransferase/transferase |
| OptiV1S30318 | GRMZM2G131266_T01 | 5.93 | 0.02 | Up | 86.15 | 0.02 | Up | Ovule development protein, putative |
| OptiV1C13062 | GRMZM2G322047_T01 | 5.92 | 0.03 | Down | 11.01 | 0.02 | Down | Exostosin family protein |
| OptiV1S28227 | AC197143.3_FGT007 | 4.53 | 0.02 | Down | 4.68 | 0.05 | Down | ATPRB1 |
| OptiV1S24670 | GRMZM2G073969_T01 | 3.93 | 0.02 | Up | 15.62 | 0.03 | Up | POUF |
| OptiV1C01853 | GRMZM2G055178_T01 | 3.66 | 0.02 | Down | 265.51 | 0.02 | Up | PTAC5 (PLASTID TRANSCRIPTIONALLY ACTIVE5) |
| OptiV1C08761 | GRMZM2G004748_T01 | 3.44 | 0.02 | Up | 5.72 | 0.00 | Up | PGP2 (P-GLYCOPROTEIN2) |
| OptiV1N40004 | AC213612.3_FGT001 | 3.20 | 0.02 | Up | 17.85 | 0.05 | Up | Gly-rich cell wall structural protein |
| OptiV1S24539 | GRMZM2G034471_T01 | 3.19 | 0.03 | Up | 4.28 | 0.03 | Up | Cytochrome P450 CYP2 subfamily protein |
| OptiV1S21266 | GRMZM2G006806_T01 | 2.56 | 0.02 | Down | 2.35 | 0.04 | Down | Ssu72-like family protein |
| OptiV1S18426 | GRMZM2G108153_T01 | 2.55 | 0.03 | Down | 74.54 | 0.04 | Up | Peroxidase12 (PER12, P12, PRXR6) |
| OptiV1C01437 | GRMZM2G459841_T01 | 2.48 | 0.02 | Up | 2.38 | 0.05 | Down | POUF |
| OptiV1C03164 | GRMZM2G127581_T01 | 2.41 | 0.03 | Up | 2.48 | 0.04 | Up | Tubulin-specific chaperone B |
| OptiV1S26447 | GRMZM2G019993_T01 | 2.03 | 0.02 | Up | 6.94 | 0.03 | Up | MADS-TF (ZCN21) |
Table III. Candidate FTi regulatory genes differentially regulated during the floral transition in leaves of the maize lines B77 and E2558W.
The table shows the identifier of the oligonucleotide on the microarray chip, transcript identifier (MaizeSequence version 4a.53) of the coding sequence from which the oligonucleotide was derived, and, for both lines, fold change (FC), P value, regulation, and annotation. Values were calculated between transcriptome data obtained from leaf samples at V3 and at 1 d before the transition of the SAM from vegetative to flowering meristem. MADS box TFs (MADS-TF) are indicated in boldface. POUF, Protein of unknown function.
| OPTIMAS Identifier | 4a.53 Coding Sequence | B77 |
E2558W |
Annotation | ||||
|---|---|---|---|---|---|---|---|---|
| FC | P | Regulation | FC | P | Regulation | |||
| OptiV1S24287 | GRMZM2G159105_T01 | 222.58 | 0.03 | Down | 56.78 | 0.02 | Down | Protein kinase family protein |
| OptiV1C11764 | GRMZM2G109627_T01 | 87.32 | 0.04 | Up | 6.34 | 0.02 | Up | PPR repeat-containing protein |
| OptiV1C02818 | GRMZM2G168898_T01 | 35.29 | 0.03 | Down | 8.82 | 0.04 | Down | HB2 (Hemoglobin2) |
| OptiV1N40037 | GRMZM2G006210_T01 | 11.87 | 0.04 | Down | 32.18 | 0.02 | Down | POUF |
| OptiV1S32916 | GRMZM2G061751_T01 | 10.30 | 0.05 | Down | 16.07 | 0.04 | Down | UNE1 (Unfertilized Embryo Sac1) |
| OptiV1S26715 | GRMZM2G052461_T01 | 8.95 | 0.03 | Up | 3.14 | 0.02 | Up | LHT2 (LYS-HIS TRANSPORTER2) |
| OptiV1C17237 | GRMZM2G171365_T01 | 7.07 | 0.05 | Up | 6.63 | 0.04 | Up | MADS-TF (MADS1) |
| OptiV1S31605 | GRMZM2G052034_T02 | 6.85 | 0.05 | Down | 8.50 | 0.04 | Down | Zinc finger (C3HC4-type RING finger) family protein |
| OptiV1S21916 | GRMZM2G046885_T02 | 6.29 | 0.03 | Up | 7.01 | 0.03 | Up | MADS-TF (M26) |
| OptiV1S19217 | GRMZM2G009475_T01 | 6.20 | 0.04 | Down | 4.71 | 0.02 | Down | POUF |
| OptiV1N35188 | AC233893.1_FGT001 | 5.66 | 0.05 | Down | 4.82 | 0.03 | Down | Homo-Cys S-methyltransferase |
| OptiV1C06771 | GRMZM2G179264_T01 | 5.60 | 0.03 | Up | 43.90 | 0.04 | Up | ZCN8 |
| OptiV1S20894 | GRMZM2G134502_T01 | 5.22 | 0.03 | Down | 5.02 | 0.03 | Down | Fiber annexin |
| OptiV1S29802 | GRMZM2G066291_T01 | 4.97 | 0.04 | Up | 3.84 | 0.02 | Up | POUF |
| OptiV1S33653 | GRMZM2G370777_T04 | 4.02 | 0.03 | Up | 3.72 | 0.04 | Up | MADS-TF (M19) |
| OptiV1C05996 | GRMZM2G370777_T05 | 3.87 | 0.04 | Up | 4.24 | 0.04 | Up | MADS-TF (M19) |
| OptiV1C09078 | GRMZM2G046885_T01 | 3.68 | 0.03 | Up | 3.59 | 0.05 | Up | MADS-TF (M26) |
| OptiV1C04175 | GRMZM2G128466_T02 | 3.30 | 0.05 | Up | 3.85 | 0.03 | Up | NQR (NADPH:QUINONE OXIDOREDUCTASE) |
| OptiV1S18918 | GRMZM2G107839_T01 | 2.72 | 0.04 | Down | 6.54 | 0.04 | Down | Putative LTP |
| OptiV1C03183 | GRMZM2G469898_T01 | 2.59 | 0.04 | Down | 4.99 | 0.03 | Down | GDSL motif lipase/hydrolase family protein |
| OptiV1S29722 | GRMZM2G101634_T01 | 2.39 | 0.03 | Down | 5.30 | 0.02 | Down | Ser/Thr protein phosphatase 2A, regulatory subunit |
| OptiV1C10635 | AC233935.1_FGT005 | 2.37 | 0.04 | Down | 5.41 | 0.04 | Down | OBP3 (OBF-BINDING PROTEIN3) |
| OptiV1S19635 | GRMZM2G055447_T01 | 2.32 | 0.03 | Up | 3.47 | 0.04 | Up | POUF |
| OptiV1C00462 | GRMZM2G174942_T02 | 2.21 | 0.03 | Up | 2.70 | 0.04 | Up | Eukaryotic translation initiation factor SUI1, putative |
| OptiV1C01938 | GRMZM2G443715_T01 | 2.06 | 0.04 | Up | 3.00 | 0.04 | Down | Mannan synthase |
Figure 2.
