The regulator protein OsNPR1 inhibits rice growth by enhancing the OsGH3.8 amido synthase of rice.
Abstract
Systemic acquired resistance is a long-lasting and broad-spectrum disease resistance to pathogens. Our previous study demonstrated that overexpression of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (OsNPR1), a master gene for systemic acquired resistance in rice (Oryza sativa), greatly enhanced resistance to bacterial blight caused by Xanthomonas oryzae pv oryzae. However, the growth and development of the OsNPR1 overexpression (OsNPR1-OX) plants were restrained, and the mechanism remained elusive. In this study, we dissected the OsNPR1-induced growth inhibition. We found that the OsNPR1-OX lines displayed phenotypes mimicking auxin-defective mutants, with decreases in root system, seed number and weight, internode elongation, and tiller number. Whole-genome expression analysis revealed that genes related to the auxin metabolism and signaling pathway were differentially expressed between the OsNPR1-OX and wild-type plants. Consistently, the indole-3-acetic acid (IAA) content was decreased and the auxin distribution pattern was altered in OsNPR1-OX plants. Importantly, we found that some GH3 family members, in particular OsGH3.8 coding IAA-amido synthetase, were constitutively up-regulated in OsNPR1-OX plants. Decreased OsGH3.8 expression by RNA interference could partially restore IAA level and largely rescue the restrained growth and development phenotypes but did not affect the disease resistance of OsNPR1-OX plants. Taken together, we revealed that OsNPR1 affects rice growth and development by disrupting the auxin pathway at least partially through indirectly up-regulating OsGH3.8 expression.
Rice (Oryza sativa) is a major staple food crop for more than half of the world’s population. However, diseases are a major challenge to rice production. Thus, breeding for disease resistance has been a long-lasting goal of rice breeding programs. Application of major disease resistance (R) genes and quantitative trait loci has contributed greatly to increasing rice resistance against diverse pathogens (McDonald and Linde, 2002; Kou and Wang, 2010). However, due to the arms race between pathogens and plants, plants frequently lose their resistance against specific pathogen strains (Jones and Dangl, 2006; Dangl et al., 2013). Therefore, breeding for varieties with durable and broad-spectrum disease resistance is critical to sustainable agricultural development (Zhang, 2007).
Recent progress in plant defense signaling and rice functional genomics provides new approaches for developing rice varieties with broad-spectrum resistance. It is well recognized that during long-term coevolution between plant hosts and pathogens, plants have developed a complicated immune system composed of both preformed and inducible defense mechanisms (Jones and Dangl, 2006; Spoel and Dong, 2012). The preformed defense system includes physical and chemical barriers consisting of antimicrobial molecules, such as saponins and quinines (Bednarek and Osbourn, 2009). The inducible defense mechanisms mainly include pathogen-associated molecular pattern-triggered immunity and effector-triggered immunity. Pathogen-associated molecular pattern-triggered immunity offers nonspecific basal defense and controls the extent of pathogen proliferation, while effector-triggered immunity inhibits pathogen infection and growth in a more pronounced manner (Jones and Dangl, 2006). In addition to restricting pathogens locally, effector-triggered immunity also can initiate systemic acquired resistance (SAR) via the production of signaling molecules, such as salicylic acid (SA) and methyl salicylic acid (Fu and Dong, 2013; Shah and Zeier, 2013). The initiation of SAR leads to systemic expression of pathogenesis-related (PR) genes in the uninoculated distal tissues to protect the rest of the plant against future pathogen attacks. SAR not only provides broad-spectrum resistance against various pathogens but also lasts for days to weeks/months, possibly extending through the entire growing season (Fu and Dong, 2013). Because of the nature of its long-lasting and broad-spectrum resistance, SAR is promising for breeding disease-resistant crops through genetic modifications of the signaling pathway (Wally and Punja, 2010).
Over the past two decades, significant progress has been made in understanding the SAR signaling pathway. A milestone is the cloning of Arabidopsis (Arabidopsis thaliana) NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1; herein referred to as AtNPR1), also known as NONIMMUNITY1, a key regulator of SAR (Cao et al., 1994; Ryals et al., 1997). To date, NPR1 orthologs have been characterized from many plant species, and overexpression of AtNPR1 or other orthologs has resulted in increased broad-spectrum resistance to pathogens in a variety of plant species, which functions as a transcriptional coactivator to activate defense gene expression (Cao et al., 1998; Friedrich et al., 2001; Lin et al., 2004; Malnoy et al., 2007; Wally et al., 2009; Parkhi et al., 2010; Zhang et al., 2012). Similarly, the rice ortholog of AtNPR1 (OsNPR1; also known as NH1) also has been cloned and functionally identified (Chern et al., 2005; Yuan et al., 2007). Therefore, NPR1 provides a promising candidate for genetic engineering of broad-spectrum disease resistance in crops.
It is well documented that cross talk between defense and development fine-tunes disease resistance and growth, indicating the fitness cost of disease resistance (López et al., 2008; Spoel and Dong, 2008; Robert-Seilaniantz et al., 2011; Yang et al., 2013). In our previous work, we found that transgenic rice constitutively expressing OsNPR1 greatly increased disease resistance to Xanthomonas oryzae pv oryzae (Xoo; Yuan et al., 2007). However, these OsNPR1 overexpression lines (OsNPR1-OX) displayed a dwarf phenotype in the paddy field. Another group observed a similar dwarf phenotype of OsNPR1/NH1 overexpression plants in a growth chamber (Chern et al., 2005). Apparently, this defect limits the application of OsNPR1 in improving elite rice varieties for enhancing disease resistance. Therefore, it is necessary to dissect the underlying mechanism of OsNPR1-mediated growth retardation. In this study, we investigated the OsNPR1-induced dwarf phenotype and documented that OsNPR1 regulates rice growth and development through repressing auxin signaling, including OsGH3.8 activation, shedding light on the cross talk between the SA-mediated defense pathway and growth.
