OsMADS1 binds to many downstream target loci with distinct sequence and biophysical characteristics and likely in combination with other factors to ensure rice floret development.
Abstract
OsMADS1 controls rice (Oryza sativa) floral fate and organ development. Yet, its genome-wide targets and the mechanisms underlying its role as a transcription regulator controlling developmental gene expression are unknown. We identify 3112 gene-associated OsMADS1-bound sites in the floret genome. These occur in the vicinity of transcription start sites, within gene bodies, and in intergenic regions. Majority of the bound DNA contained CArG motif variants or, in several cases, only A-tracts. Sequences flanking the binding peak had a higher AT nucleotide content, implying that broader DNA structural features may define in planta binding. Sequences for binding by other transcription factor families like MYC, AP2/ERF, bZIP, etc. are enriched in OsMADS1-bound DNAs. Target genes implicated in transcription, chromatin remodeling, cellular processes, and hormone metabolism were enriched. Combining expression data from OsMADS1 knockdown florets with these DNA binding data, a snapshot of a gene regulatory network was deduced where targets, such as AP2/ERF and bHLH transcription factors and chromatin remodelers form nodes. We show that the expression status of these nodal factors can be altered by inducing the OsMADS1-GR fusion protein and present a model for a regulatory cascade where the direct targets of OsMADS1, OsbHLH108/SPT, OsERF034, and OsHSF24, in turn control genes such as OsMADS32 and OsYABBY5. This cascade, with other similar relationships, cumulatively contributes to floral organ development. Overall, OsMADS1 binds to several regulatory genes and, probably in combination with other factors, controls a gene regulatory network that ensures rice floret development.
MADS-domain transcription factors are a class of DNA-binding proteins, found in animals, fungi, and plants where they regulate gene expression in diverse contexts. The prototypic members of this family, MCM1 (Saccharomyces cerevisiae), AGAMOUS (Arabidopsis thaliana), DEFICIENS (Antirrhinum majus), and SRF (Homo sapiens) share an ancient and highly conserved DNA-binding domain encoded by the MADS-domain. These proteins form two main lineages, Type I (SRF-like) and Type II (MEF2-like), probably due to an ancestral gene duplication before the divergence of plants and animals (Alvarez-Buylla et al., 2000).
Many plant MADS-domain transcription factors are central regulators of flower development. The pattern of organs in the whorls of a flower is explained by the ABCDE model, which states that the A, B, C, D, and E genes, most of which encode MADS-domain proteins, act in a combinatorial manner to specify organ identity (Coen and Meyerowitz, 1991; Pelaz et al., 2000; Pinyopich et al., 2003; Ditta et al., 2004; Immink et al., 2010). The role of B-function and C-function genes in regulating the development of second, third, and fourth whorl organs is more or less conserved between rice (Oryza sativa), a model grass species, and Arabidopsis (Arabidopsis thaliana), a model eudicot (Nagasawa et al., 2003; Prasad and Vijayraghavan, 2003; Krizek and Fletcher, 2005; Dreni et al., 2011). In contrast, E-class (SEPALLATA) members perform both conserved and diversified functions (Ditta et al., 2004; Malcomber and Kellogg, 2005; Ciaffi et al., 2011). Phylogenetically, E-class genes are divided into SEPALLATA3 (SEP3) and LOFSEP major clades (Malcomber and Kellogg, 2005). Among the five SEP-like genes in rice, OsMADS7 and OsMADS8 belong to the SEP3 clade, whereas OsMADS1, OsMADS5, and OsMADS34 are members of the LOFSEP clade (Malcomber and Kellogg, 2005). The LEAFY HULL STERILE1/OsMADS1 genes of the LOFSEP clade have been speculated to play a central role in the origin and diversification of grass spikelets (Malcomber and Kellogg, 2004, 2005; Christensen and Malcomber, 2012). Rice OsMADS1 is the best-studied monocot SEP gene whose transcripts are detected throughout the very young floret meristem (FM), in the developing lemma, palea, and weakly in carpels (Prasad et al., 2001, 2005). Functional studies show both redundant and distinct roles during FM specification and organ development. Several loss-of-function mutants or transgenic lines where OsMADS1 is down-regulated are known (lhs1, OsMADS1-RNAi, Osmads1-Tos17 insertion, nsr). They share the phenotypes of malformed and glume-like lemma and palea, lodicules transformed to glume-like organs, as well as reduced stamen and increased carpel numbers. Some strong alleles show a complete conversion of all organs to glume-like structures with an indeterminate FM (Jeon et al., 2000; Agrawal et al., 2005; Prasad et al., 2005; Chen et al., 2006). Thus OsMADS1 is required for the formation of all floret organs and for a determinate FM.
MADS-domain transcription factors bind in vitro to a consensus DNA sequence known as CArG-box (Huang et al., 2000; de Folter and Angenent, 2006). The most common of these are the SRF-type element with the CC[A/T]6GG consensus sequence and the MEF2-type element with the consensus sequence C[A/T]8G; Shore and Sharrocks, 1995; Kaufmann et al., 2009). However, plant MADS-domain proteins show a broad range of DNA-binding preferences and bind to SRF-type, MEF2-type, as well as intermediate motifs (West et al., 1998; Tang and Perry, 2003; de Folter and Angenent, 2006). Much progress has been made in understanding the molecular mechanisms underlying the combinatorial activities of MADS-domain proteins in flower development. Homo- and hetero-dimerization and the formation of multimeric complexes provide mechanisms for specific target gene recognition (Riechmann et al., 1996; Egea-Cortines et al., 1999; Melzer and Theissen, 2009; Immink et al., 2010). In addition to forming complexes among themselves, MADS-domain proteins interact with other proteins, such as transcription factors, chaperones, kinases, and chromatin modifiers in higher order complexes (Messenguy and Dubois, 2003; Smaczniak et al., 2012b). These additional interacting partners may be essential to determine net target gene output, be it activation or repression, and may also aid in target binding.
Studies on the genome-wide binding of Arabidopsis MADS-domain proteins identify mechanisms underlying their role during flower development (Kaufmann et al., 2009, 2010; Immink et al., 2012; Deng et al., 2011; Zheng et al., 2009; Wuest et al., 2012; Ó’Maoiléidigh et al., 2013; Gregis et al., 2013). These studies showed them to bind a multitude of downstream target genes and regulate their expression. However, such genome-wide approaches have not been frequently employed in grasses. Although rice mutant analyses have provided considerable insights into the regulation of floret organ patterning by OsMADS1, the molecular mechanisms and its downstream effects as a transcription regulator are less explored. Because OsMADS1 is a central regulator in the broad program of rice flower development from meristem inception to organ differentiation and meristem termination, identifying its direct downstream target genes is essential to our better understanding of flower development. Our previous studies using candidate gene approaches have revealed that OsMADS1 regulates the expression of transcription factors belonging to different families as well as genes in hormone signaling pathways (Khanday et al., 2013). To further extend our understanding of OsMADS1 function, we have employed ChIP-seq to globally identify its downstream target genes. We provide a comprehensive genome-wide binding map of OsMADS1, a detailed analysis of the structural properties of its binding sites and a snapshot of its gene regulatory network, including likely downstream genes in some floral differentiation pathways. We thus bring, to our knowledge, new insights into the mechanism of OsMADS1-mediated gene regulation during floret development.
RESULTS
Genome-Wide Identification of Target Genes Bound by OsMADS1
To understand the role of rice OsMADS1 as a regulator of rice FM specification and floral organ development, we mapped its genome-wide chromatin occupancy using chromatin immunoprecipitation (ChIP) followed by deep sequencing (ChIP-seq). To carry out ChIP, we used an affinity-purified OsMADS1-specific antibody described previously in Khanday et al. (2013). Chromatin was prepared from developing wild-type panicle tissues (2 to 200 mm length), which contained a range of young FMs and florets with developing organs (see “Materials and Methods”). As a control, ChIP was carried out in parallel with the same chromatin samples treated with beads without antibodies. The quality of the DNA in the ChIP and in mock-precipitated control samples was first qualitatively assessed by testing OsMADS1 occupancy at some target genes that were identified in prior functional studies (Khanday et al., 2013; Supplemental Fig. S1A). These samples were deep-sequenced to derive approximately 23 million high-quality, uniquely mapping reads that have 100% identity to the rice genome (Supplemental Fig. S1B). Probabilistic Inference for ChIP-seq (PICS), a peak-finding algorithm from Strand Genomics (formerly Avadis NGS), was used to estimate sequence enrichments/peaks at loci reflecting OsMADS1 chromatin association throughout the genome (Supplemental Fig. S2A). This genome-wide overview revealed that enriched regions were distributed throughout the rice genome with a higher density of peaks found on chromosomes 1, 2, and 3 (Supplemental Fig. S2B).
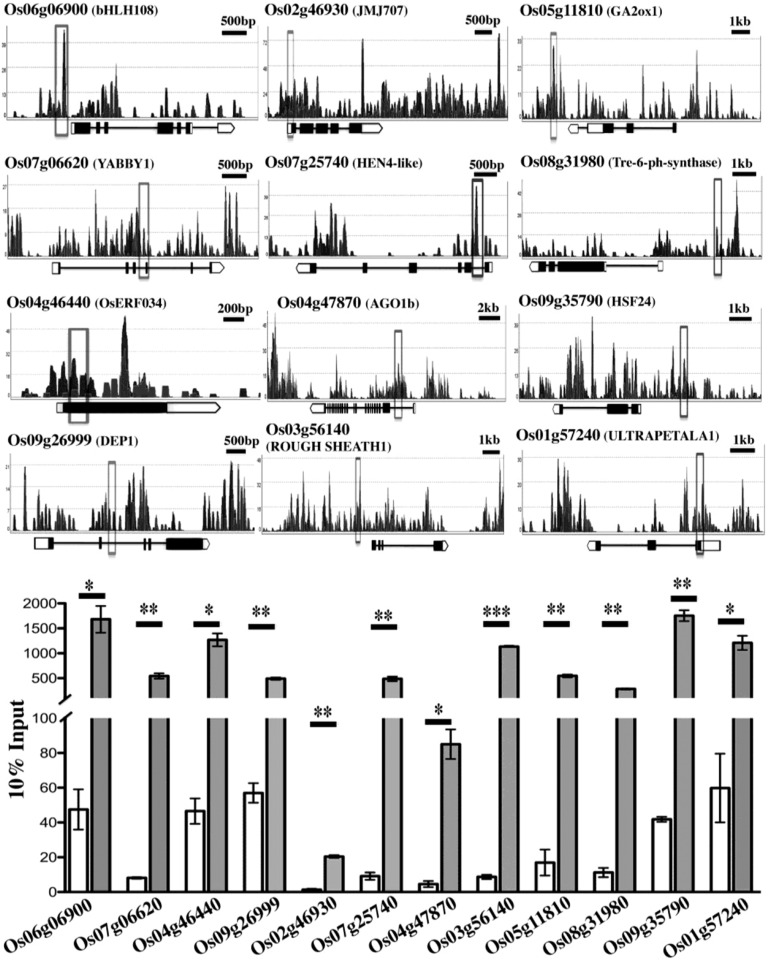
The centers of the binding peaks, with an FDR of ≤0.055, were used to search for the respective nearest annotated gene within 4 kb of the peak. A total of 3,112 gene-associated binding sites was identified (Supplemental Data Set S1). To validate these binding sites, we tested 12 and 3 loci, representing varying ChIP peak scores and binding densities, by ChIP-qPCR experiments and semiquantitative PCR, respectively, on two independent chromatin preparations. The loci chosen for ChIP-PCR validation correspond to genes with varied functions including transcription factors, chromatin modifiers, RNA processing, and metabolism (Fig. 1; Supplemental Fig. S3). These data for selected loci validated the OsMADS1 genome-wide binding-site enrichment obtained by ChIP-sequencing.
