Highly conserved genes that regulate the identity of single flowers in conventional plant models regulate the unique inflorescence architecture of the evolutionarily successful Asteraceae plant family.
Abstract
The evolutionary success of Asteraceae, the largest family of flowering plants, has been attributed to the unique inflorescence architecture of the family, which superficially resembles an individual flower. Here, we show that Asteraceae inflorescences (flower heads, or capitula) resemble solitary flowers not only morphologically but also at the molecular level. By conducting functional analyses for orthologs of the flower meristem identity genes LEAFY (LFY) and UNUSUAL FLORAL ORGANS (UFO) in Gerbera hybrida, we show that GhUFO is the master regulator of flower meristem identity, while GhLFY has evolved a novel, homeotic function during the evolution of head-like inflorescences. Resembling LFY expression in a single flower meristem, uniform expression of GhLFY in the inflorescence meristem defines the capitulum as a determinate structure that can assume floral fate upon ectopic GhUFO expression. We also show that GhLFY uniquely regulates the ontogeny of outer, expanded ray flowers but not inner, compact disc flowers, indicating that the distinction of different flower types in Asteraceae is connected with their independent evolutionary origins from separate branching systems.
In flowering plants, inflorescences are the branched structures that bear flowers. Their architecture in terms of number and arrangement of flowers shows enormous variation in nature and plays a central role in angiosperm reproductive adaptation and success. Most of our knowledge of the molecular regulation of inflorescence architecture is based on studies of three major inflorescence types: racemes in Arabidopsis (Arabidopsis thaliana) or snapdragon (Antirrhinum majus), cymes in Solanaceae species such as petunia (Petunia hybrida) or tomato (Solanum lycopersicum), and panicles in grasses (Prusinkiewicz et al., 2007; Park et al., 2014; Teo et al., 2014). In the model plant Arabidopsis, endogenous and exogenous flowering-inducing signals convert the vegetative shoot meristem into an inflorescence meristem (IM) that initiates determinate flower meristems (FMs) on its flanks. The inflorescence forms a simple, indeterminate (monopodial) raceme that elongates and never forms a terminal flower due to maintenance of the stem cells in the central zone of the meristem. In Solanaceae, the cymous IM always terminates in a flower but forms new axillary IMs that continue growth, leading to a zig-zag-like sympodial branching pattern. Panicles in grasses show more complex lateral branching, and both apical and lateral meristems may form flowers. Using mathematical modeling, Prusinkiewicz et al. (2007) showed that a single developmental model (the so-called transient model) can generate the distinct inflorescence types (racemes, cymes, and panicles) found in nature.
In all basic inflorescence types, flower meristem identity (FMI) is controlled by homologs of at least three functionally conserved proteins, LEAFY (LFY), UNUSUAL FLORAL ORGANS (UFO), and SEPALLATA3 (SEP3), that diverge in their spatiotemporal expression domains, leading to differences in IM patterning (Weigel et al., 1992; Lippman et al., 2008; Rebocho et al., 2008; Souer et al., 2008). In Arabidopsis, LFY is uniformly expressed in floral buds, where it specifies FMI (Weigel et al., 1992). By interacting with the key coregulators UFO (Chae et al., 2008) and SEP3 (Liu et al., 2009), LFY initiates the floral program by activating flower organ identity genes. Constitutive expression of LFY is sufficient to convert both apical and lateral meristems into terminal flowers (Weigel and Nilsson, 1995), while loss-of-function alleles result in partial loss of FMI, converting flowers into shoots (Weigel et al., 1992). Studies in petunia and tomato indicate that functionally similar proteins regulate patterning in cymose inflorescences, although in an opposite manner compared with Arabidopsis (Lippman et al., 2008; Rebocho et al., 2008; Souer et al., 2008; Park et al., 2014). For example, the LFY homologs ABERRANT LEAF AND FLOWER (ALF) in petunia and FALSIFLORA in tomato show more ubiquitous expression during vegetative growth (Molinero-Rosales et al., 1999; Souer et al., 2008). Moreover, constitutive expression of ALF does not affect the inflorescence architecture in transgenic petunia (Souer et al., 2008). In fact, in Solanaceae, the UFO homologs DOUBLE TOP (DOT) in petunia and ANANTHA (AN) in tomato are specifically expressed in FMs, and they are both necessary and sufficient to specify FMI (Lippman et al., 2008; Souer et al., 2008).
In the Asteraceae, the inflorescence forms a pseudanthium, or false flower. While it superficially resembles a solitary flower, the Asteraceae inflorescence is actually a tightly packed, compressed head (capitulum) composed of morphologically and functionally different types of flowers. In the sunflower (Helianthus annuus), for example, showy ray flowers are formed at the capitulum periphery, while smaller disc flowers appear at the center. Individual flowers emerge in left- and right-turning spirals, the numbers of which follow the famous Fibonacci series. The entire structure is surrounded by involucral bracts that perform sepal-like, protective functions. The rapid tribal radiation of the Asteraceae family, the largest among flowering plants, may correlate with this complex architecture (Bremer, 1994; Funk et al., 2009). Nonetheless, the evolutionary origin and patterning of the head-like inflorescences has been heavily debated, with some proposing that the capitulum represents a single highly compressed raceme or cyme (Cronquist, 1977) or a condensed structure combining both cymose and racemose branching orders accounting for the evolution of floral polymorphy (Pozner et al., 2012). The latter hypothesis is based on morphological studies of the closest relatives of Asteraceae (Menyanthaceae, Goodenicaceae, and Calyceraceae), which show complex inflorescences in which the main axis shows racemose branching while the basal first-order branches follow a cymose pattern (Endress, 2010; Pozner et al., 2012). Pozner et al. (2012) proposed that a major change during the evolution of the capitulum has been the suppression of the cymose patterning of these peripheral branches. Additionally, it has been proposed that the capitulum has evolved from a single, determinate, and expanding meristem that, through subdivision, gave rise to the multiflowered head (Claßen-Bockhoff and Bull-Hereñu, 2013).
We are using gerbera (Gerbera hybrida) as a model to explore the molecular control of flower type differentiation and inflorescence development in Asteraceae. Gerbera represents the basal Mutisieae tribe within Asteraceae (Panero and Funk, 2002) and harbors heterogamous inflorescences consisting of morphologically and functionally different types of flowers. The large and showy marginal ray flowers are female, as are the smaller, intermediate trans flowers, whereas the central disc flowers are hermaphroditic and produce functional pollen. We conducted functional analyses with the two key FMI genes GhLFY and GhUFO in gerbera to test the compelling hypotheses for the evolutionary origin of the capitulum-type inflorescence. We hypothesized that if the capitulum arose from a single meristem, genetically induced loss of FMI would show similar phenotypic changes in both flower types. On the other hand, a condensation of racemose and cymose units could be reflected by distinct phenotypes that correlate with alterations of Arabidopsis-like and Solanaceae-like expression domains, respectively, for these floral regulators. Our experiments, however, reveal a novel expression pattern and function for GhLFY, suggesting that neither of these hypotheses alone can explain the early patterning and evolution of capitulum architecture. Instead, GhLFY imparts a homeotic, FM-like identity to the entire gerbera IM.
RESULTS
GhLFY and GhUFO Show Specific Expression Patterns during Capitulum Development
Gerbera orthologs of the key FMI genes LFY and UFO were identified, cloned, and characterized (Supplemental Fig. S1). Their expression was absent from vegetative tissues (leaves, floral scape, or stem) and was shown to be restricted to young capitula with emerging flower primordia by quantitative reverse transcription (RT)-PCR (Supplemental Fig. S2). In addition, we found that the biochemical functions of GhLFY and GhUFO proteins are conserved in planta and in vitro. As found previously for Arabidopsis AtLFY (Weigel and Nilsson, 1995), ectopic expression of the orthologous gene GhLFY in transgenic Arabidopsis converted both the apical and lateral meristems into terminal flowers (Supplemental Data S1; Supplemental Fig. S3). Moreover, overexpression of AtUFO and GhUFO, respectively, led to the formation of supernumerary petals (Lee et al., 1997; Supplemental Fig. S3). GhLFY and GhUFO proteins also were shown to interact physically with each other in a yeast two-hybrid assay (Supplemental Data S1; Supplemental Fig. S4). As in the case of AtUFO (Chae et al., 2008), LFY interaction only occurred when the F-box region was removed from GhUFO.