RESULTS
OsNPR1 Overexpression Inhibits Rice Growth and Development
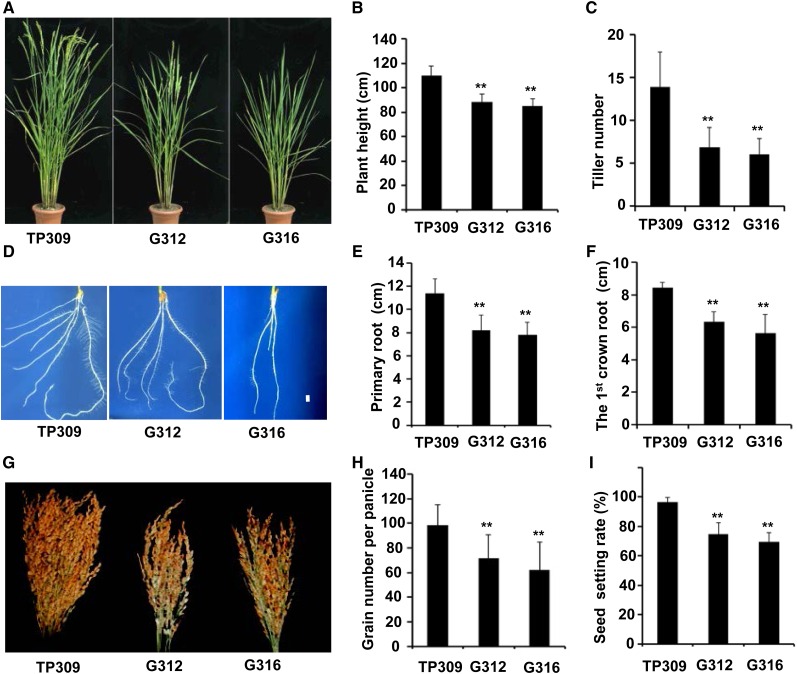
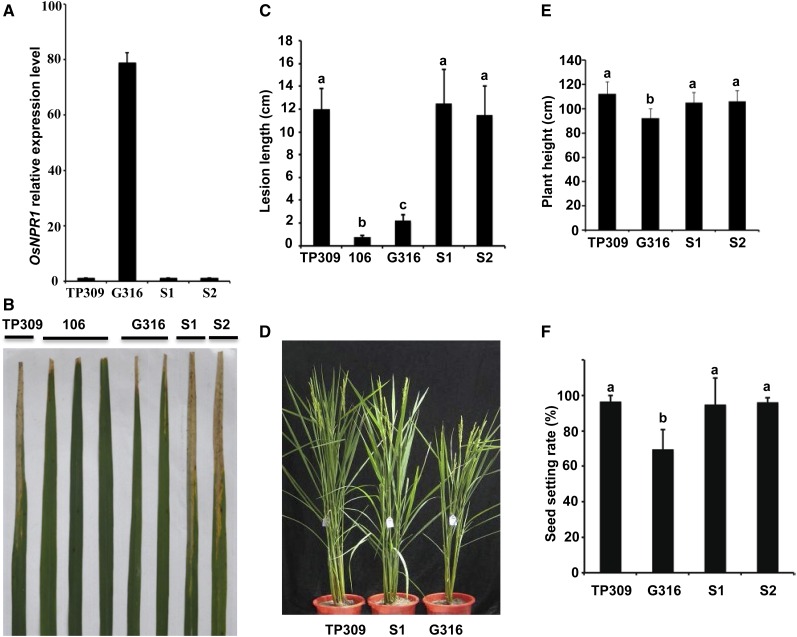
We functionally characterized the rice ortholog OsNPR1 and showed that OsNPR1 overexpression increased disease resistance to Xoo in our previous study (Yuan et al., 2007). However, we observed that growth and development were strongly inhibited in the OsNPR1-OX transgenic plants. Compared with the wild-type Taipei 309 (TP309), the two representative OsNPR1-OX lines G312 and G316 were smaller in both aboveground and belowground morphology through their entire lifetimes, with decreases in plant height, tiller number (Fig. 1, A–C), root system (Fig. 1, D–F), and grain number and seed setting (Fig. 1, G–I). To determine the OsNPR1 overexpression-mediated phenotypes, we screened for spontaneous transgene-silenced plants in a progeny population and found that transgene silencing occurred in some individuals in the progeny of the OsNPR1-OX line (Fig. 2A; Supplemental Fig. S1). Consequently, those transgene-silenced plants lost disease resistance against Xoo (Fig. 2, B and C). Simultaneously, the transgene-silenced plants also restored growth phenotypes (Fig. 2, D–F). The results further supported the hypothesis that OsNPR1 overexpression was responsible for growth inhibition.
Figure 1.
OsNPR1 overexpression plants exhibited growth and developmental defects. A, Morphological phenotypes of adult OsNPR1-OX (G312 and G316) and wild-type (TP309) plants during the heading stage, grown at the Hainan station. Note that G312 and G316 were dwarf and delayed in flowering. B and C, Plant heights and tiller numbers of OsNPR1-OX and wild-type plants. Data shown are means ± sd (n = 20). D, Root architecture of OsNPR1-OX and wild-type plants. E and F, Lengths of primary (E) and lateral (F) roots. Root lengths were measured at 21 d after seed germination. Data shown are means ± sd (n ≥ 24). G, Mature panicles of OsNPR1-OX and wild-type plants as the harvest of each plant. H and I, Grain number per panicle (H) and seed-setting rate (I) were decreased significantly in OsNPR1-OX plants compared with wild-type plants. Data were collected from all panicles of whole plants (n ≥ 5). Asterisks indicate significant differences in comparison with the wild-type TP309 (**, P < 0.01, Student’s t test).
Figure 2.
Transgene silence of OsNPR1-OX restored growth and development. A, Expression levels of OsNPR1 measured by qRT-PCR in wild-type TP309, G316, and two transgene-silenced G316 plants (S1 and S2) with the rice UBIQUITIN gene (Os03g13170) as an internal control, using primers listed in Supplemental Table S2. B and C, Decreased disease resistance of the transgene-silenced plants to Xoo (strain PXO99A) in comparison with TP309, G316, and the fully resistant Xa21 line 106 control (Song et al., 1995). D, Normal morphological phenotype of the transgene-silenced line (S1) compared with the wild type and G316 during the heading stage. E and F, Statistical analysis of plant heights (E) and seed-setting rates (F) of TP309, G316, S1, and S2 plants grown at the Shanghai station. Data were collected from adult plants grown in the field. Different letters indicate significant differences at P < 0.05 (n = 20; ANOVA followed by Duncan’s multiple range test).
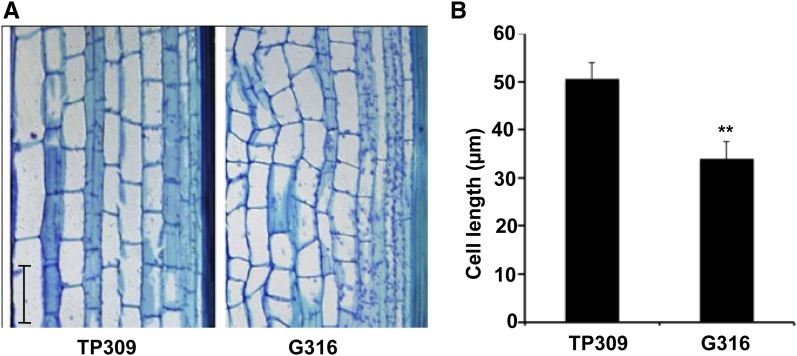
To further shape the effect of OsNPR1 overexpression on morphological changes, we performed cellular observations with sections of the uppermost internodes and observed that cells were smaller and abnormally shaped in the OsNPR1-OX plants in comparison with the wild-type control (Fig. 3). Therefore, OsNPR1 likely inhibited cell expansion when overexpressed.
Figure 3.
Cell development was repressed in OsNPR1-OX plants. A, Cell morphology in the longitudinal section of panicle necks of TP309 and G316 at the heading stage. Sections were stained with Toluidine Blue and observed with a light microscope (Leica). Bar = 50 µm. B, Statistical analysis of cell lengths in the parenchyma near the sclerenchyma. More than 100 cells from five individual stem segments were measured for both TP309 and G316. Asterisks indicate significant differences (**, P < 0.01, Student’s t test).
OsNPR1 Overexpression Alters the Expression of Hormone-Related Genes
To investigate the mechanism of OsNPR1-mediated growth retardation, we first compared the global gene expression of OsNPR1-OX and wild-type plants at heading stage by microarray analysis using the Affymetrix oligonucleotide chip GeneChip Rice Genome Array with three biological replicates. Reproducible differentially expressed probe sets were selected (fold change ≥ 2 and P < 0.05) from total normalized data using Data Mining Tool version 3.0. A total of 895 genes showed differential expression between the two plant groups, of which 729 and 166 were up- and down-regulated, respectively (Supplemental Table S1). Functional classification of the genes was performed according to the Rice Genome Annotation Project Release 7.0 (Kawahara et al., 2013; http://rice.plantbiology.msu.edu). Microarray data were validated for some genes using qRT-PCR analysis (Supplemental Fig. S2).
