Oxygen-regulated stability of the ERF-VII transcription factor RAP2.12 regulates central metabolic processes to sustain growth, development, and anoxia responses.
Abstract
Subgroup-VII-ethylene-response-factor (ERF-VII) transcription factors are involved in the regulation of hypoxic gene expression and regulated by proteasome-mediated proteolysis via the oxygen-dependent branch of the N-end-rule pathway. While research into ERF-VII mainly focused on their role to regulate anoxic gene expression, little is known on the impact of this oxygen-sensing system in regulating plant metabolism and growth. By comparing Arabidopsis (Arabidopsis thaliana) plants overexpressing N-end-rule-sensitive and insensitive forms of the ERF-VII-factor RAP2.12, we provide evidence that oxygen-dependent RAP2.12 stability regulates central metabolic processes to sustain growth, development, and anoxic resistance of plants. (1) Under normoxia, overexpression of N-end-rule-insensitive Δ13RAP2.12 led to increased activities of fermentative enzymes and increased accumulation of fermentation products, which were accompanied by decreased adenylate energy states and starch levels, and impaired plant growth and development, indicating a role of oxygen-regulated RAP2.12 degradation to prevent aerobic fermentation. (2) In Δ13RAP2.12-overexpressing plants, decreased carbohydrate reserves also led to a decrease in anoxic resistance, which was prevented by external Suc supply. (3) Overexpression of Δ13RAP2.12 led to decreased respiration rates, changes in the levels of tricarboxylic acid cycle intermediates, and accumulation of a large number of amino acids, including Ala and γ-amino butyric acid, indicating a role of oxygen-regulated RAP2.12 abundance in controlling the flux-modus of the tricarboxylic acid cycle. (4) The increase in amino acids was accompanied by increased levels of immune-regulatory metabolites. These results show that oxygen-sensing, mediating RAP2.12 degradation is indispensable to optimize metabolic performance, plant growth, and development under both normoxic and hypoxic conditions.
Plants experience low oxygen concentrations in specific tissues during their development (Geigenberger 2003; Licausi et al., 2011a; Geigenberger 2014; van Dongen and Licausi 2015) and in response to decreased oxygen availability in their environment due to waterlogging and flooding (Voesenek and Bailey-Serres 2015). Under these conditions, oxygen deprivation leads to the induction of a specific set of genes, which is involved in carbohydrate mobilization and fermentation to sustain substrate-level ATP production and promote hypoxic acclimation (van Dongen et al., 2009). Different members of subgroup VII ethylene-response factor (ERF) transcription factors have been identified to be involved in the regulation of hypoxic gene expression in Arabidopsis (Arabidopsis thaliana; Papdi et al., 2008; Hinz et al., 2010; Licausi et al., 2010, 2011b; Gasch et al., 2016). One of these members, RAP2.12, has been found to be regulated by intracellular relocalization and protein stability in response to low oxygen concentrations (Gibbs et al., 2011, Licausi et al., 2011c; Kosmacz et al., 2015). Upon hypoxia, RAP2.12 moves from the plasma membrane into the nucleus to specifically activate the expression of anaerobic genes, such as PYRUVATE DECARBOXYLASE1 (PDC1) and ALCOHOL DEHYDROGENASE1 (ADH1) involved in ethanol fermentation, SUC SYNTHASE1 (SUS1) and SUS4 involved in carbohydrate degradation, and HEMOGLOBIN1 (HB1) involved most probably in scavenging of nitric oxide to promote conversion of NADH to NAD+ (Licausi et al., 2011c).
In normoxia (21% v/v oxygen) and after reoxygenation, RAP2.12 is targeted to proteasome-mediated proteolysis via the oxygen-dependent branch of the N-end-rule pathway to down-regulate the hypoxic response (Licausi et al., 2011c, Kosmacz et al., 2015). The presence of molecular oxygen leads to oxidation of the penultimate Cys in the N terminus of RAP2.12 by specific plant Cys oxidases, targeting RAP2.12 to proteasomal degradation and leading to its destabilization (Weits et al., 2014). This is prevented in Arabidopsis plants overexpressing Δ13RAP2.12, which lacks the first 13 amino acids from the N terminus including the regulatory Cys and representing an N-end-rule-insensitive version of RAP2.12 (Licausi et al., 2011c). RAP2.12 and Cys oxidases therefore act as an oxygen-sensing system in Arabidopsis plants, allowing expression of hypoxic genes to be strictly regulated in response to oxygen supply (van Dongen and Licausi 2015). When oxygen falls to concentrations below 10% (v/v), RAP2.12 is stabilized and relocated to the nucleus to induce hypoxic gene expression (Kosmacz et al., 2015). This is fine-tuned by an antagonistic interplay between RAP2.12 and the trihelix transcription factor HYPOXIA RESPONSE ATTENUATOR1 enabling a flexible response to fluctuating hypoxia (Giuntoli et al., 2014).