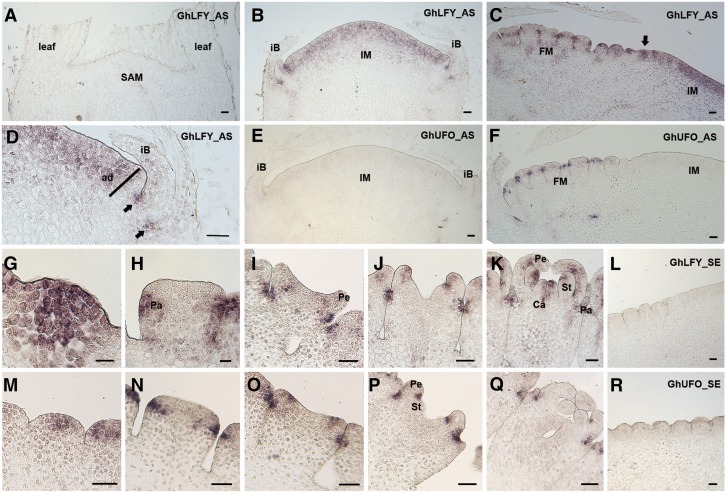
The expression domains of GhLFY and GhUFO were further investigated by in situ hybridization in gerbera. Transcripts of both genes were absent from the vegetative shoot apical meristem as well as the leaf primordia (Fig. 1, A and E). In Arabidopsis, AtLFY expression is localized strictly to the determinate FM (Weigel et al., 1992). However, we show that, after the reproductive transition, GhLFY is expressed uniformly in the naked, dome-shaped IM, an early expression domain, suggesting that the capitulum is, in fact, a determinate structure (Fig. 1B). GhLFY expression in capitula persisted throughout floral primordia development (Fig. 1, B and C) and localized to the emerging involucral bract primordia, where it appeared exclusively in their adaxial domain and, later on, in the axils of each involucral bract (Fig. 1D). During flower primordia initiation, GhLFY expression was visible in the incipient flower primordia already before their outgrowth (Fig. 1, C and G). In contrast, GhUFO was not expressed in the naked IM (Fig. 1E). Its expression initiated later and was restricted solely to the emerging floral primordia (Fig. 1, F and M). During the patterning of individual flowers, both GhLFY and GhUFO show conserved expression patterns comparable to those found in Arabidopsis and petunia (Ingram et al., 1995; Lippman et al., 2008; Souer et al., 2008). During early stages, the expression of both GhLFY and GhUFO localizes to the central region of FMs (Fig. 1, G and M), to the marginal pappus bristle (sepal) primordia (Fig. 1, H and N), and to the boundaries of petal primordia (Fig. 1, I and O). Later on, GhUFO and GhLFY show complementary expression domains. GhUFO is restricted to the bases of petals (Fig. 1, P and Q), while GhLFY transcripts are found in all four whorls of floral organs (Fig. 1, J and K).
Figure 1.
Expression domains of GhLFY and GhUFO in the IM and FM of wild-type gerbera. A, Expression of GhLFY is absent from the vegetative shoot apical meristem (SAM) and leaves. B, GhLFY shows uniform expression in the young, naked, dome-shaped IM after reproductive transition. C, GhLFY expression marks the incipient flower primordia before their outgrowth (arrow). D, GhLFY first localizes to the adaxial side of incipient bract primordia and later to the axil (arrows) of the elongated involucral bract (iB). E, Expression of GhUFO is lacking from the IM. F, GhUFO expression correlates with FM initiation but occurs later than GhLFY expression (C). G to K, Expression domain of GhLFY during early floral developmental stages. M to Q, Expression domain of GhUFO during early floral developmental stages. L and R, Negative controls were hybridized with sense probes for GhLFY (L) and GhUFO (R). ad, Adaxial; AS, antisense; Ca, carpel; Pa, pappus (sepals of individual flowers); Pe, petal; SE, sense; St, stamen. Bars = 50 µm.
GhLFY Is Required to Maintain the Determinacy of the Capitulum IM
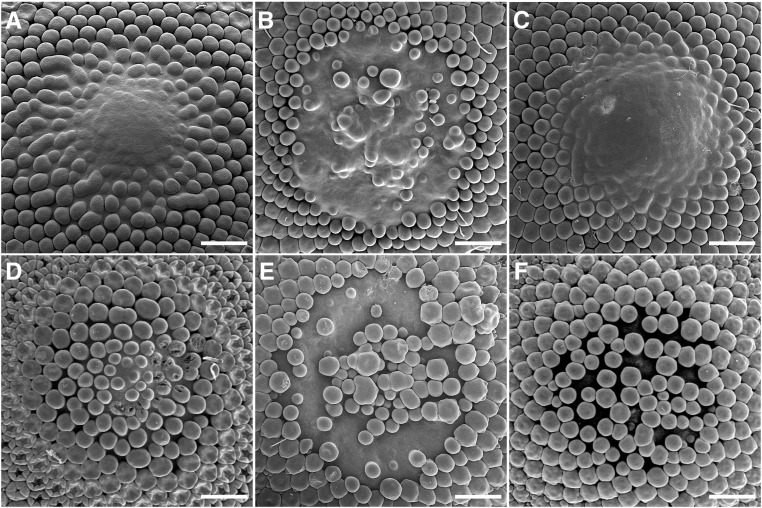
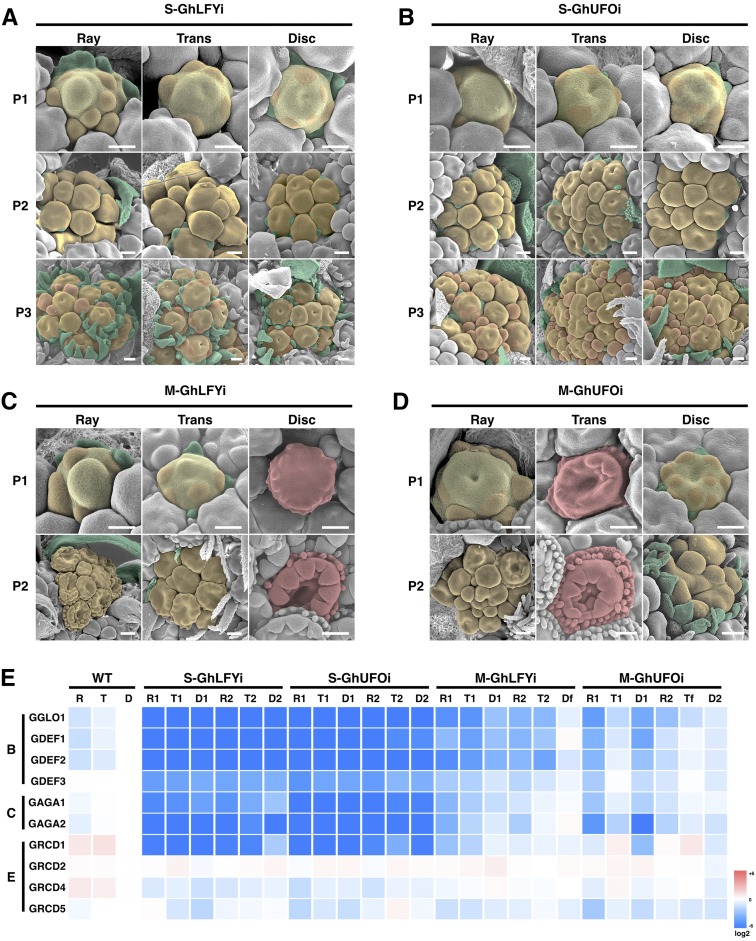
We created transgenic gerbera lines to explore the detailed functions of FMI genes in gerbera. Transformation of RNA interference (RNAi) constructs efficiently suppressed GhLFY and GhUFO expression (Fig. 2, G and H) and led to highly modified inflorescence phenotypes (Fig. 2). As expected, in the most severe transgenic lines, we observed loss of floral organ identities and conversion into green, leaf-, or bract-like organs (Fig. 2, C and E). We also identified milder phenotypes with only partially reduced expression of GhLFY and GhUFO and, accordingly, less pronounced loss of organ identities (Fig. 2, D and F). Scanning electron microscopy (SEM) analyses of the early stages of inflorescence patterning in the transgenic lines uncovered that GhLFY has evolved a novel and specific role in Asteraceae in defining the determinacy of the IM (Fig. 3). In wild-type gerbera, the capitulum produces a fixed number of flowers until the center of the meristematic area is fully consumed with disc flower primordia (Fig. 3, A and D; Uimari et al., 2004; Teeri et al., 2006). In transgenic lines with suppressed GhUFO expression, IM was similarly consumed as in the wild type (Fig. 3, C and F), but, in contrast, the capitula of GhLFY RNAi lines remained undifferentiated (Fig. 3, B and E). In these plants, disc flower primordia first develop regularly from the margins toward the center, as in the wild type, but their emergence ceases at a certain stage, whereupon they start to initiate in a random manner in the center of the IM (Fig. 2, B and E). We did not detect any signs of consumption of capitulum growth in these lines, as new primordia kept initiating constantly even when the inflorescence was fully open and most of the flowers had reached anthesis.