Figure 1.
Validation of ChIP-seq reads. ChIP-qPCR was carried out for 12 genes with different functional annotations. Binding profile of sequenced read coverage across genic structure of selected targets is depicted. Each gene structure is shown schematically on the x axis and the scale bar on the top right indicates gene length. Black boxes are exons and black lines indicate introns. White boxes represent UTRs and the white pentagon (3′UTR) shows the gene orientation. Vertical (y axis) gray open boxes depict the region assessed for enrichment by ChIP-qPCR. The histogram bars represent enrichment measures after normalization to 10% of the input chromatin in each case. Open bars show levels in Mock (no antibody controls) and the solid bars quantitate data from OsMADS1 ChIP. Error bars represent se from two independent ChIP replicates. Asterisk indicates significant enrichment in IP over mock at * = P < 0.05, ** = P < 0.01, and *** = P < 0.001, respectively.
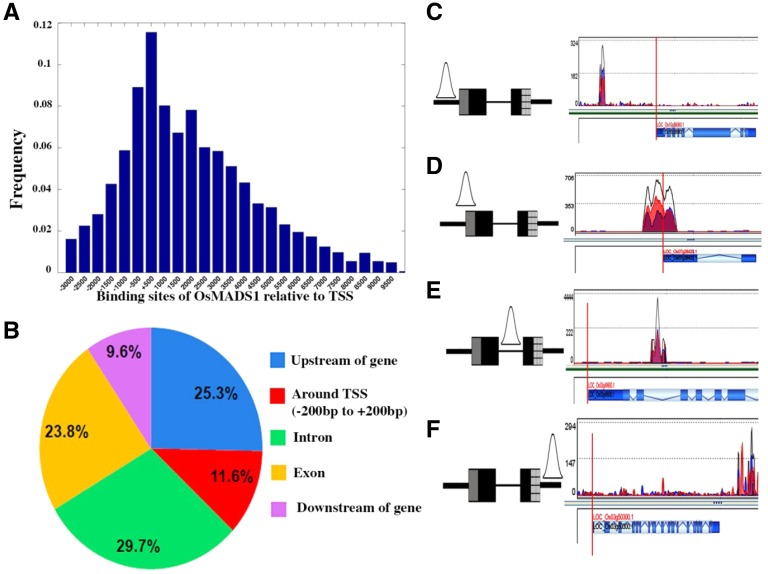
The data set of 3,112 OsMADS1-bound gene-associated sites were examined for the position of the binding site relative to the transcription start sites (TSSs) and other structural elements of the associated genes. We observed that binding sites can lie up to 4 kb upstream of the TSS (Fig. 2A; Supplemental Data Set S1). Roughly 25% of all OsMADS1-bound sites mapped themselves to regions upstream of the TSS of annotated genes (Fig. 2B). Binding sites in the vicinity of the TSS (−250 to +250) of target genes accounted for approximately 12% of the globally bound sites. Binding within the transcribed sequence, referred to henceforth as “gene body”, accounted for approximately 54% (exons and introns) of the sites, whereas approximately 10% of the global binding sites aligned with sequences downstream of the gene body (Fig. 2B). These classes of binding sites are schematically represented in Figure 2, C to F. Regardless of the position of the binding site, most binding peaks covered approximately 150 to 200 bp of DNA while 34% of the sites covered 300 to 500 bp of genomic sequences (Supplemental Fig. S4A). Because transcription factor binding sites tend to be small in size (Yu et al., 2016), these data are in agreement with OsMADS1 acting as a transcriptional regulator of gene expression.
Figure 2.
Genome-wide binding sites of OsMADS1 and the distribution of binding sites in different regions of annotated genes. A, Histogram showing the normalized frequency of occurrence of binding sites of OsMADS1 mapped to the promoters (TSSs) of annotated genes. The binding sites are widely distributed from −3,000 to +10,000, with a peak near the TSSs. B, Pie chart illustrating the location of OsMADS1 binding sites. A majority of the sites lie within the gene body (exons and introns). Approximately 12% of events overlap the TSS (−250 bp to +250 bp) while the remaining events are positioned either further upstream or are downstream of the gene body. Examples of OsMADS1 binding in the up-stream sequences (C), around the TSS (D), to an intronic region (E), and at downstream sequences (F). The vertical red line on schematics marks the TSS. C to F, The schematics to the left illustrate the binding events. Gray box represents the 5′ UTR, black boxes are exons, and the striped box the 3′ UTR. The thin black line between the exons represents the intron and thick black lines mark upstream and downstream sequences. The vertical red line on schematics marks the TSS.
Occurrence of CArG-Boxes in OsMADS1 Binding Sites
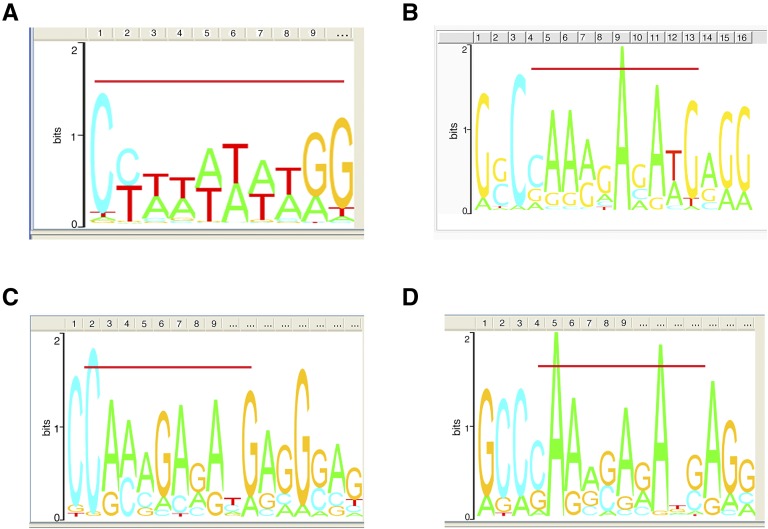
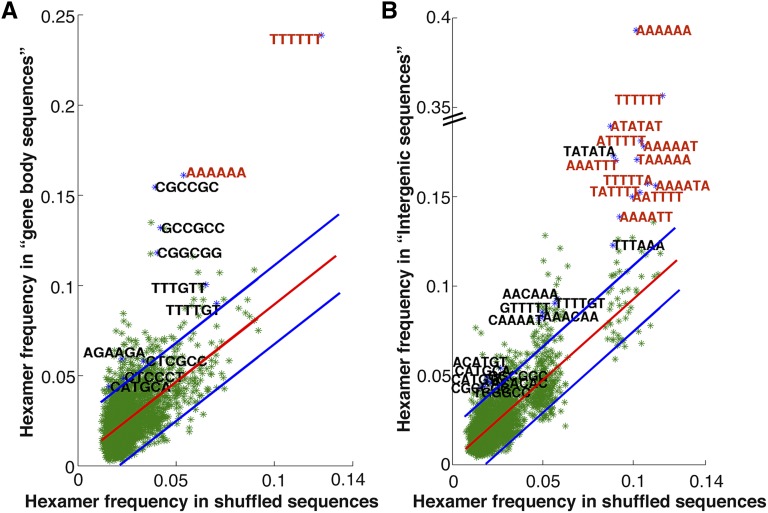
MADS-domain transcription factor binding sites are relatively degenerate with occupancy at SRF-type, MEF2-type, variant CArG-boxes, at intermediate motif types, and at half sites (Shore and Sharrocks, 1995; West et al., 1998; Tang and Perry, 2003; de Folter and Angenent, 2006; Kaufmann et al., 2009). We searched for consensus CArG elements among the global OsMADS1-bound sequences using the GADEM algorithm from Strand Genomics. We observed that OsMADS1 preferentially bound to four consensus motifs, which resemble the SRF-type (Fig. 3A), the MEF2-type (Fig. 3B), and the intermediate types (Fig. 3, C and D). Using a stringent search of all these motifs for types where A/T residues are flanked by C and G, we noted that CC(A/T)6GG, C(A/T)8G, C(A/T)7G, C(A/T)6G, C(A/T)5G, CC(A/T)3G, and CC(A/T)3GG together constituted 67.1% of all OsMADS1 CArG-box types. Only these were henceforth considered as CArG-boxes. For further analyses we grouped the whole data set of OsMADS1-bound sequences into two subsets, based on the position of the binding peak relative to the transcribed regions of the associated genes. One subset comprised sequences called “intergenic” (Fig. 2, C and F) and the other consisted of sequences that correspond to the “gene body” (transcribed sequences including the TSS as represented schematically in Fig. 2, D and E). In the rice genome, the GC content is approximately 6% higher in gene bodies as compared to intergenic regions (Morey et al., 2011). To account for this baseline effect in our analysis, we considered these two data sets separately. We found that 74.2% of the binding sites (893 out of 1,204) in the intergenic data set clearly contained CArG-boxes, whereas 62.6% of the sequences (1,195 out of 1,908) in the gene body data set contained CArG-boxes. In the remaining 1,024 sequences, CArG-boxes or its variants were not found; yet 866 of them contained A-tract sequences (see “Materials and Methods”). This set of binding sites was named “A-tract” and was identified as three or more consecutive A:T bp that are devoid of a TpA step (Strahs and Schlick, 2000; McConnell and Beveridge, 2001; Stefl et al., 2004). We found that 24.8% of the A-tract sites (299 out of 1,204) were in the intergenic and 29.7% (567 out of 1,908) in the gene body data set of OsMADS1-bound sites, respectively.
Figure 3.
CArG motifs present in the OsMADS1 binding sites as revealed by GADEM analysis. A, SRF-type. B, MEF2-type. C and D, Intermediate type. The red line above the motif indicates the sequence that corresponds to the CArG element; sequence outside the line corresponds to neighboring nucleotides that may also be essential for target recognition.
Structural Properties of OsMADS1 Binding Sites
Genome-wide binding studies with the Arabidopsis SEP3 and APETALA1 (AP1) floral organ identity MADS-domain proteins show majority target sites that do not contain perfect CArG motifs, implicating other parameters influencing the functional properties of these factors (Kaufmann et al., 2009, 2010; Muiňo et al., 2014). As our OsMADS1 ChIP-seq data set was of high quality with reads mapping to different regions of the genome (Supplemental Fig. S2), we analyzed the binding sites for their structural properties. This would characterize intrinsic DNA properties that may contribute to the energetics of target site recognition. The genome-wide OsMADS1-bound sequences, which were subgrouped into three data sets (intergenic [1] and gene body [2] binding sites containing a CArG-box; and [3] binding sites sequences with A-tracts).