Transgenic studies have been used to investigate the role of RAP2.12 in hypoxic stress acclimation; however, the results were partly contradicting. While Arabidopsis lines overexpressing RAP2.12 showed increased survival of anoxic stress conditions, lines overexpressing the N-end-rule-insensitive Δ13RAP2.12 showed the opposite (Licausi et al., 2011c). This is puzzling, because Δ13RAP2.12 overexpressors showed a constitutive hypoxic gene expression, which was stronger than the low-oxygen-inducible one in RAP2.12 lines (Licausi et al., 2011c). Moreover, studies with N-end-rule knock-out mutants such as deficient in arginyl-tRNA protein transferases (ate1ate2) and deficient in the ubiquitin ligase PROTEOLYSIS6 (prt6) showed that constitutive expression of RAP2.12 in the context of these mutants leads to increased anoxic survival rates (Gibbs et al., 2011), which is contradicting the results of Licausi et al. (2011c). Several options were suggested to explain the discrepancy between the studies, such as differences in the developmental stage and growth conditions such as the day/night cycle, air humidity, and the presence/absence of Suc in the medium (Sasidharan and Mustroph, 2011; Bailey-Serres et al., 2012; Riber et al., 2015). It remained unclear, however, how regulated and constitutive expression of RAP2.12 effect plant metabolism under normoxic and hypoxic conditions, and how this works out on low-oxygen tolerance of these plants.
In this work, we investigated the influence of oxygen-regulated RAP2.12 stabilization on plant metabolism under normoxic and hypoxic conditions, and how this affects plant growth and survival of anoxic conditions. To do this, we analyzed Arabidopsis plants overexpressing either the N-end-rule-sensitive RAP2.12 or the N-end-rule-insensitive Δ13RAP2.12, which is not targeted to proteasome-mediated degradation in response to oxygen (Licausi et al., 2011c). Under normoxic conditions, stabilization of RAP2.12 led to constitutive expression of hypoxic marker genes, increased activities of fermentative enzymes such as lactate dehydrogenase (LDH), PDC, ADH, and Ala aminotransferase (AlaAT), and accumulation of fermentation products such as lactate, ethanol, and Ala, resulting in decreased levels of starch and adenylate energy states. This was accompanied by decreased plant growth and developmental progression under normoxia and decreased survival after anoxia. In addition to this, stabilization of RAP2.12 also affected respiration rates, tricarboxylic acid (TCA) cycle intermediates, and amino acid levels, indicating a role of oxygen sensing mediated by RAP2.12 stabilization in controlling the flux-modus of the TCA cycle.
RESULTS
Overexpression of a Stabilized RAP2.12 Leads to Reduced Biomass and Delayed Developmental Progression under Normoxic Conditions
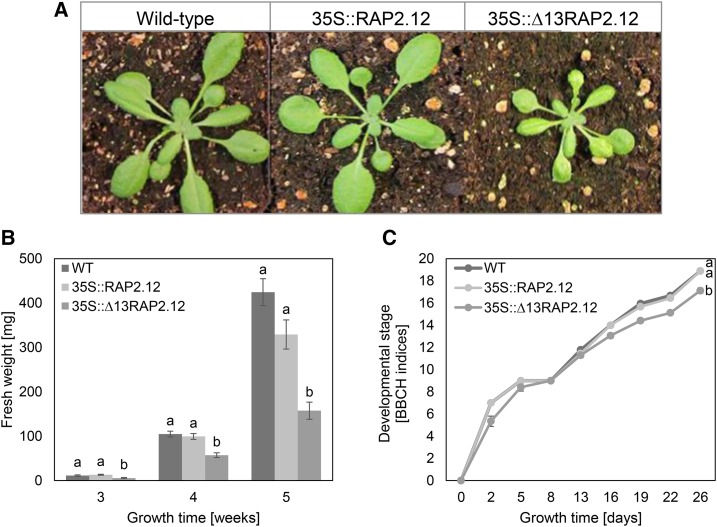
To investigate the effect of a constitutive expression of RAP2.12 on plant growth and development, 35S::Δ13RAP2.12 Arabidopsis plants growing for 5 weeks under normoxic conditions (21%) were analyzed in comparison to 35S::RAP2.12 and the wild type. Overexpression of the stabilized Δ13RAP2.12 led to a decrease in rosette size (Fig. 1A), fresh-weight (Fig. 1B), and development progression (Fig. 1C), while plants overexpressing the oxygen-regulated RAP2.12 were similar to wild type. This confirms previous studies, showing decreased growth of 35S::Δ13RAP2.12 Arabidopsis plants under normoxic conditions (Licausi et al., 2011c).
Figure 1.
Overexpression of the N-end-rule-insensitive Δ13RAP2.12 transcription factor under the control of the 35S promoter leads to decreased growth and delayed development under normoxic conditions. A, Rosette size; B, fresh weight; C, development of 35S::Δ13RAP2.12 plants in comparison to 35S::RAP2.12 and the wild type (WT). Plants were grown for 4 weeks (A) or for the times indicated in the respective figures (B and C). Results in B and C are means ± se (n = 5–6 biological replicates for fresh weight determination or 18 for development analysis) with significant differences indicated by different letters (according to one-way ANOVA for fresh weight determination and two-way ANOVA for development analysis, P < 0.05). Developmental stages were determined according to the BBCH-scale (Hess et al., 1997). FW, Fresh weight.
