Arabidopsis mat3 pollen overaccumulates methionine, germinates poorly, and has short tubes.
Abstract
S-Adenosylmethionine is widely used in a variety of biological reactions and participates in the methionine (Met) metabolic pathway. In Arabidopsis (Arabidopsis thaliana), one of the four S-adenosylmethionine synthetase genes, METHIONINE ADENOSYLTRANSFERASE3 (MAT3), is highly expressed in pollen. Here, we show that mat3 mutants have impaired pollen tube growth and reduced seed set. Metabolomics analyses confirmed that mat3 pollen and pollen tubes overaccumulate Met and that mat3 pollen has several metabolite profiles, such as those of polyamine biosynthesis, which are different from those of the wild type. Additionally, we show that disruption of Met metabolism in mat3 pollen affected transfer RNA and histone methylation levels. Thus, our results suggest a connection between metabolism and epigenetics.
S-Adenosylmethionine synthetase (SAMS), also known as methionine adenosyltransferase (MAT), is the only enzyme that synthesizes S-adenosylmethionine (SAM) from ATP and l-Met (Binet et al., 2011). SAM is used in a wide variety of biological reactions and is the major hub of Met metabolism (Shen et al., 2002; Martínez-López et al., 2008). Over 80% of the Met is metabolized into SAM, of which 90% is used for transmethylation reactions (Giovanelli et al., 1985). SAM, as a universal methyl group donor, participates in numerous enzyme-catalyzed reactions. For example, many iron-sulfur cluster-containing enzymes (radical SAM enzymes) use SAM in two ways: as a source of a methyl group transferred to a conserved Cys and as a source of 5′-deoxyadenosyl radicals (Fujimori, 2013). In eukaryotes, many radical SAM enzymes function in rRNA modifications (Kaminska et al., 2010; Yan et al., 2010), tRNA modifications (Pierrel et al., 2004; Hernández et al., 2007), and lipid metabolism (Hinson and Cresswell, 2009; Duschene and Broderick, 2010). SAM also serves as the precursor for ethylene and polyamines (Roje, 2006). Polyamines were reported to be important for pollen germination and pollen tube growth (Wolukau et al., 2004; Aloisi et al., 2016).
SAMS in Arabidopsis (Arabidopsis thaliana) is encoded by four genes. The nomenclature varies in different publications (Lindermayr et al., 2005; Mao et al., 2015); here, we will use the MAT1 to MAT4 designations. MAT1, MAT2, and MAT4 are expressed in nearly all tissues (Gómez-Gómez and Carrasco, 1998; Mao et al., 2015), whereas MAT3 is expressed predominantly in pollen (Loraine et al., 2013). MAT1 and MAT2 are most similar in sequence and expression patterns, and the double mutant mat1;mat2 exhibits decreased ethylene (Mao et al., 2015). The mat4 mutant accumulates less lignin due to the reduced supply of SAM (Shen et al., 2002).
Pollen tubes are the fastest growing plant cells (Selinski and Scheibe, 2014). Pollen tubes grow by transferring chemical energy from stored starch and newly assimilated sugars into ATP (Rounds et al., 2011). Transcriptome studies (Becker et al., 2003; Honys and Twell, 2003; Pina et al., 2005, Loraine et al., 2013) identified a large number of mRNAs in pollen that encode enzymes belonging to various metabolic pathways, and proteome profiling of pollen grains documented proteins present in pollen (Holmes-Davis et al., 2005; Noir et al., 2005; Sheoran et al., 2006). However, metabolomics studies during pollen grain maturation and pollen tube growth, and the connection between metabolism and cellular processes during tube growth, are not well established.
Given that SAM is the universal methyl donor, enzymes that control SAM levels play a critical role in determining the extent of methylation. For example, a knockdown mutant of SAMS globally reduced histone methylation in rice (Oryza sativa; Li et al., 2011; Chen et al., 2013) and in Caenorhabditis elegans (Towbin et al., 2012). Met metabolism generates the major methyl donor SAM for histone methylation. Reduced levels of some enzymes involved in Met metabolism in Drosophila melanogaster affected histone methylation and resulted in loss of viability (Liu et al., 2015). Moreover, it was reported that radical SAM enzymes function in tRNA modification (Pierrel et al., 2004; Hernández et al., 2007). The methylation of tRNA is important for correct and efficient protein synthesis (Hori, 2014). It was suggested that tRNA is the only RNA methylated in pollen tubes (Steffensen, 1971)
Here, we present evidence that MAT3, a SAMS expressed in pollen, plays an important role during pollen germination and pollen tube growth. Homozygous mat3/mat3 pollen overaccumulates Met. Metabolic profiling showed that several metabolic pathways are altered in mat3/mat3 pollen grains and pollen tubes. We used immunofluorescence assays and ultra-performance liquid chromatography-mass spectrometry to show that both histone methylation and tRNA methylation were altered in mat3/mat3 pollen These results support the idea that MAT3 is required for pollen germination and pollen tube growth by affecting metabolic pathways and for maintaining histone and tRNA methylation.
RESULTS
A mat3 Knockdown Mutant Exhibits Defects in the Male Gametophyte
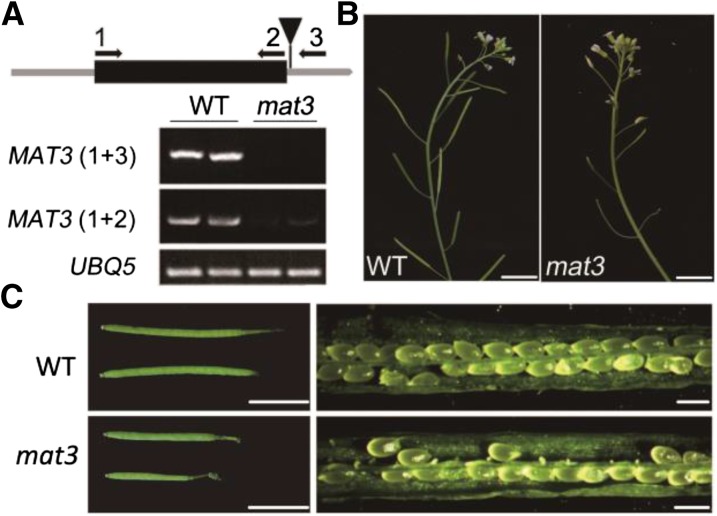
Arabidopsis has four SAMSs that are 94% identical at the amino acid level. According to RNA sequencing data (Loraine et al., 2013), MAT3 (525 reads per kilobase per million mapped reads [RPKM]) is more highly expressed in pollen than are the other three MAT family members (2, 12.5, and 47 RPKM for MAT1, MAT2, and MAT4, respectively). Therefore, it was reasonable to speculate that MAT3 is important for pollen function. We obtained a T-DNA insertion line, SALK_019375, which has a T-DNA inserted in the 3′ untranslated region. Two pairs of primers were designed to assess the levels of MAT3 transcripts in the mat3 mutant. Full-length MAT3 cDNA was very weakly detected in mat3 plants (Fig. 1A), indicating that this insertion line is a knockdown mutant (Supplemental Fig. S1). The mutant had shorter siliques and reduced seed set (Fig. 1, B and C).
Figure 1.
mat3 is a knockdown mutant and exhibits shorter siliques and reduced seed set. A, Structure of MAT3 (At2g36880). The exon is indicated by a black box, and untranslated regions are indicated by gray lines. The T-DNA in the 3′ untranslated region is indicated by the black triangle. UBIQUITIN5 (UBQ5) was used as an internal loading control. The numbers 1, 2, and 3 indicate primers used for RT-PCR. B, Representative 10-week-old plants of the wild type (WT) and mat3. Bars = 1 cm. C, Intact and opened siliques of the wild type and mat3. Bars = 0.5 cm (left) and 500 μm (right).