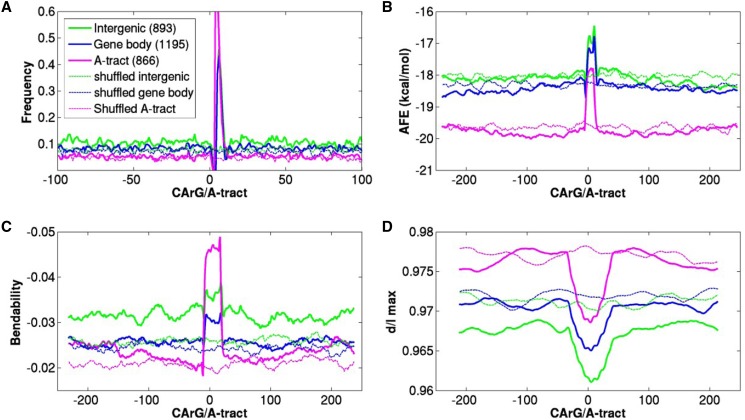
DNA duplex stability, A-tract content, and bendability and curvature properties of each of these three data sets were compared to those in their respective control (shuffled) sequences (see “Materials and Methods”). Structural properties characteristic of the bound sites are common for the gene body, intergenic, and A-tract data sets (Fig. 4). Interestingly, approximately 45% sequences with CArG motifs also contain internal A-tracts (Fig. 4A) and this is the case for binding sites in the gene body or in intergenic regions. The DNA stability (average free energy, AFE) profiles showed that intergenic regions in general have a somewhat lower stability than sequences in the gene body, but regions with A-tracts are more stable (Fig. 4B). Yet, all three data sets showed a low stability peak in the vicinity of the motif (CArG or A-tract) as compared to their flanking regions (Fig. 4B). An estimate of DNA flexibility can be made by a DNaseI-based bendability measure (Brukner et al., 1995). This model predicts the bending propensity of trinucleotides toward the major groove, based on experimentally derived DNaseI cleavage data. Because several MADS-domain protein homo- and heterodimers bend DNA in vitro (Riechmann et al., 1996; West et al., 1997), we examined all three classes of OsMADS1-bound sequences for this feature. Our data showed OsMADS1-bound DNA to be less bendable in the vicinity of motif when compared to the upstream and downstream sequences (Fig. 4C). The sequences with A-tract cis-elements showed much greater differences in their bendability around the element as compared to their flanking sequences. Importantly, the respective shuffled sequences do not have such properties. The intrinsic curvature of DNA in the overall OsMADS1 binding site sequences was also calculated. DNA curvature is a signature reported to reflect the presence of periodic A-tracts in DNA (Nagaich et al., 1994; Slama-Schwok et al., 2000; Nair, 2010). Among CArG- and A-tract containing sequences, the former are more curved in the vicinity of the motif (Fig. 4D). Overall, OsMADS1 binding sites are characterized by low stability and flexibility, while having a greater intrinsic curvature.
Figure 4.
Structural properties of 2954 OsMADS1-bound sequences in three data sets (intergenic, gene body, and A-tract). Sequences were aligned by positioning the CArG or A-tract motif at 0. Structural properties for sequences spanning 250 bp upstream and 250 downstream were calculated. Profiles for shuffled sequences for each data set are presented as controls in the same color as the respective OsMADS1-bound data set, but with a dashed line. A, The frequency of occurrence of A-tracts in all data sets shown only for the −100 to +100 region for clarity. B, DNA double helix stability was represented by AFE in kcal/mol. Smaller values indicate greater stability. C, Bendability or flexibility was measured by predicting DNaseI sensitivity. Smaller values suggest less bendability. D, Distribution of curvature around the motif was calculated by using BMHT dinucleotide parameters for all three data sets and their randomized controls. Smaller values indicate higher curvature.
Co-Occurrence of Other cis-Regulatory Motifs with CArG Boxes
Recent studies with Arabidopsis floral MADS-domain proteins show that SEP family transcription factors aid in the formation of higher order complexes and that such complexes impart distinct functional specificities (Melzer and Theissen, 2009; Smaczniak et al., 2012a). Upon knockdown of OsMADS1, the expression of its downstream genes is positively and negatively affected (Khanday et al., 2013). This points to a partnership of OsMADS1 with other transcription factors to achieve such regulatory outcomes. Hence, in addition to probing for CArG-boxes and for A-tract elements in OsMADS1-bound ChIP-seq data sets, we also computed hexamer motif frequencies for binding sites of other transcription factors. We curated hexamers that deviated > 2σ in OsMADS1-bound sequences as compared to their frequency in control data sets of randomized sequences with same nucleotide composition (see “Materials and Methods”; Supplemental Data Set S2). The hexamer searches were performed separately for data set of bound sequences that map to gene bodies (Fig. 5A) and intergenic regions (Fig. 5B), respectively. We observe that various hexameric sequence motifs, which match or are part of predicted or experimentally derived consensus binding sequences for different transcription factor families, are overrepresented in the OsMADS1-bound as compared to the randomized sequences (Table I; Fig. 5).
Figure 5.
Frequency of occurrence of all hexamers in approximately 200 bp of OsMADS1-bound DNA. The position of CArG or A-tract motif is taken as zero (and −100 to +100 regions were extracted) for hexamer analyses. A, Hexamer frequency in the gene body data set of 1908 sequences is compared with the frequency in the corresponding shuffled sequences. B, The hexamer frequencies for the 1204 sequences in intergenic data set are compared with the frequencies in shuffled sequences. Here, 130 and 99 hexamers were found to be 2σ deviated in the two data sets, respectively. Hexamers that form a part of the consensus sequences of known transcription factors are labeled in black. Other hexamers with frequencies ≥0.14 are in red.
Table I. Core hexamers enriched in gene body sequences and intergenic sequences.
The hexamers found at >2σ abundance in OsMADS1-bound chromatin sequences (as in Fig. 5) were queried for match to consensus binding sequences for 92 known transcription factor classes in PLACE (http://www.dna.affrc.go.jp/PLACE/). We show the core hexamers (column 3), and their presence in the larger consensus binding sites of which they are a part are listed (column 2, underlined nucleotides). The sequences that contain the larger consensus motif, for the respective transcription factor, are given within parentheses in column 2. The transcription factor class that binds the consensus sequence is given below where it is known. The number of OsMADS1-bound sequences containing the core hexamer sequence and the CArG element is listed in column 5, while column 4 lists the numbers in all OsMADS1-bound data. Hexamers co-occurring with CArG motif, in statistically insignificant numbers, are indicated in boldface in column 5; these bound sites likely contain A-tracts.
| cis-Element | Consensus Binding Sequence and Transcription Factor Class |
Core Hexamer Motif |
No. of Sequences with Core Hexamer Motif |
No. of Sequences with CArG Element and Core Hexamer Motif |
|---|---|---|---|---|
| Core hexamers found in gene body sequences compared with the shuffled sequences | ||||
| GCrichrepeatII | CGCCGCGC (41) | CGCCGC | 274 (P < 0.0001) | 58 (P < 0.0001) |
| AGCBOXNPGLB | AGCCGCC (28) ERF-domain | GCCGCC | 243 (P < 0.0001) | 54 (P < 0.0001) |
| A-T rich binding motif for AP2R2 or GARE | TTTGTT/AACAAA (314) euAP2 with double Ap2 domain | TTTGTT | 314 (P < 0.0001) | 246 (P < 0.0001) |
| REGION1OSOSEM | CGGCGGCCTCGCCACG bZIP | CGGCGG/CTCGCC | 316 (P < 0.0001) | 63 (P = 0.6251) |
| GLUEBOX2/OSGT3 | CTTTTGTGTACCTA | TTTTGT | 283 (P = 0.0001) | 233 (P < 0.0001) |
| RGATAOS | CAGAAGATA (2) | AGAAGA | 192 (P < 0.0001) | 112 (P < 0.0001) |
| GCrichrepeatIV | GTCTCCCT (7) | CTCCCT | 152 (P < 0.0001) | 79 (P = 0.0002) |
| RYREPEATVFLE B4/ RYREPEAT4 | CATGCATG (21) B3 domain | CATGCA | 150 (P < 0.0001) | 109 (P < 0.0001) |
| PROLAMINBOXO SGLUB1 | TGCAAAG (37) | TGCAAA | 175 (P < 0.0001) | 133 (P < 0.0001) |
| TATAboxIV | TATATACA (23) | TATATA | 178 (P = 0.3517) | 132 (P = 0.0269) |
| GCAAmotif | SCAAAATGA (11) | AAAATG | 209 (P < 0.0001) | 170 (P < 0.0001) |
| Core hexamers found in intergenic sequences compared with the shuffled sequences | ||||
| TATAboxII | TATTTAAA (14) | TTTAAA | 223 (P = 0.004) | 184 (P = 0.5465) |
| GCrichrepeatII | CGCCGCGC (8) | CGCCGC | 61 (P = 0.0002) | 26 (P = 0.8415) |
| A-T rich binding motif for AP2R2 or GARE | TTTGTT/AACAAA (183) euAP2 with double Ap2 domain | AACAAA | 183 (P < 0.0001) | 145 (P < 0.0001) |
| REGION1OSOSEM | CGGCGGCCTCGCCACG bZIP | GGCGGC | 62 (P < 0.0001) | 29 (P = 0.2109) |
| RYREPEATVFLE B4/ RYREPEAT4 | CATGCATG (22) B3 domain | CATGCA | 106 (P < 0.0001) | 68 (P < 0.0001) |
| GLUEBOX2/OSGT3 | CTTTTGTGTACCTTA | TTTTGT | 182 (P < 0.0001) | 149 (P < 0.0043) |
| DirectRepeat | GGTTTTTAAGTT | GTTTTT | 173 (P < 0.0001) | 138 (P = 0.0001) |
| GCAAmotif | SCAAAATGA (1) | CAAAAT | 172 (P < 0.0001) | 147 (P < 0.0001) |
| PROLAMINBOX | CACATGTGTAAAGGT | ACATGT | 119 (P < 0.0001) | 96 (P < 0.0001) |
| MYCATERD1 | CATGTG (109) Myc domain | CATGTG | 109 (P < 0.0001) | 76 (P < 0.0001) |
| GLUTEBP1OS | AAGCAACACACAAC | ACACAC | 77 (P < 0.0001) | 55 (P = 0.0003) |
| Element II Os region Element II OsPCNA-2 | TGGGCCCGT Class II TCP domain | TGGGCC | 74 (P < 0.0001) | 48 (P < 0.0001) |
| PYRIMIDINEBOXOSRAMY1A | CCTTTT (121) | CCTTTT | 121 (P < 0.0001) | 99 (P < 0.0001) |
The sequence GCCG(C/A)C is known as GCC-box, AGCNOXPGLB, or DRE (Hart et al., 1993; Table I), and is a binding site for AP2/ERF transcription factors with a single AP2/ERF-domain (Mizoi et al., 2012). The hexamer GCCGCC representing this consensus sequence was significantly enriched in OsMADS1-bound sequences when present in the gene body of the target locus. Many of these sites also contained a CArG-box, i.e. 54 of the 243 sites (Table I). Other members of the large AP2/ERF transcription factor family have two AP2-domains and form the euAP2-subclade, many members of which serve developmental functions (Kim et al., 2006). The TTTGTT or AACAAA element is an in vitro determined the binding site for the R2 domain of Arabidopsis APETALA2 (AP2; Dinh et al., 2012). Our data on OsMADS1-bound DNA indicated that either one of these hexameric sequences was greatly overrepresented in the gene body and in intragenic data sets. Importantly, nearly all sites with either of these two elements also contained a CArG-box. The AACAAA element is closely related to the gibberellin (GA)-responsive element (GARE) with the consensus site c/tAACc/gg/aa/cc/a. In the data set of OsMADS1-bound sequences that lie in intergenic regions, this motif could thus represent interactions with AP2-domain factors or with regulators of the GA pathway such as GAMYB (Gubler et al., 1995).