Putative leaf genes involved in regulation of the floral transition of the SAM. A, Genes differentially expressed in the upper most fully expanded leaf comparing developmental stage V3 and each stage at 1 d before transition of the SAM in the maize lines 4F-240 BX 16 (green), 4F-350 CN 2 (red), B77 (yellow), and E2558W (blue). Two genes (ZmMADS1 and ZMM26) are differentially expressed in all lines according to the expected profile shown in Figure 1B. B, Expression profiles of the putative FTi regulators ZmMADS1 (light blue) and ZMM26 (gray). Red lines indicate the SAM transition.
MADS box TFs are widely distributed throughout the plant kingdom and contribute to a variety of processes controlling development and reproduction in plants (Gramzow and Theissen, 2010). To identify proteins homologous to the identified candidate MADS box TFs and to gain first ideas about their functions, a BLASTP search was performed against the plant genomes of maize, rice (Oryza sativa), barley, wheat, and Arabidopsis. ZmMADS1 and ZMM26 homologous proteins were found in all five analyzed plant genomes (Supplemental Table S2). They all contain a MADS domain, a highly conserved C-terminal SOC1 motif (Zhao et al., 2014), as well as the K domain, thus representing MIKC-type (MEF2-like) MADS domain proteins (Becker and Theissen, 2003). In maize, two homologous proteins of ZmMADS1 (ZmMADS56 and GRZM2G070034; Supplemental Fig. S2) and three homologous proteins of ZMM26 (ZMM19, ZmMADS47, and ZMM21; Supplemental Fig. S3) were identified. A phylogenetic tree was generated to reveal proteins most closely related to the identified candidate FTi activators (Fig. 3). Within this tree, ZmMADS1 clusters together with ZmMADS56 of maize and the flowering activator OsMADS50 of rice (Tadege et al., 2003; Lee et al., 2004). ZMM26 appears homologous to ZMM19 of maize and OsMADS22, which was shown to be involved in spikelet development of rice (Sentoku et al., 2005). Moreover, it also clusters with HvBM10, an antagonist of the floral transition in barley (Trevaskis et al., 2007), and the wheat MADS box TFs TaWM22A/B and TaAGL11. The Arabidopsis homolog of ZmMADS1 is AtSOC1, whereas AtSVP and AtAGL24 (AGAMOUS-LIKE24) are homologous to ZMM26. All three proteins were shown to represent important FTi regulators in Arabidopsis (Hartmann et al., 2000; Lee et al., 2000; Yu et al., 2002), substantiating the assumption that the identified candidate genes act in regulating FTi processes in maize.
Figure 3.
Phylogenetic tree of ZmMADS1, ZMM26, and homologous proteins in different plant species (for details, see Supplemental Table S2). The subgroups containing ZmMADS1 and ZMM26 are marked by the red and green bars, respectively. ZmMADS1 and its close homolog ZmMADS56 are shaded in red, and ZMM26 and its close homolog ZMM19 are shaded in green. AtSOC1 and AtSVP are framed in black. The bar represents the number of substitutions per amino acid site.
ZmMADS1 and ZMM19 Show a Diurnal Expression Pattern
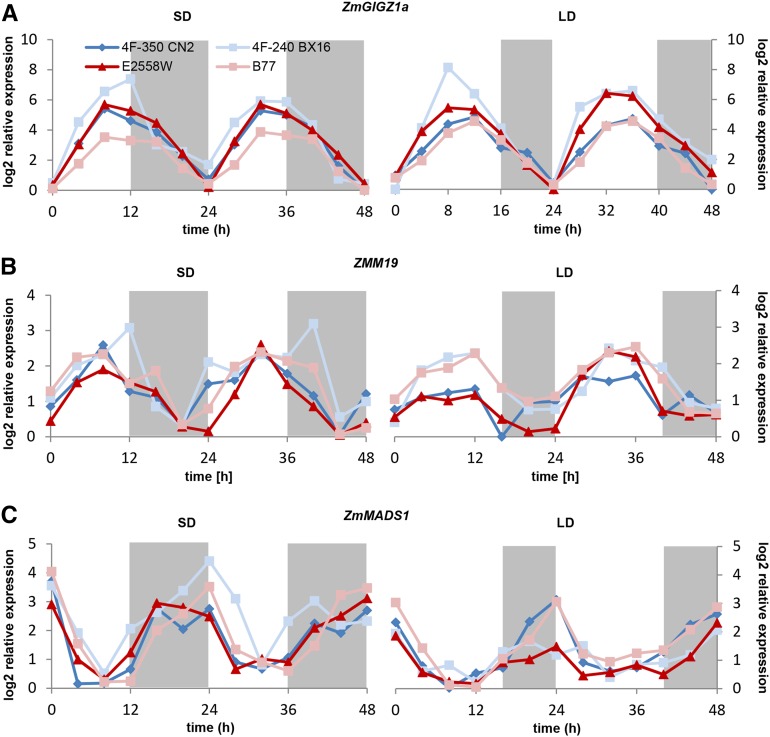
Many photoperiodic FTi genes are expressed in a diurnal manner (Park et al., 1999; Suárez-López et al., 2001). To test the regulation of the identified candidate MADS box protein genes and their homologous genes during daily periods, leaf samples of all four maize inbred lines were harvested at intervals of 4 h in a day-course experiment of 48 h, and transcript level was compared with that of ZmGIGZ1a. ZmGIGZ1a is homologous to the photoperiodic FTi regulator GI of Arabidopsis and is known to be regulated in a diurnal manner (Miller et al., 2008). We could confirm its periodic gene activity in all four maize lines, with expression maximum after 8 to 12 h in light under SD and LD conditions and complete down-regulation during the dark periods (Fig. 4A). A significant diurnal expression pattern was not visible for the AtSVP homologous candidate gene ZMM26 (data not shown). Nevertheless, its maize homolog ZMM19 showed gene activity that was regulated comparable to that of ZmGIGZ1a (Fig. 4B). In contrast to this expression pattern, ZmMADS1 showed a decrease in transcript abundance in light periods, with nearly complete down-regulation after approximately 8 h, followed by an increase of expression during dark periods at both SD and LD conditions (Fig. 4C). Compared with ZmGIGZ1a, this pattern appears inverted, indicating that ZmGIGZ1a might represent a negative regulator of the diurnal ZmMADS1 gene expression pattern. To test this hypothesis, we studied its expression pattern in the GI mutant gi1-mi (Bendix et al., 2013). In the mutant, the diurnal expression pattern of ZmMADS1 was not altered (Supplemental Fig. S4). A significant diurnal regulation could not be observed for the ZmMADS1 maize homologs ZmMADS56 and GRZM2G070034 (data not shown). Detailed expression values and sd values of ZmGIGZ1a, ZmM19, and ZmMADS1 in all four lines analyzed under LD and SD conditions are provided in Supplemental Tables S3 to S8.
Figure 4.
Diurnal regulation of ZmMADS1 and ZMM19 compared with ZmGIGZ1a in SD and LD conditions. Relative expression levels of ZmGIGZ1a (A), ZMM19 (B), and ZmMADS1 (C) are shown in the selected lines 4F-350 CN 2 (dark blue), 4F-240 BX 16 (light blue), E2558W (dark red), and B77 (light red) over a period of 2 d at 4-h intervals under SD (left) and LD (right) conditions. Means of three biological replicates are shown. The relative expression values are log2 transformed; sd values are not depicted (for details, see Supplemental Tables S3–S8). Gray areas indicate dark periods.
