Genome-scale metabolic model for Chlorella vulgaris UTEX 395 accurately predicts phenotypes under different growth conditions.
Abstract
The green microalga Chlorella vulgaris has been widely recognized as a promising candidate for biofuel production due to its ability to store high lipid content and its natural metabolic versatility. Compartmentalized genome-scale metabolic models constructed from genome sequences enable quantitative insight into the transport and metabolism of compounds within a target organism. These metabolic models have long been utilized to generate optimized design strategies for an improved production process. Here, we describe the reconstruction, validation, and application of a genome-scale metabolic model for C. vulgaris UTEX 395, iCZ843. The reconstruction represents the most comprehensive model for any eukaryotic photosynthetic organism to date, based on the genome size and number of genes in the reconstruction. The highly curated model accurately predicts phenotypes under photoautotrophic, heterotrophic, and mixotrophic conditions. The model was validated against experimental data and lays the foundation for model-driven strain design and medium alteration to improve yield. Calculated flux distributions under different trophic conditions show that a number of key pathways are affected by nitrogen starvation conditions, including central carbon metabolism and amino acid, nucleotide, and pigment biosynthetic pathways. Furthermore, model prediction of growth rates under various medium compositions and subsequent experimental validation showed an increased growth rate with the addition of tryptophan and methionine.
Photosynthetic microorganisms have gained attention for their potential utility in various biotechnology applications due to their polytrophic metabolism, including photoautotrophy, heterotrophy, and mixotrophy, enabling them to take advantage of different energy and carbon sources for growth. These organisms are capable of fixing CO2 with light as an energy source to produce biomass and oxygen, thus having a significant impact on the global oxygen and CO2 budget. Algae, as well as microalgae, play a profound role in the carbon cycle and are responsible for 50% of the atmospheric oxygen release and carbon fixation on the planet (Tabatabaei et al., 2011).
The unicellular microalga Chlorella vulgaris of the phylum Chlorophyta has long served as a model organism. Chlorella spp. strains exhibit substantial metabolic flexibility in response to environmental perturbations (Mitra et al., 2012; Liu et al., 2013). Their robust metabolic capabilities suggest the potential for industrial production of desired biomass components, most notably lipids, to serve as precursors for biofuel production as well as other value-added products (Perez-Garcia et al., 2011). In addition, Chlorella spp. strains are capable of using nutrients (i.e. organic carbon and minerals) directly from wastewater for growth, making them attractive cell factories for biosustainable production processes (Liu et al., 2013). Although algal biodiesel production is technically feasible and environmentally desirable, several challenges remain to be overcome for the process to successfully compete with fossil fuel-derived products (Miao and Wu, 2006; Xiong et al., 2008).
Species such as Chlorella protothecoides, Chlorella minutissima, and C. vulgaris can accumulate lipids up to 50%, 56%, and 60% of their dry weight, respectively (Wang et al., 2008; Guarnieri et al., 2013; Espinosa-Gonzalez et al., 2014). Microalgae with an oil content of around 50% could reach a productivity of 86,515 kg biodiesel hectare−1 year−1, compared with 170 and 340 kg biodiesel hectare−1 year−1 from soy (Glycine max) and canola (Brassica napus; Savage, 2011). Among microalgae, Chlorella spp. strains possess significant advantages, such as higher photosynthetic efficiency over other photosynthetic organisms (Doucha and Lívanský, 2006). Due to this potential, efforts have been made to characterize the biomass production of Chlorella spp. strains in large-scale bioreactors. It has been reported that the synthesis of lipids in Chlorella spp. is substantially increased by deficiencies of nitrogen and phosphorus in the culture medium, with nitrogen having the most important effect on lipid accumulation. Other studies have shown that lower nitrogen-to-carbon ratios result in higher lipid storage by C. vulgaris (Levering et al., 2015).
Industrial microorganisms, including C. vulgaris, have become targets for engineering strategies to improve their performance in biotechnological applications. Genome-scale models have proven to be useful tools for the targeted engineering of such organisms (Monk et al., 2014). These genome-scale network reconstructions can help us understand the compartmental organization of metabolism within an organism, discover new metabolic gene functions, guide adaptive evolution approaches, and optimize the production of value-added compounds, such as pigments and lipids (Espinosa-Gonzalez et al., 2014). Several genome-scale reconstructions for photosynthetic organisms have been generated previously. The most comprehensive of these, in terms of coverage of the genome, are for the plants Zea mays, Brassica napus, and Arabidopsis thaliana, for the microalgae Chlamydomonas reinhardtii, Ostreococcus spp., and Tisochrysis lutea, and for the bacteria Synechocystis sp. PCC6803 and Cyanothece spp. (Kim et al., 2012; Baroukh et al., 2015). Additionally, smaller networks have recently emerged for Chlorella sp. FC2 IITG, C. protothecoides, Chlorella variabilis, and Chlorella pyrenoidosa. These smaller networks are limited in scope and contain only core carbon and photosynthetic metabolism, with an average of 500 reactions and 390 metabolites (Yang et al., 2000; Muthuraj et al., 2013; Wu et al., 2015; Juneja et al., 2016). Here, we report the reconstruction of a genome-scale metabolic network for C. vulgaris UTEX 395 (iCZ843) based on the recently assembled genome sequence.
RESULTS
Reconstruction of the C. vulgaris UTEX 395 Metabolic Network
Using the genome annotation for C. vulgaris UTEX 395, as well as an existing and manually curated genome-scale model for C. reinhardtii (iRC1080; Chang et al., 2011), a preliminary draft reconstruction for C. vulgaris UTEX 395 based on protein homology was assembled.
This draft reconstruction was subsequently subjected to an iterative manual curation process as depicted in Supplemental Figure S1. During the conversion of the curated reconstruction to a mathematical model, quality control/quality assessment tests were performed according to established standards (Thiele and Palsson, 2010). We identified and resolved thermodynamically infeasible reactions in the model that were capable of producing energy in the form of ATP, NADPH, or NADH without allowing carbon or photon input into the system. To guide this step, we visualized flux-carrying reactions using the Web application ESCHER for building, sharing, and embedding data-rich visualizations of biological pathways (King et al., 2015). In most cases, the addition of protons or cofactors was required to prevent these thermodynamically infeasible energy-producing reactions.
Nonenzymatic reactions were included in the model following the work flow for high-quality reconstructions described previously (Thiele and Palsson, 2010) and were associated with Kyoto Encyclopedia of Genes and Genomes (KEGG) identifiers. A mechanistic representation of photosynthesis in C. reinhardtii (Chang et al., 2011), a closely related species, was adapted for the model for C. vulgaris UTEX 395 (iCZ843).
Once all the reactions were introduced into the model, mass and charge balances were checked computationally. All metabolic and transport reactions in the final model are mass balanced, except for the poorly studied reactions related to metarhodopsin hydrolase and rhodopsin retinyltransferase. In those cases, an unknown part of the metabolite formula is present, which causes an imbalance in the reaction. However, the presence of this unknown moiety does not appreciably affect simulations of iCZ843. Complete lists of reactions and metabolite annotations are given in Supplemental Tables S1 and S2.
Refinement and Gap Analysis
Model refinement and gap analysis were performed after the conversion to a mathematical model to preserve metabolic pathway connectivity. Reactions identified during the gap-filling process were categorized into different classes (non-gene/protein/reaction [GPR]-associated, nonenzymatic or spontaneous, and multistep incomplete reactions; Supplemental Table S3). A detailed protocol used for gap filling is described in Supplemental Text S1 and Supplemental Figure 1. Most of the non-GPR-associated reactions were added in subsystems related to central carbon metabolism. Bioinformatics tools were used to predict the cellular compartmental location of lipid metabolism reactions (Table I). Retinol metabolism was placed in the chloroplast based upon the predicted subcellular localization of proteins and the lack of an eyespot in C. vulgaris. We identified gene associations for 35% of the approximately 350 transport reactions. Non-gene-associated reactions were included in the model using standard mechanisms, such as passive transport, transport linked to energy consumption, and salt efflux and influx, that were associated with the transport of particular compounds. The transport reactions for monocarboxylates, dicarboxylates, tricarboxylates, amino acids, and nucleic acids were included based on previous reports (Hanson, 1985). For more details, see Supplemental Text.
Table I. Online resources used during the reconstruction and curation process.
Model Properties
The model for C. vulgaris UTEX 395 (iCZ843) consists of six compartments: the cytoplasm, mitochondrion, chloroplast, thylakoid, glyoxysome, and extracellular space. The model was tested for the ability to grow under known physiological growth conditions of C. vulgaris.