Overexpression of a Stabilized RAP2.12 Leads to Induced Expression of Anoxic Marker Genes and Fermentative Enzyme Activities under Normoxic Conditions
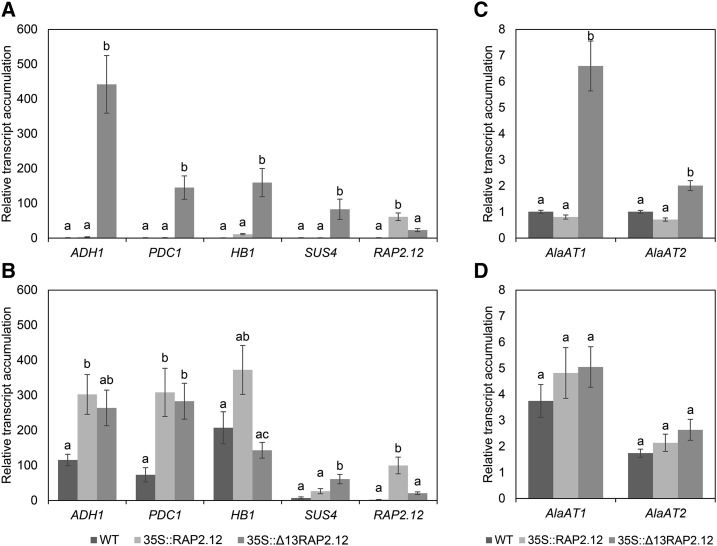
To investigate the effects on mRNA expression levels of anoxic marker genes, 5-week-old plants were exposed for 16 h to 21% or 1% (v/v) oxygen in the dark, before they were harvested for RT-PCR analyses (Fig. 2). When plants were compared at normoxic conditions (21% v/v oxygen), 35S-driven expression of Δ13RAP2.12 resulted in elevated levels of ADH1, PDC1, HB1, SUS4, AlaAT1, and AlaAT2 transcripts, while those in 35S::RAP2.12 and wild-type plants were close to or below the detection limit (Fig. 2, A and C), confirming previous studies (Licausi et al., 2011c). Hypoxic treatment (1% v/v oxygen) led to a strong increase in ADH1, PDC1, HB1, SUS4, AlaAT1, and AlaAT2 transcript levels in wild-type and 35S:RAP2.12 plants (the increase being more marked in 35S::RAP2.12 than in the wild type), while there was no further increase upon hypoxic treatment in 35S::Δ13RAP2.12 plants (Fig. 2, B and D).
Figure 2.
Overexpression of the N-end-rule-insensitive Δ13RAP2.12 transcription factor under the control of the 35S promoter leads to constitutive expression of anoxic marker genes. After 16 h incubation in normoxia (A and C) or hypoxia (B and D) in the absence of external sugars in the dark, 5-week-old wild-type, RAP2.12, and Δ13RAP2.12-overexpressing plants were sampled to analyze the mRNA expression levels of ADH1, PDC1, HB1, SUS4, and RAP2.12 (A and B) as well as AlaAT1 and AlaAT2 (C and D) in whole rosettes. Results are means ± se (n = 8–10 biological replicates) normalized to the normoxic wild type. Values of the different genotypes that significantly differ from each other are indicated by different letters (according to one-way ANOVA test, P < 0.05). WT, Wild type.
To put these data into context, RAP2.12 expression levels were analyzed by using primers for RT-PCR that do not distinguish between RAP2.12 and Δ13RAP2.12. Compared to wild type, the 35S::RAP2.12 and 35S::Δ13RAP2.12 plants revealed strongly elevated levels of RAP2.12 mRNA under normoxic (Fig. 2A) and hypoxic conditions (Fig. 2B). Intriguingly, under both conditions RAP2.12 mRNA levels were more strongly induced in 35S::RAP2.12 than in 35S::Δ13RAP2.12 lines.