ZmMADS1 Can Complement the Late-Flowering Mutant soc1-2 of Arabidopsis
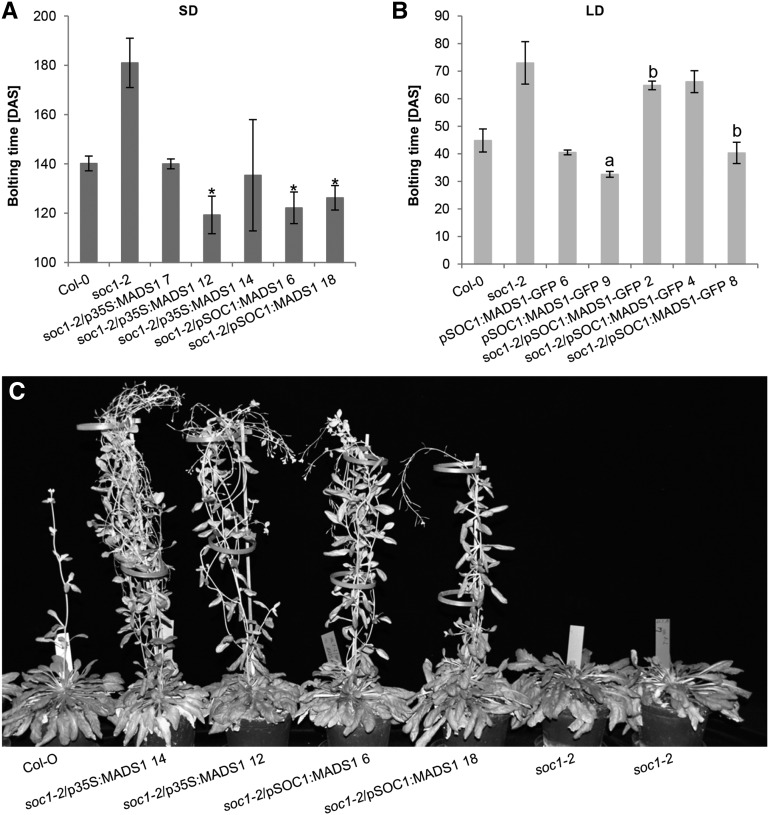
To examine whether ZmMADS1 is indeed involved in FTi regulation pathways, we first applied complementation assays using Arabidopsis soc1-2 T-DNA insertion mutants (Lee et al., 2000; Moon et al., 2003). ZmMADS1 overexpression constructs were generated both under the control of the cauliflower mosaic virus 35S promoter (Odell et al., 1985) and the AtSOC1 promoter. In SD conditions, bolting occurred in Col-0 wild-type Arabidopsis plants at about 140 DAS, whereas bolting in soc1-2 mutants was strongly delayed for approximately 40 d. In all analyzed mutant lines, both overexpression constructs could rescue the late-flowering phenotype at least to the wild-type level (Fig. 5A). Furthermore, some lines even showed a significant early-flowering phenotype. Notably, complemented mutant plants exhibited a different plant architecture compared with wild-type plants by containing more and longer shoots, indicating a higher activity of the SAM (Fig. 5C). Taken together, these data indicate that ZmMADS1 represents a functional ortholog of the FTi transcription activator AtSOC1. Furthermore, a construct generating a MADS1-GFP fusion protein under the control of the AtSOC1 promoter was generated and shown to be functional by complementing the soc1-2 mutant under LD conditions (Fig. 5B). In the complemented plant leaves as well as in tobacco (Nicotiana benthamiana) leaf epidermis cells transiently transformed with the overexpression construct, the ZmMADS1-GFP fusion protein was detected to localize exclusively in the nucleus (Supplemental Fig. S5), supporting its proposed function as a transcriptional regulator.
Figure 5.
Complementation analysis of the Arabidopsis soc1 mutant expressing ZmMADS1 in SD and LD conditions. Expression of ZmMADS1 is under the control of the Arabidopsis SOC1 or the cauliflower mosaic virus 35S promoter. A, Expression of ZmMADS1 could rescue the late-flowering phenotype of the soc1-2 mutation under SD conditions. Asterisks mark plant lines showing even reduced bolting time compared with the Columbia-0 (Col-0) wild type (P ≤ 0.05). B, ZmMADS1-GFP fusion constructs were tested for functionality under LD conditions, where complementation is less effective. Plants significantly different from Col-0 (P ≤ 0.05) are marked with a, and plants significantly different from soc1-2 (P ≤ 0.05) are marked with b (n = 2–7); mean values are indicated, and error bars depict sd. C, Comparison of growth behavior/plant architecture of Col-0, soc1-2, and selected complementation lines as indicated. All plants grew for a period of 148 d under SD conditions.
Overexpression of ZmMADS1 Results in an Early-Flowering Phenotype
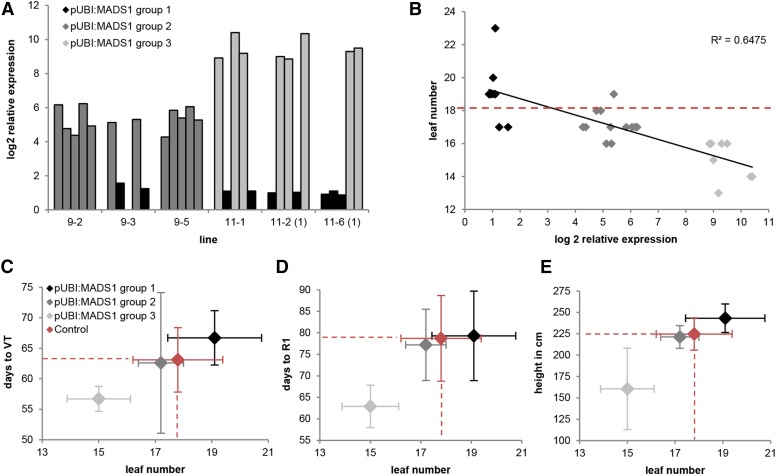
To demonstrate that ZmMADS1 acts as an FTi activator in maize, overexpression constructs under the control of the maize ubiquitin (UBI) promoter were generated and used for Agrobacterium tumefaciens-mediated stable transformation of maize plants. Three offspring lines of two independent transformation events were first analyzed for ZmMADS1 transcript levels. Samples were always taken at the same time in the morning, at 10 am (after a 4-h light period). Compared with wild-type control plants, overexpression plants showed a wide range of transcript levels and, thus, were clustered into three different groups (Fig. 6A). Plants of group 1 showed an increase of log2 relative ZmMADS1 expression levels of up to 3.3, group 2 plants showed expression levels between 3.3 and 6.6, while group 3 plants showed expression levels above log2 of 6.6 (up to maximum log2 = 10.4).
Figure 6.
Characterization of transgenic maize plants overexpressing ZmMADS1 (MADS1) using a ubiquitin promoter (pUBI). A, Relative expression levels of five T1 progeny plants of six independent transgenic lines quantified by qRT-PCR. Log2 expression values were set in relation to the average expression level of ZmMADS1 in eight control plants. Based on log2 relative expression values (log2 REV), plants were clustered into group 1 (log2 REV < 3.3), group 2 (3.3 < log2 REV < 6.6), and group 3 (6.6 < log2 REV). B, Correlation of leaf number and log2 REV of relative ZmMADS1 transcript levels of individual plants of groups 1 to 3 (A). Regression line, correlation coefficients (R), and coefficients of determination (R2) are indicated. The red dashed line indicates the mean value of eight control plants. C to E, Correlations between leaf number and time to reach developmental stage VT (C) or R1 (D) as well as plant height (E) of ZmMADS1 overexpression plants. VT represents the last vegetative stage at which all tassels are no longer enclosed by the upper leaf. R1 represents the stage at which the silks start to emerge. Mean and sd of total leaf number are indicated on the x axis and those of the second trait on the y axis. Mean values of the control group are projected as dashed lines toward the axis. Colors show expression levels of group 1 (black), group 2 (dark gray), group 3 (light gray), and control (red) plants.