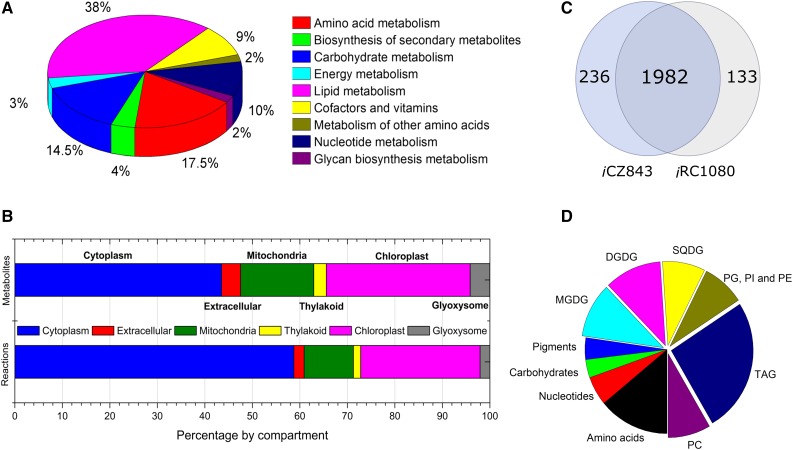
The detailed features of iCZ843 are given in Figure 1. The model contains 843 out of 7,100 annotated genes (around 12%), delineating 1,770 metabolites and 2,294 reactions. The largest subsystems in the reconstruction are amino acid metabolism and lipid metabolism. Other pathways with annotated GPRs include autotrophic metabolism, such as thiamine and biotin metabolism, and amino acid degradation, as well as ubiquinone, terpenoid, α-linolenic acid, and brassinosteroid biosynthesis. As shown in Table II, core models of various complexities have been generated for three Chlorella spp. (Chlorella sp. FC2 IITG, C. protothecoides, and C. pyrenoidosa), and a genomic scale model has been generated for C. variabilis. A detailed comparison by subsystem with other photosynthetic models is shown in Supplemental Table S4; the corresponding metabolic reactions in iRC1080 and iCZ843 are given in Figure 1C.
Figure 1.
Overview of network properties. A, Metabolic reactions of the reconstruction by subsystems (KEGG metabolic pathway diagrams). B, Distribution of metabolites and reactions in the reconstruction. Most metabolites (44%) and reactions (59%) are located in the cytoplasm, while the chloroplast contains 30% of all metabolites and 25% of the reactions. The reconstruction has 1,770 metabolites, 1,180 of which are unique. Metabolites present in all five organelles are hydrogen peroxide, water, hydrogen, NADP, NADPH, and oxygen. C, Unique and shared metabolic reactions between C. vulgaris (iCZ843) and C. reinhardtii (iRC1080). The exchange, demand, sink, and transport reactions were excluded. D, Distribution of the BOF in iCZ843. The BOF (140 metabolites) contains 20 amino acids, eight nucleotides, five carbohydrates, six pigments, and 101 lipids: 35 TAG, eight PG, four PI, four PE, 10 sulfoquinovosyldiacylglycerol (SQDG), 15 MGDG, 16 digalactosyldiacylglycerol (DGDG), and nine phosphatidylcholine (PC).
Table II. Properties of the genome-scale model for C. vulgaris (iCZ843) and other prokaryotic and eukaryotic organisms shown for comparisona.
| Microorganism | Synechocystis sp. PCC6803 | Z. mays | C. reinhardtii | C. pyrenoidosa | Chlorella sp. FC2 IITG | C. protothecoides | C. variabilis | C. vulgaris UTEX 395 |
|---|---|---|---|---|---|---|---|---|
| Reconstruction | Genomic | Genomic | Genomic | Biochemical | Biochemical | Genomic | Genomic | Genomic |
| Genome size | 3.57 Mb | 2.4 Gb | 100 Mb | ND | 46.2 Mb | 22.9 Mb | 46.2 Mb | 63 Mb |
| Genes in the model (total genes/proteins) | 678 (3,575) | 1,563 (32,540) | 1,073 (14,354) | 0 | 0 | 461 (7,039) | 526 (9,791) | 843 (7,100) |
| Total reactions | 863 | 1,985 | 2,190 | 67 | 159 | 272 | 1,455 | 2,286 |
| Metabolites | 790 | 1,825 | 1,707 | 61 | 114 | 144 | 1,236 | 1,770 |
| Compartments | 3 | 5 | 10 | 1 | 1 | 4 | 6 | 6 |
| Reference | Nogales et al. (2012) | Saha et al. (2011) | Chang et al. (2011) | Yang et al. (2000) | Muthuraj et al. (2013) | Wu et al. (2015) | Juneja et al. (2016) | This work |
aBased on the genome, but the gene association is not shown in the model. ND, Not determined.
Defining the Biomass Objective Function
The biomass reaction accounts for all known biomass constituents in terms of their fractional abundance per gram of biomass. The biomass objective function (BOF) in iCZ843 contains the stoichiometric coefficients for 140 metabolites, 39 measured and 101 estimated from the fatty acid profile.
The lipids produced by C. vulgaris can be grouped in the esters: triacylglycerol (TAG), phosphatidylglycerol (PG), phosphatidylinositol (PI), phosphatidylethanolamine (PE), sulfoquinovosyldiglycerol, monogalactosyldiglycerol (MGDG), digalactosyldiglycerol, and phosphatidylcholine, where TAG can accumulate to 20% to 60% of dry cell weight (Guarnieri et al., 2011, 2013; Bellou et al., 2014).
Fatty acids in these esters commonly fall into seven major groups: C16:0, C16:1, C16:2, C16:3, C18:1, C18:2, and C18:3 (Nakamura and Imamura, 1985; Bellou et al., 2014). We also measured 14:0 and 18:0 fatty acid contents for C. vulgaris UTEX 395 under the experimental conditions used in this study. The metabolic model encompasses pathways to synthesize each of these lipids. Using the experimentally determined fatty acid values, a theoretical categorization of the groups of lipids was conducted according to a previous study (Nichols et al., 1967).
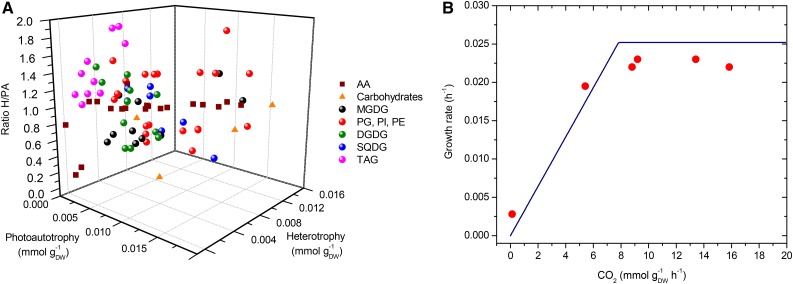
The abundances of the groups of metabolites defined in the BOF are shown in Figure 1D. The light and dark conditions produce a difference in the requirements of biomass precursor metabolites. Although metabolites specified in the BOF are the same under photoautotrophic and heterotrophic growth, experimental data revealed differences between the stoichiometric coefficients under these conditions (Fig. 2A). A ratio for each metabolite between both conditions was calculated. While the amino acid content is almost identical in light and dark, carbohydrates are accumulated under light conditions and excess carbohydrates are stored as starch.
Figure 2.
Network evaluation under different conditions. A, Experimental coefficients in the biomass objective function under photoautotrophic and heterotrophic growth. The ratio between the values in heterotrophy and photoautotrophy (H/PA) was calculated and is displayed in the y axis. AA, Amino acids; DGDG, digalactosyldiacylglycerol; SQDG, sulfoquinovosyldiacylglycerol. B, Specific growth rate simulated for photoautotrophic growth at different CO2 uptake rates shown in blue. The predicted photon input flux was 646 µE m−2 s−1. Red circles indicate experimental values.
Another useful way to constrain a metabolic model is to block the flux through enzymes that are not expressed under a certain condition. RNA sequencing data revealed high expression of genes encoding ribulose bisphosphate carboxylase (EC 4.1.1.39), phosphoribulokinase (EC 2.7.1.19), and NADP-dependent phosphorylating glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.13) under light but not under growth in the dark (Supplemental Table S5). This information was used to modify the flux bounds of related reactions depending on the experimental condition.
Network Evaluation and Robustness
The uptake rates used to constrain the model were calculated using our experimental data (Supplemental Text; Supplemental Fig. S2; Supplemental Table S6). iCZ843 is able to simulate photoautotrophic, heterotrophic, and mixotrophic conditions according to Supplemental Tables S7 to S10. These condition-specific models are provided in Supplemental Data Sets S1–S3 (SBML format).