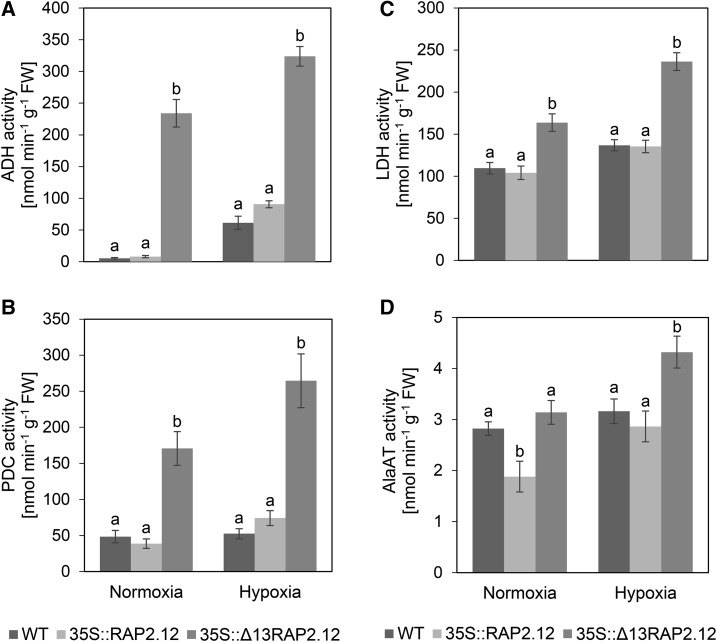
To investigate whether the induction in the mRNA levels of anoxic genes is leading to corresponding increases in fermentative enzyme activities, we analyzed the activities of ADH, PDC, LDH, and AlaAT in samples taken in parallel. Under normoxic conditions, 35S::Δ13RAP2.12 plants showed strongly increased activities of ADH (Fig. 3A), PDC (Fig. 3B), LDH (Fig. 3C), and AlaAT (Fig. 3D), compared to 35S::RAP2.12 or (with the exception of AlaAT) wild-type plants. Hypoxic treatment led to increased activities of these enzymes also in wild-type and 35S::RAP2.12 plants, but the values remained below those reached by 35S::Δ13RAP2.12 plants (Fig. 3, A–D).
Figure 3.
Overexpression of the N-end-rule-insensitive Δ13RAP2.12 transcription factor under the control of the 35S promoter leads to a constitutive activation of fermentative enzymes. After 16 h incubation in normoxia or hypoxia in the absence of external sugars in the dark, 5-week-old wild-type, RAP2.12, and Δ13RAP2.12 overexpressing plants were sampled to analyze activities of A, ADH; B, PDC; C, LDH; and D, AlaAT in whole rosettes. Results are means ± se (n = 8–10 biological replicates for activity assay of ADH and 6 for PDC, LDH, and AlaAT). Values of the different genotypes that significantly differ from each other are indicated by different letters (according to one-way ANOVA test, P < 0.05). Growth of plants and rosette stage analyzed was similar as in Fig. 2. WT = wild type.
Overexpression of a Stabilized RAP2.12 Leads to Accumulation of Fermentation Products and Decreased Respiration Rates under Normoxic Conditions
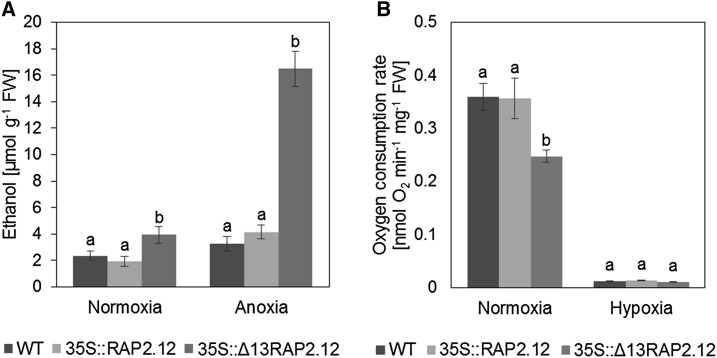
The above results demonstrate that under normoxic conditions, overexpression of the N-end-rule-insensitive Δ13RAP2.12 leads to increased expression of anaerobic proteins and increased activities of fermentative enzymes. To investigate whether this is accompanied by an induction of fermentation, we analyzed the levels of ethanol (Fig. 4A), lactate, Ala, and γ-amino butyrate (GABA; Fig. 5; Supplemental Table S1) as end-products of fermentative metabolism. Because ethanol is volatile, it is very difficult to measure in plant material of soil-grown plants. We therefore analyzed ethanol in a separate experiment using 1-week-old hydroponic seedlings, where ethanol could be easily measured in the solution in which seedlings were immersed. Under normoxic conditions, ethanol, lactate, Ala, and GABA were markedly increased in 35S::Δ13RAP2.12 plants, while 35S::RAP2.12 plants were similar to wild type (Figs. 4A and 5, B and C; Supplemental Table S1). Hypoxic treatment led to an increase in ethanol, lactate, Ala, and GABA levels also in 35S::RAP2.12 plants; however, the values obtained were not reaching those of 35S::Δ13RAP2.12 plants.
Figure 4.
Overexpression of the N-end-rule-insensitive Δ13RAP2.12 transcription factor under the control of the 35S promoter leads to increased ethanol production and decreased respiration rates under normoxic conditions. A, To analyze ethanol levels, seedlings were grown in liquid culture containing 1/2 MS medium and 30 mm Suc for 1 week under long-day conditions. After 10 h into the photoperiod, the medium was exchanged to 1/2 MS containing 90 mm Suc and the seedlings were subjected to normoxia or anoxia for further 16 h in the dark, before ethanol levels were measured. B, To analyze respiration rates, whole rosettes of 5-week-old plants were sampled at the end of the night and leaf material representing the whole rosette was introduced into the measuring chamber of an oxygen electrode to analyze oxygen consumption rates in normoxic or hypoxic conditions in the absence of external Suc in the dark. Results are means ± se (n = 25–35 different sets of plants for ethanol assay and seven biological replicates for respiration rates). Values of the different genotypes that significantly differ from each other are indicated by different letters (according to one-way ANOVA test, P < 0.05). WT, Wild type.