A set of metabolism, transcriptional regulation, kinase, calcium signaling, and hormone genes are differentially expressed (Supplemental Table S1). Because plant hormones play vital roles in cross talk between defense and growth in rice (Yang et al., 2013) and those genes are likely related to diverse hormone pathways, we focused on the genes involved in hormone signaling pathways. We found that 20 genes involved in hormone signaling pathways that regulate development and growth, including auxin, cytokinin, brassinolides (BRs), and gibberellic acids (GAs), were differentially expressed in OsNPR1-OX plants compared with wild-type plants (Table I). Five differentially expressed genes related to cytokinin, including two putative cytokinin-O-glucosyl transferase genes (Os02g51930 and Os06g11720; Kawahara et al., 2013), were up-regulated. These genes encode enzymes that can convert active cytokinin to its O-glucoside (Pineda Rodo et al., 2008; Kudo et al., 2012). The other three cytokinin response genes (Os02g35180, Os04g36070, and Os11g04720; Jain et al., 2006b; Kawahara et al., 2013) were down-regulated. This finding suggested that cytokinin levels or signaling might decrease in OsNPR1-OX plants compared with wild-type plants. In fact, cytokinin-deficient mutants displayed increased root mass and branching but also shorter stature, thinner stems, narrower leaves, and smaller meristems compared with wild-type plants (Werner et al., 2003; Pineda Rodo et al., 2008; Kudo et al., 2012). Based on the OsNPR1-OX phenotypes of decreased root system and branching (Fig. 1D), the possible disruption of cytokinin signaling might not be responsible for the OsNPR1-inhibited development and growth. Three genes possibly involved in BR signaling were differentially expressed in OsNPR1-OX plants, including Os06g16300 (annotated to encode putative BRASSINOSTEROID INSENSITIVE1-ASSOCIATED RECEPTOR KINASE1 PRECURSOR; Kawahara et al., 2013), Os06g12120 (OsSERL6/OsBAK1-3; Singla et al., 2009; Khew et al., 2015), and Os07g40630 (annotated to encode putative BRASSINOSTEROID INSENSITIVE1 precursor). The three genes were up-regulated in OsNPR1-OX plants, likely indicating enhanced BR signaling. However, we did not observe enhanced BR phenotypes that should display increases in leaf length and angle (Tong et al., 2012) and twisted leaves (Khew et al., 2015).
Table I. Differentially expressed genes related to hormone signaling in OsNPR1-OX compared with the wild type.
R1, R2, and R3 indicate three replicates.
| Hormone | Locus Identifier | Gene Product Name | Signal Log2 Ratio |
||
|---|---|---|---|---|---|
| R1 | R2 | R3 | |||
| Auxin | LOC_Os01g56240 | OsSAUR2, auxin-responsive SAUR gene family member, expressed | 5.5 | 4.0 | 2.9 |
| LOC_Os03g58350 | OsIAA14, auxin-responsive Aux/IAA gene family member, expressed | −2.1 | −4.7 | −3.4 | |
| LOC_Os04g03980 | OsYUCCA7, flavin monooxygenase, putative, expressed | −2.7 | −5.9 | −1.8 | |
| LOC_Os07g08460 | OsIAA24, auxin-responsive Aux/IAA gene family member, expressed | −3.1 | −3.8 | −1.8 | |
| LOC_Os07g40290 | OsGH3.8, probable IAA-amido synthetase, expressed | 3.8 | 2.4 | 2.0 | |
| LOC_Os09g37330 | OsSAUR39, auxin-responsive SAUR gene family member, expressed | −3.4 | −1.5 | −2.3 | |
| LOC_Os11g14190 | ARGOS, putative, expressed | −1.5 | −1.1 | −2.2 | |
| Cytokinin | LOC_Os02g35180 | OsRR2 type A response regulator, expressed | −1.1 | −1.2 | −1.9 |
| LOC_Os02g51930 | Cytokinin-O-glucosyltransferase2, putative, expressed | 1.2 | 2.7 | 1.0 | |
| LOC_Os04g36070 | OsRR1 type A response regulator, expressed | −1.7 | −1.1 | −2.4 | |
| LOC_Os06g11720 | Cytokinin-O-glucosyltransferase2, putative, expressed | 5.0 | 2.3 | 3.2 | |
| LOC_Os11g04720 | OsRR10 type A response regulator, expressed | −1.1 | −1.0 | −1.4 | |
| GA | LOC_Os03g57640 | GA receptor GID1L2, putative, expressed | 1.6 | 1.9 | 2.8 |
| LOC_Os06g15620 | GASR7, GA-regulated GASA/GAST/Snakin family protein precursor, expressed | 1.4 | 2.3 | 4.3 | |
| LOC_Os07g06830 | GA receptor GID1L2, putative, expressed | 3.3 | 1.3 | 1.9 | |
| LOC_Os07g36170 | Chitin-inducible GA-responsive protein, putative, expressed | 1.9 | 1.5 | 1.9 | |
| LOC_Os07g39470 | GA response modulator protein, putative, expressed | 1.9 | 1.3 | 1.7 | |
| Brassinosteroid | LOC_Os06g12120 | BRASSINOSTEROID INSENSITIVE1-ASSOCIATED RECEPTOR KINASE1 | 2.7 | 1.2 | 1.1 |
| LOC_Os06g16300 | BRASSINOSTEROID INSENSITIVE1-ASSOCIATED RECEPTOR KINASE1 | 1.3 | 1.4 | 1.0 | |
| LOC_Os07g40630 | BRASSINOSTEROID INSENSITIVE1 precursor | 3.1 | 2.1 | 4.9 | |
At first glance, the dwarf phenotype of OsNPR1-OX plants might imply that OsNPR1 overexpression could suppress the GA signaling pathway. Intriguingly, five GA signaling genes were up-regulated in OsNPR1-OX plants, of which Os03g57640 and Os07g06830 were predicted to belong to the GA receptor GID1 L2 family (Kawahara et al., 2013; Jiang et al., 2014); Os06g15620 was annotated to encode a GA-regulated GASA/GAST/Snakin family protein, Os07g36170 was annotated to encode a chitin-inducible GA-responsive protein, and Os07g39470 was annotated to encode a GA response modulator protein (Kawahara et al., 2013). However, OsNPR1-OX plants showed the same sensitivity to GA as wild-type plants (Supplemental Fig. S3A). Furthermore, not much similarity was found between the OsNPR1-OX and the GA-deficient EUI-OX plants that were dwarf with more tillers compared with wild-type plants (Zhu et al., 2006; Zhang et al., 2008a, 2011; Supplemental Fig. S3, B–D). Therefore, we concluded that the stunted phenotypes caused by OsNPR1 overexpression were unlikely to be caused by disruption in cytokinin, BR, or GA signaling and that these signaling pathways might be somewhat affected in OsNPR1-OX plants.
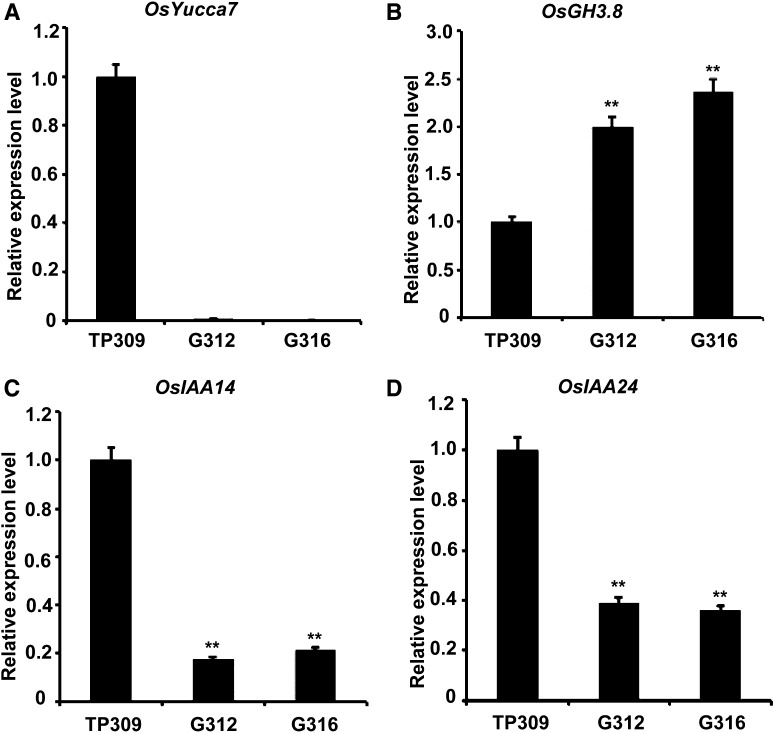
Interestingly, among the 20 differentially expressed hormone-related genes, seven genes were predicted to involve the auxin pathway (Table I); among these candidates, a putative auxin synthesis gene, OsYUCCA7 (Os04g03980; Yamamoto et al., 2007), was down-regulated (Fig. 4A), and an auxin catabolism gene, OsGH3.8 (Os07g40290), whose function in plant defense and growth and development had been identified by Ding et al. (2008), was up-regulated (Fig. 4B). The changes in the two genes might result in decreased free auxin levels. Consequently, it was indeed found that the auxin response genes OsIAA14 and OsIAA24 were down-regulated (Fig. 4, C and D). Based on the auxin-related phenotypes of OsNPR1-OX plants (Fig. 1), the results implied that OsNPR1 overexpression likely suppressed auxin signaling by disturbing auxin metabolism.
