Photoperiod-H1 controls leaf size by influencing the duration of leaf growth in barley.
Abstract
Leaf size is a major determinant of plant photosynthetic activity and biomass; however, it is poorly understood how leaf size is genetically controlled in cereal crop plants like barley (Hordeum vulgare). We conducted a genome-wide association scan for flowering time, leaf width, and leaf length in a diverse panel of European winter cultivars grown in the field and genotyped with a single-nucleotide polymorphism array. The genome-wide association scan identified PHOTOPERIOD-H1 (Ppd-H1) as a candidate gene underlying the major quantitative trait loci for flowering time and leaf size in the barley population. Microscopic phenotyping of three independent introgression lines confirmed the effect of Ppd-H1 on leaf size. Differences in the duration of leaf growth and consequent variation in leaf cell number were responsible for the leaf size differences between the Ppd-H1 variants. The Ppd-H1-dependent induction of the BARLEY MADS BOX genes BM3 and BM8 in the leaf correlated with reductions in leaf size and leaf number. Our results indicate that leaf size is controlled by the Ppd-H1- and photoperiod-dependent progression of plant development. The coordination of leaf growth with flowering may be part of a reproductive strategy to optimize resource allocation to the developing inflorescences and seeds.
Leaf size is a major determinant of plant photosynthetic activity and performance and contributes to yield in crops (Zhang et al., 2015). The leaf of cereal crops is strap shaped and organized in two main regions: the proximal sheath encloses the stem, while the distal blade projects out of the stem axis to optimize light interception and photosynthesis. The blade-sheath boundary is marked by the ligule and two auricles, epidermal structures that hold the stem. Leaf growth in grasses is initiated by the division of cells at the base of the leaf (Esau, 1977; Kemp, 1980a, 1980b). Leaves grow in a linear process as cells are displaced in parallel longitudinal files by the continuous production and expansion of cells (MacAdam et al., 1989). This creates a clearly defined spatial pattern of cell development along the longitudinal axis, with a basal division zone, where meristematic cells divide and elongate, and a distal elongation-only zone, where cells undergo postmitotic elongation (Skinner and Nelson, 1995). The location where cells stop expanding marks the end of the leaf growth zone and the initiation of the differentiation zone. Leaves are initiated at the flanks of the shoot apical meristem in a regular spatial pattern, so-called phyllotaxy, and the time interval between the emergence of two successive leaves on a culm is called phyllochron (Wilhelm and McMaster, 1995).
The final size of leaves is tightly controlled by genetic factors that coordinate cell proliferation and cell expansion. Mutant screens in rice (Oryza sativa) and maize (Zea mays) have identified a number of genes required for leaf development, for which loss-of-function mutations result in extreme mutant phenotypes (Scanlon, 2003; Chuck et al., 2007; Fujino et al., 2008; Qi et al., 2008; Zhang et al., 2009). Genes encoding transcription factors such as KNOTTED-LIKE HOMEOBOX factors, proteins involved in hormone biosynthesis and response, and microRNAs have been shown to play a role in leaf organogenesis across different species, including maize and the model dicot plant Arabidopsis (Arabidopsis thaliana; Hay and Tsiantis, 2010; González and Inzé, 2015; Sluis and Hake, 2015).
Large genetic variation in leaf size has been identified in natural populations of rice and maize (Tian et al., 2011; Li et al., 2012; Yang et al., 2015; Zhang et al., 2015). Quantitative trait locus (QTL) studies in both species revealed a complex genetic basis for leaf size variation (Peng et al., 2007; Farooq et al., 2010; Jiang et al., 2010; Tian et al., 2011; Wang et al., 2011). The majority of the identified QTLs did not coincide with any of the major known leaf development genes as identified in mutant screens (Scanlon, 2015). Consequently, different genes might underlie leaf size variation between and within species.
While natural differences in leaf size have been well characterized in rice and maize, natural variation in leaf size and its genetic basis are still poorly understood in temperate cereal crop plants such as barley (Hordeum vulgare ssp. vulgare). Barley is characterized by two major growth types, as determined by natural variation at the two vernalization genes Vrn-H1 and Vrn-H2 (Yan et al., 2003, 2004; Trevaskis et al., 2006). Winter types accelerate flowering after a prolonged period of cold (vernalization), whereas spring barley does not respond to vernalization. Winter barley usually shows a strong promotion of flowering in response to long days (LDs; Turner et al., 2005). The photoperiod response, or rapid flowering under LDs, is determined by natural variation of the PHOTOPERIOD-H1 (Ppd-H1) gene (Turner et al., 2005). The wild-type allele is prevalent in winter barley, while a natural mutation in the conserved CCT domain of Ppd-H1 causes a delay in flowering under LDs and is predominant in spring barley from cultivation areas with long growing seasons (Turner et al., 2005; von Korff et al., 2006, 2010; Jones et al., 2008; Wang et al., 2010). While the genetic basis of flowering time variation in response to vernalization and photoperiod is well characterized in barley, it is not known if variation in reproductive development affects leaf growth and size.
The aim of this study was to identify genomic regions and genes controlling natural variation in leaf size in a diverse collection of winter barley cultivars. By combining a genome-wide association scan (GWAS) analysis and detailed phenotyping of introgression lines (ILs), we establish a novel link between reproductive development and leaf size in barley.
RESULTS
Phenotypic Variation in the Field Experiments
To characterize natural variation in leaf size and its correlation to variation in reproductive development, we examined flowering date (FD), leaf width (LW), and leaf length (LL) in a diverse collection of winter barley cultivars grown in the field at two different locations in Italy and Iran (Table I). In both locations, large phenotypic variances were observed for FD, LW, and LL. In Italy, plants flowered between 202 and 230 d after sowing (DAS), with a mean of 209 DAS. In Iran, the number of days from sowing to flowering varied from a minimum of 175 DAS to a maximum of 192 DAS, with a mean of 181 DAS. LW was on average 17.8 mm in Italy, with a minimum of 12.7 mm and a maximum of 24.5 mm. In Iran, LW varied between 8.3 and 19.3 mm, with an average of 13 mm. LL, scored only in Iran, varied between 130 and 236 mm, with a mean of 177 mm.
Table I. Mean, minimum, maximum, and heritability of FD, LL, and LW scored in Italy and Iran.
h2, Heritability; n.d., not determined.
FD, LL, and LW showed high heritability values of 89%, 96%, and 82%, respectively. ANOVA demonstrated that the genotype accounted for 82%, 80%, and 31% of the total phenotypic variance for FD, LL, and LW, respectively (Supplemental Table S1). Consequently, the genetic components accounted for a large proportion of the total phenotypic variation for each trait. Positive correlations were found between FD and LW (0.32; P = 0.0001) and between FD and LL (0.34; P = 0.0001). A correlation coefficient of 0.77 (P < 2 × 10−16) was observed between LW and LL. Taken together, our analysis revealed a high genetic variation for leaf size parameters, and these were positively correlated with FD across both locations.
Population Structure, Linkage Disequilibrium, and GWAS
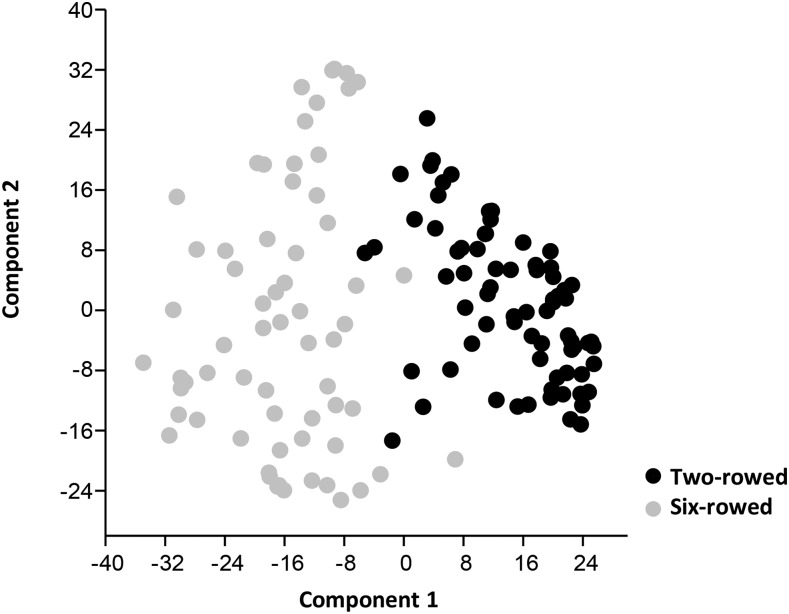
To identify the genetic basis of leaf size variation in the winter barley cultivar collection, we analyzed population structure and performed a genome-wide association study with 2,532 iSELECT single-nucleotide polymorphisms (SNPs) and three diagnostic markers in Vrn-H1, Vrn-H2, and Ppd-H1. Principal component analysis indicated the existence of two major subpopulations, which separated the two-rowed and six-rowed barley cultivars (Fig. 1). The two-rowed barley cultivars showed a higher genetic diversity, with a mean correlation coefficient of 0.39, compared with the six-row barley genotypes, with a mean genetic correlation coefficient of 0.48.
Figure 1.
Principal component analysis plot of 138 barley cultivars based on the first two principal axes (component 1 = 12% and component 2 = 8%). Two-rowed barley cultivars are indicated in black, and six-rowed cultivars are indicated in gray.