Figure 5.
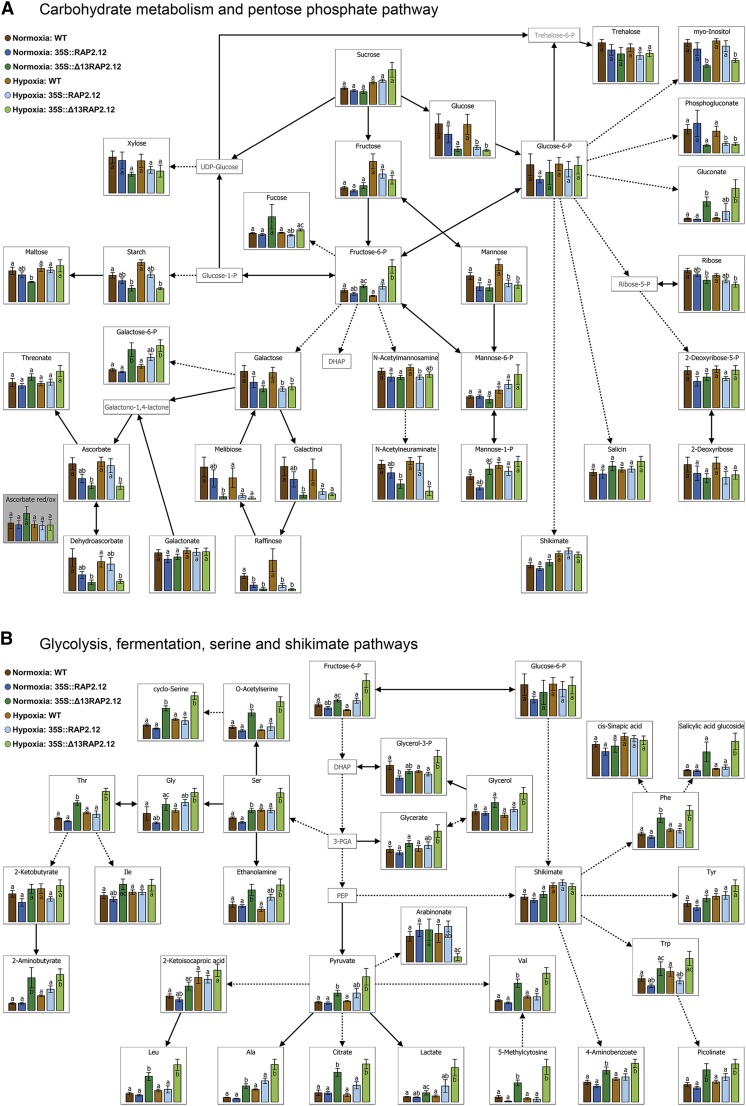
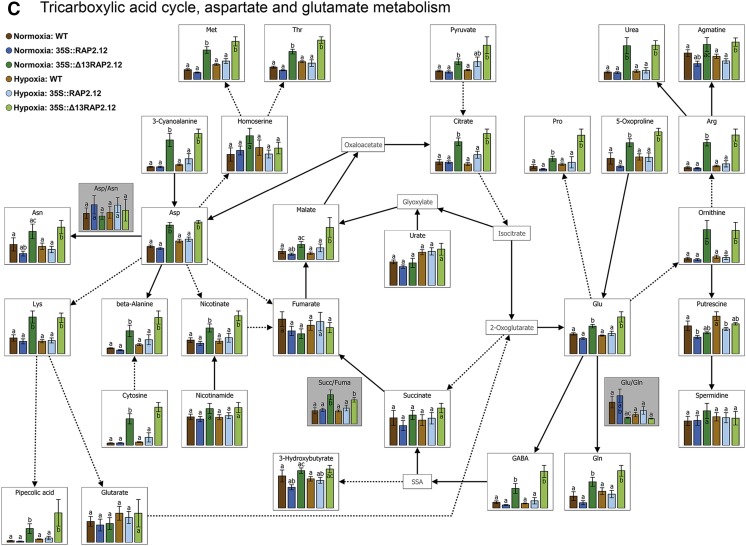
Overexpression of the N-end-rule-insensitive Δ13RAP2.12 transcription factor under the control of the 35S promoter leads to severe changes in metabolite profiles. Metabolite levels from whole rosettes of 5-week-old wild-type, RAP2.12, and Δ13RAP2.12-overexpressing plants sampled after 16 h incubation in normoxia or hypoxia in the absence of sugars in the dark are visualized using the following VANTED diagrams: A, Carbohydrate metabolism and pentose P-pathway; B, glycolysis, fermentation, Ser, and shikimate pathways; and C, tricarboxylic acid cycle, Asp, and Glu metabolism. Results are means ± se (n = 8–10 biological replicates) and are expressed as arbitrary values; the scaling of the y axis was therefore omitted. Values of the different genotypes that significantly differ from each other within one oxygen treatment are indicated by different letters (according to one-way ANOVA test, P < 0.05). Metabolite ratios are shown in boxes with gray background color. In Supplemental Table S1, the complete metabolite profile is shown together with a statistical analysis of the data. Growth of plants and rosette stage analyzed was similar as in Fig. 2. WT, Wild type.