Self-pollinated heterozygous plants (+/mat3) exhibited a distorted segregation ratio that differed significantly from the expected Mendelian ratio of 1:2:1 (Table I). To test for potential defects in male or female gametophyte transmission, we performed reciprocal crosses using +/mat3 heterozygotes as either the male or female parent. Male transmission was more affected than female transmission (Table I). However, it is curious that homozygous mutants were rarely recovered from self-pollinations, suggesting some unknown difference between manual and self-pollinations. We also performed reciprocal crosses between mat3/mat3 homozygotes and the wild type. When wild-type or mat3/mat3 plants were pollinated with mat3 pollen, approximately 40% to 60% seed set was observed. In contrast, when the wild type was used as pollen donor on mat3 plants, there was 100% seed set.
Table I. Segregation analysis of the T-DNA in mat3/+ mutants and in reciprocal crosses with mat3/+ and wild-type plants.
| Female × Male | Genotype | Expected | Observed | Transmission Efficiency |
|---|---|---|---|---|
| mat3/+ × mat3/+ | +/+; mat3/+; mat3/mat3 | 1:2:1 | 78:79:4 | |
| mat3/+ × wild type | +/+; mat3/+ | 1:1 | 93:79 | 84% |
| Wild type × mat3/+ | +/+; mat3/+ | 1:1 | 158:0 | 0% |
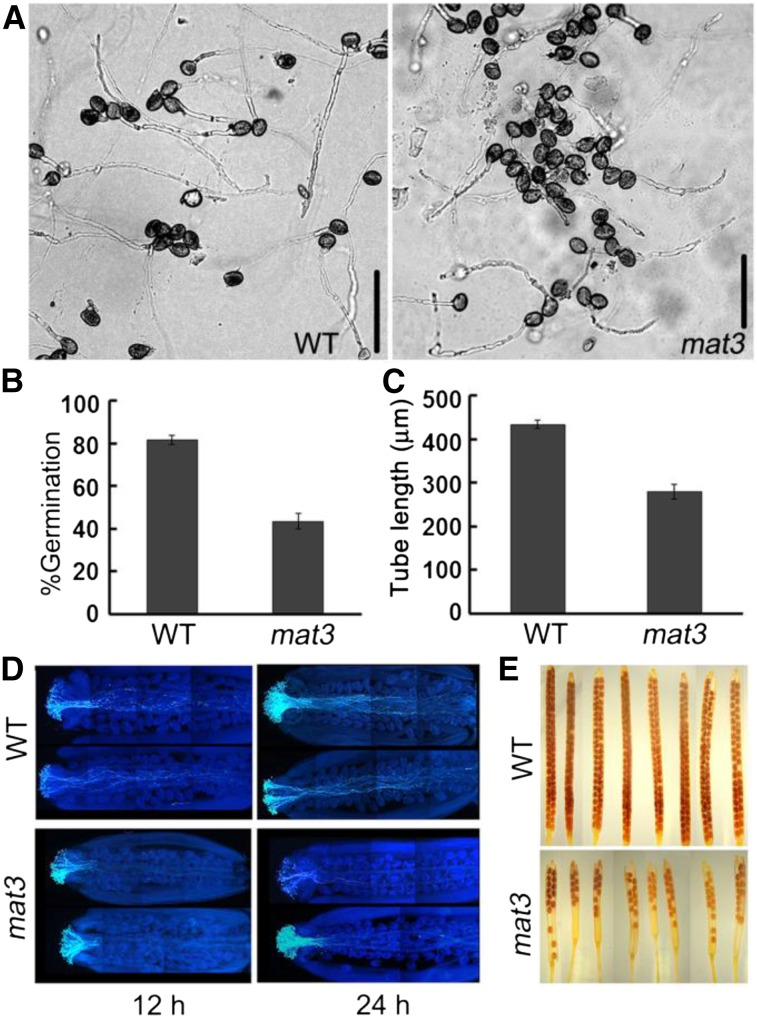
To check whether mat3 pollen had defects in pollen germination, we examined both in vitro and in vivo germination. For in vitro pollen germination, we measured germination percentage and pollen tube lengths. mat3 pollen showed reduced pollen germination and had shorter pollen tubes than wild-type pollen (Fig. 2 A–C). For in vivo pollination, we used wild-type pollen on wild-type stigmas or mat3 pollen on mat3 stigmas and then observed pollen tubes 12 h after pollination. Pollen tubes of the wild type had penetrated through the style and reached close to the end of the transmitting tract, while in mat3, pollen tubes had just begun to penetrate into the style. After 24 h, pollen tubes of the wild type had reached the end of the transmitting tract, but only a few mat3 pollen tubes had reached the middle of the transmitting tract (Fig. 2D). These findings are consistent with the seed set analyses, as most seeds on mat3 plants were located at the top of the siliques (Fig. 2E).
Figure 2.
mat3 exhibits defects in the male gametophyte. A, Representative images of pollen from wild-type (WT) and mat3 plants after in vitro germination for 6 h. Bars = 100 μm. B, Pollen germination percentage at 6 h. Data are means ± se of three experiments. For each independent replicate, 400 to 500 pollen grains were analyzed. C, Pollen tube lengths of the wild type and mat3 after germination for 6 h. Data are means ± se of three experiments. D, Semi in vivo pollen tube growth. Shown are wild-type and mat3 homozygous pistils 12 h (left) and 24 h (right) after pollination stained with decolorized Aniline Blue. E, Whole-mount images of mature siliques after clearing in a 0.2 n NaOH and 1% SDS solution.