Figure 2.
Phenotypes and expression analysis of wild-type gerbera and transgenic lines with suppressed FMI gene expression. A, Mature inflorescence of nontransgenic gerbera. B, Nontransgenic gerbera from the abaxial side. The inflorescence is surrounded by green involucral bracts. C, Phenotype of a strong transgenic GhLFY RNAi line. D, Phenotype of a mild transgenic GhLFY RNAi line. E, Phenotype of a strong transgenic GhUFO RNAi line. F, Phenotype of a mild transgenic GhUFO RNAi line. G, Expression analysis of three independent transgenic lines with suppressed GhLFY expression compared with wild-type (WT) gerbera. H, Expression analysis of two independent transgenic lines with suppressed GhUFO expression compared with wild-type gerbera. Bars = 1 cm (A–F).
Figure 3.
IM phenotypes in transgenic lines with suppressed FMI gene expression. A, Wild-type IM. The center of the expanding IM has not yet been consumed by emerging flower primordia. B, The IM in the GhLFY RNAi line shows random initiation of flower primordia. C, The IM in the GhUFO RNAi line develops similar to that in the wild type (A). D, Later stage of wild-type IM. The IM is fully consumed with disc flower primordia. E, The IM in the GhLFY RNAi line is never consumed with flower primordia. F, Later stage of IM in the GhUFO RNAi line. The IM is similarly consumed with flower primordia as in the wild type (D). Bars = 500 μm.
Ectopic Expression of GhUFO Confers Floral Fate to the Capitulum
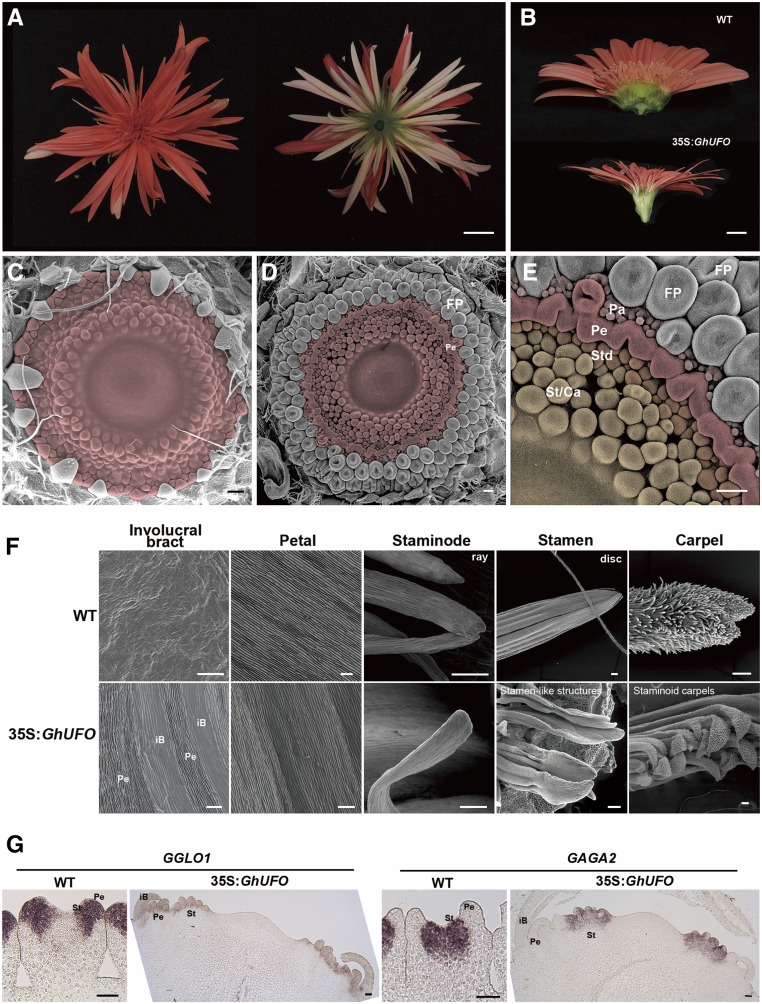
The flower primordia-specific expression of GhUFO suggests that it may be a master gene that defines FMI. We produced transgenic lines with precocious, ectopic expression of GhUFO under the control of the cauliflower mosaic virus 35S promoter (Supplemental Fig. S5) and found no effect on vegetative growth, while reproductive structures were highly modified (Fig. 4, A and B). The emerging young capitulum was more elongated than expanded, and it retained its small size throughout development (Fig. 4B). The involucral bracts surrounding the capitula were arranged in a spiral order as in the wild type, but they gained increasing petaloid identity toward the center of the capitulum. This phenotypic change correlated with the up-regulation of B class MADS box genes (Supplemental Fig. S5). However, instead of forming flower primordia in spiral phyllotaxis, the meristem showed whorled phyllotaxis and initiated floral organ primordia in concentric rings (Fig. 4, C–E). Inward of the petaloid bracts, true petals initiated from a fused, ring-like meristem (Fig. 4, D–F), followed by several whorls of staminodes or stamen-like structures (Fig. 4, D–F) that occupied the center of the capitulum. SEM was used to verify the identities of these organs (Fig. 4F). Furthermore, RNA in situ hybridization indicated expected expression patterns of MADS box organ-identity genes in the modified organs (Fig. 4G). Expression of the B class gene GGLO1 (Broholm et al., 2010) marked the petaloid involucral bracts and the petal whorl of the transgenic capitulum, and overlapping expression of both GGLO1 and the C function gene GAGA2 (Yu et al., 1999) was confined to staminodes and stamens (Fig. 3G).
Figure 4.
Constitutive expression of GhUFO confers a floral fate to the capitulum. A, General phenotype of the transgenic 35S:GhUFO inflorescence. B, The young capitulum is elongating in 35S:GhUFO rather than expanding as in the wild type. C, Ectopic GhUFO leads to highly modified floral structures with organ primordia emerging in a whorled phyllotaxis. In this line with a strong phenotype, instead of single flowers, floral organs are initiated from the margins toward the center of the capitulum. D and E, A mild phenotype (D) and closeup of the mild phenotype (E) showing that normal flower primordia (FP) are first initiated at the capitulum periphery. The fused ring-like meristem produces petals (Pe) surrounded by pappus bristles (Pa). The innermost whorls are occupied by staminode-like organs (Std) and a mixture of stamen-like structures and staminoid carpels (St/Ca). F, Epidermal cell structures of floral organs in wild-type (WT) and 35S:GhUFO plants. In 35S:GhUFO, the petaloid involucral bracts (iB) show mixtures of petal- and bract-like cell types; the fused ring-like meristem shows petal identity: the sterile staminodes are as in wild-type ray flowers, and functional stamens are as in wild-type disc flowers. We also found variants showing staminoid carpels in the center. G, Expression of GGLO1 (B function MADS box gene) and GAGA2 (C function MADS box gene) in wild-type FM and 35S:GhUFO meristem. B gene expression is confined to petaloid bracts and petals, while both B and C genes are expressed in the staminode/stamen-like organs (St). Bars = 1 cm (A and B), 100 µm (C–F), and 50 µm (G).