Maize plants produce leaves until the transition of the SAM from vegetative to generative growth; thus, the leaf number is strictly dependent on the time of the floral transition. By comparing the leaf number of overexpression plants with the log2-transformed relative expression values depicted in Figure 6A, a negative correlation (r2 = 0.6475) between leaf number and ZmMADS1 transcript levels could be detected (Fig. 6B). To examine this effect in more detail, additional phenotypic traits were used as indicators for FTi. These included day numbers until plants reached developmental stage VT (the last vegetative stage at which all tassels are no longer enclosed by the leaves [Abendroth et al., 2011]; Fig. 6C), until they reached stage R1 (the first reproductive stage at which the silks emerge [Abendroth et al., 2011]; Fig. 6D), as well as plant height (Fig. 6E). Detailed values as well as an additional trait defined as days until anthesis are given in Supplemental Table S9. To ensure maximal comparability between overexpression and control plants, plants that underwent the transformation procedure but did not show increased expression levels were chosen as a control group. In summary group 2, plants with medium overexpression showed phenotypes comparable to the control group regarding days to VT, R1, anthesis, and plant height. In contrast, group 3 plants with strong ZmMADS1 overexpression showed a clear reduction in numbers of days to reach VT, R1, and anthesis and exhibited a smaller plant habitus. On average, flowering occurred about 1 week early, silk emergence happened more than 2 weeks early, but plant size was reduced significantly (on average about 60 cm; Supplemental Table S9). Notably, group 1 plants with minimal overexpression of ZmMADS1 showed slight increases of days to anthesis and plant height, indicating a delayed FTi phenotype. Taken together, these results demonstrate that ZmMADS1 acts as an FTi activator if it is strongly overexpressed. The observation that slight increases of expression levels result in a contrary phenotype with slightly delayed flowering may indicate that ZmMADS1 levels are tightly regulated, that ZmMADS1 functions as an integrator of various FTi signals, but it is also possible that these observation are associated with the genetic background, as the plants investigated are the segregating progeny of hybrid plants.
Down-Regulation of ZmMADS1 Causes a Delay in FTi
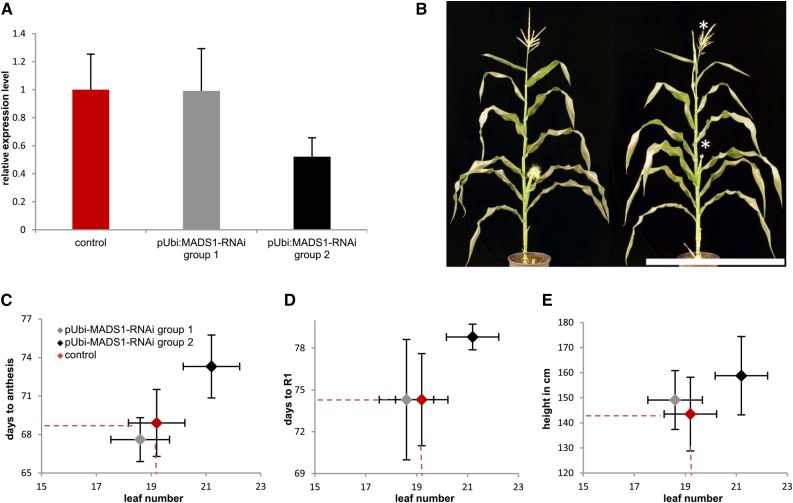
Transgenic maize lines with RNA interference (RNAi)-mediated down-regulation of ZmMADS1 under the control of the maize UBI promoter were created to further analyze its role for FTi regulation. RNAi plants were clustered into two groups based on their relative ZmMADS1 expression levels, with group 1 lacking a significant change in transcript abundance compared with the wild-type group and group 2 exhibiting down-regulation of about 50% (Fig. 7A). Samples were collected when control plants showed maximum expression levels. On average, group 2 plants contained two additional leaves and FTi occurred about 5 d delayed compared with wild-type control plants and plants lacking a reduction in ZmMADS1 expression (Fig. 7B). In detail, we investigated the number of days until plants reached anthesis (Fig. 7C) and developmental stage R1 (Fig. 7D) as well as plant height (Fig. 7E). These traits were used in correlation to leaf number as indicator traits to measure differences in the FTi point. Detailed values are given in Supplemental Table S10. For all three traits, a delay of flowering could be observed in group 2 with reduced ZmMADS1 expression level.
Figure 7.
Characterization of RNAi transgenic maize plants with down-regulated ZmMADS1 (MADS1) expression levels using a ubiquitin promoter (pUBI). A, Relative expression levels of ZmMADS1 in two transgenic groups and a wild-type control group quantified by qRT-PCR (control group, n = 12; group 1, n = 9; and group 2, n = 14). Group 1 showed ZmMADS1 expression levels comparable to wild-type plants. Samples were collected in the morning, when control plants showed maximum expression levels. B, Representative image of segregating wild-type (left) and transgenic progeny (right) plants. Delayed development of tassels and silks (right) compared with the control (left) is indicated by asterisks. Bar = 50 cm. C to E, Correlations between leaf number and time to reach developmental stages of anthesis (C) and R1 (D) and plant height (E) compared between control and the two RNAi plant groups. Means and sd of total leaf number are indicated on the x axis and those of the second trait on the y axis. Mean values of the control group are projected as dashed lines toward the axis. Colors show expression levels of group 1 (gray), group 2 (black), and control (red) plants.
DISCUSSION
Expression of Putative FTi Regulators Is Distinctly Controlled in Different Maize Lines
Flowering in plants is a precisely regulated process in which the SAM switches from vegetative growth to generative growth. Environmental factors like temperature and daylength influence the time of flowering in different plant species with varying degrees of intensity. As one of the most important agricultural crops, maize is widely distributed all over the world, growing in a wide variety of geographical regions and climate conditions. The historical center of maize domestication is Mexico, with around 13 h of daylight in summer. Thus, the ancestors of modern temperate maize as well as tropical modern maize lines were adapted to SD conditions. However, nowadays, temperate maize is generally considered a day-neutral plant, as thousands of years of domestication and breeding appear to have made this crop less sensitive to the photoperiod and enabled it to flower at higher latitude and in LD conditions (Liu et al., 2003). By analyzing the SAM, we determined the time of the floral transition of temperate and tropical maize lines under LD greenhouse conditions. Plants with a tropical background exhibited delayed FTi (31–40 DAS) compared with lines adapted to temperate climate, which showed a strong variation of FTi between 18 and 29 DAS.
To identify new regulators involved in FTi variation in maize, two pairs of related maize lines with a strong difference in FTi were used for leaf transcriptome analysis shortly before the meristem switch occurred. A number of novel and several known regulators could be identified using this approach. The FTi activator ZCN8, for example, represents a maize homolog for Arabidopsis FT (Danilevskaya et al., 2008a; Lazakis et al., 2011; Meng et al., 2011) and was found to be up-regulated in both temperate U.S. dent lines B77 and E2558W but not in lines 4F-240 BX 16 and 4F-350 CN 2 originating from Argentina. Its expression is influenced by the autonomous pathway via the main maize floral regulator ID1 as well as the photoperiodic pathway (Mascheretti et al., 2015). Another gene that was up-regulated only in the lines from Argentina was the MADS box TF ZMM19. Mutations in this gene result in the formation of pod corn displaying dose-dependent spikelet formation defects in ears and tassels with foliaceous elongated glumes (Han et al., 2012; Wingen et al., 2012). Although its role in FTi remained unknown, ZMM19 was classified as a putative homolog of the negative FTi regulator protein SVP from Arabidopsis (Hartmann et al., 2000). In contrast, the yet uncharacterized putative MADS box TF with the identifier GRMZM2G070034 was demonstrated to be up-regulated only in the lines 4F-240 BX 16 and 4F-350 CN 2. Although there is no evidence about its function, it codes for a protein homologous to the Arabidopsis FTi regulator SOC1. Additional genes with differential expression during the floral transition exclusively in these lines are ZmRAP2.7 and ZCN21. ZmRAP2.7 represents an AP2-like TF encoded by a gene downstream of the cis-regulatory element Vgt1, a major FTi-regulating quantitative trait locus (Salvi et al., 2007). ZmRAP2.7 is homologous to AtRAP2.7, a TF that is involved in the floral switch regulation of Arabidopsis (Okamuro et al., 1997; Aukerman and Sakai, 2003). ZCN21, a member of the FT-like II group, is suggested to be involved in FTi regulation in maize (Danilevskaya et al., 2008a; Bouchet et al., 2013) and also was up-regulated in these two lines.