Figure 4.
Expression of differentially expressed genes related to the auxin pathway. RNA was extracted from young uppermost internodes at the heading stage. The expression levels of OsYUCCA7 (A), OsGH3.8 (B), OsIAA14 (C), and OsIAA24 (D) were analyzed by qRT-PCR. Note that OsGH3.8 was activated in G312 and G316. The oligonucleotide sequences used for qRT-PCR are listed in Supplemental Table S2. The rice OsACTIN1 gene (Os03g50885) was used as an internal control. Asterisks indicate significant differences in comparison with TP309 (**, P < 0.01, Student’s t test).
OsNPR1 Overexpression Changes Auxin Homeostasis
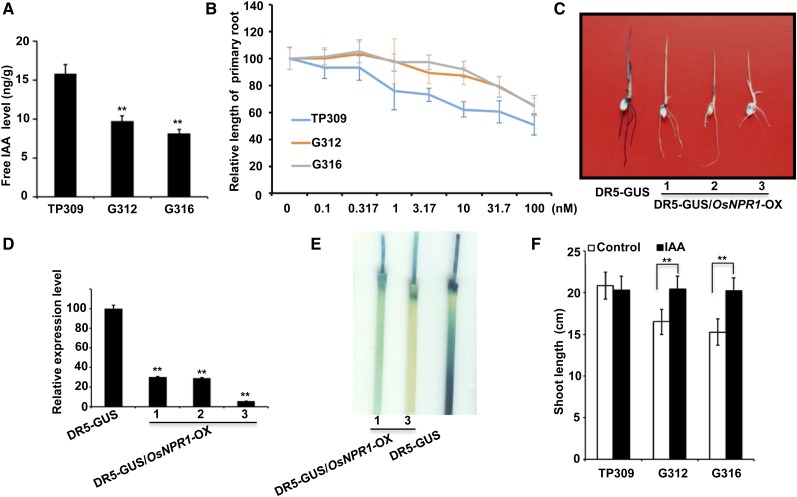
The final output of auxin signaling is determined by its levels, distribution, and perception system. To determine whether the auxin pathway indeed contributes to the OsNPR1-OX phenotypes, the free indole-3-acetic acid (IAA) levels were measured using an Accurate-Mass Q-TOF LC/MS device (Agilent 6520) with IAA-[α,α-D2] as an internal standard. The free IAA levels were significantly lower in OsNPR1-OX G312 and G316 plants than in TP309 plants (Fig. 5A), consistent with changes in the expression levels of the two auxin genes, OsYUCCA7 and OsGH3.8 (Fig. 4, A and B). Therefore, we concluded that OsNPR1 overexpression indeed decreased auxin levels through either down- or up-regulating the genes responsible for auxin biosynthesis and metabolism, respectively. To determine whether auxin perception, signaling, and transport systems also were affected by OsNPR1 overexpression, a growth inhibition assay with exogenous auxin application was conducted using root length as a parameter. The OsNPR1-OX overexpression lines exhibited lower sensitivity to naphthaleneacetic acid (NAA) than the wild-type plants (Fig. 5B). Because NAA is a substrate of GH3-encoded IAA-amido synthases, OsGH3.8 up-regulation might partially contribute to the reduced sensitivity to NAA in the OsNPR1-OX lines (Staswick et al., 2005).
Figure 5.
OsNPR1-OX changed auxin biosynthesis and response. A, OsNPR1 overexpression decreased free IAA levels in G312 and G316. Young uppermost internodes at the heading stage were collected for IAA extraction. B, Root growth inhibition by NAA in OsNPR1-OX and wild-type plants recorded and analyzed (n = 24) at day 7 after treatment. C, GUS staining and qRT-PCR analysis of GUS expression levels in the wild type (ZH11) and three DR5-GUS/OsNPR1-OX lines. Seedlings at 5 d after germination were incubated in GUS staining buffer for 4 h, indicating the decrease in GUS staining in DR5-GUS/OsNPR1-OX plants. D, Expression levels of GUS detected by qRT-PCR in RNA extracted from 3-week-old seedlings. The rice OsACTIN1 gene (Os03g50885) was used as an internal control. E, GUS staining of young uppermost nodes at the heading stage. Tissues were incubated in GUS staining buffer for 12 h. Note that GUS staining was decreased in the internodes and nodes of DR5-GUS/OsNPR1-OX. F, Exogenous application of IAA (1 nm) could partially restore the growth of OsNPR1-OX plants. Asterisks indicate significant differences in comparison with TP309 (A and F) or DR5-GUS (D; **, P < 0.01, Student’s t test).
To further examine auxin homeostasis in OsNPR1 overexpression plants, we took advantage of the DR5-GUS reporter, which is often used to detect active auxin levels and distribution in Arabidopsis (Nakamura et al., 2003) and also works well in rice (Zhao et al., 2009). We introduced the OsNPR1-OX transgene into the DR5-GUS background (variety Zhonghua 11 [ZH11]; Supplemental Fig. S4A). The ZH11 OsNPR1-OX plants also exhibited developmental defects and dramatically enhanced disease resistance (Supplemental Fig. S4, A and B). Histochemical staining of GUS activity revealed that both shoots and roots showed dramatically weaker DR5-GUS activity in OsNPR1-OX plants compared with the DR5-GUS control plants at the seedling stage (Fig. 5C). The weaker GUS activity caused by OsNPR1 overexpression was attributed to the lower GUS transcript level (Fig. 5D), indicating auxin deficiency in OsNPR1-OX plants. Consistent with the smaller adult plant height (Fig. 1, A and B), the uppermost node and the two internodes were more weakly stained in OsNPR1-OX plants compared with DR5-GUS control plants (Fig. 5E). The evidence of the exogenous auxin response and DR5-GUS reporter activity allowed us to conclude that OsNPR1 overexpression changed auxin homeostasis most likely through auxin metabolism, which resulted in decreased auxin levels in OsNPR1-OX plants. Consistent with our hypothesis, exogenous application of IAA could largely restore the growth of G316 plants (Fig. 5F).