In order to verify the growth habit of cultivars in our germplasm set, all lines were genotyped with diagnostic markers for Vrn-H1, Vrn-H2, and Ppd-H1 (Supplemental Table S2). The germplasm set revealed three different Vrn-H1 haplotypes. The majority of cultivars were characterized by winter Vrn-H1 alleles, with 117 cultivars (56 six-rowed and 61 two-rowed) carrying the full-length W-1A allele and 14 cultivars (12 six-rowed and two two-rowed) carrying the winter allele W-5C, which is characterized by a deletion of 486 bp in the first intron (Cockram et al., 2009). Seven cultivars (five six-rowed and two two-rowed) were characterized by the spring Vrn-H1 allele (Cockram et al., 2009). A full deletion of the Vrn-H2 locus, which is typical for spring barley, was identified in seven of the 138 cultivars, including five carrying a winter Vrn-H1 allele. The Vrn-H1 and Vrn-H2 spring alleles had a low frequency but were distributed equally between the two-rowed and six-rowed varieties. Consequently, seven of the 138 genotypes were characterized as spring types, while five genotypes were identified as facultative cultivars, which are characterized by a deletion of Vrn-H2 and the winter allele at Vrn-H1 (Supplemental Fig. S1A; von Zitzewitz et al., 2005). Genotyping with the diagnostic marker in the CCT domain of Ppd-H1 showed that the mutated ppd-H1 allele was present in approximately 25% of the winter barley lines and was detected preferentially in two-rowed genotypes (Supplemental Table S2). However, barley genotypes with Ppd-H1 or ppd-H1 haplotypes did not form separate clusters in the principal component analysis (Supplemental Fig. S1B). In summary, only a low number of genotypes, two of 138, carried spring alleles at Vrn-H1 and Vrn-H2, while 25% of the germplasm set was characterized by a mutated ppd-H1 allele that has been associated with late flowering under LDs (Turner et al., 2005).
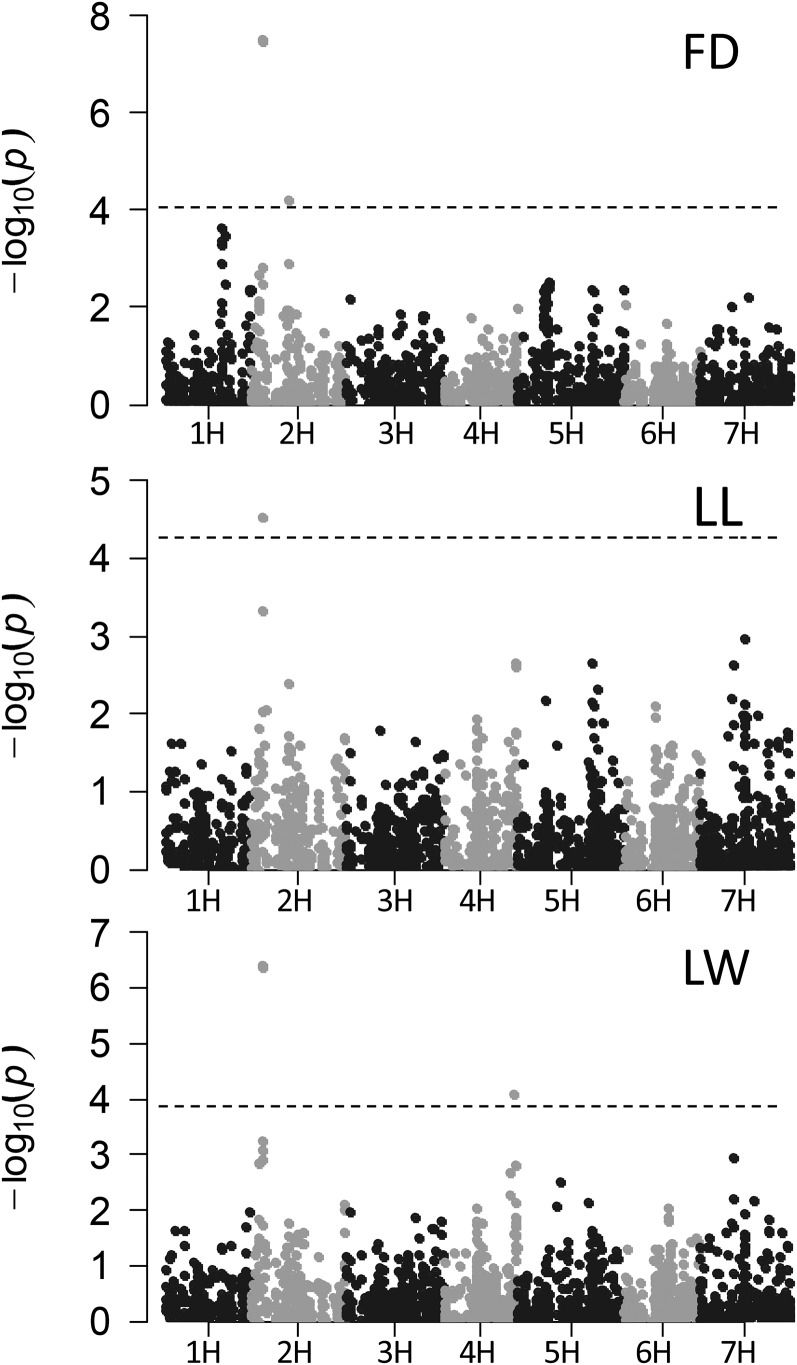
The average linkage disequilibrium decay in the population was determined at 5.5 centimorgan (cM) based on the r2 between all intrachromosomal pairs of loci (Supplemental Fig. S2). The GWAS across both locations and for each location separately revealed two significant genomic regions on chromosome 2HS associated with FD, one region on 2HS for LL, and two genomic regions on 2HS and 4HL for LW (Fig. 2; Table II; Supplemental Fig. S3). On chromosome 2HS, seven linked SNP markers at position 19.9 cM on the POPSEQ reference map (Mascher et al., 2013) were associated with FD, LW, and LL (Table II): SNP22, BK_12, BK_14, BK_15, BK_16, BOPA2_12_30871, and BOPA2_12_30872 designed on the genic sequence of Ppd-H1. All seven of these markers were in complete linkage disequilibrium and included the diagnostic marker SNP22, which was proposed by Turner et al. (2005) to be responsible for flowering time variation under LDs (Supplemental Table S2).
Figure 2.
Manhattan plots of GWAS for FD, LL, and LW in the barley cultivar collection. The −log10 (P values) from the association scans are plotted against the SNP marker positions on each of the seven barley chromosomes. The dashed horizontal lines indicate the genome-wide significance threshold at a false discovery rate < 0.05.
Table II. Summary of significant marker-trait associations identified by GWAS.
Chr, Chromosome arm; MAF, minimum allele frequency; n.s., not significant.
| Locus | Candidate Gene | Chr | Position | MAF |
FD |
LL |
LW |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −log10 | Δma | Effectb | −log10 | Δma | Effectb | −log10 | Δma | Effectb | |||||
| cM | P | % | d | P | % | mm | P | % | mm | ||||
| SNP22c | Ppd-H1 | 2HS | 19.9 | 0.27 | 7.5 | 38 | 2.1 | 4.5 | 39 | 8 | 6.4 | 25 | 1 |
| BOPA1_ConsensusGBS0008-1 | HvCEN | 2HS | 58.78 | 0.19 | 4.2 | 27 | 1.7 | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. |
| SCRI_RS_157866 | 4HL | 110.2 | 0.36 | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | 4.1 | 8 | 0.7 | |
Difference between r2 of the model with and without the marker.
Effect of a minor allele.
Marker designed on the Ppd-H1 gene, cosegregates with BK_12, BK_14, BK_15, BK_16, BOPA1_12_30871, and BOPA_12_30872.
Genetic variation at Ppd-H1 was associated with an average difference of 2 d in flowering time in the winter barley population. In addition, genetic variation at Ppd-H1 caused an estimated variation of 8 mm in LL and of 1 mm in LW (Table II). Variation at Ppd-H1 accounted for 23%, 6%, and 5% of the genetic variation for FD, LL, and LW, respectively (Supplemental Table S1). A second association on 2HS, at position 58.78 cM close to the CENTRORADIALIS (HvCEN)/EPS2 locus (Comadran et al., 2012), was identified for FD but not for LL or LW. Furthermore, for LW, a significant association was found with the marker SCRI_RS_157866 at 110.2 cM on chromosome 4H, which caused an average difference in LW of 0.7 mm. Variation at Vrn-H1 and Vrn-H2 was not associated with FD, LL, or LW. In summary, variation at the photoperiod response gene Ppd-H1 exhibited pleiotropic effects on FD, LL, and LW in our barley germplasm collection under different field conditions.
Since a flowering time gene was colocated with a QTL for leaf size, we recalculated the GWAS with FD as a covariate to test if leaf size was controlled primarily by overall plant development (Supplemental Fig. S4). LW was still significantly influenced by the Ppd-H1 locus and the marker SCRI_RS_157866 at 110.2 cM on chromosome 4H. However, the associations for LL fell below the significance threshold, possibly because LL was measured only in one environment.