ZmMADS1 and ZMM26 Showing a Diurnal Expression Pattern Are Strongly Up-Regulated during the Floral Transition
In contrast to the aforementioned genes, the genes ZmMADS1 and ZMM26 were the only candidates found to be up-regulated in all four analyzed maize lines correlated with the FTi switch in early- and late-flowering genotypes. ZmMADS1 is homologous to the Arabidopsis floral inductive pathway integrator SOC1 and the FTi regulators OsMADS50 and OsMADS56 of rice. ZmMADS1 is up-regulated in leaves during the floral transition similar to OsMADS50 in the whole shoot (Tadege et al., 2003) and AtSOC1 in the SAM (Lee et al., 2000). Just like ZMM19 (see above), the second candidate regulator ZMM26 represents a protein homologous to Arabidopsis SVP. In a phylogenetic comparison, they cluster together with OsMADS22, OsMADS47, and OsMADS55, three proteins involved in flowering processes in rice. Ectopic expression of OsMADS22 was shown to alter the meristem indeterminacy in rice spikelets (Sentoku et al., 2005) and leads to abnormal flower formation in Arabidopsis (Lee et al., 2012), but its role in FTi remained unclear. Overexpression of OsMADS47 and OsMADS55 results in abnormal flower formation in Arabidopsis and, in the case of OsMADS55, also in a delay of flowering induction (Fornara et al., 2008; Lee et al., 2012). All of these genes show a different expression pattern in leaves. Whereas OsMADS22 expression stays constant, the transcript level of OsMADS47 decreases while the level of OsMADS55 increases (Lee et al., 2008) like ZMM26, which we showed to be up-regulated during FTi. The expression pattern and overexpression phenotypes thus indicate that SVP homologs in grasses likely have developed different/additional functions that remain to be elucidated. Notably, we found that the ZMM26 homolog ZMM19 displays a diurnal expression pattern similar to the circadian clock-associated GI gene ZmGIGZ1a, both peaking 8 to 12 h after light induction in SD and LD conditions. At the end of the dark period, both genes are almost silent. Considering that both genes show almost identical expression peaks, it is unlikely that ZmGIGZ1a can act as a direct upstream regulator of ZMM19. Genes that appear as targets of ZmGIGZ1a display either delayed peaks at the end of the light period (CONZ1) or an inverted expression pattern (ZCN8; Bendix et al., 2013). Although ZmMADS1 also shows an inverted expression pattern, it is not a target of ZmGIGZ1a. Whether its regulation depends on the second maize GI homolog ZmGIGZ1b (Khan et al., 2010) remains to be shown in further experimentation.
ZmMADS1 Is Functionally Conserved to SOC1 and Acts as an FTi Regulator in Maize
Therefore, ZMM26 and ZmMADS1 and their closest homologs in maize were selected as promising candidates for line-independent FTi regulators, and their photoperiod-dependent expression pattern in leaves was studied in more detail. Expression of the SVP homolog ZMM26 was not observed to be influenced by a day rhythm. SVP of Arabidopsis is controlled by the GA3 and the autonomous pathways and acts independently from the photoperiodic pathway (Li et al., 2008); therefore, it is likely that ZMM26 also is not involved in photoperiodic flowering control if representing the functional equivalent in maize. The rice SVP-like proteins OsMADS22 and OsMADS55 can interact directly with the Arabidopsis flowering activators AGL24, AP1, and, in the case of OsMADS55, also with FLC in yeast two-hybrid screens. Furthermore, OsMADS55 can complement the early-flowering phenotype of Arabidopsis svp mutants, indicating that it has indeed an evolutionarily conserved function in flowering comparable to SVP (Lee et al., 2012). The genes coding for the homologous proteins HvBM1, HvBM10, and HvVRT2 in barley also are not diurnally expressed. They do not seem to be involved in the control of FTi, but HvBM1 and HvBM10 overexpression can lead to the inhibition of floral development and floral reversion (Trevaskis et al., 2007). It is possible, therefore, that ZMM26 also is involved in other processes than FTi. Expression of its homologous gene ZMM19, however, was controlled by a day rhythm, indicating that it may encode a maize SVP homolog. In contrast to ZMM26, the expression of ZmMADS1 in leaves was regulated by daylength, with an expression maximum at dawn under both SD and LD conditions. This pattern was in accordance with the expression profiles of the homologous rice FTi regulators OsMADS50 and OsMADS56, which also were demonstrated to be diurnally regulated (Ryu et al., 2009). ZmMADS1 was shown recently to induce an early-flowering phenotype when ectopically overexpressed in Arabidopsis under LD conditions, and it leads to up-regulation of the TFs AtLEAFY and AtAP1. This observation indicates that it can promote flowering in an evolutionarily conserved pathway (Zhao et al., 2014), as both LEAFY and AP1 control the expression of a set of target genes with a role in the onset of flowering (Winter et al., 2015). Furthermore, we could demonstrate that ZmMADS1 expression under the control of the SOC1 promoter of Arabidopsis is able to fully rescue the late-flowering phenotype of the Arabidopsis soc1-2 mutant under SD conditions and in some lines also under LD conditions. In LD conditions, we tested ZmMADS1:GFP fusion proteins that may not always be as effective as ZmMADS1 lacking the GFP reporter. Arabidopsis and tobacco leaves expressing ZmMADS1-GFP showed localization of the fusion protein in the nucleus, supporting the idea of ZmMADS1 acting as a TF. AtSOC1 as well as its rice homologs OsMADS50 and OsMADS55 also have been demonstrated to localize in the nucleus (Liu et al., 2009; Ryu et al., 2009). Taken together, this supported the idea of ZmMADS1 acting as a TF promoting flowering. Variation of FTi indicates that ZmMADS1 might act in a dosage-dependent manner, as already reported for AtSOC1 (Borner et al., 2000).
To prove the role of ZmMADS1 as an FTi regulator, we next analyzed transgenic lines with down-regulation and overexpression of ZmMADS1 under LD conditions. Plants with ZmMADS1 activity reduced to about 50% compared with the wild-type level showed an about 5-d delayed flowering phenotype regarding the number of days to anthesis and stage R1. Plants were higher and contained on average two additional leaves, supporting the idea of ZmMADS1 functioning as a positive FTi regulator. Interestingly, overexpression of ZmMADS1 caused a flowering phenotype depending on the expression level of ZmMADS1. An increase of expression intensity of less than 10-fold resulted in a slight delay of flowering, whereas plants overexpressing ZmMADS1 more than 100-fold flowered up to 2 weeks earlier compared with control plants and contained, on average, three fewer leaves. This observation indicates that ZmMADS1-regulated flowering behavior is complex, and we assume that it not only represents an FTi activator but fulfills additional roles in the complex network of flowering control and also may play a role in other developmental processes, as it is also expressed in egg cells, zygotes, and developing flowers (Heuer et al., 2001). Some MADS box TFs involved in specification of the floral meristem identify of Arabidopsis also have genetically separable functions; for example, AP1 also functions in the determination of sepal and petal identity, while FRUITFUL also is involved in fruit valve specification (for review, see Kater et al., 2006).