Model functionality was validated by simulating growth under different conditions using flux balance analysis (Orth et al., 2010). Growth is dependent on the carbon and nitrogen sources (organic as well as inorganic) and on the presence or absence of light. The model accurately predicts the consumption and release of metabolites from or into the medium. For example, simulated nitrate uptake rates under photoautotrophic and heterotrophic conditions were 0.068 and 0.044 mmol h−1, respectively, agreeing well with the experimental data (0.063 and 0.049 mmol h−1). The simulated growth rates for photoautotrophic (0.025 h−1), heterotrophic (0.016 h−1), and mixotrophic (0.0419 h−1) growth are consistent with our experimental data. Table III shows a comparison between experimental and predicted data for C. vulgaris and C. reinhardtii.
Table III. Experimental and predicted growth rates reported for C. vulgaris and C. reinhardtii.
| Model |
iCZ843 |
iRC1080 |
||
|---|---|---|---|---|
| Predicted Growth Rate | Experimental | Predicted Growth Rate | Experimental | |
| h−1 | ||||
| Photoautotrophy | 0.0252 | 0.014–0.025a | 0.1538 | 0.035–0.09b |
| Heterotrophy | 0.0168 | 0.018–0.025a | 0.0299c | 0.059–0.084b |
| Mixotrophy | 0.0407 | 0.02–0.03a | 0.1817c | 0.066b |
Data from this study and Van Baalan et al. (1973). bData from Boyle and Morgan (2009). cAcetate used as organic carbon source. The model was constrained according to Chang et al. (2011).
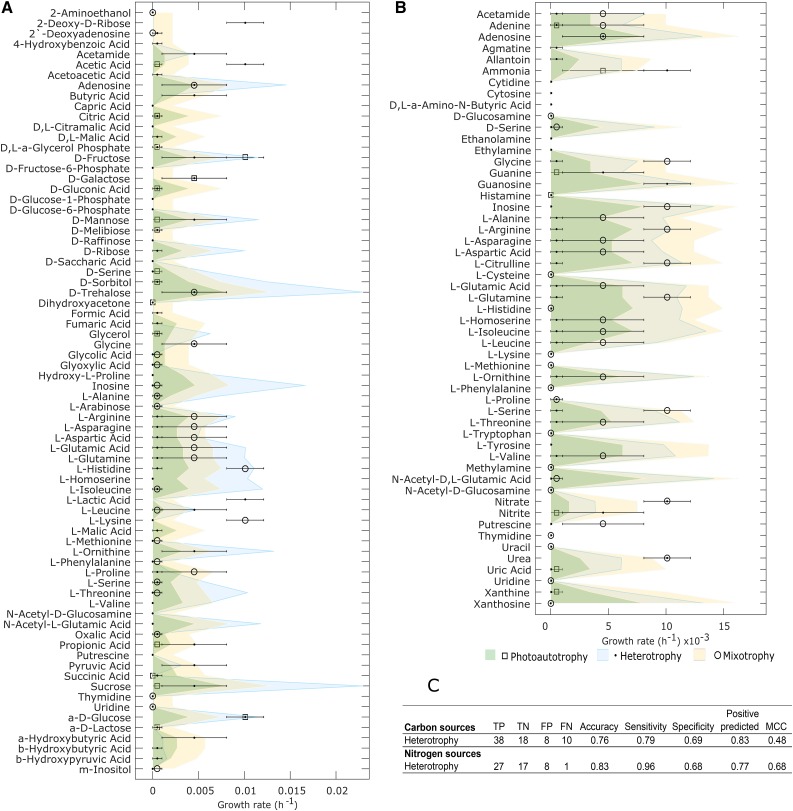
The model demonstrated good accuracy for growth/no-growth prediction on 74 carbon sources and 53 nitrogen sources. Matthews correlation coefficient (MCC) values were estimated for each condition (Fig. 3). Across carbon sources, the highest prediction accuracy was observed for photoautotrophy (MCC = 0.51), followed by heterotrophy; the positive predicted values ranged from 0.77 to 0.85 for all conditions (Supplemental Tables S11 and S12). iCZ843 was able to accurately predict the presence or absence of growth across several nitrogen sources as well, with an MCC of 0.68. The false-negative values occur primarily within particular subsystems, such as amino acid metabolism in charge of the degradation of l-homo-Ser, agmatine, and putrescine, where the GPR associations were uncertain. Additionally, some of the false-positive values under mixotrophic conditions appear to be due to the CO2 uptake rate allowed to the model; true and false negatives were not predicted under mixotrophy due to the supply of CO2 and light into the model.
Figure 3.
Model benchmarking through different carbon and nitrogen sources. A, Predicted growth rate in photoautotrophy (light + compound), heterotrophy (compound), and mixotrophy (CO2 + light + compound) for the 74 carbon sources. The Glc uptake rate was used to constrain the model for all the metabolites; markers represent experimental data for each condition. B, Comparison of predicted growth rates using 53 nitrogen sources. The model was constrained using the urea uptake rate and 1% Glc uptake rate. C, Statistics of the predictions under heterotrophy: true positive (TP), true negative (TN), false positive (FP), false negative, and MCC. The full data set and statistics under the three conditions are shown in Supplemental Tables S11 and S12.
It was found that certain enzymes, such as mitochondrial cytochrome c peroxidase (EC 1.11.1.5), ferredoxin-NADP+ reductase (EC 1.18.1.2), ethanolamine kinase (EC 2.7.1.82), and formate dehydrogenase (EC 1.2.1.2), have a high impact on the flux through the BOF and are essential for a functional model (Supplemental Fig. S3).
Robustness analysis for cytochrome c peroxidase shows an inverse relationship between the reaction flux increase and the growth rate. Here, a flux of 2 mmol g−1 dry weight h−1 results in a lethal phenotype under heterotrophic growth. According to our analysis, the enzymes ethanolamine kinase, formate dehydrogenase, and ferredoxin-NADP+ reductase show a similar sensitivity; in all cases, lower or higher fluxes result in a lethal phenotype.
Flux Distributions
Carbon and Amino Acid Metabolism
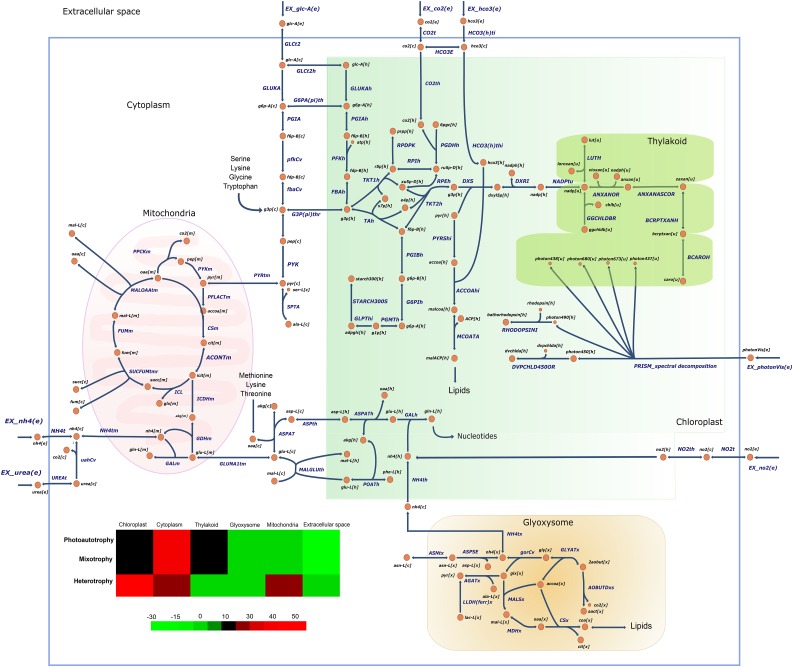
A general overview of the reconstruction, the connections, and the transport of compounds is shown in Figure 4. The heat map shows the percentage of total flux by compartment. Negative and positive values represent the total mass shuttled outside or inside of the compartments, respectively; cytoplasm is considered as a reference in order to stablish connections between the extracellular space and the rest of the organelles. Fluxes showed different behavior in light and dark conditions: cytoplasm and extracellular space are highly active in light conditions (photoautotrophy and mixotrophy), while under heterotrophy, mitochondria, cytoplasm, and chloroplast have similar activity. The chloroplast has significant metabolic activity even in the dark due to the presence of some lipid and starch synthesis pathways in this compartment.
Figure 4.
Model overview and metabolite exchange between different compartments. Color-coded values (heat map) refer to total flux by compartment (mmol g−1 dry weight h−1); the color map shows a negative value when the input mass is higher than output and vice versa. The arrows show how the reactions are carried out: arrows with two points indicate reversible reactions, which can carry flux in both directions; arrows with just one point indicate irreversible reactions and point to the products. Each reaction and metabolite contains in its name a location identifier: C, cytoplasm; E, extracellular space; H, chloroplast; M, mitochondria; U, thylakoid; and X, glyoxysome. For the naming and abbreviation of reactions and metabolites, see Supplemental Tables S1 and S2.