Variation for Leaf Size in Ppd-H1 ILs
In order to further dissect the effect of natural variation at Ppd-H1 on leaf size, LL and LW were analyzed under LDs in three pairs of spring barley cultivars and their respective ILs (Supplemental Fig. S5). The spring barley genotypes and ILs differed for the SNPs associated with variation in FD, LW, and LL in the barley germplasm collection: Scarlett, Triumph, and Bowman were homozygous ppd-H1, while their respective ILs were homozygous Ppd-H1 (Supplemental Table S3).
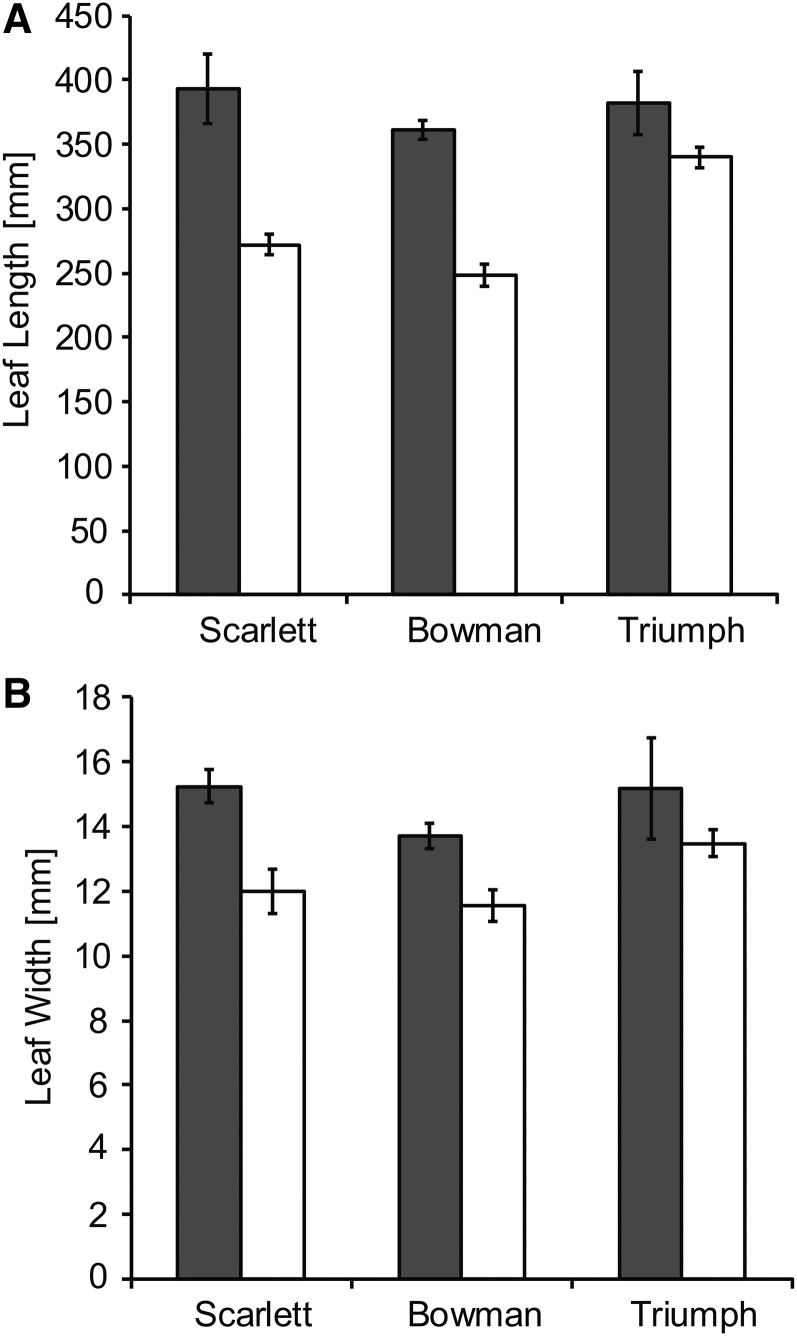
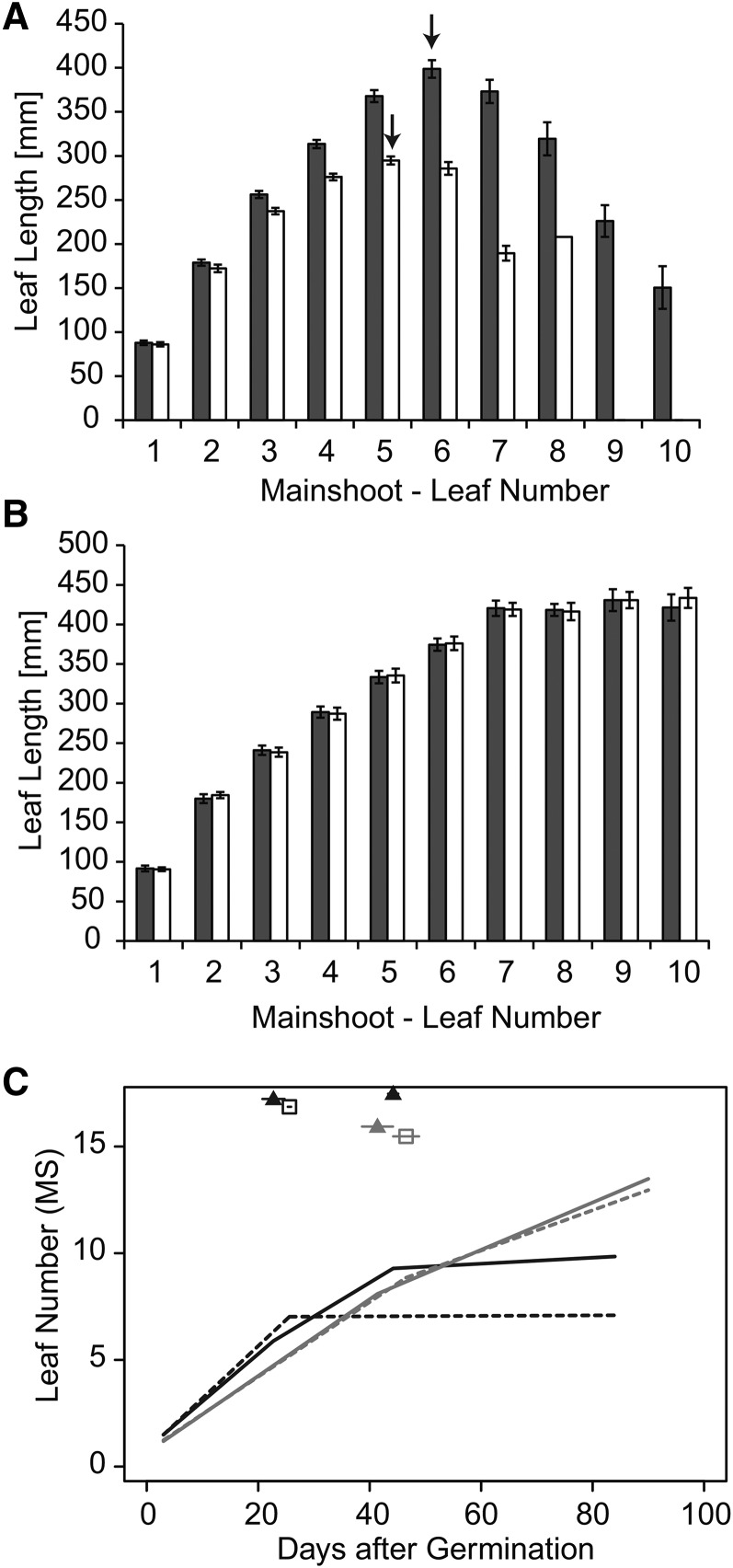
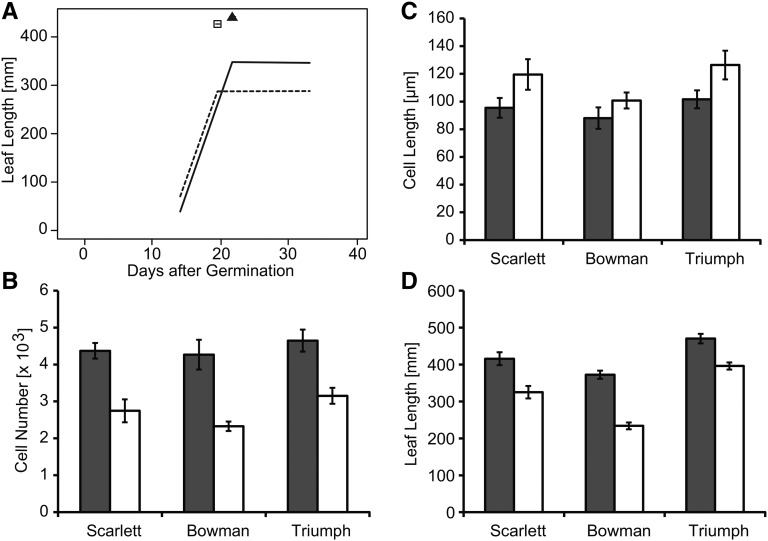
The spring barley genotypes flowered significantly later than their respective ILs under LDs (Supplemental Fig. S6). In addition, the spring barley cultivars exhibited significant increases in LW and LL of the largest leaf compared with their respective ILs under LDs (Fig. 3). Genetic differences in LL varied from an average of 121 mm between Scarlett and S42-IL107 to 113 mm between Bowman and BW281 and 42 mm between Triumph and Triumph-IL. Genetic differences in LW varied from an average of 3.2 mm between Scarlett and S42-IL107 to 2.2 mm between Bowman and BW281 and 1.7 mm between Triumph and Triumph-IL. Differences in LL were larger and less variable between the ILs and recurrent parents; therefore, we concentrated our further analyses on the effect of Ppd-H1 on LL and leaf elongation rates (LERs). Leaf blade size was dependent on the leaf position on the main shoot in all genotypes. Successive leaves exhibited a continuous increase in LL, with each node producing a leaf of gradually increasing length until a plateau was reached and, under LD conditions, leaf blade size declined again (Fig. 4, A and B; Supplemental Fig. S7, A–D). Under LD conditions, the maximum leaf size was reached with leaves 5 and 6 in the spring barley cultivars and with leaves 4 and 5 in the ILs and, thus, was dependent on the total number of leaves per main culm (eight to 11 in spring barleys versus seven to nine in ILs). Leaf blades were significantly longer in the spring barley cultivars as compared with their respective ILs, starting from the third developing leaf in Scarlett/S42-IL107 and Bowman/BW281 and starting from the fifth or sixth leaf in Triumph/Triumph-IL under LDs (Fig. 4A; Supplemental Fig. S7, A and C).