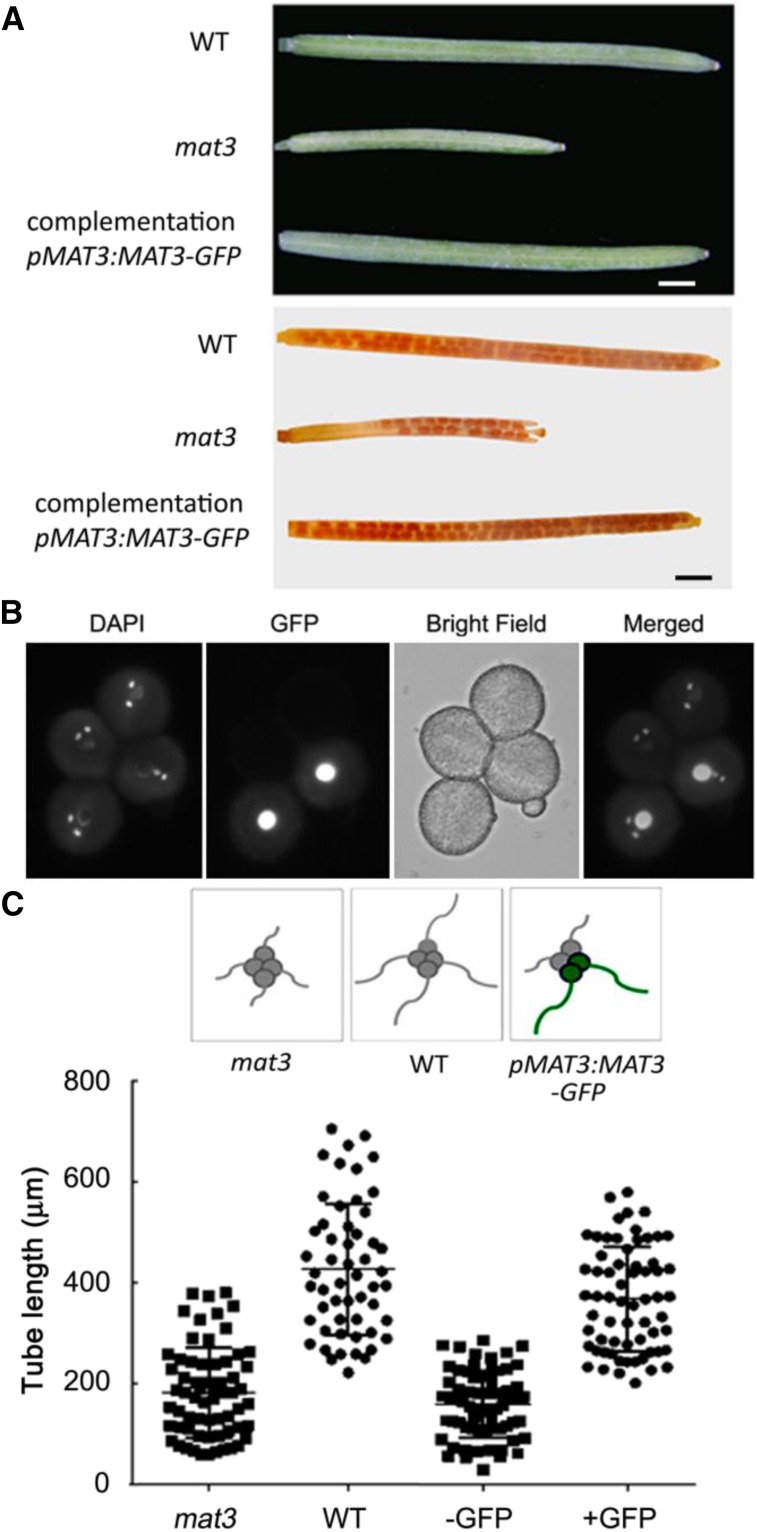
To determine if these phenotypes were due to the disruption in MAT3, a pMAT3::MAT3-GFP construct was transformed into the mat3 mutant. The complemented lines rescued silique length and seed set (Fig. 3A). The same construct also was transformed into mat3/mat3; qrt1/qrt1. qrt1/qrt1 is a sporophytic recessive mutation in which the four products of microsporogenesis remain fused so that pollen grains are released as tetrads (Preuss et al., 1994). In mat3/mat3; qrt1/qrt1 plants carrying the pMAT3::MAT3-GFP transgene, mutant and complemented pollen can be distinguished (green pollen contains the pMAT3::MAT3-GFP transgene). After germination, the green pollen had pollen tube lengths similar to those of wild-type pollen tubes, indicating that the pollen defects are indeed caused by the loss of MAT3 expression (Fig. 3C). Furthermore, with this construct, we could determine the subcellular location of MAT3-GFP. MAT3-GFP was localized in both the cytoplasm and the vegetative nucleus (Fig. 3B), with several bright spots in the vegetative nucleus (Supplemental Fig. S2).
Figure 3.
Complementation of the mat3 mutant and subcellular localization of MAT3. A, Representative wild-type (WT), mat3, and complemented immature siliques and mature siliques after clearing. Bars = 1 mm. B, Subcellular localization of MAT3 in a quartet, in which two of the four pollen grains are transgenic. Images (from left to right) show the 4′,6-diamino-2-phenylindole (DAPI) channel, GFP channel, bright field, and merged. C, Pollen tube lengths of qrt1/qrt1;mat3/mat3, qrt1/qrt1, and mat3/mat3 carrying pMAT3::MAT3-GFP measured after 6 h of germination. Data are means ± se of three experiments. For each independent replicate, 100 to 150 pollen tubes were analyzed.
Overaccumulation of Met Inhibits Pollen Tube Growth
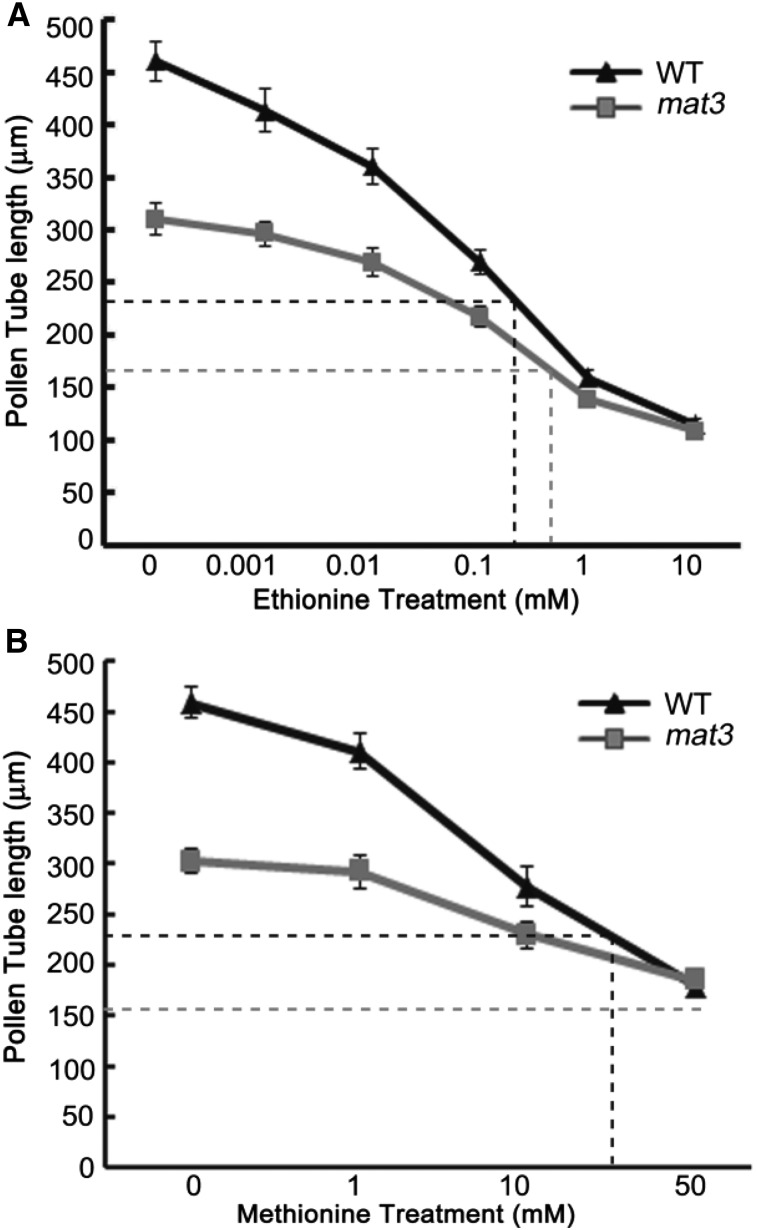
SAMS synthesizes SAM from ATP and l-Met. If the levels of SAMS were reduced, the product also might be reduced. If so, adding SAM might rescue the silique length and the seed set problem. We used an in vitro inflorescence culture system (Lardon et al., 1993; Magnard et al., 2001) to attempt to deliver SAM, and also added SAM to pollen germination medium to test if germination or tube growth was improved. However, in neither case was there an improvement (Supplemental Fig. S3). Therefore, we hypothesized that the pollen phenotype might be caused by increased amounts of Met. Ethionine is a toxic analog of Met (Inaba et al., 1994), and cells that overaccumulate free Met exhibit increased resistance to ethionine (Shen et al., 2002). Therefore, we added ethionine to pollen germination medium and measured pollen tube lengths in the wild type and mat3. Inhibition was dose dependent and was more obvious in the wild type than in mat3 (Fig. 4A): wild-type 50% inhibition (I50) = 0.3 mm, while mat3 I50 = 0.8 mm, suggesting that free Met accumulates in mat3 pollen. To verify if Met overaccumulation indeed affected pollen tube growth, exogenous Met was added to in vitro pollen germination medium. The pollen tube lengths were shorter in the wild type than in mat3, and the effect was again dose dependent (Fig. 4B): wild-type I50 = 26 mm, while mat3 I50 > 50 mm. These data support the idea that Met can inhibit pollen tube growth and that the short mat3 pollen tubes might be caused by Met overaccumulation. It is also possible that the reduction in SAM indirectly caused the phenotype (i.e. that some or all of the SAM methyl donor needs in the cell were impaired). To address these possibilities, we carried out metabolomics and assessed histone methylation and tRNA modifications.