In transgenic lines with milder phenotypes, the change from an inflorescence program to floral organ formation occurred later, suggesting that posttranscriptional output or a response threshold for GhUFO/GhLFY determines the switch. In the weaker lines, the peripheral primordia were still patterned as single flowers, but the inner whorls formed floral organs following an order normally found in single flowers: pappus bristles (sepals; whorl 1) followed by petals (whorl 2), stamen-like structures (whorl 3), and staminoid carpels (whorl 4; Fig. 4, D and E). Our data show that ectopic expression of GhUFO alone is able to trigger the floral fate of the meristem and to establish a whorled pattern of floral organ development. The transgenic phenotypes indicate that the meristem recapitulates the floral prepattern found in single flowers of wild-type gerbera.
Ray Flowers Show a Distinct Ontogenetic Pattern Associated with GhLFY Function
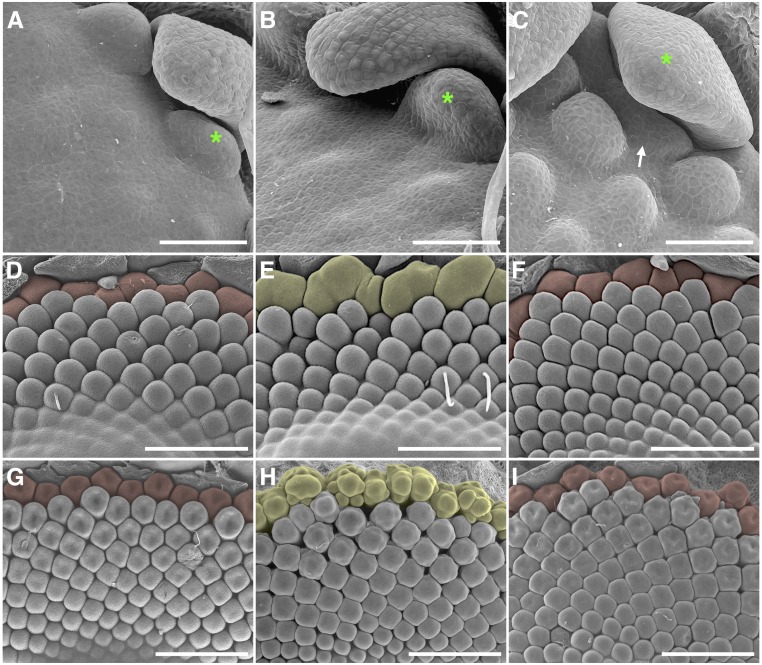
The key feature of the Asteraceae capitulum is the presence of distinct flower types. Close studies of the early ontogeny of distinct flower types in gerbera indicate that ray flower initiation deviates from that of the other flower types (Fig. 5; Supplemental Fig. S6). It is temporally delayed, and in fact, the first visible flowers are the trans flower primordia, which continue the phyllotactic pattern established by involucral bracts (Fig. 5, A–C). We also show that ray flowers develop exclusively in the axils of the last series of involucral bracts (Fig. 5C). Transgenic gerbera lines with suppressed FMI gene expression formed modified capitula with leaf- or bract-like organs. However, detailed phenotypic analysis revealed unique responses in these lines. While ray flower initiation occurred similarly in wild-type and GhUFO RNAi lines, the suppression of GhLFY expression showed a specific effect at the early initiation stage of the ray primordia (Fig. 5, D–F). The capitulum periphery in GhLFY RNAi plants initiated oval-shaped primordia, considerably larger than the single ray primordia of wild-type or GhUFO RNAi lines. These large primordia were not patterned as single flowers but, instead, were further branched into two to three primordia clearly distinguished from the neighboring trans flowers (Fig. 5E). In addition, in both the wild-type and GhUFO RNAi lines, organogenesis in ray flower primordia was delayed in comparison with trans flowers, whereas patterning of the ray primordia in GhLFY RNAi lines occurred without any delay, again suggesting an indispensable role for GhLFY in early ray flower development (Fig. 5, G–I). Intriguingly, this ontogenetic pattern of peripheral primordia development shares similarities with the branched, peripheral cymose units found in the immediate sister family of Asteraceae, Calyceraceae (Pozner et al., 2012), or the syncephalium subunit primordia described in species of Asteraceae with higher order capitulum structures (i.e. those bearing capitula within capitula; Harris, 1994, 1999). Our data suggest that ray primordia in Asteraceae may have evolved through the suppression of branching of the peripheral cymose units still found in Calyceraceae capitula and by the gain of floral fate defined by GhLFY.
Figure 5.
Early ontogeny of ray primordia initiation in wild-type and transgenic gerbera with suppressed FMI gene functions. A to C, Three consecutive developmental stages of early capitulum development in wild-type gerbera. Trans flowers initiate earlier than ray primordia (arrow) that emerge in the axils of the last series of involucral bracts (green asterisks). D to F, SEM images show that the ray flower initials (shaded in yellow) of GhLFY RNAi plants (E) are distinct from the solitary ray primordia (shaded in red) found in wild-type (D) and GhUFO RNAi (F) plants. G to I, In contrast to wild-type (G) and GhUFO RNAi (I) plants, the marginal ray flower primordia in GhLFY RNAi (H) plants show faster organogenesis compared with nearby trans flower primordia. Bars = 50 µm (A–C) and 500 µm (D–I).
Floral Patterning Is Regulated by GhLFY and GhUFO in a Flower Type-Dependent Manner
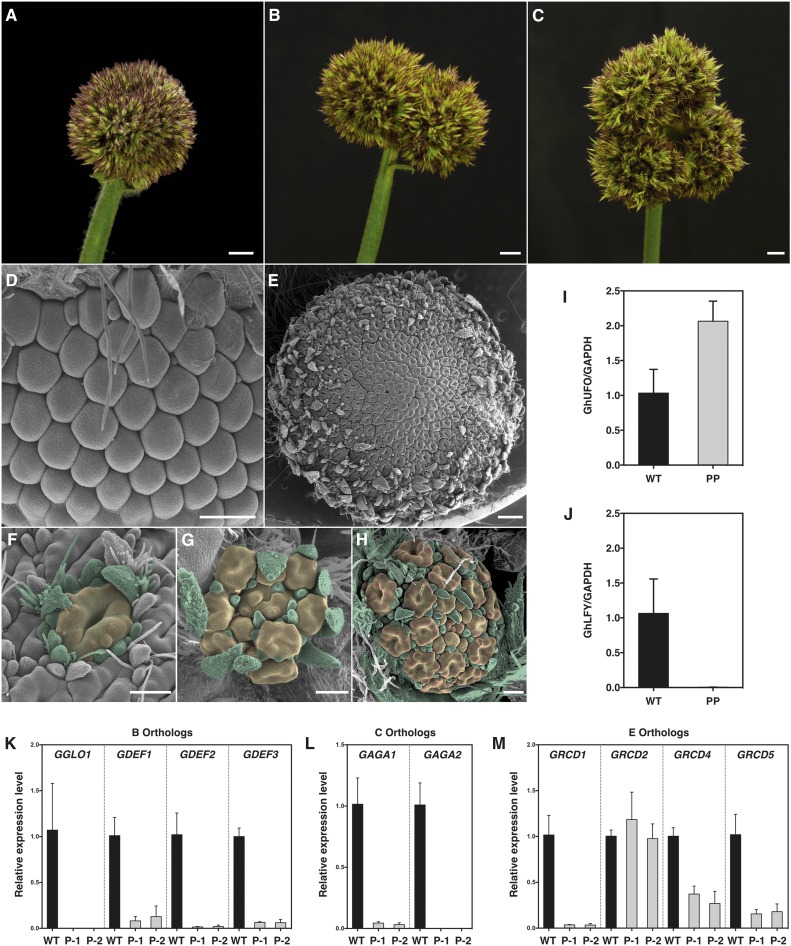
After flower primordia initiation, at the level of single flowers, loss of FMI in both GhLFY and GhUFO RNAi lines was obvious. We conducted SEM analyses in order to characterize the phenotypic changes more closely. In the most severe transgenic lines, suppression of either GhLFY or GhUFO expression led to complete loss of floral organs, and emerging primordia repeatedly initiated secondary and tertiary flower primordia subtended by bract-like structures (Fig. 6, A and B). A similar strong phenotype was observed in a mutant gerbera cultivar, Pingpong, which lacks GhUFO expression (Fig. 7, A, I, and J). This cultivar was identified from a breeder’s collection, as its inflorescence resembles the transgenic GhLFY and GhUFO phenotypes (Fig. 7A). In cv Pingpong, the marginal ray primordia initiate as in wild-type and GhUFO RNAi lines and the capitulum is fully packed with flower primordia (Fig. 7, D and E). Furthermore, the single primordium continuously produces secondary and tertiary primordia surrounded by bract-like organs (Fig. 7, F–H), leading to extensive proliferation of the inflorescences (Fig. 7, B and C). Altogether, our data reveal the necessary and evolutionarily conserved roles of both GhLFY and GhUFO functions for proper patterning of individual flowers.