Figure 3.
The size of the leaf blade is increased in spring barley lines with the mutated ppd-H1 allele. Maximal leaf blade length (A) and leaf blade width (B) are shown for the largest leaf in spring barley lines with a mutated ppd-H1 allele (gray bars) and the derived ILs (white bars) carrying the dominant Ppd-H1 under LDs. Bars represent means with 95% confidence intervals.
Figure 4.
Leaf blade length and phyllochron of Scarlett and S42-IL107 grown under different photoperiods. A and B, Leaves emerging from the main shoot in Scarlett (gray bars) and S42-IL107 (white bars) under LD (A) and SD (B) conditions. Arrows indicate the longest leaf for Scarlett and S42-IL107. Bars represent means with 95% confidence intervals. C, Number of leaves emerging on the main shoot (MS) per time unit after germination in Scarlett (solid lines, triangles) and S42-IL107 (dashed lines, squares) under LDs (black) and SDs (gray). Breakpoints of the regression model are indicated for the different genotypes and conditions above the regression curves with their 95% confidence intervals.
To further support our findings that variation at Ppd-H1 controls leaf shape, we also examined LL under short days (SDs). Since Ppd-H1 is only functional and controls development under LDs, we predicted that the ILs would not differ from their parental lines in leaf shape under SDs. Under SDs, plants of all six genotypes did not flower, as the inflorescences of the main shoots were aborted before reaching the flowering stage, and leaf emergence stopped before the flag leaf became visible (data not shown). In general, under SDs, more leaves were produced on the main culm as compared with LDs. However, as expected for the LD-dependent function of Ppd-H1, no differences in leaf size were detected between ILs and the respective recurrent parents under SDs (Fig. 4B; Supplemental Fig. S7, B and D).
In order to further understand how Ppd-H1 affects leaf growth and size, we examined the phyllochron in the spring barley lines and ILs. Under LDs, the spring barley genotypes showed an increased phyllochron compared with their respective ILs. A difference in phyllochron was observed for all leaves in S42-IL107/Scarlett and for leaves 4 to 8 in BW281/Bowman and leaves 6 to 11 in Triumph-IL/Triumph (Fig. 4C; Table III; Supplemental Fig. S7, E and F; Supplemental Table S4). The rate of leaf emergence, and thus phyllochron, was constant for all leaves in the three ILs under LDs. In contrast, the phyllochron was dependent on the position of the leaf on the main shoot in the spring barley genotypes under LDs and in both the spring barley cultivars and the ILs under SDs (Table III; Supplemental Table S4).
Table III. Variation at Ppd-H1 affects the phyllochron of Scarlett and S42-IL107.
| Photoperiod | Genotype | Leaf No. | Phyllochrona | 95% Confidence Interval |
|---|---|---|---|---|
| d | ||||
| LD | Scarlett | 1–6 | 4.5 | 4.3–4.7 |
| 7–10 | 6.3 | 6.0–6.6 | ||
| S42-IL107 | 1–8 | 4.1 | 4.0–4.2 | |
| SD | Scarlett | 1–9 | 5.7 | 5.5–5.8 |
| 10–14 | 9.0 | 8.6–9.4 | ||
| S42-IL107 | 1–9 | 5.7 | 5.6–5.9 | |
| 10–14 | 10.6 | 9.9–11.2 |
Phyllochron was calculated as the leaf emergence rate from the slopes of the linear segments of the regression lines presented in Figure 4C.
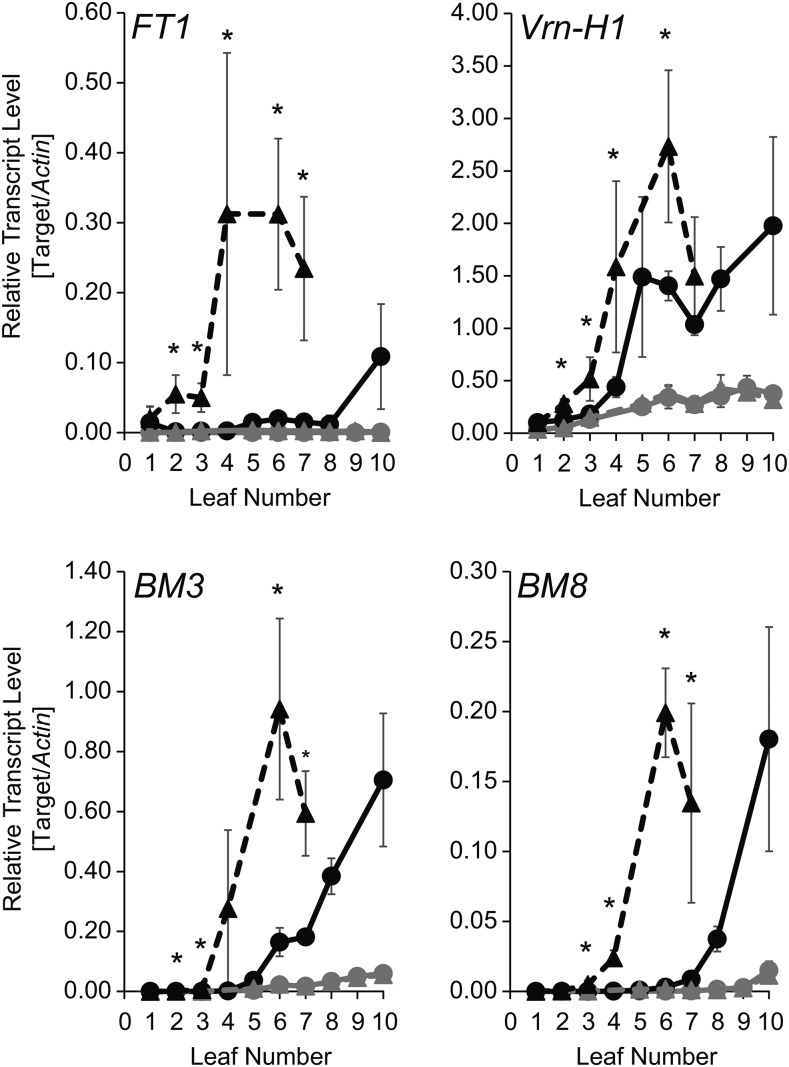
In order to examine whether an increased phyllochron was associated with a decreased LER or a delayed termination of leaf elongation (i.e. leaf growth), we scored LER in Scarlett and S42-IL107 of all leaves on the main shoot under LDs and SDs (Fig. 5A; Supplemental Fig. S8; Supplemental Table S5). Genetic variation between Scarlett and the IL did not affect LER, but the duration of leaf growth as exemplified for leaf 5 in Figure 5A. Under LDs, leaf growth terminated earlier in S42-IL107 than in Scarlett, whereas under SDs, no significant difference in the duration of leaf growth was observed between genotypes. However, leaf growth stopped earlier under LDs than SDs, as observed for leaf 8 (Supplemental Fig. S8). Under LDs, the delayed termination of leaf growth in the spring barley genotypes with the mutated ppd-H1 allele was associated with a reduced interstomatal cell size and an increased number of cells per leaf along the proximal/distal axis (Fig. 5, B–D). Under SDs, leaf cell size was not affected by allelic variation at Ppd-H1 (Supplemental Fig. S9). This suggested that the effect of Ppd-H1 on leaf size is based on differences in cell proliferation and possibly linked to its effects on floral transition and/or inflorescence development. Consistent with this view, we detected differences in the expression of the floral integrator homologs and targets of Ppd-H1 in the leaf, FLOWERING LOCUS1 (FT1), and the AP1/FUL-like genes BARLEY MADS BOX3 (BM3), BM8, and Vrn-H1 (Fig. 6). FT1 expression increased with increasing leaf number under LDs and was significantly higher in the leaves of the ILs than in the spring barley lines. While FT1 expression was higher in the IL in all leaves harvested during development compared with Scarlett, expression of the AP1/FUL-like genes BM3, BM8, and Vrn-H1 was up-regulated in the IL starting from leaf 2 or 3, when also the leaf size differences between genotypes became apparent.
Figure 5.