OsNPR1 Overexpression Also Enhances OsGH3.8 Expression at the Seedling Stage
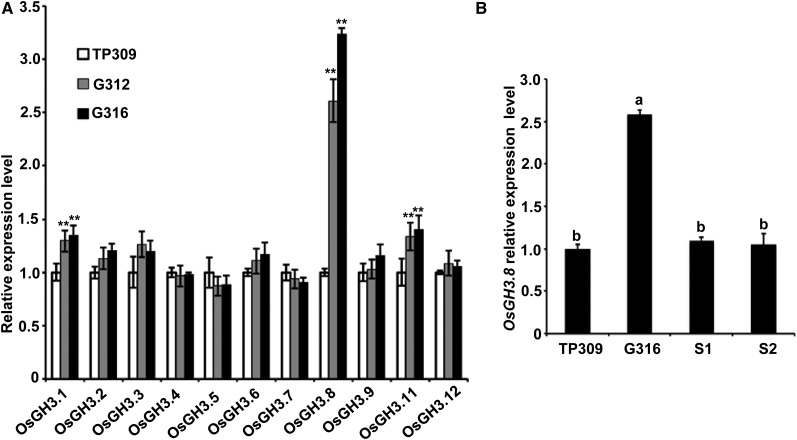
We have shown that OsYUCCA7 and OsGH3.8, genes involved in auxin biosynthesis and metabolism, respectively, were differentially regulated in OsNPR1-OX at the heading stage, suggesting that OsNPR1 might regulate the expression of the two auxin genes. Because OsNPR1 overexpression repressed rice growth and development over the plant’s entire lifetime, we then compared the expression levels of the two genes between OsNPR1-OX and wild-type plants at the seedling stage. We found that OsGH3.8 also was significantly up-regulated at the seedling stage (Fig. 6A); however, OsYUCCA7 expression was not changed at this stage (Supplemental Fig. S5). We also analyzed other members of the rice GH3 family (Jain et al., 2006a) at the seedling stage and found that two other GH3 members, OsGH3.1 and OsGH3.11, also were significantly but not dramatically up-regulated in the OsNPR1-OX lines compared with the wild type (Fig. 6A). Moreover, we found that the expression level of OsGH3.8 was correlated with that of OsNPR1, since the transgene-silenced plants in G316 reduced the expression of OsGH3.8 (Fig. 6B). Therefore, we propose that OsNPR1 overexpression indirectly up-regulates some GH3 genes, in particular OsGH3.8, triggering a defense cost process that links with the auxin signaling pathway. Consistent with this hypothesis, OsGH3.8 overexpression also caused abnormal plant morphology and retarded growth and development (Ding et al., 2008).
Figure 6.
OsNPR1 up-regulated OsGH3.8. A, Expression levels of GH3 family genes in the wild type and OsNPR1 overexpression lines G312 and G316. RNA was extracted from 3-week-old seedlings. qRT-PCR was performed with the OsACTIN1 gene (Os03g50885) as an internal control, and expression levels of the genes in wild-type TP309 were set as 1. Asterisks indicate significant differences in comparison with wild-type TP309 (**, P < 0.01, Student’s t test). B, Expression levels of OsGH3.8 in OsNPR1 overexpression line G316 and two transgene-silenced G316 plants (S1 and S2). Different letters indicate significant differences (P < 0.05, ANOVA followed by Duncan’s multiple range test).
Decrease of OsGH3.8 Expression Partially Restores OsNPR1-OX Growth But Not Disease Resistance
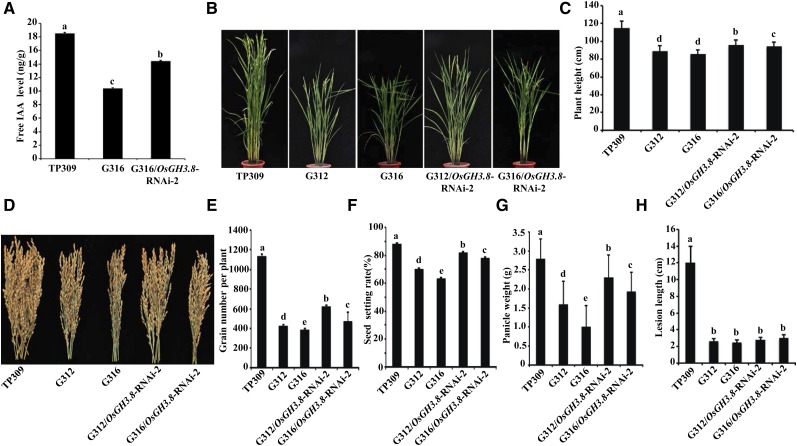
To further confirm whether the activation of OsGH3.8 was responsible for the stunted phenotypes of OsNPR1-OX plants, we first developed OsGH3.8 RNA interference lines (OsGH3.8-RNAi) in the TP309 background (Supplemental Fig. S6A), which exhibited a significantly lower seed-setting rate compared with wild-type plants, as reported (Supplemental Fig. S6B; Yadav et al., 2011). The stable OsGH3.8-RNAi line was crossed with OsNPR1-OX plants to generate OsNPR1-OX/OsGH3.8-RNAi plants. However, we could not obtain lines with strong RNA interference; rather, plants exhibited only partially reduced OsGH3.8 expression in the OsNPR1-OX background, using an unknown mechanism (Supplemental Fig. S6, C and D). We found that the decrease in OsGH3.8 expression partially restored IAA contents in the OsNPR1-OX (G316)/OsGH3.8-RNAi plants (Fig. 7A). As expected, knockdown of OsGH3.8 also rescued the OsNPR1-OX growth phenotypes, including plant height (Fig. 7, B and C) and grain productivity (Fig. 7, D–G). Importantly, the OsNPR1-mediated disease resistance was not attenuated by the OsGH3.8 knockdown (Fig. 7H). Therefore, we genetically confirmed that OsGH3.8 is an important downstream target of the OsNPR1-mediated defense signaling pathway, which regulates auxin signaling to shape rice growth and development, providing a link between defense and fitness cost. However, the mechanism by which OsNPR1 activates OsGH3.8 remains elusive, and we could not provide direct molecular evidence for a causal link between NPR1 protein activity and OsGH3.8 gene expression.
Figure 7.
Decreased OsGH3.8 expression partially rescued OsNPR1-OX developmental defects. A, Free IAA levels measured in young uppermost internodes at the heading stage. B, Morphological phenotypes of adult plants at the heading stage, grown at the Hainan station. C, Statistical analysis of plant heights. Note that OsNPR1-OX/OsGH3.8-RNAi-2 partially restored the phenotype of OsNPR1-OX in comparison with wild-type TP309. D, Mature panicles of the indicated genotypes. All panicles were harvested for individual plants. E to G, Statistical analysis of grain number per plant (E), seed-setting rate (F), and panicle weight (G). Data were collected from more than five adult plants of each genotype grown in the field. H, Statistical analysis of lesion lengths after inoculation with Xoo PXO99A. Different letters (A, C, and E–H) indicate significant differences at P < 0.05 (ANOVA followed by Duncan’s multiple range test).
DISCUSSION
OsNPR1 Overexpression Represses Rice Growth and Development by Disturbing Auxin Homeostasis
Plant defense and growth are largely governed by diverse phytohormones. Auxins, with IAA as a main active form in higher plants, can regulate many aspects of plant development, such as gravitropism, root, leaf, and flower formation, plant vasculature development, and seed germination (Kieffer et al., 2010; Zhao, 2010; Liu et al., 2013). This study reveals that the auxin signaling pathway impacts the OsNPR1-mediated disease resistance in rice partly through the IAA-amido synthetase gene OsGH3.8, which has been reported to play important roles in rice growth and basal defense through modifying cell wall structure (Ding et al., 2008). Our study provides another genetic model indicating that the plant defense machinery interacts with growth and development and further supports our previous claim that rice defense is usually prioritized over growth when defense hormones encounter growth hormones in the cereal model crop (Yang et al., 2008, 2012, 2013).