To investigate whether overexpression of Δ13RAP2.12 also affects respiration rates, oxygen consumption of leaf material from rosettes of 5-week-old normoxic plants was measured using oxygen electrodes in normoxic or hypoxic conditions in the dark. When respiration was measured under normoxic conditions, oxygen consumption rates were markedly lower in 35S::Δ13RAP2.12 plants than in 35S::RAP2.12 or wild-type plants, while 35S::RAP2.12 plants were similar to wild type (Fig. 4B). Hypoxic conditions resulted in decreased oxygen consumption rates when compared to normoxia, dropping to similar levels in all three genotypes. Overall, these data show that overexpression of a deregulated RAP2.12 leads to an induction of ethanol, lactate, and Ala fermentation in normoxic plants, while respiration is inhibited.
Overexpression of a Stabilized RAP2.12 Leads to Decreased Adenylate Energy States under Normoxic Conditions, while NAD and Ascorbate Redox-States Were Not Substantially Altered
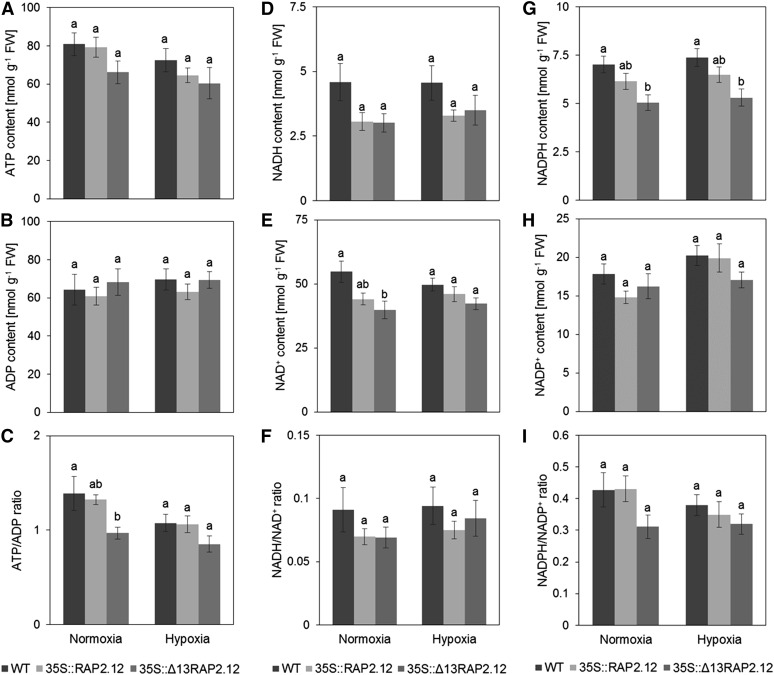
The results described above show that 35S::Δ13RAP2.12 overexpression leads to an induction of fermentation and an inhibition of respiration in normoxic plants. To investigate whether this is accompanied by changes in cellular energy states, ATP (Fig. 6A) and ADP levels (Fig. 6B) as well as ATP/ADP ratios (Fig. 6C) were analyzed in 5-week-old plants. Under normoxia, 35S::Δ13RAP2.12 plants showed a significant decrease in the ATP/ADP ratio, while 35S::RAP2.12 plants were similar to wild type. After hypoxic treatment, the ATP/ADP ratio dropped in all three genotypes, reaching lower levels in 35S::Δ13RAP2.12 than in 35S::RAP2.12 and wild-type plants.
Figure 6.
Overexpression of the N-end-rule-insensitive Δ13RAP2.12 transcription factor under the control of the 35S promoter leads to a decrease in the adenylate energy state under normoxic conditions. After 16 h incubation in normoxia or hypoxia in the absence of sugars in the dark, 5-week-old wild-type, RAP2.12, and Δ13RAP2.12-overexpressing plants were sampled to analyze the levels of A, ATP; B, ADP; C, ATP/ADP; D, NADH; E, NAD+; F, NADH/NAD+; G, NADPH; H, NADP+; and I, NADPH/NADP+ in whole rosettes. Results are means ± se (n = 9–10 biological replicates). Values of the different genotypes that significantly differ from each other are indicated by different letters (according to one-way ANOVA test, P < 0.05). Growth of plants and rosette stage analyzed was similar as in Fig. 2. WT, Wild type.
In addition to ATP and ADP, the pyridine nucleotides NAD(P)H and NAD(P)+, were also analyzed; these are important cofactors of energy metabolism. As shown in Figure 6, there were only minor changes in the levels of NADH (Fig. 6D), NAD+ (Fig. 6E), NADH/NAD+ ratio (Fig. 6F), NADPH (Fig. 6G), NADP+ (Fig. 6H), and NADPH/NADP+ ratio (Fig. 6I) across the three genotypes and the conditions. Specifically in 35S::Δ13RAP2.12 plants, there were significant decreases in NAD+ (Fig. 6E) and NADPH (Fig. 6G) under normoxic conditions. Interestingly, hypoxic manipulation did not lead to any substantial increases of the NADH/NAD+ ratio in all three genotypes, indicating that mechanisms to balance the NADH/NAD+ redox state were still effective under these conditions (Fig. 6F). Also the redox state of another redox couple, ascorbate/dehydroascorbate, was analyzed, but no significant changes were observed across either genetic or low-oxygen manipulations (Fig. 5A; Supplemental Table S1).