Ppd-H1 affects the duration of leaf elongation, cell number, cell size, and LL under LD conditions. A, LERs were determined by measuring the LL every 3 d under LDs in Scarlett (solid line, triangle) and S42-IL107 (dashed line, square) in the fifth leaf of the main shoot. Breakpoints of the regression model and their 95% confidence intervals are indicated for the different genotypes and conditions above the regression curves. B to D, Cell length (B), estimated number of interstomatal cells (C), and final LL (D) of the fifth fully expanded leaf on the main shoot. Barley genotypes with the mutated ppd-H1 allele and ILs with the photoperiod-responsive Ppd-H1 allele are represented by gray and white bars, respectively. Bars represent means with 95% confidence intervals (n = 5).
Figure 6.
Expression patterns of FT1 and AP1/FUL-like MADS box transcription factors in successive barley leaves. Quantification of gene expression levels by quantitative real-time (qRT)-PCR is shown for leaf samples harvested from successive leaves emerging from the main shoot of Scarlett (circle) and S42-IL107 (triangle) plants grown under SD (gray) and LD (black) conditions. Expression levels are demonstrated relative to the transcript abundance of the ACTIN housekeeping gene. Error bars represent sd over three biological and two technical replicates. Asterisks indicate significant differences (P < 0.05) in transcript abundance between S42-IL107 and Scarlett when plants were grown under LD conditions.
Taken together, allelic variation at Ppd-H1 had significant effects on LW, and in particular on LL, under LDs but not under SDs. Variation at Ppd-H1 affected phyllochron and leaf elongation ceased later in Scarlett than in the IL. In contrast, LER was not affected by variation at Ppd-H1. Differences in cell number along the proximal/distal leaf axis between genotypes were only partially compensated by the increased cell length of the ILs with the photoperiod-responsive Ppd-H1 allele. Thus, variation at Ppd-H1 affected LL by affecting the cell number, cell size, and duration of leaf blade elongation. In addition, leaf size differences correlated with the Ppd-H1-dependent expression difference of BM-like genes in the leaf.
DISCUSSION
Leaf size is an important agronomic trait, as it relates to radiation use efficiency and transpiration rate, directly affecting photosynthesis and response to water limitations. Variation in these traits is of key importance for winter barley varieties cultivated in Mediterranean areas with terminal drought. The first goal of this study was to explore natural genetic variation for leaf size in winter barley and identify genomic regions associated with leaf size variation in the field through an association mapping approach.
Most association studies in barley were carried out on panels of spring accessions or mixed panels of winter and spring accessions (Rostoks et al., 2006; Cockram et al., 2008; Stracke et al., 2009; Pasam et al., 2012; Tondelli et al., 2013). In this study, we used an autumn-sown panel of winter cultivars where two-rowed and six-rowed types were equally represented, as reflected by the analysis of population structure. A strong genetic differentiation between two- and six-rowed barley genotypes has already been detected in other studies (Rostoks et al., 2006; Cockram et al., 2010; Comadran et al., 2012; Pasam et al., 2012; Muñoz-Amatriaín et al., 2014) and derives from modern breeding practices: contemporary European spring and winter varieties descend from a small number of successful European landraces selected around 100 years ago (Fischbeck, 2003). In our panel, the two-rowed barley subgroup showed a relatively higher genetic diversity compared with the six-rowed group. This may be due to the use of spring two-rowed varieties in the breeding of winter two-rowed varieties potentially increasing genetic diversity (Fischbeck, 2003). Based on genotyping of Vrn-H1 and Vrn-H2, seven (five six-rowed and two two-rowed cultivars) of the 138 genotypes were characterized as spring types, while five genotypes were identified as facultative cultivars, which are characterized by a deletion of Vrn-H2 and the winter allele at Vrn-H1 (von Zitzewitz et al., 2005). However, these cultivars clearly clustered with winter barley genotypes in a principal component analysis of a comprehensive panel of spring and winter varieties (data not shown). A reduced vernalization response in winter barley might offer advantages in autumn-sowing areas with mild winters, which are not as cold as those in more northern or continental climates (Casao et al., 2011). The mutated ppd-H1 allele was present in approximately 25% of the winter barley lines and was preferentially detected in two-rowed genotypes used for malt production. This might be a consequence of the introgression of malting-related traits from spring genotypes into winter cultivars. Whether the mutation in Ppd-H1 has an impact on grain characteristics and, ultimately, on malting quality has yet to be demonstrated.
The association analysis demonstrated that, under vernalizing conditions, the Ppd-H1 locus had the strongest effect on flowering time in our barley germplasm panel, with the recessive ppd-H1 delaying flowering time and causing an increase in leaf size. While variation at the Ppd-H1 locus affected both flowering time and leaf size, a second locus on 2H harboring the floral repressor HvCEN (Comadran et al., 2012) was only associated with flowering time but not leaf size. These results suggested that Ppd-H1, but not HvCEN, influenced leaf size, possibly because these genes act at different developmental stages and in different tissues. While Ppd-H1 is expressed primarily in the leaf, where it controls photoperiod-dependent flowering, TERMINAL FLOWER1, the Arabidopsis homolog of HvCEN, acts as a repressor of floral development in the shoot apical meristem (Bradley et al., 1997; Ohshima et al., 1997).
The effects of the Ppd-H1 locus on leaf size were confirmed in three pairs of ILs, where the ppd-H1 allele increased leaf blade size under LDs but not SDs. Therefore, the effect of Ppd-H1 on leaf growth was correlated with its LD-specific effect on reproductive development. Differences in leaf size were already evident at the floral transition (three-leaf stage in ILs), suggesting that processes linked to phase transition affected leaf size. Ontogenetic changes in leaf size and morphology (heteroblasty) have been associated with the transition from vegetative to reproductive development (Goebel, 1900; Jones, 1999). In Arabidopsis and grasses such as maize and rice, early flowering correlates with a reduction in leaf size, suggesting that the fate of existing leaf primordia is changed with the transition to flowering (Poethig, 1990, 2010; Hempel and Feldman, 1994). The microRNAs miR156 and miR157 and their direct targets, the SQUAMOSA PROMOTER-BINDING PROTEIN family of transcription factors, have been identified as major regulators of vegetative phase change in a range of plants (Wu and Poethig, 2006, Chuck et al., 2007, Fu et al., 2012; Xie et al., 2012). A decrease in the expression of miR156 is linked to the juvenile-to-adult transition and heteroblastic change in leaf shape, resulting in two or multiple, discrete leaf types. However, in our experiments, Ppd-H1-dependent differences in leaf size were cumulative and increased with leaf number on the main stem. While Ppd-H1 does not have a strong effect on the vegetative-to-reproductive phase change, it strongly controls inflorescence development and stem elongation (Digel et al., 2015). Differences in the rate of leaf emergence, in the duration of leaf growth, and in the final leaf cell number suggested that Ppd-H1 affected leaf size by influencing the rate of age-dependent progression of leaf development. Pleiotropic effects of flowering time regulators might be a consequence of changes in source-sink relationships triggered by the transition from vegetative to reproductive growth or inflorescence growth. On the other hand, flowering time genes may play dual roles to control and coordinate leaf development and phase transitions at the shoot apical meristem, as indicated by studies in dicots. For example, a recent study demonstrated that natural variation of leaf shape in Cardamine hirsuta is controlled by the vernalization gene FLOWERING LOCUS C, which is known to contribute to variation in flowering time in the Brassicaceae (Cartolano et al., 2015). In addition, SINGLE FLOWER TRUSS, the tomato (Solanum lycopersicum) ortholog of FT, affected flowering time, leaf maturation, and compound leaf complexity (Lifschitz et al., 2006; Shalit et al., 2009). These studies focused on species with compound leaves, whose shape and size arise from spatiotemporal regulation of morphogenetic activity within the leaf primordium, as determined by prolonged expression of meristem-related gene functions (Sluis and Hake, 2015). However, to our knowledge, no link between specific flowering time regulators and leaf size had been reported in simple-leaf grasses.
Interestingly, in our study, the expression of FT1, and the putative downstream targets Vrn-H1, BM3, and BM8 in the leaf, negatively correlated with the duration of leaf growth and final leaf blade size. In particular, the AP1/FUL-like genes BM3 and BM8 were up-regulated in the IL (Ppd-H1) compared with Scarlett (ppd-H1) starting from leaf 3, when leaf size differences between genotypes also became apparent. In Arabidopsis, FT and its targets, the AP1/FUL MADS box genes, are best known for their role in the floral transition and floral development (Turck et al., 2008). However, studies in Arabidopsis have suggested that FT and the downstream AP1/FUL targets also control leaf size and shape (Teper-Bamnolker and Samach, 2005). Indeed, FT restricted leaf size via up-regulation of the MADS box gene FUL, and a ful loss-of-function mutation suppressed leaf size reduction caused by FT overexpression (Teper-Bamnolker and Samach, 2005). Similarly, variations in BM expression correlated with the genetic differences in the timing of leaf development, suggesting that the duration of cell proliferation and leaf maturation was controlled by Ppd-H1 and possibly downstream variation in the expression of FT1 and BM genes. To our knowledge, our work provides the first link between specific flowering time genes and the regulation of leaf growth in grasses, suggesting that monocots and dicots may share common genetic modules to coordinate leaf development and reproductive timing. This information will be important for the targeted manipulation and optimization of individual plant organs in plant breeding programs.