Figure 6.
Patterning of the individual flower primordia in transgenic GhLFY and GhUFO RNAi lines. A and B, Transgenic GhLFY RNAi (A) and GhUFO RNAi (B) plants with severe phenotypes form primary primordia (P1; shaded in yellow) that repeatedly initiate secondary (P2) and tertiary (P3) primordia (shaded in orange) in all flower types (ray, trans, and disc). C and D, Patterning of flower primordia in GhLFY RNAi (C) and GhUFO RNAi (D) transgenic plants with milder phenotypes shows flower type-specific responses. In both lines, the ray flower primordia uniformly initiate secondary primordia and, consequently, secondary flowers. The response in trans and disc flowers shows opposite effects: in GhLFY RNAi lines, the disc primordia, and in GhUFO RNAi lines, the trans primordia, pattern as normal flowers (shaded in red). Bars = 100 µm. E, Heat map of quantitative RT-PCR results shows expression profiles of the B, C, and E function MADS box genes in developing primary (R1, T1, and D1) and secondary (R2, T2, and D2) primordia in different flower types. D, Disc flower primordia; M, mild phenotype; R, ray flower primordia; S, severe phenotype; T, trans flower primordia; WT, wild type.
Figure 7.
Phenotypes and expression analysis of the gerbera mutant cv Pingpong. A to C, The inflorescence of cv Pingpong shows similarity to the transgenic GhLFY and GhUFO RNAi lines (A). Extensive proliferation of the inflorescences (B and C) causes splitting of the head. D, Patterning of the capitulum of cv Pingpong. The marginal primordia develop as in wild-type gerbera. E, The inflorescence of cv Pingpong is fully consumed by emerging flower primordia at later developmental stages. F to H, Patterning of the single primordium in cv Pingpong. The single primordium (F) produces bract-like structures surrounding the secondary primordia (G) that further initiate tertiary primordia (H). Bars = 1 cm (A–C) and 100 µm (D–H). I, Relative expression levels of GhUFO in young inflorescences of the wild type (WT) and cv Pingpong (PP). J, Relative expression levels of GhLFY in young inflorescences of the wild type and cv Pingpong. K to M, Relative expression levels of B class (K), C class (L), and SEP-like (M) MADS box genes in primary (P-1) and secondary (P-2) primordia dissected from cv Pingpong in comparison with the wild-type stage 3 flower primordia. Error bars indicate sd calculated from three biological replicates.
We also explored floral patterning in GhUFO and GhLFY RNAi lines with milder phenotypes (Supplemental Fig. S7). In these weak lines, the ray flowers responded independently from the other flower types and produced secondary flowers as in the more severe lines. In the mild GhUFO RNAi lines, the disc flower primordia also initiated secondary primordia, while the trans flower primordia were patterned as single flowers (Fig. 6D; Supplemental Fig. S7). The mild GhLFY RNAi lines instead showed an opposite effect: the disc flower primordia were patterned as single flowers, and the trans flower primordia included secondary primordia (Fig. 6C; Supplemental Fig. S7). These phenotypic changes correlated with the expression levels of the downstream B, C, and E function MADS box homeotic genes, the expression of which was lacking in the most severe transgenic lines and in the mutant cv Pingpong, while it was still visible in disc flower primordia of milder GhLFY RNAi lines and trans flowers of GhUFO RNAi lines (Figs. 6E and 7, K–M). Altogether, these data suggest that both GhUFO and GhLFY form a functional gradient across the capitulum, with GhLFY showing a more pronounced role in the development of peripheral ray and trans flowers and GhUFO in the patterning of disc flowers.
DISCUSSION
The unique feature of the Asteraceae plant family is that their inflorescences mimic solitary flowers. This homeotic transference of flower-like identity to the inflorescence was likely the key innovation for the diversification of this largest family of flowering plants. Here, using the model plant gerbera, we provide functional data suggesting that the capitulum not only resembles a single flower at the morphological level but also at the molecular level. Our data further demonstrate that transitions in plant reproductive evolution use conserved genetic modules and pathways that can be readily co-opted for adaptive success. We show a conserved function for GhUFO in specifying FMI, while the LFY homolog GhLFY has evolved novel functions in defining the determinacy of the IM and in regulating the development of the marginal ray flowers. Our data suggest that distinct flower types have independent evolutionary origins.
The Capitulum Is a Determinate Structure That Confers Floral Fate upon Ectopic GhUFO Expression
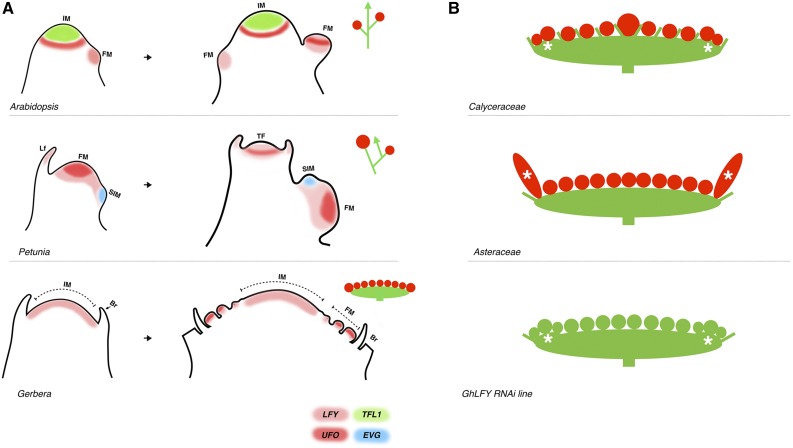
We identified the key FMI genes in gerbera in order to understand the patterning of the capitulum architecture and get insight into its evolutionary origin. As is common to most angiosperms, the FMI genes GhLFY and GhUFO were found as single-copy genes in gerbera. Their functions in a heterologous Arabidopsis background as well as their capacity to interact physically with each other indicate that their protein functions have been conserved between rosids and asterids. However, we show that both spatial and temporal modifications in the expression domains of these regulators have played a major role in the evolution of capitulum architecture (Fig. 8A). Our data indicate that the expression pattern of GhLFY has diversified, showing uniform, early expression in the dome-shaped IM before the initiation of flower primordia. This stands in contrast to the indeterminate Arabidopsis raceme, where TERMINAL FLOWER1 (TFL1) activity in both the apical and lateral IMs inhibits the expression of the FMI genes LFY and AP1 and, consequently, flower initiation (Weigel et al., 1992; Bradley et al., 1997). Similar to petunia with its cymose inflorescences, the UFO ortholog of gerbera (GhUFO) defines FMI (Fig. 8A). In petunia, the WOX homeodomain protein EVERGREEN (EVG) specifies lateral IM (sympodial IM) development and is needed to activate DOT (Rebocho et al., 2008). Whether the WOX-like proteins in gerbera play a role in meristem patterning remains to be studied.