Overexpression of a Stabilized RAP2.12 Causes Deep Effects on Metabolite Levels under Both Normoxic and Hypoxic Conditions
To investigate whether the impaired balance between respiration and fermentation caused by overexpression of a deregulated RAP2.12 leads to alterations in the levels of metabolites of central metabolic pathways, gas chromatography (GC)-mass spectrometry (MS)-based metabolite profiling was performed in 5-week-old plants. Figure 5, A–C, and Supplemental Table S1 show marked changes in metabolite levels in 35S::Δ13RAP2.12 plants relative to 35S::RAP2.12 and wild-type plants under normoxic conditions. Several sugars, such as Glc, ribose, raffinose, melibiose, and maltose and the carbon storage reserve starch were decreased (Fig. 5A), while glycolytic metabolites such as Fru-6-P and pyruvate were increased (Fig. 5B), a scenario that is in line with increased fermentation rates under these conditions. Interestingly, there were also significant changes in the levels of intermediates of the TCA cycle, leading to increased levels of citrate and malate and an increased succinate/fumarate ratio (Fig. 5C). These changes in TCA cycle intermediates have been found to be a characteristic response to hypoxic conditions in previous studies (Geigenberger et al., 2000; van Dongen et al., 2009; Narsai et al., 2011; António et al., 2016), indicating an inhibition of succinate dehydrogenase (SDH; also known as complex II of the mitochondrial electron transport chain), bifurcation of the TCA cycle, and inhibition of respiration (Rocha et al., 2010; António et al., 2016.). This was accompanied by a strong increase in the levels of many amino acids, such as Val, Ala, Ser, Thr, Met, Leu, Ile, Asp, Lys, Cyt, Pro, Orn, Glu, Gln, GABA, Arg, Phe, and Trp (Fig. 5, B and C), a response that has also been found to be a characteristic of hypoxic conditions in the past (van Dongen et al., 2009; Narsai et al., 2011; António et al., 2016). Interestingly, there was also an increase in important metabolites deriving from amino acids, such as salicylic acid glucoside, picolinic acid (Fig. 5B), and pipecolic acid (Fig. 5C), involved in hormone, defense, and immune responses (Zhang et al., 2004; Návarová et al., 2012; Bernsdorff et al., 2016). Moreover, gluconic acid, a precursor of the pentose-P pathway, was strongly increased (Fig. 5A).
Under hypoxic conditions, 35S::Δ13RAP2.12 overexpression led to similar changes in metabolite levels as in normoxia (Fig. 5, A–C; Supplemental Table S1), indicating that the role of oxygen-regulated RAP2.12 in controlling TCA cycle regulation, amino acid metabolism, and immune-regulatory metabolite levels will also extend to hypoxic conditions.
Overexpression of a Stabilized RAP2.12 Leads to Impaired Anoxic Resistance, Which Can Be Prevented by External Feeding of Suc
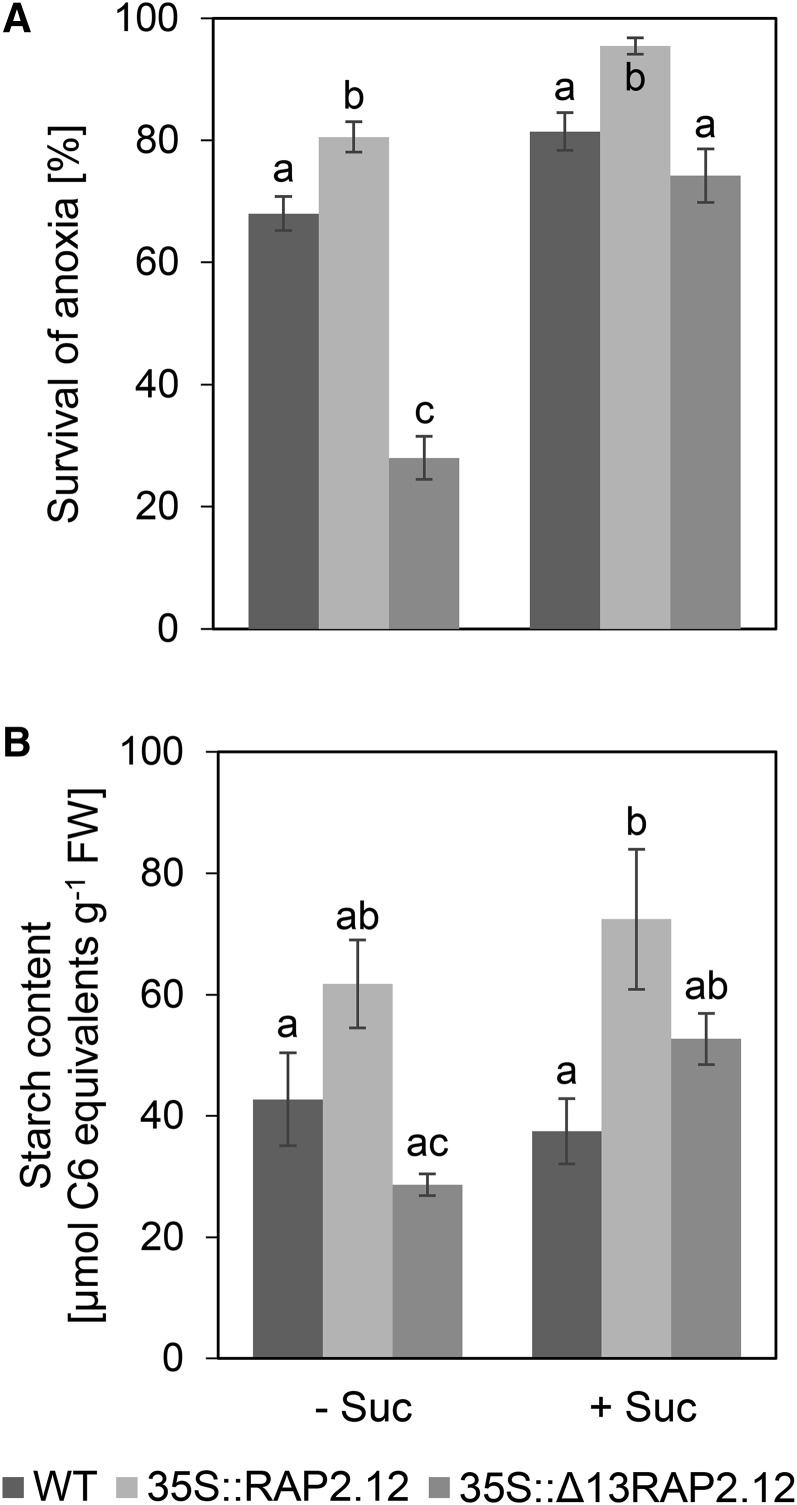