CONCLUSION
In this study, we revealed several putative FTi regulators in maize that are activated in leaves associated with the SAM transition. Differences in their expression during the floral transition in various maize lines indicate that the gene regulatory network controlling FTi in maize is very complex and not universal in all lines. Studies that aimed to identify, for example, targets of the leaf-restricted TF ID1 regulating the transition to flowering support this assumption, as the identified up- and down-regulated genes (Coneva et al., 2007, 2012) do not overlap with the genes found in this study. Thus, we conclude that, during adaption to a range of climates, maize lines have evolved distinct severities of the roles of different floral control key players. Besides identifying candidate genes with differential expression in groups of inbred lines, we could further demonstrate that the MADS box TF ZmMADS1 represents a dosage-dependent regulator of FTi in maize and shares a conserved function with its homologous protein SOC1 of Arabidopsis. Further characterization of this transcriptional regulator, which appears to be present and similarly regulated in all investigated lines, will now help us understand how FTi pathways are integrated and how the strong variation of FTi is regulated in a day-neutral and important grass crop species.
MATERIALS AND METHODS
Bioinformatics Tools
Information about gene and protein sequences was derived from databases of the National Center for Biotechnology Information (http://ncbi.nlm.nih.gov/), Gramene (http://www.gramene.org/), The Arabidopsis Information Resource (http://arabidopsis.org/), and the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu). Proteins homologous to ZmMADS1 and ZMM26 were identified using candidate protein sequences as query for BLASTP searches with default settings against the genomes of maize (Zea mays), rice (Oryza sativa), and Arabidopsis (Arabidopsis thaliana), available at the PlantGDB platform (http://plantgdb.org). For the genomes of barley (Hordeum vulgare) and wheat (Triticum aestivum), BLASTP search was performed at the National Center for Biotechnology Information with an E value cutoff of 1e−70. Protein sequence alignments were generated using the muscle algorithm of SeaView version 4.3.5 (Gouy et al., 2010) and were visualized by the program GeneDoc version 2.7.000 (Nicholas et al., 1997). A phylogenetic tree based on the alignments was constructed using the neighbor-joining method of SeaView. To test the relationship, the bootstrap method with 1,000 replicates was applied.
Plant Growth Conditions
All maize lines used in this study are publicly available and were obtained from the Maize Genetics Cooperation Stock Center (maizecoop.cropsci.uiuc.edu), except A188 (Rgbg.) and B73 (Rgbg.), which were grown and propagated exclusively in greenhouses at the University of Regensburg. Lines HiII A and HiII B were derived from the Plant Transformation Facility of Iowa State University, and the inbred line A632 and the gi1-mi−/− (CB51xsib) mutant was kindly provided by Frank G. Harmon. In the greenhouse, plants were grown under LD conditions (16 h of light) at 25°C during light and 20°C during dark periods. In climate chambers, plants were grown under both LD and SD (12 h of light) conditions at 26°C during light and 18°C during dark periods and a constant air humidity of 60%. Maize plants were illuminated using Philips MASTER HPI-T Plus 400W/645 E40 1SL and Philips MASTER GreenPower CG T 400W E40 lighting. Arabidopsis plants were grown in climate chambers under LD conditions (16 h of light) at 20°C and 70% air humidity during light and 18°C and 65% air humidity during dark periods. Under SD conditions (8 h of light), temperature and air humidity were kept constantly at 22°C and 70%. Arabidopsis plants were illuminated using Sylvania Luxline Plus F30W/840 cool-white deluxe and Sylvania Luxline Plus F30W/830 lighting. Nicotiana benthamiana plants were grown in the greenhouse under the same conditions as maize plants.
Extraction of Total RNA and Generation of cDNA
Microarray leaf samples as well as leaf samples of transgenic maize lines were taken for RNA extraction in the morning at 10 am (after a 4-h light period). Sample collection for the day-course experiment was started with the beginning of the light period at 6 am. Extraction of total RNA of 100 to 200 mg of the leaf material was performed as described (Logemann et al., 1987). RNA samples were treated with RNase-free DNase I, and cDNA was synthesized using SuperScript III reverse transcriptase, oligo(dT)18 primer, and RiboLock RNase inhibitor, according to the manufacturer’s protocol (all chemicals derived from Thermo Fisher Scientific).
Microarray Analysis
The quality of the RNA used for hybridization was analyzed using a 2100 Bioanalyzer (Agilent Technologies). The synthesis of cDNA and cRNA was performed according to the one-color microarray-based gene expression analysis protocol by Agilent Technologies. A total of 1.65 mg of the RNA sample was loaded on one-color microarrays with custom-designed oligonucleotide probes (Agilent 025271). Microarray data were normalized to the 75th percentile within each array and were analyzed using the Agilent GeneSpring software package (according to Pick et al. [2011]).
Quantitative Real-Time PCR Analysis
Quantitative real-time PCR was performed using the KAPA SYBR Fast QPCR Master Mix Universal Kit (Peqlab Biotechnologie) and a Master Cycler realplex2 (Eppendorf). The optimal annealing temperature was determined for each primer pair by gradient PCR (for primer sequences, see Supplemental Table S11). For PCR products smaller than 200 bp, a two-step qRT-PCR, and for larger fragments, a three-step qRT-PCR with 40 cycles, was performed, followed by a melting curve analysis to test product specificity. qRT-PCR data were analyzed using the ΔΔCT method (Livak and Schmittgen, 2001). GLYCERALDEHYDE-3-PHOSPHATASE DEHYDROGENASE (GRMZM2G046804) and FOLYLPOLY-GLUTAMATE SYNTHASE (GRMZM2G393334) of maize (Manoli et al., 2012) were used as reference genes for normalization.
Generation of Constructs
To design constructs for ZmMADS1 and ZmMADS1-GFP overexpression in maize and Arabidopsis, the coding sequence of ZmMADS1 (GRMZM2G171365) was amplified using the primer pair CDS MADS1 5′ pENTR/CDS MADS1 3′ for introduction of a stop codon and primer pair CDS MADS1 5′ pENTR/CDS MADS1 3′ degenBamHI without the stop codon (for primer sequences, see Supplemental Table S11). Fragments were cloned into the pENTR/D-TOPO vector using the pENTR Directional TOPO Cloning Kit (Thermo Fisher Scientific). These entry vectors were used for recombination reactions mediated by the Gateway LR Clonase II enzyme mix (Thermo Fisher Scientific). For maize transformation, the entry vector with ZmMADS1 including a stop codon was combined with the destination vector pUbi:Gate, creating the vector pUbi:ZmMADS1. The pUbi:Gate vector is based on the pNOS-AB-M vector (DNA Cloning Service) and contains a 2-kb fragment of the maize POLYUBIQUITIN promoter (GRMZM2G409726; Christensen et al., 1992) and a Gateway cassette (M. Gahrtz, unpublished data). The intermediate vector pUbi:ZmMADS1 was digested with BamHI and XhoI, and the fragment containing the promoter, the coding sequence, and the NOS terminator was cloned into pTF101.1 (Paz et al., 2004). To generate the construct for RNAi-controlled down-regulation of ZmMADS1, a 163-bp fragment from the 3′ end of the ZmMADS1 coding sequence was amplified using the primer pair M5RNAi-s-BsrGI/M5RNAi-s-MluI for sense orientation and primer pair M5RNAi-a-BamH/M5RNAi-a-EcoRI for an antisense construct. These PCR products were introduced into the ZmEAL1 RNAi construct (Krohn et al., 2012), replacing the ZmEAL1 sequences using the restriction enzymes BsrGI and MluI for sense orientation and BamHI and EcoRI for antisense orientation. The RNAi cassette containing the maize UBI promoter, the ZmMADS1 RNAi stem loop, and the NOS terminator was amplified with the primer pair M5RNAi1-XmaI-f/M5RNAi1-HindIII-r and cloned into the vector pCR-Blunt II-TOPO vector of the Zero Blunt TOPO PCR Cloning Kit (Thermo Fisher Scientific). Using the restriction enzymes XmaI and HindIII, the RNAi cassette was cloned into the transformation vector pTF101.1. Constructs for the overexpression of ZmMADS1-GFP in Arabidopsis were generated by performing Clonase reactions of the entry vector containing ZmMADS1 without a stop codon with the destination vector pB7FWG2,0 (Karimi et al., 2002) and a modified version of pB7FWG2,0. In this modified version, the 35S promoter was replaced by a SOC1 promoter fragment of 1,951 bp upstream of the SOC1 gene (AT2G45660) using SacI and SpeI restriction sites. The resulting vectors were named pUbi:MADS1-GFP and pSOC1:MADS1-GFP.