Figure 4.
Wild type (WT) pollen tubes are more sensitive to ethionine and Met. A, In vitro pollen tube lengths after 6 h, with various concentrations of ethionine added to pollen germination medium. Data are means ± se of triplicate experiments. B, In vitro pollen tube lengths after 6 h, with various concentrations of Met added to pollen germination medium. Data are means ± se of triplicate experiments. Dashed lines mark I50 of pollen tube length, black for the wild type and gray for mat3.
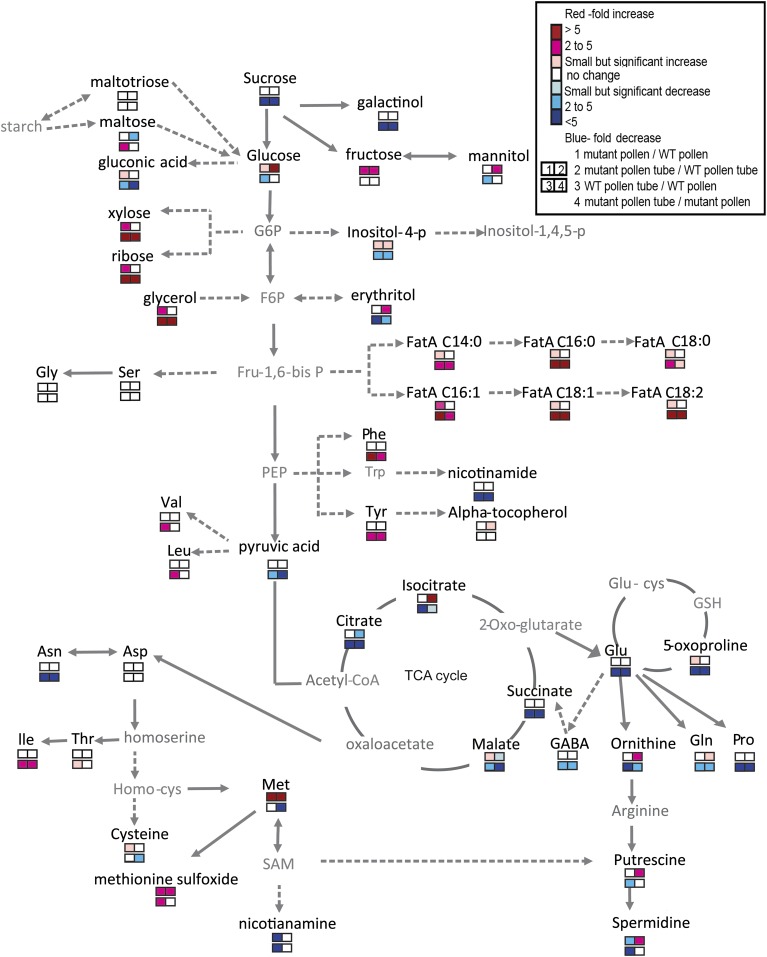
mat3 Pollen and Pollen Tubes Exhibit Alterations in the Tricarboxylic Acid Cycle and Polyamine Biosynthetic Pathways
Met biosynthesis is part of a complex pathway with multiple regulatory controls (Hesse and Hoefgen, 2003). A previous study of three Met overaccumulation (mto) mutants found changes in several metabolic pathways (Kusano et al., 2010). Most relevant for our study, mto3-1 (i.e. a mutant in MAT4, which encodes a homolog expressed in seedlings) showed changes in asparate biosynthesis, in isocitrate and citrate levels in the tricarboxylic acid cycle, in γ-aminobutyrate, and in levels of the polyamine putrescine. To determine whether there was any overlap of these changes in mat3 pollen and pollen tubes, we performed metabolomics analysis on wild-type and mat3/mat3 mature pollen and pollen tubes after 3 h of germination using gas chromatography-time of flight-mass spectrometry. We obtained mass spectra for 185 metabolites, including sugars, amino acids, polyamines, and fatty and organic acids. As expected, Met in mat3/mat3 pollen was 33-fold higher than in wild-type pollen, and in pollen tubes, Met was 9-fold higher than in the wild type.
We compared pollen tubes of mat3 and the wild type. As found for mto3-1 in seedlings, in mat3 pollen tubes, the most obvious differences involved the Met-related pathways, such as Met sulfoxide (2.3-fold higher in mat3), and polyamines, such as putrescine (2-fold higher) and spermidine (2.1-fold higher). Additionally, four intermediates in the tricarboxylic acid cycle were altered in mat3: malate (2-fold lower), fumaric acid (1.6-fold lower), and citrate (2.6-fold lower) were decreased, while isocitrate (7.1-fold higher) was increased (Fig. 5). Amino acids derived from oxoglutarate, such as Gln (1.8-fold) and Pro (1.6-fold), also were higher (Fig. 5; Supplemental Table S3). In starch and Suc metabolism, maltose was lower (3.7-fold) in mat3 pollen tubes (Supplemental Table S3), coupled with more Glc (5.8-fold) and Fru (3.4-fold), consistent with the expected need for starch breakdown to provide an immediate source of soluble sugars during pollen tube growth.
Figure 5.
Metabolite profiles of mat3 and wild-type (WT) mutant pollen and pollen tubes. Changes are represented as fold change between the level of a metabolite in mat3 and in the wild type and are listed in Supplemental Data S1. Solid arrows represent a single-step reaction between two metabolites, and dashed arrows indicate multiple steps. Metabolites in gray were not detected. FatA C14:0, Myristic acid; FatA C16:0, palmitic acid; FatA C18:0, stearic acid; FatA C16:1, palmitoleic acid; FatA C18:1, oleic acid; FatA C18:2, linoleic acid; F6P, fructose6-phosphate; Fru-1,6-bisP, fructose 2,6-bisphosphate; G6P, glucose 6-phosphate; GABA, γ-aminobutyric acid; Glu-cys, glutamyl-l-cysteine; Homo-cys, homocysteine; PEP, phosphoenolpyruvate; SAM, S-adenosyl-methionine.
Next, we compared pollen of mat3 and the wild type. Spermidine levels were lower (4.3-fold) in mat3 pollen, consistent with the report that spermidine stimulated Arabidopsis in vitro pollen germination (Rodriguez-Enriquez et al., 2013). In contrast to the differences seen when mat3 and wild-type pollen tubes were compared, intermediates in the tricarboxylic acid cycle were not different in mat3 and wild-type pollen, but many metabolites in lipid metabolism were higher in mat3 pollen: five unsaturated fatty acids: arachidic acid (3.4-fold), linolenic acid (1.8-fold), linoleic acid (1.6-fold), palmitoleic acid (4.7-fold) and oleic acid (1.7-fold); three saturated fatty acids: myristic acid (1.9-fold), palmitic acid (1.7-fold), and stearic acid (1.8-fold); and glycerol (2.2-fold; Supplemental Table S3).