Auxin, as a negative regulator of plant immunity, has long been known to actively modify plant defense (Navarro et al., 2006; Chen et al., 2007). A set of evidence has demonstrated that auxin plays roles in balancing plant defense responses and growth in both monocotyledonous and dicotyledonous plants. For example, auxin signaling repression through negatively regulated mRNAs for the F-box receptor was reported to contribute to antibacterial resistance in Arabidopsis (Navarro et al., 2006). Interestingly, increased auxin levels even could counteract the effect of activated SA signaling on disease resistance (Zhang et al., 2007). Wang et al. (2007) demonstrated that auxin might down-regulate the defense response by inhibiting the full induction of SA-mediated signaling and that inhibition of auxin signaling is part of the SA-mediated disease resistance in Arabidopsis (Wang et al., 2007). In rice, reducing auxin content by constitutively expressing OsGH3.8 and OsGH3.1 enhanced disease resistance to both fungal and bacterial pathogens (Ding et al., 2008; Domingo et al., 2009). Therefore, auxin signaling is likely subjected to strict monitoring and modification during plant-pathogen interactions.
OsGH3.8 Is a Downstream Target of OsNPR1
The intertwining relationships between the SA and auxin pathways have been widely reported (Navarro et al., 2006; Wang et al., 2007; Zhang et al., 2007, 2008b). Some members of the GH3 gene family are actively involved in fine-tuning plant defense and growth. Our previous study with the GH3.5 activation-tagged mutant gh3.5-1D documented that GH3.5 acts as a bifunctional modulator in both SA and auxin signaling pathways during pathogen infection in Arabidopsis. GH3.5 overexpression in gh3.5-1D plants resulted in elevated SA accumulation and high levels of free IAA after pathogen infection, simultaneously augmenting the SA and auxin pathways and impairing different R gene-mediated resistance (Zhang et al., 2007). Interestingly, GH3.5 also modulated abiotic stress adaptation by changing auxin homeostasis (Park et al., 2007). Another Arabidopsis GH3 gene, GH3.3, was up-regulated using benzothiadiazole, an SA analog (Wang et al., 2007). In rice, reducing auxin levels by constitutively expressing OsGH3.8 (Ding et al., 2008) and OsGH3.1 (Domingo et al., 2009) also enhanced disease resistance to fungal and bacterial pathogens. Intriguingly, however, the enhanced disease resistance conferred by OsGH3.8 up-regulation is suggested to be independent of the SA pathway, given that PR1 gene induction and SA accumulation were slightly suppressed in OsGH3.8-overexpressing plants (Ding et al., 2008).
NPR1 is a key regulator of the SA-mediated pathway and acts as a transcriptional coactivator in plant innate immunity (Cao et al., 1998; Friedrich et al., 2001; Malnoy et al., 2007; Wally et al., 2009; Parkhi et al., 2010; Zhang et al., 2012). Our results here indicate that OsGH3.8 was up-regulated in OsNPR1-OX plants and consequently decreased IAA levels (Fig. 5). We propose that OsGH3.8 is a downstream target of OsNPR1 for modifying plant growth and development based on the following lines of evidence. First, OsGH3.8 is up-regulated in OsNPR1-OX. Second, OsGH3.8 is greatly inducible by SA (Ding et al., 2008). Third, the decrease in OsGH3.8 expression partially restored the growth and developmental defects of OsNPR1-OX plants. Because the OsGH3.8 expression level in the OsNPR1-OX/OsGH3.8-RNAi line could not be decreased to the level in the original OsGH3.8-RNAi lines (Supplemental Fig. S6, A and D), we proposed that the OsNPR1-OX stunted phenotype would be more profoundly rescued if OsGH3.8 expression could be further knocked down. Nevertheless, it is interesting that OsGH3.8 knockdown did not affect OsNPR1-enhanced disease resistance, regardless of the role that OsGH3.8 plays in basal defense in rice. These observations suggest that the strong OsNPR1-dependent enhancement of the SA defense pathway could not be compromised by the potential loss of OsGH3.8-mediated basal defense. Alternatively, as suggested previously, the functional redundancy of the GH3 family led to a nondecrease of disease resistance in GH3.8 suppression or knockout plants (Ding et al., 2008). NPR1 is a master transcription regulator that interacts with transcription factors to activate target genes. Chern et al. (2014) showed that OsNPR1 (NH1) is capable of interacting strongly with rTGA2.1, rTGA2.2, rTGA2.3, rLG2, TGAL2, and TGAL4, which belong to the TGA family of basic-region Leu zipper transcription factors involved in defense responses. Therefore, OsNPR1 likely indirectly regulates OsGH3.8 through an unrecognized transcriptional mechanism. We propose that other unknown transcription factor(s) may be regulated directly by OsNPR1 and may play an important role in OsGH3.8 activation. The transcription factor(s) could be identified through extensive screenings of OsNPR1-interacting proteins or genetic suppressors of OsNPR1-OX disease resistance or developmental phenotypes.
Potential Strategy for OsNPR1 Application in Rice Breeding for Broad-Spectrum Disease Resistance
OsNPR1 is a key regulator of innate immunity in rice and has been regarded as an ideal candidate for durable and broad-spectrum disease resistance breeding by transgenic ectopic expression (Chern et al., 2005; Yuan et al., 2007). The fact that OsNPR1 overexpression inhibits rice growth and development by repressing auxin signaling partially by activating OsGH3.8 makes it difficult to utilize this gene directly in improving disease resistance in elite rice varieties. Our results have shown that the auxin pathway is the target of OsNPR1-dependent growth and development, which likely does not affect OsNPR1-mediated disease resistance. Therefore, one strategy of OsNPR1 application in rice molecular breeding is to block the downstream auxin signal in growth and development by mutating the OsNPR1-interacting gene(s) or its promoter, such as GH3.8, using CRISPR/Cas9 technology, provided that the precise mechanism or transcription factor(s) has been recognized. Another strategy is to promote growth and development and thereby increase yield potential in OsNPR1-OX plants by introducing some auxin-related yield regulators, such as the OsSPL14 elite alleles, IPA1 and WFP (Jiao et al., 2010; Miura et al., 2010), into OsNPR1-OX plants, which would promote plant growth and development with stronger and elongated culms and higher grain productivity and likely would overcome the negative effects of OsNPR1 on yield traits.
CONCLUSION
In this study, the SAR master gene OsNPR1 was recognized to inhibit plant growth and development through repressing the auxin signaling pathway in overexpression transgenic rice. This finding provides further insight into the fine-tuned hormone network of defense and growth interactions.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The rice (Oryza sativa) varieties TP309 and ZH11 were used in rice transformation. The OsNPR1 overexpression lines (G312 and G316) and the EUI1 overexpression line all were developed in our laboratory (Zhu et al., 2006; Yuan et al., 2007). The transgene-silenced plants of OsNPR1-OX, S1 and S2, were isolated from the progeny of G316. The presence of transgene in S1 and S2 plants was confirmed by PCR using forward primer that located in LacZ on the plasmid of 1301-35SN (Yuan et al., 2007) and reverse primer that located in OsNPR1 (Supplemental Table S2). The Xa21 transgenic line 106 was provided by Dr. Pamela Ronald (University of California, Davis; Song et al., 1995). The DR5-GUS transgenic rice line was provided by Dr. Yu Zhao (Huazhong Agricultural University). The OsNPR1 overexpression line in ZH11 was developed as described below and then was crossed with DR5-GUS to obtain the DR5-GUS/OsNPR1-OX line. The OsGH3.8 RNA interference lines (OsGH3.8-RNAi) were developed as described below. The OsNPR1-OX/OsGH3.8-RNAi double transformants were obtained by crossing G312, G316, and OsGH3.8-RNAi plants. Rice plants were grown in experimental fields (the Shanghai station and/or the Hainan station) or in growth chambers under growth conditions with 12-h day, 28°C, and 80% relative humidity followed by 12-h night, 26°C, and 60% relative humidity.