Plant Transformation
Overexpression and RNAi constructs of ZmMADS1 were sent to the Plant Transformation Facility of Iowa State University for Agrobacterium tumefaciens-mediated stable transformation of HiII A × HiII B hybrids (protocol according to Frame et al. [2002]). Arabidopsis plants were stably transformed using the floral dip method (Clough and Bent, 1998). Infiltration of N. benthamiana leaves was performed according to Bartetzko et al. (2009), with resuspension of the bacteria in buffer (10 mm MgCL2, 10 mm MES-KOH, pH 5.7, and 100 µm actosyringone) before infiltration.
Microscopic Analysis
The developmental stage of dissected SAMs was determined using the binocular microscopes Zeiss Discovery.V8 and Nikon SMZ 645. Examination of infiltrated N. benthamiana leaf epidermis cells was performed using a Leica SP8 confocal microscopy system and the HC PL APO CS2 20×/0.75 Leica glycerol-immersion objective. GFP was excited by a 488-nm argon laser, and emission was detected by a hybrid detector at 500 to 550 nm.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Validation of microarray results by RT-qPCR.
Supplemental Figure S2. Protein sequence alignment of ZmMADS1 and homologous proteins.
Supplemental Figure S3. Protein sequence alignment of ZMM26 and its homologs.
Supplemental Figure S4. Diurnal expression of ZmMADS1 is not dependent on GI.
Supplemental Figure S5. Subcellular localization of ZmMADS1-GFP.
Supplemental Table S1. Time points of floral transition in SD and LD conditions.
Supplemental Table S2. Identifiers of proteins homologous to ZmMADS1 and ZMM26.
Supplemental Table S3. Log2-transformed relative expression values of ZmGIGZ1a in leaves of selected inbred lines under SD conditions.
Supplemental Table S4. Log2-transformed relative expression values of ZmGIGZ1a in leaves of selected inbred lines under LD conditions.
Supplemental Table S5. Log2-transformed relative expression values of ZmMADS1 in leaves of selected inbred lines under SD conditions.
Supplemental Table S6. Log2-transformed relative expression values of ZmMADS1 in leaves of selected inbred lines under LD conditions.
Supplemental Table S7. Log2-transformed relative expression values of ZMM19 in leaves of selected inbred lines under SD conditions.
Supplemental Table S8. Log2-transformed relative expression values of ZMM19 in leaves of selected inbred lines under LD conditions.
Supplemental Table S9. Flowering time in pUBI:MADS1 plants.
Supplemental Table S10. Flowering time in pUBI:MADS1 RNAi plants.
Supplemental Table S11. Primers used in this study for RT-qPCR.
Supplementary Material
Acknowledgments
We thank Günther Peissig, Ursula Wittmann, and Armin Hildebrand for plant care as well as Kan Wang and Frank G. Harmon for providing various maize inbred and mutant lines.
Glossary
- FTi
flowering time
- SAM
shoot apical meristem
- LD
long-day
- SD
short-day
- DAS
days after sowing
- TF
transcription factor
- RNAi
RNA interference
Footnotes
This work was supported by the German Federal Ministry of Education and Research in the frame of OPTIMAS (grant no. FKZ 0315430 to U.So. and T.D.), by the Alexander von Humboldt Foundation (postdoctoral fellowship to L.-Z.Z.), and by the University of Regensburg (to S.B.).
Articles can be viewed without a subscription.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Abendroth LJ, Elmore RW, Boyer MJ, Marlay SK (2011) Corn Growth and Development. Iowa State University Extension, Ames, IA [Google Scholar]
- Amasino R. (2004) Vernalization, competence, and the epigenetic memory of winter. Plant Cell 16: 2553–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés F, Porri A, Torti S, Mateos J, Romera-Branchat M, García-Martínez JL, Fornara F, Gregis V, Kater MM, Coupland G (2014) SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc Natl Acad Sci USA 111: E2760–E2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartetzko V, Sonnewald S, Vogel F, Hartner K, Stadler R, Hammes UZ, Börnke F (2009) The Xanthomonas campestris pv. vesicatoria type III effector protein XopJ inhibits protein secretion: evidence for interference with cell wall-associated defense responses. Mol Plant Microbe Interact 22: 655–664 [DOI] [PubMed] [Google Scholar]
- Becker A, Theissen G (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Bendix C, Marshall CM, Harmon FG (2015) Circadian clock genes universally control key agricultural traits. Mol Plant 8: 1135–1152 [DOI] [PubMed] [Google Scholar]
- Bendix C, Mendoza JM, Stanley DN, Meeley R, Harmon FG (2013) The circadian clock-associated gene gigantea1 affects maize developmental transitions. Plant Cell Environ 36: 1379–1390 [DOI] [PubMed] [Google Scholar]
- Blümel M, Dally N, Jung C (2015) Flowering time regulation in crops: what did we learn from Arabidopsis? Curr Opin Biotechnol 32: 121–129 [DOI] [PubMed] [Google Scholar]
- Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S (2000) A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J 24: 591–599 [DOI] [PubMed] [Google Scholar]
- Bouchet S, Servin B, Bertin P, Madur D, Combes V, Dumas F, Brunel D, Laborde J, Charcosset A, Nicolas S (2013) Adaptation of maize to temperate climates: mid-density genome-wide association genetics and diversity patterns reveal key genomic regions, with a major contribution of the Vgt2 (ZCN8) locus. PLoS ONE 8: e71377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AH, Sharrock RA, Quail PH (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol 18: 675–689 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colasanti J, Coneva V (2009) Mechanisms of floral induction in grasses: something borrowed, something new. Plant Physiol 149: 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Yuan Z, Sundaresan V (1998) The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603 [DOI] [PubMed] [Google Scholar]
- Colmsee C, Mascher M, Czauderna T, Hartmann A, Schlüter U, Zellerhoff N, Schmitz J, Bräutigam A, Pick TR, Alter P, et al. (2012) OPTIMAS-DW: a comprehensive transcriptomics, metabolomics, ionomics, proteomics and phenomics data resource for maize. BMC Plant Biol 12: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coneva V, Guevara D, Rothstein SJ, Colasanti J (2012) Transcript and metabolite signature of maize source leaves suggests a link between transitory starch to sucrose balance and the autonomous floral transition. J Exp Bot 63: 5079–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coneva V, Zhu T, Colasanti J (2007) Expression differences between normal and indeterminate1 maize suggest downstream targets of ID1, a floral transition regulator in maize. J Exp Bot 58: 3679–3693 [DOI] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Hou Z, Ananiev EV, Simmons CR (2008a) A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol 146: 250–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Selinger DA, Deschamps S, Hermon P, Vansant G, Gupta R, Ananiev EV, Muszynski MG (2008b) Involvement of the MADS-box gene ZMM4 in floral induction and inflorescence development in maize. Plant Physiol 147: 2054–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Danilevskaya O, Abadie T, Messina C, Coles N, Cooper M (2012) A gene regulatory network model for floral transition of the shoot apex in maize and its dynamic modeling. PLoS ONE 7: e43450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Gregis V, Pelucchi N, Colombo L, Kater M (2008) The rice StMADS11-like genes OsMADS22 and OsMADS47 cause floral reversions in Arabidopsis without complementing the svp and agl24 mutants. J Exp Bot 59: 2181–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame BR, Shou H, Chikwamba RK, Zhang Z, Xiang C, Fonger TM, Pegg SE, Li B, Nettleton DS, Pei D, et al. (2002) Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol 129: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224 [DOI] [PubMed] [Google Scholar]