Figure 8.
Functional diversification of LFY during capitulum development. A, In Arabidopsis racemes, TFL1 activity regulates the indeterminacy of the IM while LFY defines FMI. In petunia cymes, the IM terminates in a flower. FMI is defined by DOT, and the growth of the inflorescence continues from a sympodial IM (SIM) defined by EVG activity. In gerbera, GhLFY expression is uniform in the determinate IM that subdivides into single flower primordia, where FMI is defined by GhUFO. B, Suggested evolutionary pathway for capitulum development. Species representing Calyceraceae, a close relative of Asteraceae, typically show branched cymose units (marked with asterisks) in the periphery of their inflorescences (Pozner et al., 2012). LFY has evolved a specific role to suppress branching in the marginal ray flower primordia of Asteraceae, as evidenced by the GhLFY RNAi lines.
The expanding gerbera IM lacks a terminal flower. Suppression of GhLFY expression led to the loss of determinacy and random initiation of flower primordia in the center-most region of the IM. Moreover, ectopic expression of GhUFO was able convert the capitulum into a large single flower. We observed the up-regulation of both B and C genes in accordance with the identities of single organs. As the GhUFO RNAi lines showed the absence of B and C gene expression (Fig. 6), we suggest that GhUFO is needed to regulate both these organ identity genes, as also found previously in petunia (Souer et al., 2008). Altogether, GhUFO expression reestablished the whorled floral patterning and led to the development of single floral organs in the capitulum background, indicating that the gene has a conserved and indispensable role in defining FMI even when transferred to the whole-inflorescence level.
The fact that GhLFY has evolved a broader expression domain in Asteraceae suggests that it is involved in the regulation of early IM growth. It has been proposed previously that, apart from floral identity determination, the ancestral function of LFY was in promoting meristematic growth more generally, by affecting cell division (for review, see Moyroud et al., 2010; Teo et al., 2014). In legumes, LFY mutants develop simpler leaves, indicating the involvement of LFY function in leaf indeterminacy (Hofer et al., 1997; Hofer and Ellis, 2002; Dong et al., 2005). Furthermore, in grasses, LFY homologs are required to maintain the indeterminacy of the early panicle meristem affecting the branching architecture of the inflorescences (for review, see Moyroud et al., 2010; Kyozuka et al., 2014). Moreover, in Arabidopsis, LFY stimulates the growth of axillary meristems (Moyroud et al., 2010). Our findings indicate that, in Asteraceae, LFY shows further functional diversification. Uniform expression of GhLFY across the naked capitulum defines it as a determinate structure resembling the single floral meristems borne on racemes or cymes (Weigel et al., 1992; Souer et al., 2008). The capitulum can thus be seen as an analog of a single flower not just functionally or by its ontogeny but also at the molecular level. Still, as yet uncharacterized factors contribute to the compression and/or fusion of the capitulum.
LFY Has a Specific Role in Regulating the Ontogeny of Ray Flowers
It is commonly presented that flower primordia in Asteraceae heads develop acropetally, from the margins toward the center following the spiral phyllotaxis. Harris (1995) found such acropetal development to be strictly limited to species that display capitula consisting of only a single flower type. Species with true ray flowers, however, show a consistent delay in ray flower development. Furthermore, bidirectional development of flower primordia was documented for several species (Harris, 1995). For example, in Erigeron philadelphicus, the outermost disc flowers are the first to emerge, and the subsequent disc flowers initiate acropetally. The first ray flowers initiate proximally next to the oldest disc flowers (from nonperipheral starting points), and the next ones develop basipetally toward the involucral bracts, suggesting that ray flower development is not derived directly from that of disc flowers (Harris, 1991). We similarly show that the early ontogeny of peripheral ray flowers is different from that of other flower types. In gerbera, the ray primordia initiated later than the closest trans flowers. We found that GhLFY expression localized to the base of the involucral bracts already at a stage when these bracts started to initiate. These cells with localized GhLFY expression may represent suppressed axillary structures, as GhLFY silencing specifically converted the ray primordia into branched peripheral units. Furthermore, our data indicated that the temporal delay in ray flower patterning was regulated by GhLFY.
We also observed radial gradients for both GhLFY and GhUFO functions, with GhLFY function being more pronounced at the periphery and GhUFO in the center of the capitulum. Interestingly, a similar gradient along the inflorescence axis also is found in the Arabidopsis lfy mutant, which shows stronger phenotypes in early-arising meristems compared with the younger ones (Weigel et al., 1992). In gerbera, this gradient was already reflected in our previous work that investigated the differential expression of several MADS box genes across the capitulum (Yu et al., 1999; Kotilainen et al., 2000; Laitinen et al., 2006) as well as the specific expression of CYCLOIDEA2-like TCP domain transcription factor genes in ray flower primordia (Broholm et al., 2008; Tähtiharju et al., 2012), emphasizing the key role that the FMI genes play in defining flower type differentiation. However, what exactly creates this gradient and what its actual nature is remain obscure. AtLFY is known to be regulated by auxin (Yamaguchi et al., 2013), but still, the putative role of auxin in regulating the patterning of inflorescences, either racemes or capitula, remains to be studied.
Evolution of Capitulum Architecture
Condensation of the elongated inflorescence into a capitulum is an example of convergent evolution, as it has occurred several times independently during the evolution of Asteridae and other taxa (Harris, 1999). In this article, our focus is on Asteraceae primary capitula, the most basic type of head-like inflorescences combining ray and disc flowers into a single structure. We show that, in gerbera, GhLFY has a specific role in defining ray flower development. Suppression of GhLFY expression converted the ray flower primordia into large meristems that subdivided and formed branched structures. The ontogeny of these structures was similar to the so-called cymose units found in the Calyceraceae, a close relative of Asteraceae (Pozner et al., 2012; Fig. 8B). Therefore, we propose that the differential development of gerbera ray flowers is not related to their marginal position per se but instead to their distinct ontogenetic origin as separate, cymose inflorescence axes and, correspondingly, that LFY has contributed to a gain of floral fate for the peripheral branching units found in the capitula of Calyceraceae (Pozner et al., 2012). Interestingly, the ontogeny of branched primordia in GhLFY RNAi lines also shares similarity with the syncephalium subunit primordia that are found in secondary flower heads of several Asteraceae taxa (Harris, 1995). A syncephalium is a higher order aggregation of capitula (capitula within capitula) that evolved more recently than the simple capitulum, and it is often accompanied by a reduction of the primary inflorescences to a single-flowered state (Harris, 1995). Our data, therefore, suggest that LFY function may play a role in regulating the development of secondary flower heads as well.
CONCLUSION
Inflorescence architecture displays enormous variation in nature and plays a central role in the reproductive fitness and adaptation of plants, as well as in crop yield. The evolutionary success of the large Asteraceae plant family has been associated with the unique head-like inflorescences consisting of different flower types. Here, we show that the highly conserved FMI regulator LFY has evolved novel functions in regulating inflorescence development in Asteraceae. Unlike in other model species, LFY specifies the development of the naked IM and specifically regulates ray flower development. Our data support the hypothesis that flower type differentiation in Asteracae is connected to their different developmental origins.
MATERIALS AND METHODS
Plant Material
Gerbera hybrida (Asteraceae) ‘Terraregina’ and ‘Pingpong’ (Terra Nigra) were grown under standard greenhouse conditions as described previously (Ruokolainen et al., 2010b). The developmental stages for gerbera flower primordia development are described by Laitinen et al. (2006) and Tähtiharju et al. (2012).