Plasmid Construction and Rice Transformation
To develop OsNPR1-OX in the ZH11 background, the OsNPR1 transgene (Yuan et al., 2007) was transformed into ZH11 calli. To generate OsGH3.8-RNAi plants, a hairpin structure was constructed with a specific fragment of this gene coding region (nucleotides 69–700) and its partial antisense fragment (nucleotides 75–508). The sense fragment was amplified using the primers 5′-TGGATCCCGACGCCTCTTCCTCT-3′ and 5′-AGGGCCCAATGATTCATGGCCAT-3′ (BamHI and ApaI sites), and the antisense fragment was amplified using the primers 5′-AGGGCCCGGTGAGGAACTCGGAG-3′ and 5′-AGGTACCTTCCTCTCGCTACTAC-3′ (ApaI and KpnI sites). Both fragments were fused using ApaI digestion and ligation, and the resultant fragment was inserted into the expression vector pUN1301 (BamHI and KpnI sites) under the control of the maize (Zea mays) ubiquitin promoter. Rice transformation was performed using the Agrobacterium tumefaciens-mediated method to generate more than 15 independent transgenic plants.
Microarray Data Analysis
The young uppermost internodes of OsNPR1-OX G316 and wild-type TP309 plants at the heading stage were harvested and stored in liquid nitrogen for RNA isolation. Microarray analysis was performed using the oligonucleotide chip GeneChip Rice Genome Array developed by Affymetrix. Microarray experiments were performed at an Affymetrix-authorized center (GeneTech Biotechnology) following the protocol of the Affymetrix GeneChip Rice Genome Array with three biological replicates. Raw data analysis was conducted as described previously (Wang et al., 2008). Reproducibly differentially expressed probe sets were selected (fold change ≥ 2 and P < 0.05) from total normalized data using Data Mining Tool version 3.0. Functional classification of the genes was performed according to the Rice Genome Annotation Project Release 7.0 (Kawahara et al., 2013; http://rice.plantbiology.msu.edu) and some publications involved in gene function analysis, such as Jain et al. (2006a). Microarray data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO series accession number GSE76178 (the gene expression data of OsNPR1 overexpression and its wild-type TP309 plants).
Auxin and GA Response Experiments
Sterile TP309, G312, and G316 seeds were germinated in the dark for 2 d at 28°C and then were incubated in 0.5× MS liquid medium supplied with different concentrations of NAA. After 7 d, the lengths of the primary roots of seedlings were measured. Thirty seedlings of each line were analyzed with three biological repeats. For GA treatment, seeds after germination were grown in one-half-strength MS medium with different concentrations of GA3. The shoot lengths were measured at day 17 after treatment (n ≥ 24). For exogenous application of IAA, 6-d-old seedlings grown in one-half-strength MS liquid medium were incubated with 1 nm IAA for another 10 d. Shoot length was measured (n = 20).
Auxin Extraction and Quantification
The young uppermost internodes of OsNPR1-OX G316, G316/OsGH3.8-RNAi, and wild-type (TP309) plants at heading stage were harvested for IAA extraction. IAA extraction and quantification were performed according to Pan et al. (2010) using an Agilent 6520 Accurate-Mass Q-TOF LC/MS device. IAA-[α,α-D2] (International Laboratory) was used as an internal standard of IAA.
Gene Expression Analysis
Total RNA was extracted using TRIzol reagent and treated with DNase I using a DNA-free kit (Invitrogen; http://www.invitrogen.com/). cDNA was synthesized from 3 µg of total RNA using an oligo(dT) primer and SuperScript III reverse transcriptase according to the manufacturer’s instructions (Invitrogen). Real-time reverse transcription-PCR analysis was performed using SYBR Premix Ex Taq (TaKaRa; http://www.takara.com.cn/) and gene-specific primers with OsACTIN1 as the internal control. Semiquantitative reverse transcription-PCR was conducted with a rice ubiquitin gene (Os03g13170) as the internal control. All of the primer sequences are listed in Supplemental Table S2.
Histological Analysis
The elongating zones of the uppermost internodes were cut into 0.2-cm sections and then immersed in hydrofluoric acid, followed by paraffin section preparation using a previously described protocol (Yang et al., 2011). Sections were stained with 0.05% Toluidine Blue O (Chroma Gesellshaft Shaud) and observed with a light microscope (Leica).
DR5-GUS Staining
For GUS staining, the materials were placed in staining solution (50 mm NaPO4, pH 7.2, 2 mm X-gluc, 0.5 mm K3Fe[CN]6, and 0.5 mm K4Fe[CN]6), vacuum infiltrated for 5 min, and incubated at 37°C. Five-day-old whole seedlings of DR5-GUS and DR5-GUS/OsNPR1-OX were incubated in GUS staining buffer for 4 h. Young uppermost internodes at the heading stage were incubated in GUS staining buffer for 12 h.
Pathogen Inoculation
Rice plants were inoculated with Xanthomonas oryzae pv oryzae strain PXO99A using the leaf clip method (Song et al., 1995). After 14 d, the lesion lengths were recorded. More than five individual plants, with five to six leaves of each plant, were inoculated for each genotype.
Statistical Analysis
For gene expression levels detected by qRT-PCR with three replicates, and plant growth and yield components measured with 20 or 24 plants, Student’s t test or ANOVA followed by Duncan’s multiple range test was performed.
The complete set of microarray data has been deposited in a MIAME-compliant format under accession number GSE76178 (http://www.ncbi.nlm.nih.gov/geo).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Genotyping of two transgene-silenced G316 plants, S1 and S2.
Supplemental Figure S2. qRT-PCR validation of differentially expressed genes revealed by microarray assay.
Supplemental Figure S3. OsNPR1 overexpression has no effect on the GA response.
Supplemental Figure S4. Generation of OsNPR1 overexpression lines in ZH11.
Supplemental Figure S5. OsYUCCA7 expression levels at the seedling stage.
Supplemental Figure S6. Generation of OsNPR1-OX/OsGH3.8-RNAi.
Supplemental Table S1. Differentially expressed genes revealed by microarray between TP309 and G316.
Supplemental Table S2. Primers used for PCR analysis.
Supplementary Material
Acknowledgments
We thank Pieter B.F. Ouwerkerk, Pamela Ronald, Yinong Yang, and Yu Zhao for providing vectors and rice materials as well as Dr. Yining Liu for help with IAA quantification.
Glossary
- SAR
systemic acquired resistance
- SA
salicylic acid
- Xoo
Xanthomonas oryzae pv oryzae
- qRT
quantitative reverse transcription
- BR
brassinolide
- NAA
naphthaleneacetic acid
- TP309
Taipei 309
- ZH11
Zhonghua 11
- MS
Murashige and Skoog
Footnotes
This work was supported by the National Natural Science Foundation of China (grant no. 31330061 to Z.H. and grant no. 30900917 to X.L.), the Natural Science Foundation of Jiangsu (grant no. BK20150659), and the National Key Transformation Program (grant no. 2016ZX08001002 to D.-L.Y.).
Articles can be viewed without a subscription.