The two hexamers TTTTGT and CATGCA co-occur (P value < 0.0001) with CArG-boxes among the OsMADS1-bound sites. The hexamer motif CATGCA is common to RY-repeat sequence motifs (Bäumlein et al., 1992), such as RYREPEATVFLEB4 and RYREPEAT4. Notably, the exact octamer motif CATGCATG of the RY-repeat is detected in 150 and 106 cases of OsMADS1-bound sites in the gene body and intergenic data sets, respectively (Table I). The CGCCGC motif in the GCrichrepeatII element was present together with CArG-boxes among OsMADS1-bound DNA in the gene body data set. Although at a lower frequency, this hexamer is also statistically enriched in intergenic OsMADS1-bound sequences. The CGCCGC hexamer is present in the GC-box of rice Amy8 promoter and is related to the consensus sequence of a sugar-responsive element (Lu et al., 1998).
The AGAAGA motif was present only in the gene body data set, while the hexamers DirectRepeat (McElroy et al., 1990), MYCATERD1 (Simpson et al., 2003), PROLAMINBOX (Maier et al., 1987), GLUTEBP1OS, and regionII OsPCNA-2 (Kosugi et al., 1995) were only detected in OsMADS1-bound DNA of the intergenic data set (Table I). Among these, the hexamer TGGGCC (regionII OsPCNA-2) is part of the consensus sequence TGGGCCCG (Table I), the binding site of Class II TCP transcription factors (Kosugi et al., 1995; Kosugi and Ohashi, 2002). Strikingly, this element co-occurred with OsMADS1-bound CArG-boxes (P value < 0.0001; Table I). The CATCTG motif (MYCATERD1 binding site) is recognized by MYC-domain transcription factors (Abe et al., 2003; Simpson et al., 2003), several of which are well-known developmental regulators.
Network of Direct OsMADS1 Target Genes Encoding Transcription Factors and Chromatin Modifiers
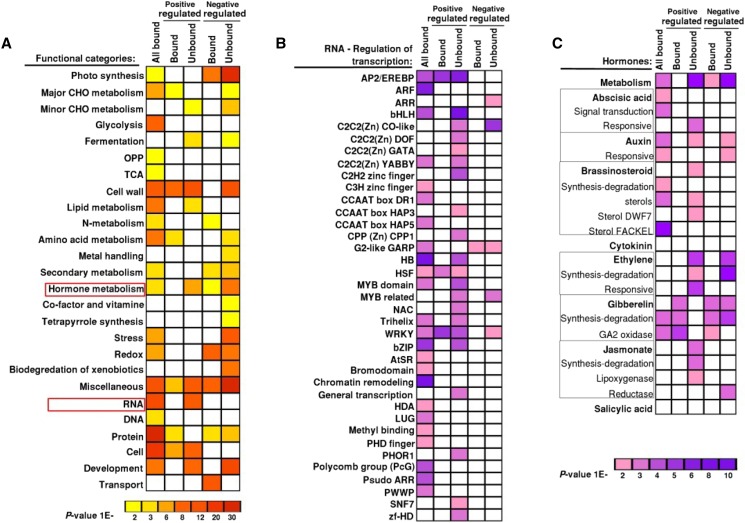
The data set of 3112 OsMADS1-bound genes were examined for their functional categories (by GO-MAPMAN) to identify pathways downstream of OsMADS1-bound genes. Overall, these gene classes were enriched for various functional categories such as RNA (transcription factor genes, RNA processing genes), protein (posttranslational regulation), development and cellular processes, hormone metabolism, and signaling pathways (Fig. 6A; Thimm et al., 2004). We have recently determined global gene expression patterns in developing wild-type panicles and florets, and compared them to similar tissues from plants where OsMADS1 was down-regulated (Khanday et al., 2013). We integrated the differential gene expression data with the data set of 3,112 OsMADS1-bound genes obtained in this study. A total of 652 genes were identified as being both bound by OsMADS1 (direct targets) and affected in their expression (>3-fold) in developing OsMADS1-RNAi florets (Supplemental Fig. S5).
Figure 6.
Functional categories of OsMADS1-bound genes. GO term enrichment for biological processes is shown in column “All Bound” for the 3112 OsMADS1-bound gene-associated ChIP-seq data set. These classified genes are juxtaposed with GO-term-classified differentially expressed genes. Both positively as well as negatively regulated gene data sets were derived based on differential expression in 2 to 20 mm OsMADS1 knockdown versus wild-type panicles (Khanday et al., 2013). Functional classifications of bound and unbound differentially expressed genes are presented in separate columns. A, All main biological functional categories in each data set are shaded from yellow to red in the order of increasing significance. B, Enrichment for various functional families within the biological process RNA regulation of transcription. C, Representation of functional groups within hormone signaling and metabolism are shaded from pink to purple based on their increasing significance.
Functional categories were assigned to both direct targets and indirectly affected genes (Fig. 6; Supplemental Data Set S3). Significant enrichment among the negatively regulated genes was found for the categories of photosynthesis, hormone metabolism, and redox signaling (Fig. 6A). Also prominent among the directly and positively regulated genes were those with functions in cell wall, other cellular processes, and protein regulation. A more detailed analysis of the subclasses of genes in the categories of RNA processes and hormone pathways was carried out. Several transcription factor genes from the AP2-ERF, HSF, and WRKY families were bound by OsMADS1 and deregulated upon OsMADS1 knockdown (Fig. 6B). Other bound loci include genes encoding transcription factors of the homeodomain (HB), bHLH, and auxin response (ARF) classes and several genes encoding chromatin modelers, such as HDAC, PcG, and LEUNIG. While OsMADS1 binds to many loci encoding factors in diverse hormone metabolism and signaling pathways, its direct effects on expression are evident for the GA pathway (Fig. 6C). Together, these results suggest that OsMADS1, besides being a master regulator of many transcription factors and chromatin modifiers, might balance other metabolic or cellular process to ensure rice floral development.
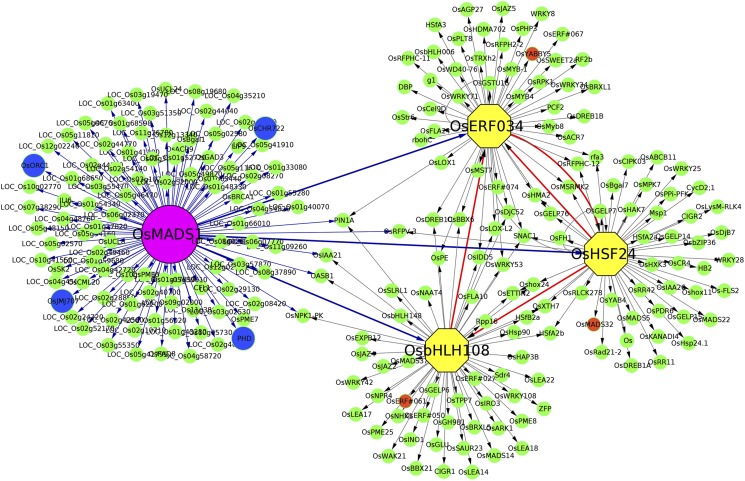
We also investigated whether transcription factors positively regulated by OsMADS1 form nodes in gene expression networks that connects to other targets of OsMADS1. Such an analysis provides a snapshot of the direct and indirect interactions between regulatory transcription factors and downstream genes. An interaction network was constructed for a selected list of 107 directly regulated targets of OsMADS1, which included transcription factors, chromatin modifiers, and factors in RNA processing. All 107 genes are positively regulated by OsMADS1, which bind to their upstream sequences (Supplemental Data Set S4A). In this snapshot of a gene regulatory network, we identified three nodal transcription factors, OsbHLH108, OsHSF24, and OsERF034, and their putative 695 downstream genes in developing florets (Fig. 7; Supplemental Data Set S4B). Among the many bHLH-domain factors regulated by OsMADS1, we found that OsbHLH108 (a predicted G-box binding protein; Li et al., 2006) was connected to 269 genes whose expression was indirectly affected in OsMADS1-RNAi tissues.
Figure 7.
Network analysis of some transcription factors and chromatin regulators that are directly and positively regulated by OsMADS1. Magenta circle represents OsMADS1; three of its directly regulated transcription factor targets, in this developmental pool of rice panicles, are in yellow hexagons. Direct targets encoding chromatin modifiers are shown in blue circles. The network shows interactions between OsMADS1 and direct targets with blue edges. To simplify the network visualization, only the well-annotated indirect targets are shown. Marked in orange circles are genes that were taken for experimental validation as indirect targets of OsMADS1.
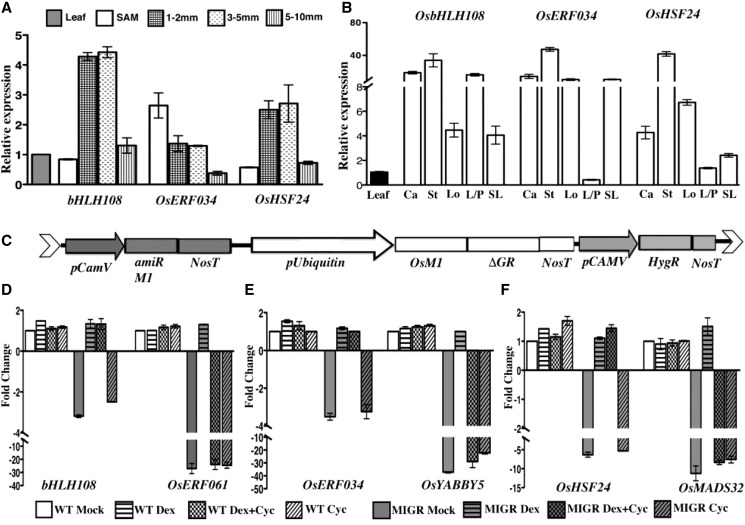
To experimentally validate these three nodal transcription factors and several other direct targets of OsMADS1 that could form nodes in other networks, we first studied expression profiles for several genes in wild-type floral tissues. By quantitative RT-PCR we find that OsHSF24 and OsbHLH108/SPATULA (SPT) are at higher levels in panicles that are predominantly with florets undergoing organogenesis. The varied expression levels of these targets of OsMADS1 in different floral tissues are suggestive of their roles in subsets of organogenesis programs (Fig. 8B).