- Gramzow L, Theissen G (2010) A hitchhiker's guide to the MADS world of plants. Genome Biol 11: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JJ, Jackson D, Martienssen R (2012) Pod corn is caused by rearrangement at the Tunicate1 locus. Plant Cell 24: 2733–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Höhmann S, Nettesheim K, Wisman E, Saedler H, Huijser P (2000) Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J 21: 351–360 [DOI] [PubMed] [Google Scholar]
- Heuer S, Hansen S, Bantin J, Brettschneider R, Kranz E, Lörz H, Dresselhaus T (2001) The maize MADS box gene ZmMADS3 affects node number and spikelet development and is co-expressed with ZmMADS1 during flower development, in egg cells, and early embryogenesis. Plant Physiol 127: 33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA (2007) FT protein acts as a long-range signal in Arabidopsis. Curr Biol 17: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Jung C, Müller AE (2009) Flowering time control and applications in plant breeding. Trends Plant Sci 14: 563–573 [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kater MM, Dreni L, Colombo L (2006) Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J Exp Bot 57: 3433–3444 [DOI] [PubMed] [Google Scholar]
- Khan MRG, Ai XY, Zhang JZ (2014) Genetic regulation of flowering time in annual and perennial plants. Wiley Interdiscip Rev RNA 5: 347–359 [DOI] [PubMed] [Google Scholar]
- Khan S, Rowe SC, Harmon FG (2010) Coordination of the maize transcriptome by a conserved circadian clock. BMC Plant Biol 10: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Krohn NG, Lausser A, Juranić M, Dresselhaus T (2012) Egg cell signaling by the secreted peptide ZmEAL1 controls antipodal cell fate. Dev Cell 23: 219–225 [DOI] [PubMed] [Google Scholar]
- Lazakis CM, Coneva V, Colasanti J (2011) ZCN8 encodes a potential orthologue of Arabidopsis FT florigen that integrates both endogenous and photoperiod flowering signals in maize. J Exp Bot 62: 4833–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Park SH, Ahn JH (2012) Functional conservation and diversification between rice OsMADS22/OsMADS55 and Arabidopsis SVP proteins. Plant Sci 185-186: 97–104 [DOI] [PubMed] [Google Scholar]
- Lee S, Choi SC, An G (2008) Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. Plant J 54: 93–105 [DOI] [PubMed] [Google Scholar]
- Lee S, Kim J, Han JJ, Han MJ, An G (2004) Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J 38: 754–764 [DOI] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H (2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Liu C, Xi W, Shen L, Tan C, Yu H (2009) Regulation of floral patterning by flowering time genes. Dev Cell 16: 711–722 [DOI] [PubMed] [Google Scholar]
- Liu K, Goodman M, Muse S, Smith JS, Buckler E, Doebley J (2003) Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165: 2117–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L (1987) Improved method for the isolation of RNA from plant tissues. Anal Biochem 163: 16–20 [DOI] [PubMed] [Google Scholar]
- Manoli A, Sturaro A, Trevisan S, Quaggiotti S, Nonis A (2012) Evaluation of candidate reference genes for qPCR in maize. J Plant Physiol 169: 807–815 [DOI] [PubMed] [Google Scholar]
- Mascheretti I, Turner K, Brivio RS, Hand A, Colasanti J, Rossi V (2015) Florigen-encoding genes of day-neutral and photoperiod-sensitive maize are regulated by different chromatin modifications at the floral transition. Plant Physiol 168: 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Muszynski MG, Danilevskaya ON (2011) The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 23: 942–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TA, Muslin EH, Dorweiler JE (2008) A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta 227: 1377–1388 [DOI] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Muszynski MG, Dam T, Li B, Shirbroun DM, Hou Z, Bruggemann E, Archibald R, Ananiev EV, Danilevskaya ON (2006) delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol 142: 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB, Deerfield DW (1997) GeneDoc: analysis and visualization of genetic variation. Embnet.news 4: 14 [Google Scholar]
- Odell JT, Nagy F, Chua NH (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313: 810–812 [DOI] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA 94: 7076–7081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582 [DOI] [PubMed] [Google Scholar]
- Paz MM, Shou HX, Guo ZB, Zhang ZY, Banerjee AK, Wang K (2004) Assessment of conditions affecting Agrobacterium-mediated soybean transformation using the cotyledonary node explant. Euphytica 136: 167–179 [Google Scholar]
- Pick TR, Bräutigam A, Schlüter U, Denton AK, Colmsee C, Scholz U, Fahnenstich H, Pieruschka R, Rascher U, Sonnewald U, et al. (2011) Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. Plant Cell 23: 4208–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu CH, Lee S, Cho LH, Kim SL, Lee YS, Choi SC, Jeong HJ, Yi J, Park SJ, Han CD, et al. (2009) OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ 32: 1412–1427 [DOI] [PubMed] [Google Scholar]
- Salvi S, Sponza G, Morgante M, Tomes D, Niu X, Fengler KA, Meeley R, Ananiev EV, Svitashev S, Bruggemann E, et al. (2007) Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad Sci USA 104: 11376–11381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentoku N, Kato H, Kitano H, Imai R (2005) OsMADS22, an STMADS11-like MADS-box gene of rice, is expressed in non-vegetative tissues and its ectopic expression induces spikelet meristem indeterminacy. Mol Genet Genomics 273: 1–9 [DOI] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Tadege M, Sheldon CC, Helliwell CA, Upadhyaya NM, Dennis ES, Peacock WJ (2003) Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotechnol J 1: 361–369 [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Tadege M, Hemming MN, Peacock WJ, Dennis ES, Sheldon C (2007) Short vegetative phase-like MADS-box genes inhibit floral meristem identity in barley. Plant Physiol 143: 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Wingen LU, Münster T, Faigl W, Deleu W, Sommer H, Saedler H, Theißen G (2012) Molecular genetic basis of pod corn (Tunicate maize). Proc Natl Acad Sci USA 109: 7115–7120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter CM, Yamaguchi N, Wu MF, Wagner D (2015) Transcriptional programs regulated by both LEAFY and APETALA1 at the time of flower formation. Physiol Plant 155: 55–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Shah T, Warburton ML, Buckler ES, McMullen MD, Crouch J (2009) Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS ONE 4: e8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Xu Y, Tan EL, Kumar PP (2002) AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc Natl Acad Sci USA 99: 16336–16341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Luo Y, Zhang Z, Xu M, Wang W, Zhao Y, Zhang L, Fan Y, Wang L (2014) ZmSOC1, a MADS-box transcription factor from Zea mays, promotes flowering in Arabidopsis. Int J Mol Sci 15: 19987–20003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.