Isolation of GhLFY and GhUFO Complementary DNAs and Genetic Transformation of Gerbera
The gerbera homologs for LFY and UFO were searched from the gerbera EST sequence database, and the full-length complementary DNAs (cDNAs) were amplified with gene-specific primers (Supplemental Table S1). Total RNA was isolated from young inflorescences (5–10 mm in size) with Trizol reagent (Invitrogen), and first-strand cDNA (Boehringer First-Strand cDNA Kit) was used as a template for PCR. The full-length cDNAs were cloned into Gateway entry vector pDONR221 (Invitrogen) and were verified by sequencing. For genetic transformation, we used the Gateway binary vectors pK7WG2D and pK7GWIWG2D(II) (Karimi et al., 2002) to generate overexpression and RNAi constructs, respectively. The gene constructs were electroporated into the Agrobacterium tumefaciens strain C58C1 harboring pGV3101. Transformation of gerbera ‘Terraregina’ with GhUFO overexpression and GhLFY and GhUFO RNAi constructs was done according to Elomaa and Teeri (2001).
Expression Analysis
To verify the efficiency of RNAi (Fig. 2), we selected three independent GhLFY RNAi lines (TR6, TR1, and TR7) and two GhUFO RNAi lines (TR9 and TR1) for expression analysis. Sampling was done by dissecting inflorescences of 2 to 3 mm in diameter for GhLFY RNAi plants and 4 to 5 mm in diameter for GhUFO RNAi plants together with wild-type controls of corresponding sizes. For the mutant cv Pingpong, inflorescences were 3 to 4 mm in diameter, but they showed a similar developmental stage of flower primordia as the 4- to 5-mm cv Terraregina inflorescences. To verify the expression of the downstream floral homeotic genes in transgenic cv Terraregina lines and cv Pingpong, individual flower primordia at different developmental stages were sampled. For GhLFY and GhUFO RNAi plants, we dissected primary and secondary primordia from three flower types according to their morphology (Fig. 6, A–D) by stereomicroscopy. For comparison, we collected stage 3 primordia of three different flower types from the wild type (Laitinen et al., 2006). Since there were no clear flower type-specific responses in cv Pingpong, we collected the center-most primary and secondary primordia for expression analysis (Fig. 7).
The expression analysis of 35S:GhUFO plants was done by semiquantitative RT-PCR. Young leaves (3–4 mm) from six independent transgenic lines (TR1, TR2, TR4, TR5, TR6, and TR10) were sampled for RT-PCR to verify the ectopic expression of GhUFO in comparison with the wild type (Supplemental Fig. S5). Furthermore, involucral bracts with petaloid identity were collected from fully opened inflorescences (lines TR1, TR2, TR5, and TR6) to check the expression of B class MADS box genes (GGLO1 and GDEF2) that define petal identity in gerbera (Yu et al., 1999; Broholm et al., 2010; Supplemental Fig. S5).
Expression analysis of GhLFY and GhUFO RNAi lines was conducted with quantitative RT-PCR. Total RNA was extracted with the CTAB method modified from Chang et al. (1993). After precipitation, the pellet was washed in 70% ethanol and dissolved in RNase-free water. DNase treatment was performed according to the instructions of the RNA Clean-Up Kit (Macherey-Nagel). cDNA synthesis (500 ng) and quantitative PCR were conducted as described previously (Tähtiharju et al., 2012). The quantitative PCR primers are shown in Supplemental Table S1. For testing GhLFY and GhUFO RNAi efficiency, we normalized GhLFY and GhUFO expression levels to the expression of the gerbera GAPDH reference gene according to the ΔΔCt method (Pfaffl, 2001). The expression levels of downstream MADS box genes were normalized against the total RNA amount used for cDNA synthesis as stated by Tähtiharju et al. (2012). Expression analysis was conducted for the B class MADS box genes GGLO1 (Yu et al., 1999), GDEF1, GDEF2, and GDEF3 (Broholm et al., 2010) as well as the C class genes GAGA1 and GAGA2 (Yu et al., 1999). We also included the SEPALLATA-like E class genes. In gerbera, GRCD1 and GRCD2 have been shown to specifically affect stamen and carpel development, respectively (Kotilainen et al., 2000; Uimari et al., 2004), while GRCD4 and GRCD5 provide redundant and general E function in gerbera (Ruokolainen et al., 2010a). The quantitative RT-PCR results were analyzed in three biological replicates. For construction of the heat map, we used log-transformed average means of relative expression levels.
In Situ Hybridization
For in situ expression analysis, the preparation of the plant samples, sectioning, and hybridization were done as described by Elomaa et al. (2003). Gene-specific probes for GhLFY (294 bp), GhUFO (299 bp), and GAGA2 (288 bp) were synthesized as described by Juntheikki-Palovaara et al. (2014) using a PCR-amplified fragment of the given gene with primers containing a few extra nucleotides (uppercase letters) and a T7 overhang (lowercase letters) (taatacgactcactataGGGAGG or CAtaatacgactcactataGGG) at the 5′ end (Supplemental Table S1) and labeled following the instructions of the DIG RNA Labeling Kit (Roche). The probe corresponding to GGLO1 (217 bp) was the same as that described by Broholm et al. (2010). Sections were examined and photographed using the Leitz Laborlux S Microscope equipped with the Leica DFC420 C Digital Camera.
SEM
For SEM analysis of wild-type and transgenic gerbera plants, a time series of capitula at early developmental stages was hand dissected under stereomicroscopy. When necessary, excessive involucral bracts were removed to expose the center IM and the developing flower primordia. Sample preparations were performed as described previously by Uimari et al. (2004), except that an automated Leica EM CP300 dryer was used for critical point drying. Samples were examined using the Quanta 250 (FEI) SEM device at the Electron Microscopy Unit at the Institute of Biotechnology, University of Helsinki. The obtained SEM images were further edited and pseudocolored in Adobe Photoshop CC 2015.
Accession Numbers
Sequence data from this article can be found in GenBank under the following accession numbers: KU554694 for GhLFY and KU554695 for GhUFO.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Amino acid sequence alignments of GhUFO and GhLFY with selected orthologous genes.
Supplemental Figure S2. Expression of GhLFY and GhUFO in diverse plant parts.
Supplemental Figure S3. Ectopic expression of GhLFY and GhUFO in transgenic Arabidopsis.
Supplemental Figure S4. Protein-protein interactions of GhLFY and GhUFO.
Supplemental Figure S5. Expression analysis and organ identities of 35S:GhUFO lines.
Supplemental Figure S6. Early ontogeny of gerbera capitulum development.
Supplemental Figure S7. Individual flowers in wild-type and GhLFY and GhUFO RNAi lines with mild phenotypes.
Supplemental Table S1. Primer sequences used in this study.
Supplemental Data S1. Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Regine Claßen-Bockhoff and Przemyslaw Prusinkiewicz for valuable discussions; the electron microscopy unit at the Institute of Biotechnology, University of Helsinki, for providing excellent facilities; Eija Takala and Anu Rokkanen for skillful technical assistance; and the greenhouse team led by Sanna Peltola for taking care of plants.
Glossary
- IM
inflorescence meristem
- FM
flower meristem
- FMI
flower meristem identity
- RT
reverse transcription
- RNAi
RNA interference
- SEM
scanning electron microscopy
- cDNA
complementary DNA
Footnotes
This work was supported by the Academy of Finland (grant no. 139092 to P.E.), by the Jenny and Antti Wihuri Foundation (grant to Y.Z.), and by the Doctoral Programme in Plant Sciences at the University of Helsinki (to Y.Z. and T.Z.).