Figure 8.
Expression levels of some OsMADS1 target genes in various floral tissues and their regulation by OsMADS1. A, Relative expression of three direct OsMADS1 target genes—OsbHLH108/SPT, OsERF034, and OsHSF24—in wild-type panicles at different developmental stages. Expression was quantified in tissues of vegetative SAM, with panicles of 1 to 2 mm (branch meristems); 3 to 5 mm (branch meristems, early spikelet, and floral meristem); and 5 to 10 mm (early floral organ differentiation). B, Relative expression of three OsMADS1-bound transcription factor genes in organs of wild-type florets. C, Schematic of the T-DNA segment in the PUbi: OsMADS1-ΔGR; P35S:OsMADS1amiR present in transgenics used for induced complementation studies with OsMADS1-GR. These transgenics were used to study transcriptional regulation of three nodal transcription factor genes and their predicted downstream genes. Wild-type or PUbi: OsMADS1-ΔGR; P35S:OsMADS1amiR (MIGR) plants were subjected to mock, dexamethasone, cycloheximide, or both dexamethasone and cycloheximide simultaneously. D, Quantitation of the fold-change in the normalized expression levels of OsbHLH108 and OsERF061 transcription factor genes in wild-type and MIGR panicles to assess direct and indirect effects of OsMADS1-GR. E, RT-qPCR analyses showing direct transcriptional regulation of OsERF034 by OsMADS1-GR and the indirect regulation of OsYABBY5. F, RT-qPCR analyses showing direct transcriptional regulation of OsHSF24 by OsMADS1-GR and indirect regulation of OsMADS32. Error bars represent se for two biological replicates. Ca, carpel; St, stamen; Lo, lodicule; L/P, lemma/palea; SL, sterile lemma.
To provide evidence for direct regulation of the three targets OsbHLH108/SPT, OsHSF24, and OsERF034 by OsMADS1, we raised transgenic lines in which the effects of the dexamethasone-inducible OsMADS1-GR fusion protein on gene expression could be examined. Transgenic rice plants with a single T-DNA (PUbi: OsMADS1-ΔGR; P35S:OsMADS1amiR as reported in Khanday et al. (2013), hereby referred to as “MIGR”) allow the knockdown of OsMADS1 by artificial miRNA and the inducible complementation by nuclear translocation of OsMADS1-GR fusion protein upon dexamethasone treatment (Fig. 8C). In planta treatment was performed using dexamethasone, cycloheximide, a combination of both chemicals, and a mock control treatment. In the harvested panicles, expression of OsbHLH108/SPT, OsHSF24, and OsERF034 was determined by RT-qPCR. As expected, transcript levels of these nodal target genes were lower in mock-treated MIGR panicle tissues than in control wild-type tissues but was restored to near wild-type levels upon induction (Fig. 8, D–F). Importantly, this rescue of gene expression occurred even in the presence of cycloheximide, indicating that nuclearly translocated OsMADS1-GR protein alone suffices to restore the expression level of the nodal target genes. To validate the predicted downstream effect of these nodal transcription factors and, therefore, indirect regulation by OsMADS1, we chose three genes that are linked in the network to each of these nodal transcription factors (Fig. 7). Low expression levels of the OsMADS32, OsYABBY5, and OsERF061 genes were detected in mock-treated MIGR panicles but this effect was reversed upon induction. However, gene expression levels were not restored when cycloheximide, blocking protein synthesis, was included (Fig. 8, D–F). Thus, OsMADS1-GR nuclear translocation alone cannot restore the altered expression levels of these genes, validating them as indirect, downstream genes.
DISCUSSION
The MADS-domain containing SEP genes of flowering plants fall into the SEP3 and LOFSEP major clades (Malcomber and Kellogg, 2005). Rice genes within the LOFSEP clade have members with both distinct and overlapping functions in panicle development (Jeon et al., 2000; Prasad et al., 2005; Gao et al., 2010; Kobayashi et al., 2010). While Arabidopsis SEP genes are largely functionally redundant in other plant species, SEP3 clade genes have specific developmental roles (Ampomah-Dwamena et al., 2002; Uimari et al., 2004; Ditta et al., 2004). In rice, interactions among SEP, LOFSEP, and other classes of MADS-domain factors have been described but how the members of a specific clade execute distinct developmental gene expression programs is yet unknown.
OsMADS1, belonging to the LOFSEP clade, is required to form a determinate FM on a short branch (spikelet) and is needed for floral organ development (Jeon et al., 2000; Agrawal et al., 2005; Malcomber and Kellogg, 2005; Prasad et al., 2005; Wang et al., 2010). Previous differential gene expression analyses showed that approximately 8,000 genes are either positively or negatively deregulated in panicles where OsMADS1 was down-regulated (Khanday et al., 2013). To elucidate the global targets of OsMADS1, understand its DNA interactions, obtain clues on likely transcription factor partners, and derive a snapshot of its gene regulatory network, we analyzed the genome-wide chromatin occupancy of OsMADS1. A notable class of OsMADS1-occupied sequences lies in the vicinity of the TSS of the associated gene; therefore, effects through the basal transcription machinery is a possible mode of regulation, as seen for other factors (Koudritsky and Domany, 2008; Kaufmann et al., 2009; Lu et al., 2013). Further, we show a majority of OsMADS1-bound sequences map to regions within gene bodies. We expect that several of these binding events are functionally relevant, especially when the target genes encode developmental regulators that often have functionally important cis-elements in their gene bodies (Busch et al., 1999; Prasad et al., 2003; Jeon et al., 2008).
In vitro DNA binding of homo- or heteromeric plant MADS-domain transcription factor complexes has been studied (West et al., 1998; de Folter and Angenent, 2006; Riechmann et al., 1996). However, sequences recognized in vivo by plant MADS-domain proteins and the links to their dynamic developmental gene expression are much less explored. Recent data on target sites bound by the Arabidopsis AP1 and SEP3 proteins during different stages of flower development and the biophysical properties of bound sequences provide some insights (Muiňo et al., 2014; Pajoro et al., 2014a). Approximately 17% of the in vivo SEP3-bound sequences contained a perfect CArG-box. In vitro studies with SEP3 show that A-tract sequences within and around the CArG-boxes influence its binding to DNA (Kaufmann et al., 2009; Muiňo et al., 2014). We observe that OsMADS1 binds in planta to consensus CArG-boxes and its variants, which may also contain embedded A-tracts (Fig. 3). More importantly, we identify that approximately 28% of the sites bound by OsMADS1 have only A-tracts (Fig. 4). Thus, the basic property of A/T-rich sequences can define the binding specificity of SEP family members. Arabidopsis AP1 and SEP3 proteins have varying affinities for subtypes of CArG-boxes at different stages of flower development (Pajoro et al., 2014a). However, although OsMADS1 can form homomeric and heteromeric complexes, in this study we cannot distinguish whether the variant CArG-boxes bound by OsMADS1 reflect the binding of different in planta complexes. Some known MADS-domain protein partners for OsMADS1 are OsMADS3, OsMADS7, OsMADS8, OsMADS13, OsMADS14, OsMADS15, OsMADS16, and OsMADS58 (Lim et al., 2000; Cui et al., 2010; Hu et al., 2015), but the binding characteristics of these other MADS proteins to CArG-box variants have not been studied. Our analysis of the larger sequence context that surrounds the core binding sites of OsMADS1 suggests that structural DNA features may also contribute to the binding site specificity of OsMADS1-containing complexes. We observe that intergenic OsMADS1-bound regions extending up to 250 bp upstream and downstream of the binding peaks have low DNA bendability and higher curvature, suggesting a pervasive role for these features in the binding of OsMADS1 to its target sites (Fig. 4, C and D).
Differences in the target site specificity of related transcription factors from two species and their downstream effects on gene expression could depend on cofactors present at different developmental stages in the different tissues. A comparison of the gene expression effects of Arabidopsis SEP3 and rice OsMADS1 shows some examples of altered gene regulatory relationships. For example, Arabidopsis SEP3 represses SVP/AGL24 to promote organ differentiation (Kaufmann et al., 2009), while we found OsMADS1 to activate the rice SVP homolog OsMADS55 (Khanday et al., 2013), done perhaps to promote early stages of FM growth. Another example would be the direct effects of Arabidopsis SEP3 on B function organ identity genes (Kaufmann et al., 2009), whereas OsMADS1 has indirect effects on rice B-function genes OsMADS16/SPW1 and OsMADS2, a PISTILLATA homolog (Khanday et al., 2013). Such altered regulatory relationships suggest OsMADS1 may interact with distinct cofactors. Proteomic analyses of Arabidopsis meristems detect MADS-domain proteins in complexes with other transcription factors such as KNAT3, BLR, and BLH1 and with chromatin modifiers (Immink et al., 2010; Smaczniak et al., 2012a, 2012b). Furthermore, Arabidopsis AP1-SEP3 heteromeric complexes can repress gene expression by interaction with SEUSS and LEUNIG transcriptional repressors (Sridhar et al., 2006). We identified bioinformatic signatures for the binding of transcription factor members of the bHLH, AP2/ERF, B3 domain, MYC, and Class II TCP families at sites bound by OsMADS1 (Table I). We suggest that certain rice proteins of these transcription factor families play a role in flower development, perhaps as partners of OsMADS1 in heteromeric complexes.
Several Arabidopsis bHLH transcription factors are activators of specific programs in organ development for example in anther differentiation and carpel development (Zhu et al., 2015; Heisler et al., 2001; Gremski et al., 2007). Interestingly, OsMADS1 binds and regulates some bHLH transcription factor genes. Among its targets are OsbHLH108 (Figs. 1, 7, and 8) and OsbHLH120 (Fig. 7; Supplemental Data Set S4A), which are related to Arabidopsis SPATULA and HECATE2, respectively. These Arabidopsis genes regulate carpel development (Heisler et al., 2001; Gremski et al., 2007). Even though their rice homologs are not functionally characterized, genes like OsbHLH018 could be involved in a pathway through which OsMADS1 contributes to carpel development.