Under photoautotrophic and mixotrophic growth, CO2 is shuttled from the cytoplasm to the chloroplast, while the flux is reversed during heterotrophy. Mitochondria provide CO2 to the cytoplasm under all the conditions, but this flux doubles when Glc is present in the culture medium. When C. vulgaris UTEX 395 grows on NO3 as a nitrogen source, the synthesis of NH4 is carried out intracellularly through six reactions. We found that NH4 is imported to the chloroplast in light conditions, where it is used in amino acid and nucleotide biosynthesis, but under heterotrophy, NH4 is released to the cytoplasm, allowing the use of NH4 in other pathways and organelles.
The 13C-tracing metabolic flux data set previously reported for C. protothecoides was used to validate the iCZ843’s flux distributions (Wu et al., 2015). The Pearson correlation (r2) in photoautotrophy was 0.85 and 0.77 for heterotrophy (Supplemental Figs. S4 and S5). The reactions were grouped by subsystem, where the flux distribution related to energy, carbon fixation, exchange, and transport reactions was predicted accurately. Some reaction fluxes in glycolysis, Fru metabolism, and tricarboxylic acid cycle were subestimated by the model. The highest complexity of iCZ843 does not allow direct comparison with the experimental data, due to either the presence of the same reaction in several compartments or the curtailment of pathways in just one experimental value. The reaction correspondence between iCZ843 and the reactions in the C. protothecoides model is shown in Supplemental Tables S13 and S14.
One of the highest flux pathways in iCZ842 was the tricarboxylic acid cycle, which contains 22 reactions. While most enzymes are highly active under heterotrophic growth, only eight reactions are active under photoautotrophic and mixotrophic growth. The cytoplasmic enzymes aconitate hydratase (EC 4.2.1.3), isocitrate dehydrogenase (EC 1.1.1.42), mitochondrial citrate synthase (EC 2.3.3.1), and fumarate hydratase (EC 4.2.1.2) as well as chloroplastic malate dehydrogenase (EC 1.1.1.37) are active under the three conditions. On the other hand, the mitochondrial isocitrate dehydrogenase (EC 1.1.1.41) and chloroplastic malate and isocitrate dehydrogenase (EC 1.1.1.37 and EC 1.1.1.42) only carry flux during heterotrophic growth.
The flux distributions of the pentose phosphate pathway (PPP) are similar under photoautotrophic and mixotrophic conditions but have lower activity under heterotrophy. Next, we examined a set of enzymes within pyruvate metabolism, namely pyruvate decarboxylase (EC 4.1.1.1) and pyruvate dehydrogenase (EC 1.2.4.1 and EC 2.3.1.12), which connect glycolysis/gluconeogenesis, carbon fixation, and Ala and Asp metabolism. iCZ843 predicts these reactions with higher activity under photoautotrophic and mixotrophic conditions compared with heterotrophic conditions. This fact is linked to greater growth efficiency in these conditions.
Phe, Tyr, and Trp biosynthesis is an active subsystem powered by phosphoenolpyruvate and acetoacetyl-CoA, key precursors in central carbon metabolism and lipid synthesis. Under mixotrophic growth, fluxes in this subsystem increased by around 25% during photoautotrophy and 60% during heterotrophy. The aminotransferase enzymes (EC 2.6.1.1) are highly active during photoautotrophic, heterotrophic, and mixotrophic growth. These enzymes catalyze the synthesis of Phe and Tyr through α-ketoglutarate/Glu or the assimilation into hydroxyphenyl pyruvate, which participates in terpenoid and secondary metabolite biosynthesis. The flux distribution within Glu and other amino acid metabolism is laid out in Supplemental Figure S6. The predicted fluxes reveal the connectivity of Glu in the cytoplasm with the synthesis of several amino acids (Pro, His, Val, and Arg) in both light and dark conditions. Chloroplastic Glu is used by amino acid transferases (EC 2.6.1.42) for the synthesis of Ile and Tyr in the chloroplast under mixotrophic and photoautotrophic conditions.
Pigment Metabolism
Certain pigments are synthesized in large quantities during heterotrophic growth. Members of the class Chlorophyceae contain α- and β-carotenes as well as the xanthophylls lutein, zeaxanthin, violaxanthin, and neoxanthin, all of which were included in the BOF of iCZ843. The model indicates that all of these pigments can be synthesized under either light or dark conditions.
The flux distribution showed that enzymes such as cryptoxanthin hydroxylase (CXHY; EC 1.14.99.45) and zeinoxanthin forming α-carotene hydroxylase (CHYA2; EC 1.14.13.-) carry flux in both light and dark. During growth conditions, lutein, loraxanthin, and zeaxanthin are produced through the enzyme CHYA2, which condenses α-carotene to cryptoxanthin using NADPH. Ultimately, zeaxanthin is converted into lutein by CXHY and eventually to loraxanthin. The flux through CHYA2 is responsible for the large concentration of lutein observed in C. protothecoides growing mixotrophically (Shi and Chen, 1999). The pigment biosynthesis pathways also revealed changes in flux activity depending on the simulated conditions (Supplemental Fig. S7).
Shadow Prices
Shadow price analysis predicts the effect that a single modification to the culture medium has on the objective function in the conditions studied, in this case growth rate. We estimated shadow prices for all 20 amino acid substitutions to the medium as well as for acetate and glycerol addition and confirmed the predictions experimentally.
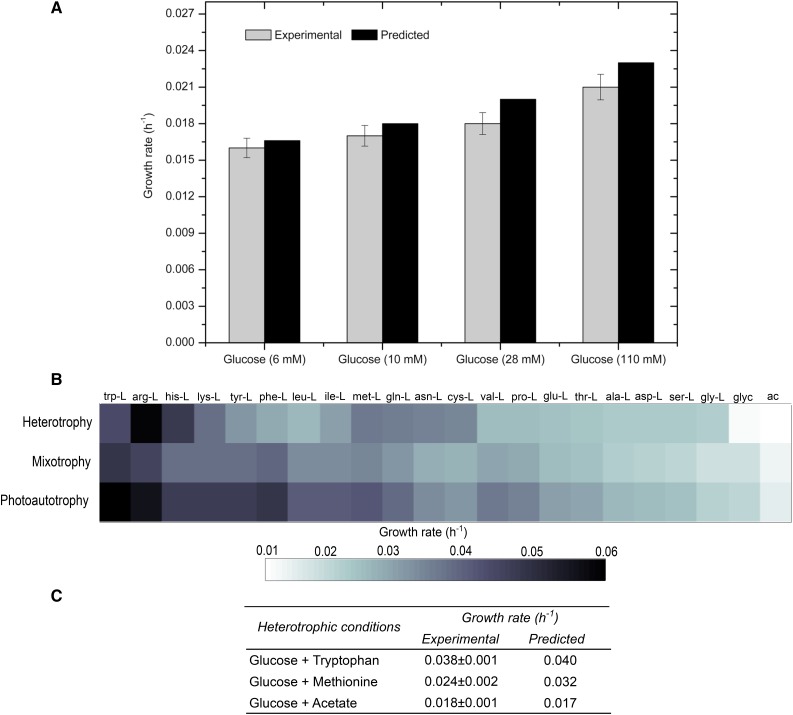
The comparison of growth rates under heterotrophy and estimated shadow prices are shown in Figure 5. Assuming an uptake rate of 1 mmol g−1 h−1, the model predicted that the addition of Trp or Met leads to an increase in growth rate (Trp, 0.040 h−1; and Met, 0.032 h−1).
Figure 5.
Predicted growth rates and shadow price analysis for heterotrophic growth. A, Growth rates were determined experimentally and simulated for four different Glc concentrations. B, The heat map displays the theoretical growth rate after the addition of 20 individual amino acids as well as glycerol (glyc) and acetate (ac) for heterotrophic, mixotrophic, and photoautotrophic conditions. The highest growth rate was achieved while adding Trp. The heterotrophy data are displayed with a conversion factor 10−1. C, Experimental and predicted growth rates for medium containing Trp or Met.
These hypotheses were tested experimentally by adding equimolar concentrations (10 mm) of Trp and Met to the medium. The growth rate improved for both Trp (0.038 h−1) and Met (0.024 h−1) addition (Fig. 5C). The addition of acetate to the medium was used as a negative control, and it had no effect on the growth rate in heterotrophic conditions.
DISCUSSION
Reconstruction
Due to recently developed tools, the creation of draft genome-scale models for bacteria is a largely automated process (Overbeek et al., 2014). However, for eukaryotes, this automation procedure often proved to be difficult, because of the complex compartmentalization present in these organisms. One tool that enables the automated generation of draft reconstructions suitable for eukaryotes is the RAVEN Toolbox (Agren et al., 2013). In contrast to what has been reported for other Chlorella spp. reconstructions (Wu et al., 2015), we were able to find gene evidence for enzymes and transporters associated with amino acid metabolism as well as steroid, porphyrin, and chlorophyll metabolism. Vitamin, ubiquinone, terpenoid, and brassinosteroid biosynthesis also have been included into the model, which, among all photosynthetic organisms, appears only in the Z. mays reconstruction (Saha et al., 2011).