References
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E (1997) Inflorescence commitment and architecture in Arabidopsis. Science 275: 80–83 [DOI] [PubMed] [Google Scholar]
- Bremer K. (1994) Asteraceae: Cladistics and Classification. Timber Press, Portland, OR [Google Scholar]
- Broholm SK, Pöllänen E, Ruokolainen S, Tähtiharju S, Kotilainen M, Albert VA, Elomaa P, Teeri TH (2010) Functional characterization of B class MADS-box transcription factors in Gerbera hybrida. J Exp Bot 61: 75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broholm SK, Tähtiharju S, Laitinen RAE, Albert VA, Teeri TH, Elomaa P (2008) A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc Natl Acad Sci USA 105: 9117–9122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E, Tan QKG, Hill TA, Irish VF (2008) An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 135: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Claßen-Bockhoff R, Bull-Hereñu K (2013) Towards an ontogenetic understanding of inflorescence diversity. Ann Bot (Lond) 112: 1523–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronquist A. (1977) The Compositae revisited. Brittonia 29: 137–153 [Google Scholar]
- Dong ZC, Zhao Z, Liu CW, Luo JH, Yang J, Huang WH, Hu XH, Wang TL, Luo D (2005) Floral patterning in Lotus japonicus. Plant Physiol 137: 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elomaa P, Teeri TH (2001) Transgenic Gerbera. In Bajaj YPS, ed, Biotechnology in Agriculture and Forestry 48. Springer-Verlag, Berlin, pp 139–154 [Google Scholar]
- Elomaa P, Uimari A, Mehto M, Albert VA, Laitinen RAE, Teeri TH (2003) Activation of anthocyanin biosynthesis in Gerbera hybrida (Asteraceae) suggests conserved protein-protein and protein-promoter interactions between the anciently diverged monocots and eudicots. Plant Physiol 133: 1831–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK. (2010) Disentangling confusions in inflorescence morphology: patterns and diversity of reproductive shoot ramification in angiosperms. J Syst Evol 48: 225–239 [Google Scholar]
- Funk VA, Susanna A, Stuessy TF, Bayer RJ, editors (2009) Systematics, Evolution, and Biogeography of Compositae. International Association for Plant Taxonomy, Vienna [Google Scholar]
- Harris EM. (1991) Floral initiation and early development in Erigeron philadelphicus (Asteraceae). Am J Bot 78: 108–121 [Google Scholar]
- Harris EM. (1994) Developmental evidence for the derivation of syncephalia in Lagascea (Heliantheae: Asteraceae). Am J Bot 9: 1139–1148 [Google Scholar]
- Harris EM. (1995) Inflorescence and floral ontogeny in Asteraceae: a synthesis of historical and current concepts. Bot Rev 61: 93–278 [Google Scholar]
- Harris EM. (1999) Capitula in the Asteridae: a widespread and varied phenomenon. Bot Rev 65: 348–369 [Google Scholar]
- Hofer J, Ellis N (2002) Conservation and diversification of gene function in plant development. Curr Opin Plant Biol 5: 56–61 [DOI] [PubMed] [Google Scholar]
- Hofer J, Turner L, Hellens R, Ambrose M, Matthews P, Michael A, Ellis N (1997) UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr Biol 7: 581–587 [DOI] [PubMed] [Google Scholar]
- Ingram GC, Goodrich J, Wilkinson MD, Simon R, Haughn GW, Coen ES (1995) Parallels between UNUSUAL FLORAL ORGANS and FIMBRIATA, genes controlling flower development in Arabidopsis and Antirrhinum. Plant Cell 7: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntheikki-Palovaara I, Tähtiharju S, Lan T, Broholm SK, Rijpkema AS, Ruonala R, Kale L, Albert VA, Teeri TH, Elomaa P (2014) Functional diversification of duplicated CYC2 clade genes in regulation of inflorescence development in Gerbera hybrida (Asteraceae). Plant J 79: 783–796 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kotilainen M, Elomaa P, Uimari A, Albert VA, Yu D, Teeri TH (2000) GRCD1, an AGL2-like MADS box gene, participates in the C function during stamen development in Gerbera hybrida. Plant Cell 12: 1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka J, Tokunaga H, Yoshida A (2014) Control of grass inflorescence form by the fine-tuning of meristem phase change. Curr Opin Plant Biol 17: 110–115 [DOI] [PubMed] [Google Scholar]
- Laitinen RAE, Broholm S, Albert VA, Teeri TH, Elomaa P (2006) Patterns of MADS-box gene expression mark flower-type development in Gerbera hybrida (Asteraceae). BMC Plant Biol 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Wolfe DS, Nilsson O, Weigel D (1997) A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr Biol 7: 95–104 [DOI] [PubMed] [Google Scholar]
- Lippman ZB, Cohen O, Alvarez JP, Abu-Abied M, Pekker I, Paran I, Eshed Y, Zamir D (2008) The making of a compound inflorescence in tomato and related nightshades. PLoS Biol 6: e288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Xi W, Shen L, Tan C, Yu H (2009) Regulation of floral patterning by flowering time genes. Dev Cell 16: 711–722 [DOI] [PubMed] [Google Scholar]
- Molinero-Rosales N, Jamilena M, Zurita S, Gómez P, Capel J, Lozano R (1999) FALSIFLORA, the tomato orthologue of FLORICAULA and LEAFY, controls flowering time and floral meristem identity. Plant J 20: 685–693 [DOI] [PubMed] [Google Scholar]
- Moyroud E, Kusters E, Monniaux M, Koes R, Parcy F (2010) LEAFY blossoms. Trends Plant Sci 15: 346–352 [DOI] [PubMed] [Google Scholar]
- Panero JL, Funk VA (2002) Toward a phylogenetic subfamial classification for the Compositae. Proc Biol Soc Wash 115: 909–922 [Google Scholar]
- Park SJ, Eshed Y, Lippman ZB (2014) Meristem maturation and inflorescence architecture: lessons from the Solanaceae. Curr Opin Plant Biol 17: 70–77 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozner R, Zanotti C, Johnson LA (2012) Evolutionary origin of the Asteraceae capitulum: insights from Calyceraceae. Am J Bot 99: 1–13 [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E (2007) Evolution and development of inflorescence architectures. Science 316: 1452–1456 [DOI] [PubMed] [Google Scholar]
- Rebocho AB, Bliek M, Kusters E, Castel R, Procissi A, Roobeek I, Souer E, Koes R (2008) Role of EVERGREEN in the development of the cymose petunia inflorescence. Dev Cell 15: 437–447 [DOI] [PubMed] [Google Scholar]
- Ruokolainen S, Ng YP, Albert VA, Elomaa P, Teeri TH (2010a) Large scale interaction analysis predicts that the Gerbera hybrida floral E function is provided both by general and specialized proteins. BMC Plant Biol 10: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruokolainen S, Ng YP, Broholm SK, Albert VA, Elomaa P, Teeri TH (2010b) Characterization of SQUAMOSA-like genes in Gerbera hybrida, including one involved in reproductive transition. BMC Plant Biol 10: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer E, Rebocho AB, Bliek M, Kusters E, de Bruin RAM, Koes R (2008) Patterning of inflorescences and flowers by the F-box protein DOUBLE TOP and the LEAFY homolog ABERRANT LEAF AND FLOWER of petunia. Plant Cell 20: 2033–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tähtiharju S, Rijpkema AS, Vetterli A, Albert VA, Teeri TH, Elomaa P (2012) Evolution and diversification of the CYC/TB1 gene family in Asteraceae: a comparative study in Gerbera (Mutisieae) and sunflower (Heliantheae). Mol Biol Evol 29: 1155–1166 [DOI] [PubMed] [Google Scholar]
- Teeri TH, Uimari A, Kotilainen M, Laitinen R, Help H, Elomaa P, Albert VA (2006) Reproductive meristem fates in Gerbera. J Exp Bot 57: 3445–3455 [DOI] [PubMed] [Google Scholar]
- Teo ZW, Song S, Wang YQ, Liu J, Yu H (2014) New insights into the regulation of inflorescence architecture. Trends Plant Sci 19: 158–165 [DOI] [PubMed] [Google Scholar]
- Uimari A, Kotilainen M, Elomaa P, Yu D, Albert VA, Teeri TH (2004) Integration of reproductive meristem fates by a SEPALLATA-like MADS-box gene. Proc Natl Acad Sci USA 101: 15817–15822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Weigel D, Nilsson O (1995) A developmental switch sufficient for flower initiation in diverse plants. Nature 377: 495–500 [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Wu MF, Winter CM, Berns MC, Nole-Wilson S, Yamaguchi A, Coupland G, Krizek BA, Wagner D (2013) A molecular framework for auxin-mediated initiation of flower primordia. Dev Cell 24: 271–282 [DOI] [PubMed] [Google Scholar]
- Yu D, Kotilainen M, Pöllänen E, Mehto M, Elomaa P, Helariutta Y, Albert VA, Teeri TH (1999) Organ identity genes and modified patterns of flower development in Gerbera hybrida (Asteraceae). Plant J 17: 51–62 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.