References
- Bednarek P, Osbourn A (2009) Plant-microbe interactions: chemical diversity in plant defense. Science 324: 746–748 [DOI] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Li X, Dong X (1998) Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc Natl Acad Sci USA 95: 6531–6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN (2007) Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci USA 104: 20131–20136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern M, Bai W, Ruan D, Oh T, Chen X, Ronald PC (2014) Interaction specificity and coexpression of rice NPR1 homologs 1 and 3 (NH1 and NH3), TGA transcription factors and Negative Regulator of Resistance (NRR) proteins. BMC Genomics 15: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern M, Fitzgerald HA, Canlas PE, Navarre DA, Ronald PC (2005) Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol Plant Microbe Interact 18: 511–520 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ (2013) Pivoting the plant immune system from dissection to deployment. Science 341: 746–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S (2008) Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20: 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo C, Andrés F, Tharreau D, Iglesias DJ, Talón M (2009) Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol Plant Microbe Interact 22: 201–210 [DOI] [PubMed] [Google Scholar]
- Friedrich L, Lawton K, Dietrich R, Willits M, Cade R, Ryals J (2001) NIM1 overexpression in Arabidopsis potentiates plant disease resistance and results in enhanced effectiveness of fungicides. Mol Plant Microbe Interact 14: 1114–1124 [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64: 839–863 [DOI] [PubMed] [Google Scholar]
- Jain M, Kaur N, Tyagi AK, Khurana JP (2006a) The auxin-responsive GH3 gene family in rice (Oryza sativa). Funct Integr Genomics 6: 36–46 [DOI] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP (2006b) Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biol 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, Xiang Y, Zhao J, Yin D, Zhao X, Zhu L, Zhai W (2014) Regulation of inflorescence branch development in rice through a novel pathway involving the pentatricopeptide repeat protein sped1-D. Genetics 197: 1395–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, et al. (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42: 541–544 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khew CY, Teo CJ, Chan WS, Wong HL, Namasivayam P, Ho CL (2015) Brassinosteroid insensitive 1-associated kinase 1 (OsI-BAK1) is associated with grain filling and leaf development in rice. J Plant Physiol 182: 23–32 [DOI] [PubMed] [Google Scholar]
- Kieffer M, Neve J, Kepinski S (2010) Defining auxin response contexts in plant development. Curr Opin Plant Biol 13: 12–20 [DOI] [PubMed] [Google Scholar]
- Kou Y, Wang S (2010) Broad-spectrum and durability: understanding of quantitative disease resistance. Curr Opin Plant Biol 13: 181–185 [DOI] [PubMed] [Google Scholar]
- Kudo T, Makita N, Kojima M, Tokunaga H, Sakakibara H (2012) Cytokinin activity of cis-zeatin and phenotypic alterations induced by overexpression of putative cis-zeatin-O-glucosyltransferase in rice. Plant Physiol 160: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Lu CF, Wu JW, Cheng ML, Lin YM, Yang NS, Black L, Green SK, Wang JF, Cheng CP (2004) Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res 13: 567–581 [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang H, Zhao Y, Feng Z, Li Q, Yang HQ, Luan S, Li J, He ZH (2013) Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc Natl Acad Sci USA 110: 15485–15490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López MA, Bannenberg G, Castresana C (2008) Controlling hormone signaling is a plant and pathogen challenge for growth and survival. Curr Opin Plant Biol 11: 420–427 [DOI] [PubMed] [Google Scholar]
- Malnoy M, Jin Q, Borejsza-Wysocka EE, He SY, Aldwinckle HS (2007) Overexpression of the apple MpNPR1 gene confers increased disease resistance in Malus × domestica. Mol Plant Microbe Interact 20: 1568–1580 [DOI] [PubMed] [Google Scholar]
- McDonald BA, Linde C (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol 40: 349–379 [DOI] [PubMed] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42: 545–549 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, Shimada Y, Yoshida S (2003) Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol 133: 1843–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X (2010) Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat Protoc 5: 986–992 [DOI] [PubMed] [Google Scholar]
- Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J, Lee YH, Park CM (2007) GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem 282: 10036–10046 [DOI] [PubMed] [Google Scholar]
- Parkhi V, Kumar V, Campbell LM, Bell AA, Shah J, Rathore KS (2010) Resistance against various fungal pathogens and reniform nematode in transgenic cotton plants expressing Arabidopsis NPR1. Transgenic Res 19: 959–975 [DOI] [PubMed] [Google Scholar]
- Pineda Rodo A, Brugière N, Vankova R, Malbeck J, Olson JM, Haines SC, Martin RC, Habben JE, Mok DW, Mok MC (2008) Over-expression of a zeatin O-glucosylation gene in maize leads to growth retardation and tasselseed formation. J Exp Bot 59: 2673–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JDG (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Ryals J, Weymann K, Lawton K, Friedrich L, Ellis D, Steiner HY, Johnson J, Delaney TP, Jesse T, Vos P, et al. (1997) The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell 9: 425–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Zeier J (2013) Long-distance communication and signal amplification in systemic acquired resistance. Front Plant Sci 4: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla B, Khurana JP, Khurana P (2009) Structural characterization and expression analysis of the SERK/SERL gene family in rice (Oryza sativa). Int J Plant Genomics 2009: 539402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, et al. (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong X (2008) Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3: 348–351 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong X (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12: 89–100 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H, Liu L, Jin Y, Du L, Yin Y, Qian Q, Zhu L, Chu C (2012) DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24: 2562–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wally O, Jayaraj J, Punja ZK (2009) Broad-spectrum disease resistance to necrotrophic and biotrophic pathogens in transgenic carrots (Daucus carota L.) expressing an Arabidopsis NPR1 gene. Planta 231: 131–141 [DOI] [PubMed] [Google Scholar]
- Wally O, Punja ZK (2010) Genetic engineering for increasing fungal and bacterial disease resistance in crop plants. GM Crops 1: 199–206 [DOI] [PubMed] [Google Scholar]
- Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X (2007) Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol 17: 1784–1790 [DOI] [PubMed] [Google Scholar]
- Wang E, Wang J, Zhu X, Hao W, Wang L, Li Q, Zhang L, He W, Lu B, Lin H, et al. (2008) Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet 40: 1370–1374 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav SR, Khanday I, Majhi BB, Veluthambi K, Vijayraghavan U (2011) Auxin-responsive OsMGH3, a common downstream target of OsMADS1 and OsMADS6, controls rice floret fertility. Plant Cell Physiol 52: 2123–2135 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T (2007) Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143: 1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DL, Li Q, Deng YW, Lou YG, Wang MY, Zhou GX, Zhang YY, He ZH (2008) Altered disease development in the eui mutants and Eui overexpressors indicates that gibberellins negatively regulate rice basal disease resistance. Mol Plant 1: 528–537 [DOI] [PubMed] [Google Scholar]
- Yang DL, Yang Y, He Z (2013) Roles of plant hormones and their interplay in rice immunity. Mol Plant 6: 675–685 [DOI] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Ren S, Zhang X, Gao M, Ye S, Qi Y, Zheng Y, Wang J, Zeng L, Li Q, et al. (2011) BENT UPPERMOST INTERNODE1 encodes the class II formin FH5 crucial for actin organization and rice development. Plant Cell 23: 661–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Zhong S, Li Q, Zhu Z, Lou Y, Wang L, Wang J, Wang M, Li Q, Yang D, et al. (2007) Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol J 5: 313–324 [DOI] [PubMed] [Google Scholar]
- Zhang JY, Qiao YS, Lv D, Gao ZH, Qu SC, Zhang Z (2012) Malus hupehensis NPR1 induces pathogenesis-related protein gene expression in transgenic tobacco. Plant Biol (Stuttg) (Suppl 1) 14: 46–56 [DOI] [PubMed] [Google Scholar]
- Zhang Q. (2007) Strategies for developing green super rice. Proc Natl Acad Sci USA 104: 16402–16409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang B, Yan D, Dong W, Yang W, Li Q, Zeng L, Wang J, Wang L, Hicks LM, et al. (2011) Two Arabidopsis cytochrome P450 monooxygenases, CYP714A1 and CYP714A2, function redundantly in plant development through gibberellin deactivation. Plant J 67: 342–353 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhu Y, Peng Y, Yan D, Li Q, Wang J, Wang L, He Z (2008a) Gibberellin homeostasis and plant height control by EUI and a role for gibberellin in root gravity responses in rice. Cell Res 18: 412–421 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li Q, Li Z, Staswick PE, Wang M, Zhu Y, He Z (2007) Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol 145: 450–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang M, Li Z, Li Q, He Z (2008b) Arabidopsis GH3.5 regulates salicylic acid-dependent and both NPR1-dependent and independent defense responses. Plant Signal Behav 3: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hu Y, Dai M, Huang L, Zhou DX (2009) The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21: 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Nomura T, Xu Y, Zhang Y, Peng Y, Mao B, Hanada A, Zhou H, Wang R, Li P, et al. (2006) ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.