Considering the genome size and number of genes in the reconstruction, as well as the detail of lipid metabolism, iCZ843 is the most comprehensive model for any eukaryotic photosynthetic organism to date. Previously, a comprehensive reconstruction of lipid biosynthesis was available only for C. reinhardtii and Synechocystis sp. PCC6803, both unsuitable candidates for biofuel production because of their inability to accumulate high concentrations of TAGs (Chang et al., 2011; Saha et al., 2011). C. vulgaris has the ability to produce high amounts of TAG, which makes it a prime model organism for elucidating biofuel production.
Furthermore, amino acid biosynthesis has been reconstructed in detail for C. vulgaris. Amino acid metabolism in iCZ843 contains 282 reactions and the total related reaction in other subsystems account for 312, compared with 34 reactions in C. protothecoides, 256 in C. reinhardtii, and 143 in Synechocystis spp. (Supplemental Table S4; Chang et al., 2011; Saha et al., 2011; Overbeek et al., 2014).
It is noteworthy that models for phototrophic bacteria, such as the cyanobacterium Synechocystis sp. PCC6803, often contain a larger fraction of genes per genome (Table II). This discrepancy between kingdoms is due to the fact that functional genome annotations for bacteria are, in general, more comprehensive. The size and metabolic scope of iCZ843 allow more detailed and complex simulations.
C. vulgaris UTEX 395 drastically adapts its metabolism in response to changes in the environment, such as growth in light or dark or the availability of a nitrogen source (e.g. nitrate). These metabolic changes affect the biomass composition, primarily consisting of carbohydrates, proteins, lipids, and nucleic acids. Therefore, we measured changes in total carbohydrates, proteins, lipids, and RNA during photoautotrophic and heterotrophic (with Glc) growth with nitrate as the nitrogen source (Supplemental Fig. S2).
In other Chlorella spp., such as Chlorella zofingiensis, starch has been found to be the main stored carbohydrate, accounting for 66.7% of total biomass (Zhu et al., 2014). Other studies with C. vulgaris showed a considerable breakdown of starch even during photosynthesis (Nakamura and Imamura, 1985).
We found a slightly higher content of TAG, PG, PI, and PE under heterotrophic growth conditions, while the MGDG content was high during photoautotrophic growth. This could be related to the production of pools of apoproteins, such as those in the PSII complex that require lipids for their function (Thompson, 1996).
Simulations Using iCZ843
Simulations confirmed that the metabolism of C. vulgaris UTEX 395 is redirected toward the synthesis of storage compounds like lipids and carbohydrates under nitrogen starvation (Guarnieri et al., 2013). The uptake of nutrients from the medium is in agreement with previously reported values as well as our experimental data, showing good accuracy in growth predictions for more than 120 carbon and nitrogen sources. The predictions on Glc confirm that the specific growth rate for mixotrophy can be represented approximately by the sum of heterotrophic and photoautotrophic growth rates (Killam and Myers, 1956; Perez-Garcia et al., 2011).
The efficiency of a microbial production process can be evaluated based on high titer, high yield, high productivity, and process robustness (Liu et al., 2013). Robustness analysis allows the evaluation of the essentiality of the function of enzymes within the network. It correlates the enzyme-pathway relationship and the overall growth rate and can identify targets for metabolic engineering. A set of reactions that have the most profound effect on growth were identified.
Flux Distribution
Genome-scale metabolic models help explain how targeted compounds can be synthesized through all possible routes. The predictions of iCZ843 provide metabolic details that correspond to the physiology of C. vulgaris and green algae.
The PPP and glycolysis have been fairly well documented in C. vulgaris (Van Baalan et al., 1973). According to the model, the oxidative phase of the PPP, responsible for the production of NADPH, is inactive under heterotrophic conditions. Glycolysis and gluconeogenesis have been studied intensively in C. vulgaris (Perez-Garcia et al., 2011). The degradation of Glc to pyruvate occurs throughout the cytoplasm, the mitochondria, and the chloroplast. It is known that central carbon metabolism is active in different compartments depending on the growth conditions (Hanson, 1985). The model shows interchanges of products and substrates from the active reactions uniquely present in the chloroplast with those functioning in the cytoplasm and mitochondria.
The model contains the reaction ACCOAth (gene associated to maker_Scaffold_645-augustus-gene-0.44), which facilitates the transport of acetyl-CoA from the cytoplasm to the chloroplast. When this reaction is deleted (knocked out) under heterotrophy, the predicted growth rate decreases by 80%. After delving into the effects of some transporters on metabolism as a whole, we found that the flux of acetate from the chloroplast to the cytoplasm correlates with the acetyl-CoA transport and is vitally important for a viable growth prediction.
Acetate transport under dark conditions plays a very important role in green algae metabolism. This metabolite can be incorporated into acetyl-CoA following two possible pathways that both require ATP: a direct conversion with acetyl-CoA synthetase or a two-step reaction involving acetate kinase and phosphate acetyltransferase (Johnson and Alric, 2013). Analyzing our predicted flux distribution under heterotrophy enabled us to conclude that, in order to get enough acetate into the cytoplasm, the same amount of acetyl-CoA needs to be transported into the chloroplast. Then, a cascade effect activates chloroplastic enzymes such as phosphate acetyltransferase and pyruvate synthase, where ferredoxin is reduced. Eventually, ferredoxin will be the precursor in the synthesis of NH4, which is one of the main precursors needed for the synthesis of amino acids. The cytoplasmic acetate is incorporated into the tricarboxylic acid cycle and lipid biosynthesis, and the ATP necessary for these reactions is produced in the mitochondria by the ATP synthase complex.
Furthermore, calculations obtained by iCZ843 agree with experimental 13C data. We found the highest flux correlation with experimental data in photoautotrophy followed by heterotrophy. The discrepancies in the flux distributions were attributed to (1) the condensed nature of the experimental data, where explicit comparison of predictions with data were not possible due to the presence of the same reaction in several compartments in the model, and (2) the presence of anaplerotic (refilling) reactions, which replenish tricarboxylic acid cycle intermediates consumed in amino acid, lipid, and nucleotide synthesis, allowing many alternative flux solutions, although the predicted and experimental data follow similar behaviors. For heterotrophy, we also found prediction errors with reactions related to carbon fixation and pentose and Fru metabolism.
Gln, Glu, Asn, and Asp contribute as building blocks for the synthesis of organic nitrogen compounds, such as other amino acids, chlorophylls, nucleotides, alkaloids, and polyamines. Alternatively, we found that Glu needs to be converted to Gln and exported from the chloroplast to the cytoplasm in the dark, where it is subsequently used for nucleic acid synthesis or incorporated in the tricarboxylic acid cycle.
It is well known that the enzyme Glu ammonium ligase (EC 6.3.1.2) uses Gln and NH3 as substrates. The direct reduction of nitrate to NH4 and the availability of Gln have been observed in chloroplasts during heterotrophy (Perez-Garcia et al., 2011). The role of the chloroplast in the dark has been traced and attributed to the qualities of the common ancestor, but a detailed experimental description of its metabolism has not been reported in either plants or green algae. Some reports have proposed a shuttling of metabolites between mitochondria, peroxisome, and chloroplast to fuel the glycolate pathway or central carbon metabolism, but the transport of amino acids between different compartments is still poorly understood (Van Baalan et al., 1973; Hanson, 1985; Ren and Paulsen, 2007). iCZ843 provides novel insights into the amino acid metabolism in Chlorella spp. Several enzymes involved in amino acid metabolism were found to be active only when carbon was redirected mainly to lipid production. The model not only enables the study of metabolism at the reaction level; additionally, the detailed compartmentalization of the model facilitates the analysis of enzyme activity in different organelles of the cell under various conditions.
The three available core models of Chlorella spp. (Table II) assume that the fluxes differ between light and dark only by the layout of the core carbon network (tricarboxylic acid, glycolysis, and PPP), and the rest of the metabolism (synthesis of amino acids, DNA, and RNA) does not vary significantly in terms of relative fluxes (Yang et al., 2000; Muthuraj et al., 2013; Wu et al., 2015). Despite previous reports that assume the lack of change in the flux distribution for nucleotide metabolism (Yang et al., 2000; Muthuraj et al., 2013; Wu et al., 2015), we found that fluxes vary widely depending on the growth condition. Our results suggest that the anabolic part of the metabolism is not independent of growth conditions, and iCZ843 provides an accurate qualitative and quantitative analysis of the metabolism.