MATERIALS AND METHODS
Plant Materials and Phenotyping
A panel of 138 European winter barley (Hordeum vulgare) cultivars (65 two-rowed and 73 six-rowed) released between 1921 and 2006 (Supplemental Table S2) was evaluated at two experimental field stations in Fiorenzuola d’Arda, Piacenza, Italy (44°55′N and 9°53′E), and at the University of Shiraz, Iran (29°50′N and 52°46′E), during the growing season 2012-2013. The experimental fields were organized in a randomized complete block design with three replicates; each plot consisted of four rows of 2 m, with 40 cm spacing between rows and 30 cm between plants within a row. Seeds were sown in mid-October and the beginning of November 2012 in Italy and Iran, respectively.
FD was recorded when 60% of spikes were at the anthesis stage (Zadoks stage 68; Zadoks et al., 1974). LW and LL were measured on three to five mature plants per plot. For each plant, the longest and widest leaf of a culm was measured for a total of three to five culms per plant. LW was measured at the widest point of the blade, and LL was measured from the ligule to the tip of the blade. In the experiment in Italy, only LW was measured, while in Iran, both LW and LL were scored.
In order to confirm the effects of the major association with leaf size parameters in the candidate gene Ppd-H1, we analyzed three pairs of barley near isogenic lines: spring cultivars with a mutated ppd-H1 allele and derived backcross lines carrying introgressions of the dominant Ppd-H1 allele. These spring barley genotypes were Scarlett, Bowman, and Triumph, and the derived ILs were S42-IL107, BW281, and Triumph-IL, respectively. S42-IL107 and BW281 carry introgressions of the dominant Ppd-H1 allele from wild barley (Schmalenbach et al., 2008; Druka et al., 2011). Triumph-IL is a BC4F2-selected IL derived from the double haploid population of a cross between Triumph and the winter barley cv Igri (Laurie et al., 1995) and was kindly provided by David Laurie (John Innes Centre). The size of the introgression segments was determined by high-resolution genotyping using the Barley Oligo Pool arrays (Illumina Golden Gate; Druka et al., 2011; Schmalenbach et al., 2011; Digel et al., 2015).
LL and LW of the largest leaf on the main shoot were scored in the three pairs of spring barley and ILs at full expansion by measuring the distance from the ligule to the tip of the fully elongated leaf blade and the widest point of the leaf, respectively, in 10 to 30 replicated plants grown under LDs. In addition, plants of Scarlett/S42-IL107 were germinated in two independent experiments under SD conditions (8 h of light/16 h of dark, PAR of 270 µmol m−2 s−1, and 22°C/18°C) in a growth room, with 40 and 12 pots per genotype in the first and second experiment, respectively. After germination, half of the pots per genotype were transferred to LD conditions (16 h of light/8 h of dark, PAR of 270 µmol m−2 s−1, and 22°C/18°C) or cultivation was continued under SDs. The number of leaves emerged from the main shoot (leaf emergence [LEM]) was recorded every 2 to 3 d under LDs and every 3 to 4 d under SDs. Leaves were scored as fully emerged as soon as the ligule was visible. If a leaf was not fully emerged, it was scored as 0.25, 0.5, or 0.75 relative to the length of the fully emerged leaf blade of the preceding leaf. In addition, LL was determined for each leaf on the main culm in Scarlett/S42-IL107. The experiment was repeated for all six genotypes, scoring heading date, LEM, and LL of five replicated plants under SDs and LDs.
In parallel to scoring LEM, three plants per genotype were dissected and the developmental stage of the main shoot apex was determined according to the scale of Waddington et al. (1983), which is based on the morphogenesis of the shoot apex and carpels.
Phyllochron was calculated from the rate of LEM on the main culms as the inverse of the leaf emergence rate (i.e. the slope of the linear segments obtained for LEM from the regression models; see “Statistical Analyses: Phenotype”).
Furthermore, LER in Scarlett/S42-IL107 was obtained from one of the experiments. During leaf blade emergence from the leaf sheath (i.e. the ligule of the emerging leaf was not yet visible), LER was determined by measuring the length from the ligule of the preceding leaf to the leaf tip of the emerging leaf every 2 to 3 d under LDs and every 3 to 4 d under SDs. After leaf blade emergence, the measurement was continued from the ligule to the tip of the expanding leaf blade.
To determine leaf cell number and leaf cell size, five plants per genotype were germinated and grown under SDs and LDs, respectively. The cell length of 50 interstomatal cells of leaf 5 was determined at 33% and 66% of the total LL, as described by Wenzel (1997). Copies of the adaxial epidermal cell layer were transferred to microscopy slides by applying a solution of cellulose acetate (5% in acetone) to the leaf surface and transferring the solidified cellulose layer to the slides using transparent duct tape. Cell length was determined on the copied epidermal surface using a light microscope (Nikon SMZ18). The number of interstomatal cells per leaf was estimated as LL (mm) × 1,000/cell length (µm).
RNA Extraction, cDNA Synthesis, and qRT-PCR
For Scarlett and S42-IL107, we harvested leaf samples from every leaf on the main shoot at the time of their complete emergence from the leaf sheath to analyze the expression of FT1, Vrn-H1, BM3, and BM8 by qRT-PCR relative to the expression of the housekeeping gene ACTIN. Extraction of total RNA, reverse transcription, and qRT-PCR on cDNA samples using gene-specific primer pairs as listed in Supplemental Table S6 were performed as described by Campoli et al. (2012a, 2012b). To estimate the concentrations of target transcripts in the cDNA samples, dilution series of plasmids containing the respective target gene amplicons also were subjected to qRT-PCR analysis. qRT-PCR assays were conducted on the LightCycler 480 (Roche; software version 1.5).
Genotyping
Genomic DNA was extracted from young leaves of the winter barley population using Qiagen DNeasy 96 or Tepnel Nucleoplex plant DNA extraction kits according to the manufacturers’ instructions (Qiagen or Tepnel Life Sciences).
Genotyping was carried out at TraitGenetics using a set of 7,864 high-confidence gene-based SNPs incorporated in the Illumina iSELECT Chip (Comadran et al., 2012). Genotype calling was performed as described by Comadran et al. (2012). A total of 6,810 SNPs were successfully assayed in the 138 winter barley genotypes. Filtering was carried out to include only 4,257 markers positioned in the POPSEQ map (Mascher et al., 2013) and to exclude SNPs with more than 5% missing data or less than 10% MAF. Finally, a total of 2,532 iSELECT SNPs that were mapped on the POPSEQ reference map (Mascher et al., 2013) were employed for all the following analyses.
Among them, SNP markers BK_12, BK_14, BK_15, BK_16, BOPA2_12_30871, and BOPA2_12_30872 are located within the Ppd-H1 genic sequence (Supplemental Table S2). The population also was genotyped for functional variation at the two vernalization genes Vrn-H1 and Vrn-H2 and at Ppd-H1 using diagnostic markers as published by Cockram et al. (2009), Karsai et al. (2005), and Turner et al. (2005), respectively (Supplemental Table S6). Functional variation at VRN-H1, VRN-H2, and Ppd-H1 was tested for association with trait variation without filtering for MAF together with the SNP panel.
Statistical Analyses
Phenotype
All statistical analyses were performed using the R software version 3.1.1 (R Development Core Team, 2008). Variance components for FD, LW, and LL including genotypes, replicates, and locations (except for LL) as factors were calculated with a mixed linear model implemented by the lmer function from the lme4 package version 1.1.7, where genotypes, replicates, and locations were considered as random factors. Broad-sense heritability (h2) values were computed according to Knapp et al. (1985): h2 = σ2g/(σ2g + σ2lg/n + σ2e/n), where σ2g is the genetic variance, σ2lg is the genotype-by-location interaction variance, σ2e is the error variance, and n is the number of locations. For LL, analyzed only in one location, heritability was calculated as h2 = σ2g/(σ2g + σ2e). In addition, the lm function was used to conduct an ANOVA based on a linear model including locations (except for LL), genotypes, and replicates. Finally, we partitioned the genotype and genotype-by-location effects using Ppd-H1 marker alleles.
Best linear unbiased estimators (BLUEs) of FD, LW, and LL were calculated as the phenotypic values estimated for each genotype in a mixed linear model implemented by the lmer function, where genotypes were set as fixed factor and locations, location-by-genotype interactions, and replicates were considered as random factors (for LL, the random factors were reduced to the replicates). Pearson correlation analyses between FD and LW were calculated based on BLUEs across environments and replicates. For correlations between LL and FD or LW, only BLUEs across replicate measurements in Iran were calculated.
LEM and LER over time were calculated with a piece-wise regression in the segmented package (version 0.2-9.5; Muggeo, 2003, 2008) implemented in the R software. The Bayesian information criterion was used to decide on the number of breakpoints in the final regression model. Point estimates and 95% confidence intervals for the slopes of each linear segment in the selected regression models were extracted using the slope function of the segmented package.
Population Structure, Linkage Disequilibrium, and GWAS Analyses
The population structure of the panel was investigated by principal component analysis based on a correlation matrix derived from 2,532 iSELECT SNP markers using the Paleontological Statistical software (Hammer et al., 2001).
In order to identify the intrachromosomal linkage disequilibrium among markers, squared allele frequency correlations (r2) were calculated between pairs of loci using the TASSEL software (Bradbury et al., 2007). Linkage disequilibrium decay was evaluated by plotting significant (P < 0.001) pairwise r2 values against genetic distances between each pair of loci and by fitting the locally weighted scatterplot smoothing curve on the graph using the R software. A critical r2 value was estimated as the 95th percentile of r2 values between pairs of unlinked loci (pairs of loci on the same chromosome with greater than 50-cM distance).