Rapid pollen tube growth requires high rates of energy flow from sugars, the citric acid cycle, and lipid biosynthesis. We thus also compared pollen with pollen tubes (i.e. wild-type pollen with wild-type pollen tubes and mat3 pollen with mat3 pollen tubes), as such comparisons might be useful for understanding the importance of particular metabolic pathways during pollen tube growth. Most noticeably, in the wild type, three metabolites decreased, putrescine (2.3-fold), spermidine (5.3-fold), and nicotiamine (33-fold), in pollen tubes, but they did not decrease in mat3 pollen tubes. Glc decreased (3.4-fold) in wild-type pollen tubes, relative to its level in pollen, but did not decrease in mat3 pollen tubes relative to its level in mat3 pollen. Lastly, maltose (2.9-fold higher) and Fuc (2.5-fold higher) levels increased in pollen tubes in wild-type but not in mat3 pollen tubes. These differences are consistent with the idea that mat3 pollen tubes are impaired in sugar metabolism.
Histone Methylation Is Decreased in the Vegetative Nucleus of mat3 Pollen
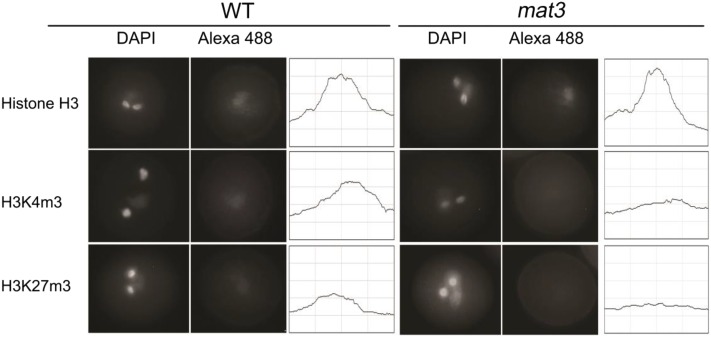
Met metabolism generates the major methyl donor SAM for histone methylation (Liu et al., 2015). In some weak MAT3-GFP expression lines, we observed some bright spots in the nucleus (Supplemental Fig. S2). Such nuclear subcompartmentalization is reminiscent of structures called chromatin hubs (Bantignies and Cavalli, 2011), which are thought to regulate the 3D chromatin state (Gómez-Díaz and Corces, 2014). As methyl groups and methyltransferase proteins are important for chromatin modifications, it was reasonable to hypothesize that histone methylation might be affected in mat3. Therefore, we assessed histone methylation levels in mat3 pollen grains using immunofluorescence with antibodies that recognize trimethyl H3K4 and trimethyl H3K27. These methylation marks were detectable in vegetative nuclei of wild-type pollen (Fig. 6) but were weaker in mat3 pollen, and especially for H3K27m3, they were not above background. The control antibody, against unmodified histone H3, showed similar intensities in both wild-type and mat3 pollen.
Figure 6.
Immunofluorescence using specific antibodies against methylated histone H3 in wild-type (WT) and mat3 pollen. Mature pollen from wild-type and mat3 plants was incubated with anti-H3 (control), anti-H3K4m3, and anti-H3K27m3 antibodies. Representative examples show histone methylation in the vegetative nucleus. Quantitation of signal intensity is shown next to each image. Each x axis represents a 5-μm distance centered on the fluorescent focus, and each y axis represents relative fluorescence intensity. All images were acquired using the same exposure time. Forty to 50 pollen grains for each genotype were examined. DAPI, 4′,6-Diamino-2-phenylindole.
Methylation of tRNA Is Reduced in mat3 Pollen
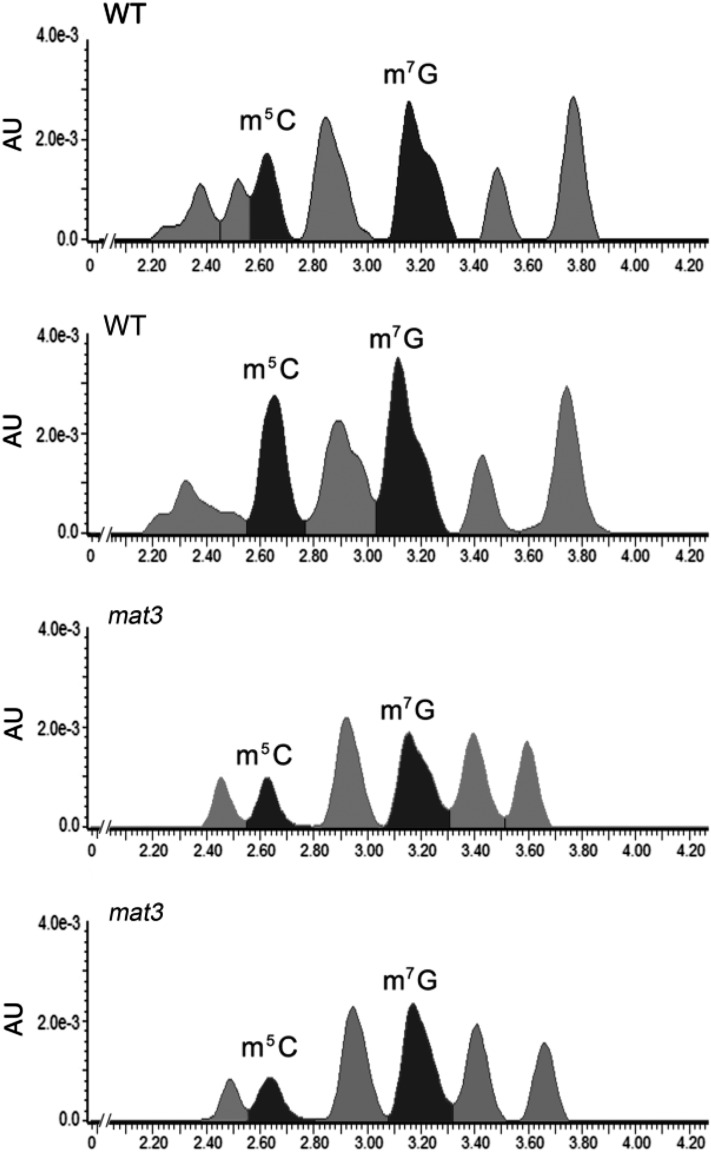
It was reported that tRNA is the only methylated RNA in pollen tubes and that, therefore, tRNA methylation might be a primary mechanism affecting protein synthesis in pollen tubes (Steffensen, 1971). More than 90 tRNA methyltransferase genes are predicted in the Arabidopsis genome, based on the homology of conserved domains with tRNA methyltransferase genes in Sacccharomyces cerevisiae and Escherichia coli (Chen et al., 2010). From these 90 tRNA methyltransferase genes, we generated a list of 27 candidate tRNA methyltransferases expressed in pollen by sorting the RNA sequencing data (Loraine et al., 2013) using greater than 1 RPKM as the threshold (Supplemental Table S2). Of these 27, 18 were annotated as responsible for ncm5U modification, four for pseudouridine modification, and five single genes for specific nucleoside modifications: dihydrouridine (At5g66180), m1A (At2g45730), m5C (At5g66180), m7G (At1g03110), and m5U (At2g28450). In order to determine if any tRNA methylated moieties were reduced in mat3, liquid chromatography-mass spectrometry was used to analyze purified tRNAs from the wild type and mat3. The retention time of each peak was compared with commercial standards, and each peak was further analyzed by mass spectrophotometry. Only two peaks matched commercial standards, those for m5C and m7G, and their identities were confirmed by mass spectrophotometry. Both m5C and m7G were reduced in mat3 pollen, more so for m5C (Fig. 7). Even though there were reductions for some of the other peaks, they did not match the retention times or mass spectral data for other modified nucleosides (Su et al., 2014) and remain unidentified.