Guided Medium Alteration
An increase in growth rate will deplete nitrogen faster, which in turn will get to a lipid accumulation state faster. The predicted and experimentally determined growth rates for C. vulgaris UTEX 395 containing amino acid additions are 2 to 3 times higher than in regular medium. The growth rate for Trp-supplemented cultures can be attributed to its degradation to formate and kynurenine, which is further metabolized to generate Ala and eventually acetyl-CoA. Trp catabolic products also can fuel the synthesis of Arg and Ser, nucleotides, as well as NAD and NADP. Met is a source of methyl groups for a number of cellular components (e.g. for S-adenosyl-Met, a purine and pyrimidine intermediary) and is used in cell wall formation. Met had been intimately associated with cell division, and some analogs of Met prevent cells of C. vulgaris from dividing but allow them to maintain other cellular activities that can lead to giant cells. Analysis of these abnormally large cells has revealed that there are increases in the rate of respiration, dry weight, and protein content (Shrift, 1960).
CONCLUSION
An in-depth understanding of metabolism is necessary to improve the production of desired products, such as nutraceuticals or biofuels, by microalgae. Genome-scale network reconstructions combine detailed biochemical and physiological information for an organism and provide new insights into growth conditions and subsequent manipulation strategies to enhance productivity. The final C. vulgaris UTEX 395 reconstruction contains 843 genes, 2,294 reactions, and 1,770 metabolites. The reconstruction was constrained during the validation process using transcriptomics and other experimental data under photoautotrophic, mixotrophic, and heterotrophic growth conditions. iCZ843 can accurately simulate the growth rates under these conditions. The model was deployed successfully to guide strategies altering the culture medium for increased growth performance.
MATERIALS AND METHODS
Draft Generation
The draft was generated using the RAVEN Toolbox (Agren et al., 2013). As input, we provided the translated genome sequence of Chlorella vulgaris UTEX 395 and the manually curated reference network of the microalga Chlamydomonas reinhardtii (iRC1080; Chang et al., 2011), which was used as a reference network. The C. vulgaris UTEX 395 genome sequence was taken from the previously deposited sequence (GenBank no. LDKB00000000).
Several resources were used during the manual curation phase, such as primary literature and the databases KEGG, EMBL-EBI, ExplorEnz, BIGG, BRENDA, MetaCyc, and SwissProt (McDonald et al., 2007; Schellenberger et al., 2010; Scheer et al., 2011; McWilliam et al., 2013; Caspi et al., 2014; Kanehisa et al., 2014; UniProt Consortium, 2015). All the resources used during the reconstruction process are highlighted in Table I. Information regarding transport proteins was obtained from TransportDB and TCDB (Ren and Paulsen, 2007). Subcellular protein localization was predicted using SignalP, ChloroP, HECTAR, and WolF PSORT (Emanuelsson et al., 1999; Horton et al., 2007; Gschloessl et al., 2008; Petersen et al., 2011; Table I).
Metabolic Network Reconstruction
The draft reconstruction was manually curated following previously published protocols (Thiele and Palsson, 2010). The reconstruction was arranged using a KEGG pathway structure (e.g. systems: amino acid metabolism; subsystems: Lys biosynthesis). Metabolites and reactions were identified using KEGG identifiers as well as EC codes for enzymes. For every metabolite, the elemental formula and charge were included in the annotation. The protonation state of each metabolite changes according to the pH of the compartment. The cytosolic pH was determined to be 7.2 at extracellular pH 6.5 (Komor and Tanner, 1974). The chloroplast and its subcompartment, the thylakoid, were assumed to share the same pH, determined for the chloroplast to be 8 in light conditions (Hogetsu and Miyachi, 1979; Goss and Garab, 2001). The pH of the mitochondrial matrix has been measured in green algae at 7.8 (Giordano et al., 2003). The glyoxysome pH was assumed to be 8.2 (Dansen et al., 2001). The extracellular pH was 7 ± 0.2 based on the growth medium used. Metabolite charges and protonation states were calculated using ChemAxon (http://www.chemaxon.com; Table I).
New pathways and their annotation were added. Names for reactions and metabolites were assigned according to the information in BIGG and SimPheny (Schilling et al., 2008; Schellenberger et al., 2010); when the reactions and metabolites were not found in any database, a new naming was provided. Each pathway was manually curated; mass and charge balance, directionality, and cofactors involved in the reactions were accounted for (Supplemental Fig. S1). Initially, reactions in the draft reconstruction were imported, keeping the same location as in iRC1080. All information for these reactions was double checked, and in some cases, the reactions were relocated to a different compartment. The manual GPR associations were set using BLAST to compare the protein sequences of C. vulgaris UTEX 395 with the corresponding sequences of Chlorella sp. NC64A, Chlorella variabilis, and Chlorella sorokiniana as well as (in some cases) with the green algae Coccomyxa subellipsoidea and Volvox carteri (Altschul et al., 1990). The phylogenic tree shows the relationship between these photosynthetic organisms (Supplemental Fig. S8).
Once the manual refinement was finished and the annotation of the model was completed, gap filling and dead-end identification were executed using the available COBRA tools (Schellenberger et al., 2011). The connectivity of the pathways was ensured by the addition of enzymes without gene associations but backed by literature evidence. When the missing transporter was detected, passive diffusion was assumed if the exact transport mechanism was unknown. Finally, quality control and assessment tests for ATP, NADPH, and NADH maintenance were performed. These tests ensured that the model cannot maximize for any of the energy cofactors (ATP, NADPH, and NADH) without any input; the objective function has an expected result of zero. Analysis of loops and plot maps were done using the ESCHER metabolic pathway visualization tool (King et al., 2015). The reconstruction was converted to a JSON model for ESCHER using COBRApy (Ebrahim et al., 2013).
Biomass Composition and Experimental Data
C. vulgaris can grow under different trophic conditions (e.g. autotrophic, heterotrophic, and mixotrophic). Each of these was represented mathematically through different BOFs (named in the model Biomass_Cvu_auto-, Biomass_Cvu_mixo-, and Biomass_Cvu_hetero-). Every equation contains the stoichiometric coefficients expressed in mmol g−1 dry weight, and all metabolites that are part of the biomass should be included. For the BOF under mixotrophy, we assumed the same data as for heterotrophy. ATP maintenance was established according to Boyle and Morgan (2009).
Lipid, protein, carbohydrate, and ribose in RNA contents were measured experimentally under photoautotrophic and heterotrophic conditions following the method described previously (Antoniewicz et al., 2007, 2011; Long and Antoniewicz, 2014). C. vulgaris UTEX 395 was grown in a 250-mL bottle with 200 mL of Bold’s basal medium (BBM) at 24°C (Guarnieri et al., 2013) and cycling of 14/10-h light/dark at10,000 lx and a gas flow of 1% CO2 (12 mL min−1). For the heterotrophic condition, C. vulgaris UTEX 395 was grown in 500 mL of BBM + 20 mm Tris and a gas flow of air (400 mL min−1) using 6, 10, 28, and 110 mm Glc as the organic carbon source at 24 h of dark. The urea uptake rate was measured in heterotrophy and mixotrophy, testing an initial concentration of 1 mm and 1% Glc.
The amino acid content was completed with literature data: Arg, Cys, Gln, and Trp under photoautotrophy were obtained from Faheed and Fattah (2008); and Cys content in heterotrophy was taken from Wu et al. (2015).
The quantified fatty acids were spread out according to previously reported lipid profiles (Nichols et al., 1967). The relation of nucleotide composition (28 RNA/DNA) was taken from Muthuraj et al. (2013). Literature and experimental data were normalized assuming an idealized size and weight for C. vulgaris (see below). The specific starch production and degradation rates were calculated from the experimental data, and the curves were adjusted to the Gompertz model using Kaleidagraph (Synergy Software; Nichols et al., 1967; Wang and Zuidhof, 2004).
The CO2 consumption rates and the chlorophylls a and b were measured under photoautotrophic conditions. Experiments were performed in a 1.25-L reactor with 500 mL of BBM supplemented with 20 mm Tris, a total gas flow of 400 mL min−1 with 0.04%, 3%, 5%, 10%, and 12.5% CO2, at an agitation rate of 200 rpm at approximately 25°C (room temperature), in light/dark cycles of 12/12 h and a light intensity of 10,000 lx (approximately 300 µE m−2 s−1). Additional compositions of pigment were taken from Safi et al. (2014).
The addition of amino acids was tested for heterotrophic conditions, adding to the culture medium equimolar concentrations (0.01 m) of Glc and either Trp or Met.
Constraints and Growth Simulations
RNA sequencing data and literature data such as specific CO2 and Glc consumption rates were used to constrain the model under growth conditions in the light and dark (Guarnieri et al., 2011). For each growth condition, the storage and consumption of starch were taken into account; we calculated the rates using experimental data (Supplemental Table S6).