GWAS was performed based on BLUEs across environments and replicates and based on BLUEs for individual environments across replicates with the GAPIT package version 2 (Lipka et al., 2012) implemented in the R software. To identify significant marker-trait associations, a mixed linear model described by the following formula was used: phenotype = M + Q + K + e, in which M and e denote the genotypes at the marker and residuals, respectively, Q is a fixed factor due to population structure, and K is a random factor due to the coancestry of individuals. Q was calculated as the first three components of the principal component analysis (Supplemental Fig. S10). The kinship matrix K represents similarities between genotypes and was calculated based on the proportion of allele mismatches at each SNP between pairs of genotypes in GAPIT with the method of VanRaden (2008). In a second mixed model, we used FD as a covariate to correct for flowering time-dependent changes in leaf size. The P values of genotype-phenotype associations were adjusted based on a false discovery rate (Benjamini and Hochberg, 1995) separately for each trait, and a threshold value for significant associations was set at 0.05. Manhattan plots displaying GWAS results were prepared with the qqman package (Turner, 2014) implemented in the R software.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Principal component analysis plot based on the first two principal axes, with spring and facultative genotypes and Ppd-H1 variants indicated in color.
Supplemental Figure S2. Intrachromosomal LD decay of marker pairs over all chromosomes as a function of genetic distance.
Supplemental Figure S3. Manhattan plots of GWAS for FD, LL, and LW calculated for Iran and Italy separately.
Supplemental Figure S4. Manhattan plots of GWAS for LL and LW with FD as a covariate.
Supplemental Figure S5. Size and flanking markers of Ppd-H1 introgressions in three independent ILs.
Supplemental Figure S6. Heading date is delayed in the presence of the mutated ppd-H1 allele under LDs.
Supplemental Figure S7. LL and leaf emergence of Bowman/BW281 and Triumph/Triumph-IL.
Supplemental Figure S8. Variation at Ppd-H1 does not affect the rate of leaf elongation.
Supplemental Figure S9. Leaf blade anatomy of the fifth leaf emerging from the main shoot of SD-grown plants.
Supplemental Figure S10. Variance explained by the first 10 principal components for the genetic diversity of the winter barley collection.
Supplemental Table S1. ANOVA for FD, LW, and LL.
Supplemental Table S2. Genetic material and genotyping information.
Supplemental Table S3. Ppd-H1 haplotypes in the ILs.
Supplemental Table S4. Variation at Ppd-H1 affects the phyllochron.
Supplemental Table S5. Variation at Ppd-H1 does not affect the rate of leaf blade elongation.
Supplemental Table S6. Primers used for genotyping and real-time qRT-PCR assays.
Supplementary Material
Acknowledgments
We thank Klaus Pillen (Martin Luther University), Robbie Waugh (James Hutton Institute), and David Laurie (John Innes Center) for seeds of barley lines; Kerstin Luxa, Caren Dawidson, and Andrea Lossow for excellent technical assistance; and A.R. Kazemeini for assistance with field management in Iran.
Glossary
- QTL
quantitative trait locus
- LD
long day
- GWAS
genome-wide association scan
- FD
flowering date
- LW
leaf width
- LL
leaf length
- DAS
days after sowing
- SNP
single-nucleotide polymorphism
- IL
introgression line
- SD
short day
- LER
leaf elongation rate
- LEM
leaf emergence
- MAF
minimum allele frequency
- BLUEs
best linear unbiased estimators
Footnotes
This work was supported by the Max Planck Society and an International Max Planck Research School fellowship to B.D., by the Deutsch Forschungsgemeinschaft (grant no. SPP1530 and Excellence Cluster EXC1028), by the ERA-PG-funded project Exbardiv and a FIRB grant from the Ministero dell’Istruzione dell’Università e della Ricerca (MIUR), Italy (grant no. RBIN047MBH), by the ERA-NET Cofund FACCE SURPLUS (grant no. 93), by the Università degli Studi di Milano (postdoctoral fellowship to E.T.), and by a PhD fellowship from MIUR to G.V.
Articles can be viewed without a subscription.
References
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E (1997) Inflorescence commitment and architecture in Arabidopsis. Science 275: 80–83 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Campoli C, Drosse B, Searle I, Coupland G, von Korff M (2012a) Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. Plant J 69: 868–880 [DOI] [PubMed] [Google Scholar]
- Campoli C, Shtaya M, Davis SJ, von Korff M (2012b) Expression conservation within the circadian clock of a monocot: natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biol 12: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartolano M, Pieper B, Lempe J, Tattersall A, Huijser P, Tresch A, Darrah PR, Hay A, Tsiantis M (2015) Heterochrony underpins natural variation in Cardamine hirsuta leaf form. Proc Natl Acad Sci USA 112: 10539–10544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casao MC, Karsai I, Igartua E, Gracia MP, Veisz O, Casas AM (2011) Adaptation of barley to mild winters: a role for PPDH2. BMC Plant Biol 11: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Cigan AM, Saeteurn K, Hake S (2007) The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet 39: 544–549 [DOI] [PubMed] [Google Scholar]
- Cockram J, Norris C, O’Sullivan DM (2009) PCR-based markers diagnostic for spring and winter seasonal growth habit in barley. Crop Sci 49: 403 [Google Scholar]
- Cockram J, White J, Leigh FJ, Lea VJ, Chiapparino E, Laurie DA, Mackay IJ, Powell W, O’Sullivan DM (2008) Association mapping of partitioning loci in barley. BMC Genet 9: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockram J, White J, Zuluaga DL, Smith D, Comadran J, Macaulay M, Luo Z, Kearsey MJ, Werner P, Harrap D, et al. (2010) Genome-wide association mapping to candidate polymorphism resolution in the unsequenced barley genome. Proc Natl Acad Sci USA 107: 21611–21616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comadran J, Kilian B, Russell J, Ramsay L, Stein N, Ganal M, Shaw P, Bayer M, Thomas W, Marshall D, et al. (2012) Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat Genet 44: 1388–1392 [DOI] [PubMed] [Google Scholar]
- Digel B, Pankin A, von Korff M (2015) Global transcriptome profiling of developing leaf and shoot apices reveals distinct genetic and environmental control of floral transition and inflorescence development in barley. Plant Cell 27: 2318–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druka A, Franckowiak J, Lundqvist U, Bonar N, Alexander J, Houston K, Radovic S, Shahinnia F, Vendramin V, Morgante M, et al. (2011) Genetic dissection of barley morphology and development. Plant Physiol 155: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. (1977) Anatomy of Seed Plants, Ed 2 John Wiley & Sons, New York [Google Scholar]
- Farooq M, Tagle AG, Santos RE, Ebron LA, Fujita D, Kobayashi N (2010) Quantitative trait loci mapping for leaf length and leaf width in rice cv. IR64 derived lines. J Integr Plant Biol 52: 578–584 [DOI] [PubMed] [Google Scholar]
- Fischbeck G. (2003) Diversification through breeding. In von Bothmer R, van Hintum T, Knüpffer H, Sato K, eds, Diversity in Barley (Hordeum vulgare). Elsevier Science, Amsterdam, pp 29–52 [Google Scholar]
- Fu C, Sunkar R, Zhou C, Shen H, Zhang JY, Matts J, Wolf J, Mann DG, Stewart CN Jr, Tang Y, et al. (2012) Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol J 10: 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K, Matsuda Y, Ozawa K, Nishimura T, Koshiba T, Fraaije MW, Sekiguchi H (2008) NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol Genet Genomics 279: 499–507 [DOI] [PubMed] [Google Scholar]
- Goebel K. (1900) Organography of Plants. Part I. General Organography. (English translation by IB Balfour.) Clarendon Press, Oxford [Google Scholar]
- González N, Inzé D (2015) Molecular systems governing leaf growth: from genes to networks. J Exp Bot 66: 1045–1054 [DOI] [PubMed] [Google Scholar]
- Hammer Ø, Ryan P, Harper D (2001) PAST: Paleontological Statistics software package for education and data analysis. Palaeontologia Electronica 4: 9 [Google Scholar]
- Hay A, Tsiantis M (2010) KNOX genes: versatile regulators of plant development and diversity. Development 137: 3153–3165 [DOI] [PubMed] [Google Scholar]
- Hempel FD, Feldman LJ (1994) Bi-directional inflorescence development in Arabidopsis thaliana: acropetal initiation of flowers and basipetal initiation of paraclades. Planta 192: 276–286 [Google Scholar]
- Jiang SK, Zhang XJ, Wang JY, Chen WF, Xu ZJ (2010) Fine mapping of the quantitative trait locus qFLL9 controlling flag leaf length in rice. Euphytica 176: 341–347 [Google Scholar]
- Jones CS. (1999) An essay on juvenility, phase change, and heteroblasty in seed plants. Int J Plant Sci (Suppl 6) 160: S105–S111 [DOI] [PubMed] [Google Scholar]
- Jones H, Leigh FJ, Mackay I, Bower MA, Smith LMJ, Charles MP, Jones G, Jones MK, Brown TA, Powell W (2008) Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the Fertile Crescent. Mol Biol Evol 25: 2211–2219 [DOI] [PubMed] [Google Scholar]
- Karsai I, Szucs P, Mészáros K, Filichkina T, Hayes PM, Skinner JS, Láng L, Bedo Z (2005) The Vrn-H2 locus is a major determinant of flowering time in a facultative × winter growth habit barley (Hordeum vulgare L.) mapping population. Theor Appl Genet 110: 1458–1466 [DOI] [PubMed] [Google Scholar]