Figure 7.
Liquid chromatography-mass spectrometry analysis of total 5-methylcytidine (m5C) and 7-methylguanosine (m7G) levels in tRNA purified from wild-type (WT) and mat3 pollen. AU represents absorbance units.
DISCUSSION
Single mutants of SAMS in Arabidopsis did not show any seedling phenotypes, presumably because of gene redundancy, as all four SAMS are expressed in seedlings (Shen et al., 2002; Mao et al., 2015). Here, we provide evidence that mat3 plants have impaired pollen tube growth and reduced seed set (Figs. 1–3). This finding raised the question of whether there is a specific requirement for MAT3 during pollen germination and pollen tube growth, because MAT1, MAT2, and MAT4 also are expressed in pollen (Loraine et al., 2013), but at much lower levels than MAT3. Microarray experiments show that MAT4 and MAT3 are equivalently expressed in pollen tubes that have first grown through pistil tissue for 1 h and then collected 3 h later (Qin et al., 2009), suggesting that MAT3 and MAT4 could be partially redundant. However, MAT4 was not expressed at 0.5- or 4-h time points of in vitro-grown pollen tubes (Qin et al., 2009). Thus, these results are consistent with the idea that MAT3 plays a more important role at the beginning stages of pollen germination.
SAM synthesis is reduced or absent in SAMS mutants, so the flux from Met into SAM is impaired. It was not possible to correct the pollen tube growth defect of mat3 by adding SAM (Supplemental Fig. S3), probably because it is rapidly degraded or poorly, if at all, taken up by cells (García-Trevijano et al., 2000). Figure 4 shows that endogenous Met is toxic to wild-type pollen and suggests that the higher levels of Met in mat3 pollen somehow obviate the effect of exogenous Met. Indeed, metabolite analysis showed that Met accumulated in mat3 pollen and pollen tubes (Fig. 5), as also seen for mto3 (SAMS3; At3g17390) rosette leaves (Kusano et al., 2010). Why would altered Met levels contribute to pollen tube growth inhibition? In mat3 pollen tubes, the most obvious metabolic changes were in the Met-related pathway, but there also were increases in sugar metabolism metabolites, particularly Glc, Fru, and mannitol. Fatty acids were increased in mat3 pollen, suggesting that there are enough sources of carbon storage in mat3 pollen (Fig. 5). Interestingly, citrate and malate, components of the tricarboxylic acid cycle, decreased in mat3 pollen tubes, consistent with the idea that storage mobilization is impaired in mat3 during pollen tube growth. Decreased spermidine in mat3 pollen was correlated with a decrease of pollen germination, consistent with the report that spermidine stimulated in vitro pollen germination (Rodriguez-Enriquez et al., 2013). We also detected polyamine biosynthesis metabolism differences in mat3 pollen, consistent with the previously described link between the Met cycle and polyamine metabolism (Bürstenbinder et al., 2010). It was suggested that changes in polyamine levels could affect DNA methylation and, consequently, gene expression (Fraga et al., 2004).
SAMS was localized in the cytosol in Arabidopsis and Catharanthus roseus (Schröder et al., 1997; Ravanel et al., 1998), but more recent studies in Arabidopsis and rice localized SAMS to both the nucleus and cytoplasm (Li et al., 2011; Mao et al., 2015), as we also showed for MAT3 (Fig. 3B). This finding raised the question of whether there might be a specific need for SAM synthesis in the vegetative nucleus. SAM is essential for the methylation of DNA and RNA (Kobayashi et al., 1990; Ying et al., 1999; Block et al., 2002; Cheng et al., 2003). GFP localization of MAT3 in nuclear subcompartments (Supplemental Fig. S2), reminiscent of chromatin hubs (Bantignies and Cavalli, 2011), suggests that MAT3 might function in DNA and/or RNA methylation in the vegetative nucleus. In D. melanogaster, SAMS is important for histone methylation (Liu et al., 2015). We showed that mat3 pollen had less trimethylated H3K4 and H3K27 (Fig. 6), modifications that are associated with transcriptional regulation. Additionally, SAMS affects the amounts of metabolites in the polyamine pathway (Larsson et al., 1996). Polyamines can bind DNA and affect chromatin (Matthews, 1993). So another possibility is that SAMS affects histone methylation by affecting polyamine content. This hypothesis is supported by our metabolomics data (Fig. 5).
Defects of tRNA modifications might cause tRNA degradation, resulting in decreased translation efficiency, increased translational errors, and altered cellular metabolism and signaling pathways (Chen et al., 2010; Tuorto et al., 2012). Thus, tRNA methylation changes could provide a possible mechanistic explanation for the defects observed in mat3. Although 27 tRNA modification genes annotated as responsible for tRNA modification are expressed in pollen, the poor sensitivity of the HPLC, and/or the overall low abundance of modified nucleosides, only allowed us to detect m5C and m7G, but mat3 pollen did have lower levels of those (Fig. 7). Thus, any or a combination of these differences might contribute to the pollen germination and pollen tube growth phenotypes.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown in the greenhouse under an 18-h-light/6-h-dark cycle at 22°C. The SALK_019375 line was obtained from the Arabidopsis Biological Resource Center (Ohio State University) by Kang Chong’s laboratory (Institute of Botany, Chinese Academy of Sciences). Columbia-0 was used as the wild type.
SAMS Nomenclature
The nomenclature for SAMS genes was as follows: MAT1 (also named SAM1, SAMS1, MTO1, and NP_849577), MAT2 (also named SAM2, SAMS2, and AAA32869), MAT3 (also named AAD31573), and MAT4 (also named SAMS3, MTO3, and AAO11581).
Constructs and Plant Transformation
All primers are listed in Supplemental Table S1. For complementation constructs, a 2-kb fragment upstream of the ATG and the full-length At2g36880 open reading frame was amplified from the wild type using primers mat3-prof/mat3-CDS-r, subcloned into the pCR8/GW-TOPO vector (Invitrogen), and then transferred into pGWB4 (Nakagawa et al., 2007). The complementation construct was transformed using the floral dip method (Clough and Bent, 1998). All PCR amplifications used Phusion hot start high-fidelity DNA polymerase with the annealing temperature and extension times recommended by the manufacturer (Thermo Scientific). The vector was sequenced using an ABI 3300 sequencer, and sequences were analyzed using Vector NTI (Invitrogen). Bioneer PCR purification and Spin miniprep kits were used for PCR product recovery and plasmid DNA extraction, respectively.
Pollen Tube Growth
Arabidopsis in vitro pollen tube germination was as described by Boavida and McCormick (2007). Decolorized Aniline Blue staining was as described by Johnson-Brousseau and McCormick (2004).
RT-PCR and Quantitative PCR
Total RNA was extracted using the RNeasy plant mini kit (Qiagen) and reverse transcribed using the SuperScript III first-strand synthesis system (Invitrogen) according to the manufacturer’s instructions. RT-PCR was performed with gene-specific primers, and runs were 18 to 25 cycles depending on the linear range of products for each gene. Quantitative PCR was performed using a MyIQ Real-Time PCR detection system (Bio-Rad) with SYBR Green PCR Master Mix (Bio-Rad). Fold change for gene expression was calculated by normalizing Ct values using the 2−ΔΔCt method. The primers used are listed in Supplemental Table S1.