The binding sites for AP2 and ERF transcription factors from the large AP2/ERF family are present in OsMADS1-enriched DNA. Significantly, our GO functional category analysis of genes differentially regulated by OsMADS1 (direct or indirect) also show an overrepresentation of the AP2/ERF family (Fig. 6B). Several Arabidopsis members of the double AP2-domain clade (e.g. AP2, AINTEGUMENTA [ANT], and PLETHORA) have functions in flower development. The pivotal role of AP2 in floral meristem development is well known (Komaki et al., 1988; Bowman et al., 1991; Jofuku et al., 1994). Among the OsMADS1 targets belonging to the double AP2-domain family, we find that a rice PLETHORA gene (OsPLETHORA8) is directly and positively regulated by OsMADS1 (Fig. 6). Its closest Arabidopsis ortholog is ANT, which plays important roles in floral meristems, organ patterning, and particularly ovule and gametophyte development (Krizek, 2009; Klucher et al., 1996; Yamaguchi et al., 2016). Moreover, Arabidopsis SEP3 also directly influences ANT expression in differentiating Arabidopsis floral organs (Pajoro et al., 2014b). Rice has five genes closely related to Arabidopsis AP2 (Tang et al., 2007), including SUPERNUMERARY BRACT and INDETERMINATE SPIKELET1, mutants of which indicate critical roles in rice FM specification and floret organ development (Lee and An, 2012). We find that the rice AP2-like factor gene Os04g55560 is directly and negatively regulated by OsMADS1. Whereas Arabidopsis AP2 locus is bound by SEP3, it is apparently not transcriptionally regulated (Pajoro et al., 2014b). We suggest that the rice AP2-like and PLETHORA-like factors are OsMADS1 targets in an intricate gene regulatory network. These genes may even be partners of OsMADS1 in higher order complexes, given the high co-occurrence of AP2 binding sites with CArG-boxes in OsMADS1-bound DNA (Table I).
The phytohormone GA is well documented for its developmental roles, particularly for the vegetative-to-reproductive transition of the shoot meristem and in flowers for anther development. In both Arabidopsis and rice developing flowers, GA levels are the highest during stamen development, where GA ensures filament elongation and the progression of microsporogenesis to form mature pollen grains (Mutasa-Göttgens and Hedden, 2009; Plackett et al., 2011). The GO enrichment analysis of OsMADS1-bound and regulated genes showed a significant enrichment of GA synthesis and degradation pathway genes (Fig. 6B). Based on the predicted functions of these rice GA pathway genes, we anticipate that in the young florets studied here (panicles of 2 to 20 mm) OsMADS1 contributes to low GA levels by directly activating OsGA2Ox1 and by directly repressing the OsGA20ox1-like and OsGA20ox3 genes encoding biosynthetic enzymes.
Large-scale gene expression changes seen in Arabidopsis mutants affecting floral MADS-domain transcription factors is a combined consequence of their direct and indirect effects (Kaufmann et al., 2009; Immink et al., 2012). The snapshot of a rice gene regulatory network we generated (Fig. 7), depicting the integration of OsMADS1 binding upstream of genes with reduced expression in OsMADS1 knockdown panicles, shows some direct OsMADS1 target genes are nodes that connect to its indirect targets. This network identified three nodal genes—OsbHLH108/SPT, OsERF034, and OsHSF24—each connected to indirect OsMADS1 targets (Figs. 7 and 8). Although the developmental functions of these nodal genes are not yet known, we show that they are highly expressed in panicles with florets at different stages of development (Fig. 8A) and that they are directly regulated by OsMADS1 (Fig. 8, D–F). These genes are likely coexpressed with OsMADS1 in specific floral organs (Fig. 8B), implicating some function in floral development. OsbHLH108 may have a role in carpel development as its homolog in Arabidopsis is SPATULA (Heisler et al., 2001). Arabidopsis AtHSF2B is required for normal gametophyte development (Wunderlich et al., 2014), hinting at plausible floral functions for OsHSF24. We show that the nodal genes are linked to other important factors such as OsMADS32/CHIMERIC FLORAL ORGANS and OsYABBY5. OsMADS32 regulates floral organ identity in rice (Sang et al., 2012), and the defective palea, internal lemma-like organs, and extra carpels in florets of the cfo mutant in OsMADS32 partly resemble floral phenotypes of osmads1 mutants (Sang et al., 2012: Prasad et al., 2005; Jeon et al., 2000). Similarly, OsYABBY5 contributes to floral lateral organ development and meristem maintenance (Tanaka et al., 2012), and its deregulation may partly explain organ identity and number defects seen in osmads1 mutants. Overall, our findings indicate that OsMADS1 directly controls the expression of key developmental transcriptional factors to ensure normal floral organ development and FM determinacy.
The interplay between floral developmental regulators and epigenetic factors such as SWI/SNF2 and PHD is well documented (Wu et al., 2012; Yang et al., 2003; Sung et al., 2006; Saiga et al., 2012; Sun and Ito, 2015). Our analysis shows that rice genes encoding Jumoji, PHD-domain, SWI/SNF, and other chromatin regulators, which themselves have no sequence-specific DNA binding activity, are targets of OsMADS1 (Figs. 1, 6B, and 7). Such genes may facilitate changes in chromatin state at other OsMADS1 target loci. The binding of OsMADS1 at loci encoding chromatin remodelers such as SWI/SNF-SWI3, JMJ707, and PHD could serve to establish and/or maintain chromatin states at OsMADS1 target genes, as is well documented in Arabidopsis (Sun and Ito, 2015; Pajoro et al., 2014a).
In this study, we identified the genome-wide targets of OsMADS1 and show that its binding sites have typical CArG-box sequences and in many cases sequences with A-tracts only. Hence, the DNA sequences and structural features of CArG-boxes and A-tracts, as well as their neighboring regions, may be essential for site selection. Using computational approaches, we provide hypotheses for plausible gene regulatory networks and for interaction of OsMADS1 with other factors. These could cumulatively regulate the broad program from FM formation to floral organ development.
MATERIALS AND METHODS
Plant Materials and ChIP
Wild-type panicles of the 2 to 20 mm stage were used to carrying out ChIP. Approximately 250 mg of 1% formaldehyde-fixed, young wild-type rice (Oryza sativa) panicles were ground in liquid nitrogen and then sonicated to yield chromatin with DNA fragments of less than 500 bp in length. ChIP was performed with affinity-purified anti-OsMADS1 antibodies, and the DNA-protein complexes were pulled-down using Protein A Dynabeads (Invitrogen) as described in Khanday et al. (2013). For sequencing, 6 ng of immunoprecipitated DNA was used. The DNA was quantified with a Qubit fluorometer (Invitrogen) as per the manufacturer’s instructions. As the yield of DNA from each ChIP experiments is very low (<1 ng), immunoprecipitated DNA from eight ChIP experiments was pooled to obtain 6 ng of total DNA. The same number of mock ChIP experiments (without the antibody) was processed in exactly the same way to serve as controls. A T-DNA construct of PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR was transformed in the TP309 variety of rice to raise transgenic plants. These plants were subjected to different chemical treatment and panicles were mixed in a specific ratio as described in Khanday et al. (2013).
ChIP-qPCR and RT-PCR
Two independent replicates of OsMADS1 immunoprecipitated chromatin, along with respective no-antibody control (Mock), was prepared as described by Khanday et al. (2013). The final ChIP was diluted 10 times and 1 μL used for qPCR with loci-specific primers (Supplemental Table S1). Enrichment was normalized to 10% of input and was calculated as described by Lin et al. (2012). For gene expression analyses, total RNA was extracted by TriZol (Sigma-Aldrich) as per manufacturer’s specification. Here, 25 ng Oligo d(T) primed cDNA (synthesized with MMLV RTase; NEB) was used for qPCRs with gene-specific primers (Supplemental Table S1). To quantify gene expression levels of genes in wild-type panicles of different stages and in floral organs, OsUBIQUITIN5 transcripts were used as an internal normalizing control. The fold change in transcript levels were estimated with respect to normalized expression levels in leaves. In wild-type and PUbi: OsMADS1-ΔGR; P35S:OsMADS1amiR panicle tissues (MIGR), the OsUBIQUITIN5-normalized Ct values for each gene in each treated sample were compared to the wild-type mock-treated sample (Khanday et al., 2013).
ChIP-Seq, Sequence Alignment, and Peak Detection
Solexa/Illumina sequencing of mock and anti-OsMADS1 immunoprecipitated DNA was carried out following Illumina’s instructions. The 34 nucleotide reads obtained after sequencing were mapped to the rice genome sequence release 6.1 (http://rice.plantbiology.msu.edu/), and only uniquely mapping reads were retained for downstream analysis. Out of approximately 30 million reads, 23 million uniquely mapped to the rice genome (Supplemental Fig. S1). For calculating the length of enriched regions, the SeqMonk program (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/) from the Babraham Institute was used. For peak calling in the OsMADS1-enriched regions, the PICS algorithm available from Strand Genomics (formerly Avadis NGS) was utilized (Zhang et al., 2011). Visualization of sequenced read profile for each loci of interest (Fig. 1) was extracted from Genic view of the Avadis NGS platform.
Data Sets for DNA Structural Analyses
The data set sizes for OsMADS1-bound sequences comprise of 893, 1195, and 866 sequences for those mapping to intergenic sites, positioned within the gene body, and with A-tract sequences, respectively. The 158 sequences bound by OsMADS1 neither contain CArG nor an A-tract motif and they were not included in analyses for DNA biophysical properties.
Analysis of Structural Features and Enrichment of A-Tract Motifs
Three different DNA structural features, AFE (Kanhere and Bansal, 2005; Morey et al., 2011), bendability (Kanhere and Bansal, 2005), and curvature (Bhattacharya and Bansal, 1988; Bansal et al., 1995) as well as frequency of A-tracts (Strahs and Schlick, 2000; McConnell and Beveridge, 2001; Stefl et al., 2004) in the binding site regions, have been analyzed. The binding sites of OsMADS1 were aligned with respect to CArG or A-tract and then structural properties of sequences were calculated for the window spanning −250 to +250 region. Three data sets—gene body, intergenic, and A-tract—were taken for all four analyses mentioned below. For each OsMADS1-bound DNA data set, we compared properties with their respective shuffled sequences. We shuffled the nucleotide positions 10,000 times to generate the randomized sequence set that served as the control sequence set. Details for methods used to calculate of A-tracts and methods for analysis of structural properties (AFE, bendability, and curvature) are described in the Supplemental Materials and Methods.
Motif Search and Analysis
For the identification of DNA motifs in the enriched regions, the genetic algorithm guided formation of spaced dyads coupled with an EM algorithm for motif discovery (GADEM) algorithm from Strand Genomics (formerly Avadis NGS) was used. For probing the percentage of OsMADS1-binding sites containing CArG-boxes and the co-occurrence of binding sites for other transcription factors, the binding regions created by PICS were taken as input. We merged the 866 sequences with an A-tract and 158 sequences without a canonical MADS-domain binding motif with the 2088 sequences containing the CArG-box. The position of the bound sequence relative to the nearest target gene (i.e. intergenic or gene body) was used as the criteria to create the two master data sets. The master gene body data set (final size 1,204) or the master intergenic data set (final size 1,908) were thus categorized based solely on their position relative to the nearest target gene.