The constraints related to mineral medium composition and the reactions that were set to zero are summarized in Supplemental Tables S7 to S10. Growth simulations were performed in the COBRA Toolbox for MATLAB (Schellenberger et al., 2011) using the flux balance analysis procedure (Orth et al., 2010). The stoichiometric coefficients in the BOFs were set according to our experimental data. The 13C data set was taken from Wu et al. (2015). Model benchmarking on carbon and nitrogen sources was performed using Biolog plates PM1 to PM3, following the previously reported protocol described by Chaiboonchoe et al. (2014), with the following modifications. C. vulgaris was grown to midlog phase in modified BBM (Guarnieri et al., 2011), pelleted via centrifugation at 4,000g for 5 min, washed, and resuspended in fresh medium to a final optical density of 0.1 (nitrate was excluded for nitrogen phenotyping). Aliquots of 100 μL were inoculated onto Biolog plates and examined for 96 h in the plate reader with readings every 15 min. Plates were housed in a plate reader with no light (heterotrophic growth). The plates for both PM1 and PM2 (carbon sources) were run at 490 nm to examine dye absorbance alterations and at 750 nm to assess optical density. A confusion matrix and various measures of quality, such as accuracy, specificity, sensitivity, and MCC, were estimated according to Matthews (1975; Supplemental Tables S11 and S12).
Robustness and Shadow Prices
The robustness analysis was performed under photoautotrophic, heterotrophic, and mixotrophic conditions. The respective analyses were executed using the same constraints. The CO2 uptake rates were plotted using MATLAB; the experimental CO2 uptake rates reported by Nascimento et al. (2015) were used to validate the simulated results.
The sensitivity of the flux balance analysis solution can be indicated by shadow prices, which represent the change of the BOF with respect to the external exchange flux for all the metabolites, computing the theoretical addition of 1 mol of every metabolite present in the model (Palsson, 2011). Negative values describe metabolites that are demanded, and positive values identify metabolites that would be excreted in order to improve the objective value. Shadow prices were calculated using the COBRA Toolbox (Schellenberger et al., 2011) in order to compare the effects of the addition of amino acids, glycerol, and acetate compounds.
Supplemental Data
The following supplemental materials are available.
Supplemental Text S1. Reconstruction process.
Supplemental Figure S1. Manual curation workflow used for the metabolic network reconstruction.
Supplemental Figure S2. Experimental data for C. vulgaris UTEX 395.
Supplemental Figure S3. Robustness analysis.
Supplemental Figure S4. Comparison of the experimental flux distribution of Chlorella protothecoides and predicted data by iCZ843.
Supplemental Figure S5. Comparison of the experimental flux distribution of Chlorella protothecoides and predicted data by iCZ843.
Supplemental Figure S6. Flux distribution associated to glutamate metabolism in the chloroplast and cytoplasm.
Supplemental Figure S7. Flux distribution of pigment biosynthesis pathways under heterotrophic growth.
Supplemental Figure S8. Arbitrarily rooted phylogenetic tree based on multiple sequence alignments of the big subunit of RuBisCO (EC 4.1.1.39) inferred using Phylogeny.fr.
Supplemental Table S1. iCZ843 model.
Supplemental Table S2. Metabolite annotation.
Supplemental Table S3. Non-gene-associated reactions.
Supplemental Table S4. Comparison of iCZ843 with the available green algae models.
Supplemental Table S5. GPR for enzymes sensitive to light.
Supplemental Table S6. Starch synthesis and degradation.
Supplemental Table S7. Bold’s mineral medium.
Supplemental Table S8. Photoautotrophic solution.
Supplemental Table S9. Heterotrophic solution.
Supplemental Table S10. Mixotrophic solution.
Supplemental Table S11. Experimental and predicted growth of C. vulgaris on various carbon sources.
Supplemental Table S12. Experimental and predicted growth of C. vulgaris on various nitrogen sources.
Supplemental Table S13. Reaction association for 13C-tracing analysis.
Supplemental Table S14. 13C-tracing analysis of C. protothecoides under photoautotrophic and heterotrophic conditions.
Supplemental Data Set S1. Model in photoautotrophy.
Supplemental Data Set S2. Model in mixotrophy.
Supplemental Data Set S3. Model in heterotrophy.
Supplementary Material
Acknowledgments
We thank Jared Broddrick and Adam M. Feist for assisting with the reconstruction process, Bernhard Palsson (University of California, San Diego) for guidance, and an anonymous reviewer for the careful evaluation of the reconstruction, which significantly improved the model.
Glossary
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GPR
gene/protein/reaction
- BOF
biomass objective function
- TAG
triacylglycerol
- PG
phosphatidylglycerol
- PI
phosphatidylinositol
- PE
phosphatidylethanolamine
- MGDG
monogalactosyldiacylglycerol
- MCC
Matthews correlation coefficient
- PPP
pentose phosphate pathway
- BBM
Bold’s basal medium
Footnotes
This work was supported by the National Science Foundation (grant no. 1332344), the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research (grant no. DE–SC0012658), and the Mexican National Research Council (fellowship no. 237897 to C.Z.).
References
- Agren R, Liu L, Shoaie S, Vongsangnak W, Nookaew I, Nielsen J (2013) The RAVEN toolbox and its use for generating a genome-scale metabolic model for Penicillium chrysogenum. PLOS Comput Biol 9: e1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G (2007) Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. Anal Chem 79: 7554–7559 [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G (2011) Measuring deuterium enrichment of glucose hydrogen atoms by gas chromatography/mass spectrometry. Anal Chem 83: 3211–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroukh C, Muñoz-Tamayo R, Steyer JP, Bernard O (2015) A state of the art of metabolic networks of unicellular microalgae and cyanobacteria for biofuel production. Metab Eng 30: 49–60 [DOI] [PubMed] [Google Scholar]
- Bellou S, Baeshen MN, Elazzazy AM, Aggeli D, Sayegh F, Aggelis G (2014) Microalgal lipids biochemistry and biotechnological perspectives. Biotechnol Adv 32: 1476–1493 [DOI] [PubMed] [Google Scholar]
- Boyle NR, Morgan JA (2009) Flux balance analysis of primary metabolism in Chlamydomonas reinhardtii. BMC Syst Biol 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A, et al. (2014) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 42: D459–D471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiboonchoe A, Dohai BS, Cai H, Nelson DR, Jijakli K, Salehi-Ashtiani K (2014) Microalgal metabolic network model refinement through high-throughput functional metabolic profiling. Front Bioeng Biotechnol 2: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang RL, Ghamsari L, Manichaikul A, Hom EFY, Balaji S, Fu W, Shen Y, Hao T, Palsson BØ, Salehi-Ashtiani K, et al. (2011) Metabolic network reconstruction of Chlamydomonas offers insight into light-driven algal metabolism. Mol Syst Biol 7: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansen TB, Pap EHW, Wanders RJ, Wirtz KW (2001) Targeted fluorescent probes in peroxisome function. Histochem J 33: 65–69 [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucha J, Lívanský K (2006) Productivity, CO2/O2 exchange and hydraulics in outdoor open high density microalgal (Chlorella sp.) photobioreactors operated in a middle and southern European climate. J Appl Phycol 18: 811–826 [Google Scholar]
- Ebrahim A, Lerman JA, Palsson BO, Hyduke DR (2013) COBRApy: constraints-based reconstruction and analysis for python. BMC Syst Biol 7: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Gonzalez I, Parashar A, Bressler DC (2014) Heterotrophic growth and lipid accumulation of Chlorella protothecoides in whey permeate, a dairy by-product stream, for biofuel production. Bioresour Technol 155: 170–176 [DOI] [PubMed] [Google Scholar]
- Faheed AF, Fattah ZA (2008) Effect of Chlorella vulgaris as bio-fertilizer on growth parameters and metabolic aspects of lettuce plant. J Agric Soc Sci 4: 165–169 [Google Scholar]
- Giordano M, Norici A, Forssen M, Eriksson M, Raven JA (2003) An anaplerotic role for mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol 132: 2126–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss R, Garab G (2001) Non-photochemical chlorophyll fluorescence quenching and structural rearrangements induced by low pH in intact cells of Chlorella fusca (Chlorophyceae) and Mantoniella squamata (Prasinophyceae). Photosynth Res 67: 185–197 [DOI] [PubMed] [Google Scholar]
- Gschloessl B, Guermeur Y, Cock JM (2008) HECTAR: a method to predict subcellular targeting in heterokonts. BMC Bioinformatics 9: 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnieri MT, Nag A, Smolinski SL, Darzins A, Seibert M, Pienkos PT (2011) Examination of triacylglycerol biosynthetic pathways via de novo transcriptomic and proteomic analyses in an unsequenced microalga. PLoS ONE 6: e25851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnieri MT, Nag A, Yang S, Pienkos PT (2013) Proteomic analysis of Chlorella vulgaris: potential targets for enhanced lipid accumulation. J Proteomics 93: 245–253 [DOI] [PubMed] [Google Scholar]