Preparation and Collection of Pollen and Pollen Tubes for Metabolite Analyses
Mature pollen grains were collected from fresh flowers using a vacuum as described (Johnson-Brousseau and McCormick, 2004). To collect pollen tubes, pollen was germinated for 3 h. The pollen/pollen tube mixture was collected and washed six times using double distilled water and then filtered using a 60-μm nylon mesh to remove ungerminated pollen grains. Pollen tube samples were collected from the mesh, checked with a microscope to confirm that ungerminated pollen grains were almost absent from the mesh, and then lyophilized in a vacuum evaporator before metabolite analyses.
Metabolite Analysis
For each analysis (three biological replicates), 50 mg of pollen or pollen tubes was used. Metabolite analysis was carried out by the West Coast Metabolomics Center (University of California Davis) using a Leco Pegasus II gas chromatograph-time of flight-mass spectrometer.
Immunofluorescence
Pollen grains were collected by vacuum (Johnson-Brousseau and McCormick, 2004), fixed in 4% paraformaldehyde in PBS for 2 h, treated with 0.5% Triton X-100 in PBS for 10 min, and incubated in blocking buffer (PBS with 5% BSA for 1 h at 37°C, then with H3 antibody (1:100 dilution) or H3K27m3 or H3K4m3 antibody (1:200 dilution) overnight at 4°C. After washing with blocking buffer, the pollen was transferred to blocking buffer containing Alexa Fluor 488 goat anti-rabbit antibody (1:200 dilution) for 6 h at room temperature. The samples were washed in PBS five times, then transferred to a microscope slide, and counterstained with 4′,6-diamino-2-phenylindole (1 μg mL−1). Images were acquired using the GFP channel with an Axiovert microscope (Zeiss) and an AxioCamRM camera and AxioVision 4.3 software. Images were processed using Adobe Photoshop 7.0.
tRNA Isolation and UPLC Analysis
tRNA was prepared from total RNA as described (Björk et al., 2001) using 150 mg of pollen for each sample. tRNA was eluted with buffer (10 mm Tris-H3PO4, pH 6.3, 15% ethanol, and 600 mm KCl), and 40 μg of tRNA was recovered. 5S RNA was not eluted from the column, as it requires at least 650 mm for elution (Björk et al., 2001). Forty micrograms of purified tRNA was digested with Nuclease P1 for 16 h at 37°C and then treated with bacterial alkaline phosphatase for 2 h at 37°C. The hydrolysate was analyzed by ultra-performance liquid chromatography with a C-18 reverse-phase column as described (Su et al., 2014). A UV light detector was used for chromatography. Peaks were collected separately and analyzed using a Perkin-Elmer AxION 2 time of flight-mass spectrometer. There were two replicates for each sample.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: MAT1 (NP_849577), MAT2 (AAA32869), MAT3 (AAD31573), and MAT4 (AAO11581).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. qPCR of MAT3 in mature pollen in the wild type and mat3.
Supplemental Figure S2. Subcellular localization of MAT3.
Supplemental Figure S3. Exogenous SAM did not correct the mat3 pollen tube growth defect.
Supplemental Table S1. Primers used for this study.
Supplemental Table S2. tRNA modification genes expressed Arabidopsis pollen.
Supplemental Table S3. Metabolites identified in this study.
Supplementary Material
Acknowledgments
We thank Binglian Zheng, Weihua Tang, and Jorge Muschietti for comments on the article; Jimmy Li, Madhur Garg, and Sumedha Ravishankar for technical assistance (they participated in the University of California Berkeley undergraduate research apprentice program); Kang Chong and Wenxuan Li (Institute of Botany, Chinese Academy of Sciences) for providing Arabidopsis mat3 seeds; Jennifer Fletcher for providing trimethyl H3K4 and trimethyl H3K27 antibodies; Delilah F. Wood and Tina Williams for help with microscopy; and Miao Zhang for help with ultra-performance liquid chromatography and mass spectrometry analyses.
Glossary
- SAM
S-adenosylmethionine
- RPKM
reads per kilobase per million mapped reads
Footnotes
This work was supported by the U.S. Department of Agriculture-Agricultural Research Service Current Research Information System (grant no. 5335–21000–030–00D to S.M.)
Articles can be viewed without a subscription.
References
- Aloisi I, Cai G, Serafini-Fracassini D, Del Duca S (2016) Polyamines in pollen: from microsporogenesis to fertilization. Front Plant Sci 7: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies F, Cavalli G (2011) Polycomb group proteins: repression in 3D. Trends Genet 27: 454–464 [DOI] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijó JA (2003) Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol 133: 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binet R, Fernandez RE, Fisher DJ, Maurelli AT (2011) Identification and characterization of the Chlamydia trachomatis L2 S-adenosylmethionine transporter. MBio 2: doi/10.1128/mBio.00051-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk GR, Jacobsson K, Nilsson K, Johansson MJ, Byström AS, Persson OP (2001) A primordial tRNA modification required for the evolution of life? EMBO J 20: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block MA, Tewari AK, Albrieux C, Maréchal E, Joyard J (2002) The plant S-adenosyl-L-methionine:Mg-protoporphyrin IX methyltransferase is located in both envelope and thylakoid chloroplast membranes. Eur J Biochem 269: 240–248 [DOI] [PubMed] [Google Scholar]
- Boavida LC, McCormick S (2007) Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J 52: 570–582 [DOI] [PubMed] [Google Scholar]
- Bürstenbinder K, Waduwara I, Schoor S, Moffatt BA, Wirtz M, Minocha SC, Oppermann Y, Bouchereau A, Hell R, Sauter M (2010) Inhibition of 5′-methylthioadenosine metabolism in the Yang cycle alters polyamine levels, and impairs seedling growth and reproduction in Arabidopsis. Plant J 62: 977–988 [DOI] [PubMed] [Google Scholar]
- Chen P, Jäger G, Zheng B (2010) Transfer RNA modifications and genes for modifying enzymes in Arabidopsis thaliana. BMC Plant Biol 10: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Xu Y, Luo W, Li W, Chen N, Zhang D, Chong K (2013) The F-box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice. Plant Physiol 163: 1673–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Sattler S, Maeda H, Sakuragi Y, Bryant DA, DellaPenna D (2003) Highly divergent methyltransferases catalyze a conserved reaction in tocopherol and plastoquinone synthesis in cyanobacteria and photosynthetic eukaryotes. Plant Cell 15: 2343–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Duschene KS, Broderick JB (2010) The antiviral protein viperin is a radical SAM enzyme. FEBS Lett 584: 1263–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Berdasco M, Diego LB, Rodríguez R, Cañal MJ (2004) Changes in polyamine concentration associated with aging in Pinus radiata and Prunus persica. Tree Physiol 24: 1221–1226 [DOI] [PubMed] [Google Scholar]
- Fujimori DG. (2013) Radical SAM-mediated methylation reactions. Curr Opin Chem Biol 17: 597–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Trevijano ER, Latasa MU, Carretero MV, Berasain C, Mato JM, Avila MA (2000) S-Adenosylmethionine regulates MAT1A and MAT2A gene expression in cultured rat hepatocytes: a new role for S-adenosylmethionine in the maintenance of the differentiated status of the liver. FASEB J 14: 2511–2518 [DOI] [PubMed] [Google Scholar]
- Giovanelli J, Mudd SH, Datko AH (1985) Quantitative analysis of pathways of methionine metabolism and their regulation in Lemna. Plant Physiol 78: 555–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Díaz E, Corces VG (2014) Architectural proteins: regulators of 3D genome organization in cell fate. Trends Cell Biol 24: 703–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Carrasco P (1998) Differential expression of the S-adenosyl-L-methionine synthase genes during pea development. Plant Physiol 117: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández HL, Pierrel F, Elleingand E, García-Serres R, Huynh BH, Johnson MK, Fontecave M, Atta M (2007) MiaB, a bifunctional radical-S-adenosylmethionine enzyme involved in the thiolation and methylation of tRNA, contains two essential [4Fe-4S] clusters. Biochemistry 46: 5140–5147 [DOI] [PubMed] [Google Scholar]