Enriched hexamer motifs were screened to trace possible binding sites for other transcription factors in regions bound by OsMADS1. The binding sites of OsMADS1 (as indicated by PICS data) were aligned by centering the motif (CArG or A-tract) and 200 nucleotide sequences flanking the motif (i.e. −100 upstream and +100 downstream) were extracted. In absence of either motif (i.e. 158 OsMADS1-bound regions), the midpoint of the PICS peak was used as 0 and sequences from −100 to +100 were extracted. Frequency of occurrence of all possible hexamers was calculated, using a 1-nucleotide sliding window for the 1,204 master gene body OsMADS1-bound data set and for the 1,908 master intergenic data set. The hexamer frequency was then compared with the frequency of occurrence in the shuffled sequences generated for each of these master data sets. Each sequence was shuffled 10,000 times to generate the randomized sequence data set. Hexamer frequency calculation and shuffled sequences were generated using a Perl script. MatLab R2011b was used to plot hexamer frequencies and to identify hexamers whose frequency in the OsMADS1-bound genomic sequence differed by >2σ from that in the shuffled sequence (Fig. 5). The Osiris database was used to screen 92 transcription factors (Morris et al., 2008; http://www.bioinformatics2.wsu.edu/Osiris) for a match between their binding sites and the enriched hexamer sequences derived from the analysis of the OsMADS1 data sets. The χ2 analysis was performed to calculate the P value for the occurrence of the enriched hexamer motifs in the sequences bound by OsMADS1 and their co-occurrence with a CArG-box if present.
Analysis of Functional Categories and Network for the Identification of Target Genes with Altered Gene Expression
Enrichment of functional categories were done by using MAPMAN for complete bound data set as well as for the overlapped genes found when combined with the gene expression data (Khanday et al., 2013). Details of deriving the OsMADS1-bound regulated and unbound regulated gene lists and details of their statistical functional categorization are in Supplemental Material and Methods. The protocol described in ChipArray (Qin et al., 2011) was used to identify the direct and indirect targets of OsMADS1 for building the network. ChIP-seq data for OsMADS1-bound DNA was combined with our previous data on the global gene expression profile in OsMADS1 knockdown panicles with developing florets. Only the genes whose expression was ≥3-fold reduced in the OsMADS1 knockdown as compared to the wild type (Data Set 1 in Khanday et al., 2013) were used for this purpose. For network development, we included only those cases where OsMADS1 binding took place between −4 kb and +200 bp of the downstream gene TSS based on the position of the Chip-seq DNA peak. Intersection of these 1151 OsMADS1-bound sites and 2147 positively regulated genes gave us the direct targets of OsMADS1. We queried genes whose expression was indirectly affected owing to their link to a direct transcription factor target of OsMADS1. To this aim, we screened for consensus binding sites for transcription factors (direct targets of OsMADS1) in promoter regions (−500 to +100 bp with regard to the TSS at position 0) of the genes with reduced expression in the gene expression data set of OsMADS1 knockdown tissue. The consensus sequences were obtained from the transcription factor database “PLACE” (http://www.dna.affrc.go.jp/PLACE/), and the network was created using Cytoscape 3.0.2. The 269 genes linked to OsbHLH108 contained a G-box (CACGTG) in their TSS proximal regions. The 183 genes downstream to OsERF034 and the 243 genes downstream to OsHSF24 contained the AGCCGCC and GAANNTCC/TTCNNGAA elements, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Quality assurance of OsMADS1 immunoprecipitated sample and mapping of the ChIP-seq reads.
Supplemental Figure S2. Genomic view of OsMADS1 peaks and density of OsMADS1 binding sites.
Supplemental Figure S3. Semiquantitative PCR validation for some additional OsMADS1 ChIP-seq binding sites.
Supplemental Figure S4. Length of OsMADS1-bound sequences in the genome.
Supplemental Figure S5. A representation of the overlap of OsMADS1-bound loci with the genes modulated by OsMADS1.
Supplemental Table S1. List of primers for annotated genes used for all ChIP validations and gene expression studies.
Supplemental Data Set S1. List of genes bound by OsMADS1.
Supplemental Data Set S2. Lists of hexamers found in OsMADS1-bound sequences.
Supplemental Data Set S3. List of important genes present in various functional categories as per MAPMAN analysis.
Supplemental Data Set S4A. List of genes positively and directly regulated by OsMADS1.
Supplemental Data Set S4B. List of genes regulated by three direct transcription factor target genes of OsMADS1.
Supplementary Material
Acknowledgments
We gratefully acknowledge the help of Drs. Ramesh Hariharan, Vamsi, Pramila Tata, and Janakiraman from Strand Life Sciences, Bangalore with mapping of raw reads and preliminary ChIP-seq data analysis.
Footnotes
This work was supported by the Indian Institute of Science and the University of Zurich and grants from the Department of Biotechnology, Government of India on Rice Functional Genomics (grant no. BT/AB/FG-I PH-II/2009 to U.V.), the Department of Science and Technology, J.C. Bose fellowship (grant no. SR/S2/JCB-49/2009 to U.V.), a J.C. Bose Fellowship (grant no. SR/S2/JCB-47/2009 to M.B.), and a Personal Exchange Programme (grant no. RF29 to I.K., U.V., and U.G.) through the Indo-Swiss Joint Research Programme.
Articles can be viewed without a subscription.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal GK, Abe K, Yamazaki M, Miyao A, Hirochika H (2005) Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol Biol 59: 125–135 [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, Ribas de Pouplana L, Martínez-Castilla L, Yanofsky MF (2000) An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc Natl Acad Sci USA 97: 5328–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampomah-Dwamena C, Morris BA, Sutherland P, Veit B, Yao JL (2002) Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol 130: 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal M, Bhattacharyya D, Ravi B (1995) NUPARM and NUCGEN: software for analysis and generation of sequence dependent nucleic acid structures. Comput Appl Biosci 11: 281–287 [DOI] [PubMed] [Google Scholar]
- Bäumlein H, Nagy I, Villarroel R, Inzé D, Wobus U (1992) Cis-analysis of a seed protein gene promoter: the conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. Plant J 2: 233–239 [PubMed] [Google Scholar]
- Bhattacharya D, Bansal M (1988) A general procedure for generation of curved DNA molecules. J Biomol Struct Dyn 6: 93–104 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Drews GN, Meyerowitz EM (1991) Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 3: 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukner I, Sánchez R, Suck D, Pongor S (1995) Sequence-dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO J 14: 1812–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch MA, Bomblies K, Weigel D (1999) Activation of a floral homeotic gene in Arabidopsis. Science 285: 585–587 [DOI] [PubMed] [Google Scholar]
- Chen ZX, Wu JG, Ding WN, Chen HM, Wu P, Shi CH (2006) Morphogenesis and molecular basis on naked seed rice, a novel homeotic mutation of OsMADS1 regulating transcript level of AP3 homologue in rice. Planta 223: 882–890 [DOI] [PubMed] [Google Scholar]
- Christensen AR, Malcomber ST (2012) Duplication and diversification of the LEAFY HULL STERILE1 and Oryza sativa MADS5 SEPALLATA lineages in graminoid Poales. Evodevo 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciaffi M, Paolacci AR, Tanzarella OA, Porceddu E (2011) Molecular aspects of flower development in grasses. Sex Plant Reprod 24: 247–282 [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Cui R, Han J, Zhao S, Su K, Wu F, Du X, Xu Q, Chong K, Theissen G, Meng Z (2010) Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J 61: 767–781 [DOI] [PubMed] [Google Scholar]
- de Folter S, Angenent GC (2006) Trans meets cis in MADS science. Trends Plant Sci 11: 224–231 [DOI] [PubMed] [Google Scholar]
- Deng W, Ying H, Helliwell CA, Taylor JM, Peacock WJ, Dennis ES (2011) FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc Natl Acad Sci USA 108: 6680–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh TT, Girke T, Liu X, Yant L, Schmid M, Chen X (2012) The floral homeotic protein APETALA2 recognizes and acts through an AT-rich sequence element. Development 139: 1978–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14: 1935–1940 [DOI] [PubMed] [Google Scholar]
- Dreni L, Pilatone A, Yun D, Erreni S, Pajoro A, Caporali E, Zhang D, Kater MM (2011) Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 23: 2850–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Cortines M, Saedler H, Sommer H (1999) Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J 18: 5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Liang W, Yin C, Ji S, Wang H, Su X, Guo C, Kong H, Xue H, Zhang D (2010) The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol 153: 728–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Andrés F, Sessa A, Guerra RF, Simonini S, Mateos JL, Torti S, Zambelli F, Prazzoli GM, Bjerkan KN, et al. (2013) Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biol 14: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremski K, Ditta G, Yanofsky MF (2007) The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134: 3593–3601 [DOI] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV (1995) Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell 7: 1879–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart CM, Nagy F, Meins F Jr (1993) A 61 bp enhancer element of the tobacco beta-1,3-glucanase B gene interacts with one or more regulated nuclear proteins. Plant Mol Biol 21: 121–131 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Atkinson A, Bylstra YH, Walsh R, Smyth DR (2001) SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128: 1089–1098 [DOI] [PubMed] [Google Scholar]
- Hu Y, Liang W, Yin C, Yang X, Ping B, Li A, Jia R, Chen M, Luo Z, Cai Q, et al. (2015) Interactions of OsMADS1 with floral homeotic genes in rice flower development. Mol Plant 8: 1366–1384 [DOI] [PubMed] [Google Scholar]
- Huang K, Louis JM, Donaldson L, Lim FL, Sharrocks AD, Clore GM (2000) Solution structure of the MEF2A-DNA complex: structural basis for the modulation of DNA bending and specificity by MADS-box transcription factors. EMBO J 19: 2615–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RG, Kaufmann K, Angenent GC (2010) The ‘ABC’ of MADS domain protein behaviour and interactions. Semin Cell Dev Biol 21: 87–93 [DOI] [PubMed] [Google Scholar]
- Immink RG, Posé D, Ferrario S, Ott F, Kaufmann K, Valentim FL, de Folter S, van der Wal F, van Dijk AD, Schmid M, Angenent GC (2012) Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol 160: 433–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Jang S, Lee S, Nam J, Kim C, Lee SH, Chung YY, Kim SR, Lee YH, Cho YG, An G (2000) leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Lee S, An G (2008) Intragenic control of expression of a rice MADS box gene OsMADS1. Mol Cells 26: 474–480 [PubMed] [Google Scholar]
- Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Bansal M (2005) Structural properties of promoters: similarities and differences between prokaryotes and eukaryotes. Nucleic Acids Res 33: 3165–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC (2009) Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muiño JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueño F, Krajewski P, Meyerowitz EM, Angenent GC, Riechmann JL (2010) Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Khanday I, Yadav SR, Vijayraghavan U (2013) Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways. Plant Physiol 161: 1970–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Soltis PS, Wall K, Soltis DE (2006) Phylogeny and domain evolution in the APETALA2-like gene family. Mol Biol Evol 23: 107–120 [DOI] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8: 137–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J (2010) PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol 51: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki MK, Kitotaka O, Nishino E, Shimura Y (1988) Isolation and characterization of novel mutants of Arabidopsis thaliana defective in flower development. Development 104: 195–203 [Google Scholar]