- Hanson J. (1985) Membrane transport systems of plant mitochondria. Encycl Plant Physiol 18: 248–275 [Google Scholar]
- Hogetsu D, Miyachi S (1979) Role of carbonic anhydrase in photosynthetic CO2. Symp A Q J Mod Foreign Lit 20: 747–756 [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Alric J (2013) Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: metabolic constraints for carbon partitioning between oil and starch. Eukaryot Cell 12: 776–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneja A, Chaplen FWR, Murthy GS (2016) Genome scale metabolic reconstruction of Chlorella variabilis for exploring its metabolic potential for biofuels. Bioresour Technol 213: 103–110 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M (2014) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42: D199–D205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killam A, Myers J (1956) A special effect of light on the growth of Chlorella vulgaris. Am J Bot 43: 569 [Google Scholar]
- Kim TY, Sohn SB, Kim YB, Kim WJ, Lee SY (2012) Recent advances in reconstruction and applications of genome-scale metabolic models. Curr Opin Biotechnol 23: 617–623 [DOI] [PubMed] [Google Scholar]
- King ZA, Dräger A, Ebrahim A, Sonnenschein N, Lewis NE, Palsson BO (2015) Escher: a web application for building, sharing, and embedding data-rich visualizations of biological pathways. PLOS Comput Biol 11: e1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor E, Tanner W (1974) The hexose-proton cotransport system of chlorella: pH-dependent change in Km values and translocation constants of the uptake system. J Gen Physiol 64: 568–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levering J, Broddrick J, Zengler K (2015) Engineering of oleaginous organisms for lipid production. Curr Opin Biotechnol 36: 32–39 [DOI] [PubMed] [Google Scholar]
- Liu CH, Chang CY, Liao Q, Zhu X, Chang JS (2013) Photoheterotrophic growth of Chlorella vulgaris ESP6 on organic acids from dark hydrogen fermentation effluents. Bioresour Technol 145: 331–336 [DOI] [PubMed] [Google Scholar]
- Long CP, Antoniewicz MR (2014) Quantifying biomass composition by gas chromatography/mass spectrometry. Anal Chem 86: 9423–9427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BW. (1975) Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta 405: 442–451 [DOI] [PubMed] [Google Scholar]
- McDonald AG, Boyce S, Moss GP, Dixon HBF, Tipton KF (2007) ExplorEnz: a MySQL database of the IUBMB enzyme nomenclature. BMC Biochem 8: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R (2013) Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 41: W597– W600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao X, Wu Q (2006) Biodiesel production from heterotrophic microalgal oil. Bioresour Technol 97: 841–846 [DOI] [PubMed] [Google Scholar]
- Mitra D, van Leeuwen J, Lamsal B (2012) Heterotrophic/mixotrophic cultivation of oleaginous Chlorella vulgaris on industrial co-products. Algal Res 1: 40–48 [Google Scholar]
- Monk J, Nogales J, Palsson BO (2014) Optimizing genome-scale network reconstructions. Nat Biotechnol 32: 447–452 [DOI] [PubMed] [Google Scholar]
- Muthuraj M, Palabhanvi B, Misra S, Kumar V, Sivalingavasu K, Das D (2013) Flux balance analysis of Chlorella sp. FC2 IITG under photoautotrophic and heterotrophic growth conditions. Photosynth Res 118: 167–179 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Imamura M (1985) Regulation of ADP-glucose pyrophosphorylase from Chlorella vulgaris. Plant Physiol 78: 601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento IA, Cabanelas-Dominguez IT, Nunes JD, Nascimento MA, Sousa L, Sansone G (2015) Biodiesel yields and fuel quality as criteria for algal-feedstock selection: effects of CO2-supplementation and nutrient levels in cultures. Algal Res 8: 53–60 [Google Scholar]
- Nichols BW, James AT, Breuer J (1967) Interrelationships between fatty acid biosynthesis and acyl-lipid synthesis in Chlorella vulgaris. Biochem J 104: 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales J, Gudmundsson S, Knight EM, Palsson BO, Thiele I (2012) Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis. Proc Natl Acad Sci USA 109: 2678–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Thiele I, Palsson BØ (2010) What is flux balance analysis? Nat Biotechnol 28: 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, et al. (2014) The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42: D206–D214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson BØ. (2011) Systems Biology: Simulation of Dynamic Network States. Cambridge University Press, Cambridge, UK [Google Scholar]
- Perez-Garcia O, Escalante FME, de-Bashan LE, Bashan Y (2011) Heterotrophic cultures of microalgae: metabolism and potential products. Water Res 45: 11–36 [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Ren Q, Paulsen IT (2007) Large-scale comparative genomic analyses of cytoplasmic membrane transport systems in prokaryotes. J Mol Microbiol Biotechnol 12: 165–179 [DOI] [PubMed] [Google Scholar]
- Safi C, Zebib B, Merah O, Pontalier PY, Vaca-Garcia C (2014) Morphology, composition, production, processing and applications of Chlorella vulgaris: a review. Renew Sustain Energy Rev 35: 265–278 [Google Scholar]
- Saha R, Suthers PF, Maranas CD (2011) Zea mays iRS1563: a comprehensive genome-scale metabolic reconstruction of maize metabolism. PLoS ONE 6: e21784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH Jr, Reddy VS, Tamang DG, Västermark A (2014) The transporter classification database. Nucleic Acids Res 42: D251–D258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage N. (2011) Algae: the scum solution. Nature 474: S15–S16 [DOI] [PubMed] [Google Scholar]
- Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Söhngen C, Stelzer M, Thiele J, Schomburg D (2011) BRENDA, the enzyme information system in 2011. Nucleic Acids Res 39: D670–D676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberger J, Park JO, Conrad TM, Palsson BØ (2010) BiGG: a Biochemical Genetic and Genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinformatics 11: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberger J, Que R, Fleming RMT, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S, et al. (2011) Quantitative prediction of cellular metabolism with constraint-based models: the COBRA toolbox v2.0. Nat Protoc 6: 1290–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling CH, Thakar R, Travnik E, Van Dien S, Wiback S (2008) SimPheny TM: A Computational Infrastructure for Systems Biology
- Shi XM, Chen F (1999) Production and rapid extraction of lutein and the other lipid-soluble pigments from Chlorella protothecoides grown under heterotrophic and mixotrophic conditions. Food/Nahrung 43: 109–113 [Google Scholar]
- Shrift A. (1960) A role for methionine in division of Chlorella vulgaris. Plant Physiol 35: 510–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabatabaei M, Tohidfar M, Jouzani GS, Safarnejad M, Pazouki M (2011) Biodiesel production from genetically engineered microalgae: future of bioenergy in Iran. Renew Sustain Energy Rev 15: 1918–1927 [Google Scholar]
- Thiele I, Palsson BØ (2010) A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc 5: 93–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GA. (1996) Lipids and membrane function in green algae. Biochim Biophys Acta 1302: 17–45 [DOI] [PubMed] [Google Scholar]
- UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res 43: D204–D212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Baalan C, Pulich WM, Brandeis MG (1973) Heterotrophic growth of the microalgae. Crit Rev Microbiol 2: 229–254 [Google Scholar]
- Wang B, Li Y, Wu N, Lan CQ (2008) CO2 bio-mitigation using microalgae. Appl Microbiol Biotechnol 79: 707–718 [DOI] [PubMed] [Google Scholar]
- Wang Z, Zuidhof MJ (2004) Estimation of growth parameters using a nonlinear mixed Gompertz model. Poult Sci 83: 847–852 [DOI] [PubMed] [Google Scholar]
- Wu C, Xiong W, Dai J, Wu Q (2015) Genome-based metabolic mapping and 13C flux analysis reveal systematic properties of an oleaginous microalga Chlorella protothecoides. Plant Physiol 167: 586–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Li X, Xiang J, Wu Q (2008) High-density fermentation of microalga Chlorella protothecoides in bioreactor for microbio-diesel production. Appl Microbiol Biotechnol 78: 29–36 [DOI] [PubMed] [Google Scholar]
- Yang C, Hua Q, Shimizu K (2000) Energetics and carbon metabolism during growth of microalgal cells under photoautotrophic, mixotrophic and cyclic light-autotrophic/dark-heterotrophic conditions. Biochem Eng J 6: 87–102 [DOI] [PubMed] [Google Scholar]
- Zhu S, Wang Y, Huang W, Xu J, Wang Z, Xu J, Yuan Z (2014) Enhanced accumulation of carbohydrate and starch in Chlorella zofingiensis induced by nitrogen starvation. Appl Biochem Biotechnol 174: 2435–2445 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.