- Hesse H, Hoefgen R (2003) Molecular aspects of methionine biosynthesis. Trends Plant Sci 8: 259–262 [DOI] [PubMed] [Google Scholar]
- Hinson ER, Cresswell P (2009) The N-terminal amphipathic α-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J Biol Chem 284: 4705–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes-Davis R, Tanaka CK, Vensel WH, Hurkman WJ, McCormick S (2005) Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics 5: 4864–4884 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H. (2014) Methylated nucleosides in tRNA and tRNA methyltransferases. Front Genet 5: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K, Fujiwara T, Hayashi H, Chino M, Komeda Y, Naito S (1994) Isolation of an Arabidopsis thaliana mutant, mto1, that overaccumulates soluble methionine: temporal and spatial patterns of soluble methionine accumulation. Plant Physiol 104: 881–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Brousseau SA, McCormick S (2004) A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J 39: 761–775 [DOI] [PubMed] [Google Scholar]
- Kaminska KH, Purta E, Hansen LH, Bujnicki JM, Vester B, Long KS (2010) Insights into the structure, function and evolution of the radical-SAM 23S rRNA methyltransferase Cfr that confers antibiotic resistance in bacteria. Nucleic Acids Res 38: 1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Ngernprasirtsiri J, Akazawa T (1990) Transcriptional regulation and DNA methylation in plastids during transitional conversion of chloroplasts to chromoplasts. EMBO J 9: 307–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M, Fukushima A, Redestig H, Kobayashi M, Otsuki H, Onouchi H, Naito S, Hirai MY, Saito K (2010) Comparative metabolomics charts the impact of genotype-dependent methionine accumulation in Arabidopsis thaliana. Amino Acids 39: 1013–1021 [DOI] [PubMed] [Google Scholar]
- Lardon A, Triboi-Blondel AM, Dumas C (1993) A model for studying pollination and pod development in Brassica napus: the culture of isolated flowers. Sex Plant Reprod 6: 52–56 [Google Scholar]
- Larsson J, Zhang J, Rasmuson-Lestander A (1996) Mutations in the Drosophila melanogaster gene encoding S-adenosylmethionine synthetase [corrected] suppress position-effect variegation. Genetics 143: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Han Y, Tao F, Chong K (2011) Knockdown of SAMS genes encoding S-adenosyl-L-methionine synthetases causes methylation alterations of DNAs and histones and leads to late flowering in rice. J Plant Physiol 168: 1837–1843 [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 137: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Barnes VL, Pile LA (2015) Disruption of methionine metabolism in Drosophila melanogaster impacts histone methylation and results in loss of viability. G3 (Bethesda) 6: 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loraine AE, McCormick S, Estrada A, Patel K, Qin P (2013) RNA-seq of Arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiol 162: 1092–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnard JL, Yang M, Chen YC, Leary M, McCormick S (2001) The Arabidopsis gene tardy asynchronous meiosis is required for the normal pace and synchrony of cell division during male meiosis. Plant Physiol 127: 1157–1166 [PMC free article] [PubMed] [Google Scholar]
- Mao D, Yu F, Li J, Van de Poel B, Tan D, Li J, Liu Y, Li X, Dong M, Chen L, et al. (2015) FERONIA receptor kinase interacts with S-adenosylmethionine synthetase and suppresses S-adenosylmethionine production and ethylene biosynthesis in Arabidopsis. Plant Cell Environ 38: 2566–2574 [DOI] [PubMed] [Google Scholar]
- Martínez-López N, Varela-Rey M, Ariz U, Embade N, Vazquez-Chantada M, Fernandez-Ramos D, Gomez-Santos L, Lu SC, Mato JM, Martinez-Chantar ML (2008) S-Adenosylmethionine and proliferation: new pathways, new targets. Biochem Soc Trans 36: 848–852 [DOI] [PubMed] [Google Scholar]
- Matthews HR. (1993) Polyamines, chromatin structure and transcription. BioEssays 15: 561–566 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Noir S, Bräutigam A, Colby T, Schmidt J, Panstruga R (2005) A reference map of the Arabidopsis thaliana mature pollen proteome. Biochem Biophys Res Commun 337: 1257–1266 [DOI] [PubMed] [Google Scholar]
- Pierrel F, Douki T, Fontecave M, Atta M (2004) MiaB protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J Biol Chem 279: 47555–47563 [DOI] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijó JA, Becker JD (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138: 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D, Rhee SY, Davis RW (1994) Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460 [DOI] [PubMed] [Google Scholar]
- Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, Vasic B, Johnson MA, Palanivelu R (2009) Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel S, Gakière B, Job D, Douce R (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci USA 95: 7805–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Enriquez MJ, Mehdi S, Dickinson HG, Grant-Downton RT (2013) A novel method for efficient in vitro germination and tube growth of Arabidopsis thaliana pollen. New Phytol 197: 668–679 [DOI] [PubMed] [Google Scholar]
- Roje S. (2006) S-Adenosyl-L-methionine: beyond the universal methyl group donor. Phytochemistry 67: 1686–1698 [DOI] [PubMed] [Google Scholar]
- Rounds CM, Winship LJ, Hepler PK (2011) Pollen tube energetics: respiration, fermentation and the race to the ovule. AoB Plants 2011: plr019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder G, Eichel J, Breinig S, Schröder J (1997) Three differentially expressed S-adenosylmethionine synthetases from Catharanthus roseus: molecular and functional characterization. Plant Mol Biol 33: 211–222 [DOI] [PubMed] [Google Scholar]
- Selinski J, Scheibe R (2014) Pollen tube growth: where does the energy come from? Plant Signal Behav 9: e977200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Li C, Tarczynski MC (2002) High free-methionine and decreased lignin content result from a mutation in the Arabidopsis S-adenosyl-L-methionine synthetase 3 gene. Plant J 29: 371–380 [DOI] [PubMed] [Google Scholar]
- Sheoran IS, Sproule KA, Olson DJH, Ross ARS, Sawhney VK (2006) Proteome profile and functional classification of proteins in Arabidopsis thaliana (Landsberg erecta) mature pollen. Sex Plant Reprod 19: 185–196 [Google Scholar]
- Steffensen DM. (1971) Ribosome synthesis compared during pollen and pollen tube development. In Heslop-Harrison J, ed, Pollen: Development and Physiology. Butterworths, London, pp 223–228 [Google Scholar]
- Su D, Chan CT, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC (2014) Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat Protoc 9: 828–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM (2012) Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150: 934–947 [DOI] [PubMed] [Google Scholar]
- Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F (2012) RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 19: 900–905 [DOI] [PubMed] [Google Scholar]
- Wolukau JN, Zhang SL, Xu GH, Chen D (2004) The effect of temperature, polyamines and polyamine synthesis inhibitor on in vitro pollen germination and pollen tube growth of Prunus mume. Sci Hortic 99: 289–299 [Google Scholar]
- Yan F, LaMarre JM, Röhrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG (2010) RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J Am Chem Soc 132: 3953–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Z, Mulligan RM, Janney N, Houtz RL (1999) Rubisco small and large subunit N-methyltransferases: bi- and mono-functional methyltransferases that methylate the small and large subunits of Rubisco. J Biol Chem 274: 36750–36756 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.