- Kosugi S, Ohashi Y (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Suzuka I, Ohashi Y (1995) Two of three promoter elements identified in a rice gene for proliferating cell nuclear antigen are essential for meristematic tissue-specific expression. Plant J 7: 877–886 [DOI] [PubMed] [Google Scholar]
- Koudritsky M, Domany E (2008) Positional distribution of human transcription factor binding sites. Nucleic Acids Res 36: 6795–6805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B. (2009) AINTEGUMENTA and AINTEGUMENTA-LIKE6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiol 150: 1916–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Fletcher JC (2005) Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet 6: 688–698 [DOI] [PubMed] [Google Scholar]
- Lee DY, An G (2012) Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J 69: 445–461 [DOI] [PubMed] [Google Scholar]
- Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, et al. (2006) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol 141: 1167–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Moon YH, An G, Jang SK (2000) Two rice MADS domain proteins interact with OsMADS1. Plant Mol Biol 44: 513–527 [DOI] [PubMed] [Google Scholar]
- Lin X, Tirichine L, Bowler C (2012) Protocol: Chromatin immunoprecipitation (ChIP) methodology to investigate histone modifications in two model diatom species. Plant Methods 8: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CA, Lim EK, Yu SM (1998) Sugar response sequence in the promoter of a rice α-amylase gene serves as a transcriptional enhancer. J Biol Chem 273: 10120–10131 [DOI] [PubMed] [Google Scholar]
- Lu L, Sun K, Chen X, Zhao Y, Wang L, Zhou L, Sun H, Wang H (2013) Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO J 32: 2575–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier UG, Brown JW, Tologcyzki C, Feix G (1987) Binding of a nuclear factor to a consensus sequence in the 5′ flanking region of zein genes from maize. EMBO J 6: 17–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA (2004) Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell 16: 1692–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA (2005) SEPALLATA gene diversification: brave new whorls. Trends Plant Sci 10: 427–435 [DOI] [PubMed] [Google Scholar]
- McConnell KJ, Beveridge DL (2001) Molecular dynamics simulations of B′-DNA: sequence effects on A-tract-induced bending and flexibility. J Mol Biol 314: 23–40 [DOI] [PubMed] [Google Scholar]
- McElroy D, Zhang W, Cao J, Wu R (1990) Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2: 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer R, Theissen G (2009) Reconstitution of ‘floral quartets’ in vitro involving class B and class E floral homeotic proteins. Nucleic Acids Res 37: 2723–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F, Dubois E (2003) Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316: 1–21 [DOI] [PubMed] [Google Scholar]
- Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819: 86–96 [DOI] [PubMed] [Google Scholar]
- Morey C, Mookherjee S, Rajasekaran G, Bansal M (2011) DNA free energy-based promoter prediction and comparative analysis of Arabidopsis and rice genomes. Plant Physiol 156: 1300–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RT, O’Connor TR, Wyrick JJ (2008) Osiris: an integrated promoter database for Oryza sativa L. Bioinformatics 24: 2915–2917 [DOI] [PubMed] [Google Scholar]
- Muiño JM, Smaczniak C, Angenent GC, Kaufmann K, van Dijk AD (2014) Structural determinants of DNA recognition by plant MADS-domain transcription factors. Nucleic Acids Res 42: 2138–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutasa-Göttgens E, Hedden P (2009) Gibberellin as a factor in floral regulatory networks. J Exp Bot 60: 1979–1989 [DOI] [PubMed] [Google Scholar]
- Nagaich AK, Bhattacharyya D, Brahmachari SK, Bansal M (1994) CA/TG sequence at the 5′ end of oligo(A)-tracts strongly modulates DNA curvature. J Biol Chem 269: 7824–7833 [PubMed] [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y (2003) SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130: 705–718 [DOI] [PubMed] [Google Scholar]
- Nair TM. (2010) Sequence periodicity in nucleosomal DNA and intrinsic curvature. BMC Struct Biol 10(Suppl 1): S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ó’Maoiléidigh DS, Wuest SE, Rae L, Raganelli A, Ryan PT, Kwasniewska K, Das P, Lohan AJ, Loftus B, Graciet E, Wellmer F (2013) Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 25: 2482–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajoro A, Biewers S, Dougali E, Leal Valentim F, Mendes MA, Porri A, Coupland G, van de Peer Y, van Dijk AD, Colombo L, Davies B, Angenent GC (2014a) The (r)evolution of gene regulatory networks controlling Arabidopsis plant reproduction: a two-decade history. J Exp Bot 65: 4731–4745 [DOI] [PubMed] [Google Scholar]
- Pajoro A, Madrigal P, Muiño JM, Matus JT, Jin J, Mecchia MA, Debernardi JM, Palatnik JF, Balazadeh S, Arif M, et al. (2014b) Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol 15: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF (2003) Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88 [DOI] [PubMed] [Google Scholar]
- Plackett ARG, Thomas SG, Wilson ZA, Hedden P (2011) Gibberellin control of stamen development: a fertile field. Trends Plant Sci 16: 568–578 [DOI] [PubMed] [Google Scholar]
- Prasad K, Kushalappa K, Vijayraghavan U (2003) Mechanism underlying regulated expression of RFL, a conserved transcription factor, in the developing rice inflorescence. Mech Dev 120: 491–502 [DOI] [PubMed] [Google Scholar]
- Prasad K, Parameswaran S, Vijayraghavan U (2005) OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J 43: 915–928 [DOI] [PubMed] [Google Scholar]
- Prasad K, Sriram P, Kumar CS, Kushalappa K, Vijayraghavan U (2001) Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev Genes Evol 211: 281–290 [DOI] [PubMed] [Google Scholar]
- Prasad K, Vijayraghavan U (2003) Double-stranded RNA interference of a rice PI/GLO paralog, OsMADS2, uncovers its second-whorl-specific function in floral organ patterning. Genetics 165: 2301–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li MJ, Wang P, Zhang MQ, Wang J (2011) ChIP-Array: combinatory analysis of ChIP-seq/chip and microarray gene expression data to discover direct/indirect targets of a transcription factor. Nucleic Acids Res 39: W430–W436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Krizek BA, Meyerowitz EM (1996) Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA 93: 4793–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiga S, Möller B, Watanabe-Taneda A, Abe M, Weijers D, Komeda Y (2012) Control of embryonic meristem initiation in Arabidopsis by PHD-finger protein complexes. Development 139: 1391–1398 [DOI] [PubMed] [Google Scholar]
- Sang X, Li Y, Luo Z, Ren D, Fang L, Wang N, Zhao F, Ling Y, Yang Z, Liu Y, He G (2012) CHIMERIC FLORAL ORGANS1, encoding a monocot-specific MADS box protein, regulates floral organ identity in rice. Plant Physiol 160: 788–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore P, Sharrocks AD (1995) The MADS-box family of transcription factors. Eur J Biochem 229: 1–13 [DOI] [PubMed] [Google Scholar]
- Simpson SD, Nakashima K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J 33: 259–270 [DOI] [PubMed] [Google Scholar]
- Slama-Schwok A, Zakrzewska K, Léger G, Leroux Y, Takahashi M, Käs E, Debey P (2000) Structural changes induced by binding of the high-mobility group I protein to a mouse satellite DNA sequence. Biophys J 78: 2543–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C, Immink RG, Angenent GC, Kaufmann K (2012a) Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139: 3081–3098 [DOI] [PubMed] [Google Scholar]
- Smaczniak C, Immink RG, Muiño JM, Blanvillain R, Busscher M, Busscher-Lange J, Dinh QD, Liu S, Westphal AH, Boeren S, et al. (2012b) Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci USA 109: 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Liu Z (2006) APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133: 3159–3166 [DOI] [PubMed] [Google Scholar]
- Stefl R, Wu H, Ravindranathan S, Sklenár V, Feigon J (2004) DNA A-tract bending in three dimensions: solving the dA4T4 vs. dT4A4 conundrum. Proc Natl Acad Sci USA 101: 1177–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahs D, Schlick T (2000) A-Tract bending: insights into experimental structures by computational models. J Mol Biol 301: 643–663 [DOI] [PubMed] [Google Scholar]
- Sun B, Ito T (2015) Regulation of floral stem cell termination in Arabidopsis. Front Plant Sci 6: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Schmitz RJ, Amasino RM (2006) A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev 20: 3244–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka W, Toriba T, Ohmori Y, Yoshida A, Kawai A, Mayama-Tsuchida T, Ichikawa H, Mitsuda N, Ohme-Takagi M, Hirano HY (2012) The YABBY gene TONGARI-BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice spikelet. Plant Cell 24: 80–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Li G, Chen M (2007) The phylogeny and expression pattern of APETALA2-like genes in rice. J Genet Genomics 34: 930–938 [DOI] [PubMed] [Google Scholar]
- Tang W, Perry SE (2003) Binding site selection for the plant MADS domain protein AGL15: an in vitro and in vivo study. J Biol Chem 278: 28154–28159 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Uimari A, Kotilainen M, Elomaa P, Yu D, Albert VA, Teeri TH (2004) Integration of reproductive meristem fates by a SEPALLATA-like MADS-box gene. Proc Natl Acad Sci USA 101: 15817–15822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Tang D, Hong L, Xu W, Huang J, Li M, Gu M, Xue Y, Cheng Z (2010) DEP and AFO regulate reproductive habit in rice. PLoS Genet 6: e1000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AG, Causier BE, Davies B, Sharrocks AD (1998) DNA binding and dimerisation determinants of Antirrhinum majus MADS-box transcription factors. Nucleic Acids Res 26: 5277–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AG, Shore P, Sharrocks AD (1997) DNA binding by MADS-box transcription factors: a molecular mechanism for differential DNA bending. Mol Cell Biol 17: 2876–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Sang Y, Bezhani S, Yamaguchi N, Han SK, Li Z, Su Y, Slewinski TL, Wagner D (2012) SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc Natl Acad Sci USA 109: 3576–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest SE, Ó’Maoiléidigh DS, Rae L, Kwasniewska K, Raganelli A, Hanczaryk K, Lohan AJ, Loftus B, Graciet E, Wellmer F (2012) Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc Natl Acad Sci USA 109: 13452–13457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich M, Gross-Hardt R, Schöffl F (2014) Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol Biol 85: 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Jeong CW, Nole-Wilson S, Krizek BA, Wagner D (2016) AINTEGUMENTA and AINTEGUMENTA-LIKE6/PLETHORA3 induce LEAFY expression in response to auxin to promote the onset of flower formation in Arabidopsis. Plant Physiol 170: 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Makaroff CA, Ma H (2003) The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 15: 1281–1295 [PMC free article] [PubMed] [Google Scholar]
- Yu CP, Lin JJ, Li WH (2016) Positional distribution of transcription factor binding sites in Arabidopsis thaliana. Sci Rep 6: 25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Robertson G, Krzywinski M, Ning K, Droit A, Jones S, Gottardo R (2011) PICS: probabilistic inference for ChIP-seq. Biometrics 67: 151–163 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE (2009) Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu E, You C, Wang S, Cui J, Niu B, Wang Y, Qi J, Ma H, Chang F (2015) The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J 83: 976–990 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.