- Kemp DR. (1980a) The location and size of the extension zone of emerging wheat leaves. New Phytol 84: 729–737 [Google Scholar]
- Kemp DR. (1980b) The growth rate of successive leaves of wheat plants in relation to sugar and protein concentrations in the extension zone. J Exp Bot 31: 1400–1411 [Google Scholar]
- Knapp SJ, Stroup WW, Ross WM (1985) Exact confidence intervals for heritability on a progeny mean basis. Crop Sci 25: 192–194 [Google Scholar]
- Laurie DA, Pratchett N, Snape JW, Bezant JH (1995) RFLP mapping of five major genes and eight quantitative trait loci controlling flowering time in a winter × spring barley (Hordeum vulgare L.) cross. Genome 38: 575–585 [DOI] [PubMed] [Google Scholar]
- Li X, Zhu C, Yeh CT, Wu W, Takacs EM, Petsch KA, Tian F, Bai G, Buckler ES, Muehlbauer GJ, et al. (2012) Genic and nongenic contributions to natural variation of quantitative traits in maize. Genome Res 22: 2436–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y (2006) The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA 103: 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: Genome Association and Prediction Integrated Tool. Bioinformatics 28: 2397–2399 [DOI] [PubMed] [Google Scholar]
- MacAdam JW, Volenec JJ, Nelson CJ (1989) Effects of nitrogen on mesophyll cell division and epidermal cell elongation in tall fescue leaf blades. Plant Physiol 89: 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M, Muehlbauer GJ, Rokhsar DS, Chapman J, Schmutz J, Barry K, Muñoz-Amatriaín M, Close TJ, Wise RP, Schulman AH, et al. (2013) Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). Plant J 76: 718–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muggeo VMR. (2003) Estimating regression models with unknown break-points. Stat Med 22: 3055–3071 [DOI] [PubMed] [Google Scholar]
- Muggeo VMR. (2008) segmented: an R package to fit regression models with broken-line relationships. R News 8: 20–25 [Google Scholar]
- Muñoz-Amatriaín M, Cuesta-Marcos A, Endelman JB, Comadran J, Bonman JM, Bockelman HE, Chao S, Russell J, Waugh R, Hayes PM, et al. (2014) The USDA barley core collection: genetic diversity, population structure, and potential for genome-wide association studies. PLoS ONE 9: e94688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima S, Murata M, Sakamoto W, Ogura Y, Motoyoshi F (1997) Cloning and molecular analysis of the Arabidopsis gene Terminal Flower 1. Mol Gen Genet 254: 186–194 [DOI] [PubMed] [Google Scholar]
- Pasam RK, Sharma R, Malosetti M, van Eeuwijk FA, Haseneyer G, Kilian B, Graner A (2012) Genome-wide association studies for agronomical traits in a world wide spring barley collection. BMC Plant Biol 12: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng MM, Yang GH, Zhang QQ, An BG, Li YS (2007) QTL analysis for flag leaf morphological traits in rice (Oryza sativa L.) under different genetic backgrounds. Chin J Rice Sci 21: 247–252 [Google Scholar]
- Poethig RS. (1990) Phase change and the regulation of shoot morphogenesis in plants. Science 250: 923–930 [DOI] [PubMed] [Google Scholar]
- Poethig RS. (2010) The past, present, and future of vegetative phase change. Plant Physiol 154: 541–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Qian Q, Bu Q, Li S, Chen Q, Sun J, Liang W, Zhou Y, Chu C, Li X, et al. (2008) Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol 147: 1947–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2008) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Rostoks N, Ramsay L, MacKenzie K, Cardle L, Bhat PR, Roose ML, Svensson JT, Stein N, Varshney RK, Marshall DF, et al. (2006) Recent history of artificial outcrossing facilitates whole-genome association mapping in elite inbred crop varieties. Proc Natl Acad Sci USA 103: 18656–18661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon MJ. (2003) The polar auxin transport inhibitor N-1-naphthylphthalamic acid disrupts leaf initiation, KNOX protein regulation, and formation of leaf margins in maize. Plant Physiol 133: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon MJ. (2015) Leaves of grass: focusing phenomics on maize leaf growth. Genome Biol 16: 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalenbach I, Körber N, Pillen K (2008) Selecting a set of wild barley introgression lines and verification of QTL effects for resistance to powdery mildew and leaf rust. Theor Appl Genet 117: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Schmalenbach I, March TJ, Bringezu T, Waugh R, Pillen K (2011) High-resolution genotyping of wild barley introgression lines and fine-mapping of the threshability Locus thresh-1 using the Illumina GoldenGate assay. G3 (Bethesda) 1: 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalit A, Rozman A, Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y, Lifschitz E (2009) The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc Natl Acad Sci USA 106: 8392–8397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner RH, Nelson CJ (1995) Elongation of the grass leaf and its relationship to the phyllochron. Crop Sci 35: 4–10 [Google Scholar]
- Sluis A, Hake S (2015) Organogenesis in plants: initiation and elaboration of leaves. Trends Genet 31: 300–306 [DOI] [PubMed] [Google Scholar]
- Stracke S, Haseneyer G, Veyrieras JB, Geiger HH, Sauer S, Graner A, Piepho HP (2009) Association mapping reveals gene action and interactions in the determination of flowering time in barley. Theor Appl Genet 118: 259–273 [DOI] [PubMed] [Google Scholar]
- Teper-Bamnolker P, Samach A (2005) The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17: 2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES (2011) Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet 43: 159–162 [DOI] [PubMed] [Google Scholar]
- Tondelli A, Xu X, Moragues M, Sharma R, Schnaithmann F, Ingvardsen C, Manninen O, Comadran J, Russell J, Waugh R, et al. (2013) Structural and temporal variation in genetic diversity of European spring two-row barley cultivars and association mapping of quantitative traits. Plant Genome 6: doi/10.3835/plantgenome2013.03.0007 [Google Scholar]
- Trevaskis B, Hemming MN, Peacock WJ, Dennis ES (2006) HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol 140: 1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Turner SD. (2014) qqman: an R package for visualizing GWAS results using Q-Q and Manhattan plots. doi/http://dx.doi.org/10.1101/005165
- VanRaden PM. (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91: 4414–4423 [DOI] [PubMed] [Google Scholar]
- von Korff M, Léon J, Pillen K (2010) Detection of epistatic interactions between exotic alleles introgressed from wild barley (H. vulgare ssp. spontaneum). Theor Appl Genet 121: 1455–1464 [DOI] [PubMed] [Google Scholar]
- von Korff M, Wang H, Léon J, Pillen K (2006) AB-QTL analysis in spring barley. II. Detection of favourable exotic alleles for agronomic traits introgressed from wild barley (H. vulgare ssp. spontaneum). Theor Appl Genet 112: 1221–1231 [DOI] [PubMed] [Google Scholar]
- von Zitzewitz J, Szucs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, Casas A, Chen THH, Hayes PM, Skinner JS (2005) Molecular and structural characterization of barley vernalization genes. Plant Mol Biol 59: 449–467 [DOI] [PubMed] [Google Scholar]
- Waddington SR, Cartwright PM, Wall PC (1983) A quantitative scale of spike initial and pistil development in barley and wheat. Ann Bot (Lond) 51: 119–130 [Google Scholar]
- Wang G, Schmalenbach I, von Korff M, Léon J, Kilian B, Rode J, Pillen K (2010) Association of barley photoperiod and vernalization genes with QTLs for flowering time and agronomic traits in a BC2DH population and a set of wild barley introgression lines. Theor Appl Genet 120: 1559–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhou G, Yu H, Yu S (2011) Fine mapping a major QTL for flag leaf size and yield-related traits in rice. Theor Appl Genet 123: 1319–1330 [DOI] [PubMed] [Google Scholar]
- Wenzel C. (1997) Characterization of the leaf epidermis of barley (Hordeum vulgare L. “Himalaya”). Ann Bot (Lond) 79: 41–46 [Google Scholar]
- Wilhelm WW, McMaster GS (1995) Importance of the phyllochron in studying development and growth in grasses. Crop Sci 35: doi/10.2135/cropsci1995.0011183X003500010001x [Google Scholar]
- Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Shen J, Hou X, Yao J, Li X, Xiao J, Xiong L (2012) Gradual increase of miR156 regulates temporal expression changes of numerous genes during leaf development in rice. Plant Physiol 158: 1382–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100: 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Guo Z, Huang C, Wang K, Jiang N, Feng H, Chen G, Liu Q, Xiong L (2015) Genome-wide association study of rice (Oryza sativa L.) leaf traits with a high-throughput leaf scorer. J Exp Bot 66: 5605–5615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14: 415–421 [Google Scholar]
- Zhang B, Ye W, Ren D, Tian P, Peng Y, Gao Y, Ruan B, Wang L, Zhang G, Guo L, et al. (2015) Genetic analysis of flag leaf size and candidate genes determination of a major QTL for flag leaf width in rice. Rice (N Y) 8: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GH, Xu Q, Zhu XD, Qian Q, Xue HW (2009) SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21: 719–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.