To investigate the effect of overexpression of a deregulated RAP2.12 on anoxic resistance, normoxic seedlings grown on agar plates containing no sugars or 30 mm Suc were subjected to anoxic conditions in the dark for 6.5 or 8 h, respectively, to analyze the percentage of surviving seedlings 1 week later. Without external sugar supply, overexpression of Δ13RAP2.12 led to decreased anoxic survival rates, while overexpression of RAP2.12 had the opposite effect (Fig. 7A). To investigate whether the impaired anoxic resistance was due to the low levels of carbon reserves prevailing in 35S::Δ13RAP2.12 plants (see Fig. 5A; Supplemental Table S1), anoxic survival experiments were performed with seedlings grown on agar plates containing 30 mm Suc. In 35S::Δ13RAP2.12 plants, external sugar supply led to a very strong increase in anoxic survival rates, reaching values similar to the wild type and close to 35S::RAP2.12 plants (Fig. 3A). The dependency of anoxic survival rates on carbohydrate availability was confirmed by analyzing starch levels in the seedlings (Fig. 7B). This indicates low levels of carbon reserves to be the most likely reason for impaired anoxic resistance in 35S::Δ13RAP2.12 plants.
Figure 7.
Overexpression of the N-end-rule-insensitive Δ13RAP2.12 transcription factor under the control of the 35S promoter leads to decreased anoxic survival rates, which can be prevented by external feeding of 30 mm Suc. A, Seedlings of 1-week-old wild-type, RAP2.12, and Δ13RAP2.12-overexpressing plants grown on MS medium with (+) or without (−) 30 mm Suc were exposed to anoxic conditions for 6.5 and 8 h, respectively, before survival rates were measured 1 week later. B, To put the data into context, starch levels were measured in the shoots of the seedlings at the time point of transfer to anoxic conditions (2 h into the photoperiod). Results are means ± se (n = 10 different sets of plants for survival rates or 5–6 for starch levels). Values of the different genotypes that significantly differ from each other are indicated by different letters (according to one-way ANOVA test, P < 0.05). WT, Wild type.
DISCUSSION
Expression of anoxic genes is regulated by proteasome-mediated proteolysis of ERF-VII transcription factors via the oxygen-dependent branch of the N-end-rule pathway (Gibbs et al., 2011, Licausi et al., 2011c). Protein stability of the ERF-VII family member RAP2.12 has been found to be strictly dependent on the oxygen concentration, being increased when oxygen is decreasing from 21 to below 10% (v/v; Kosmacz et al., 2015). While research into RAP2.12 mainly focused on its role to regulate anoxic gene expression, little is known on the impact of this oxygen-sensing system in regulating plant metabolism and growth. By comparing plants overexpressing N-end-rule-sensitive and -insensitive forms of RAP2.12, this article provides evidence that oxygen-regulated RAP2.12 stabilization plays important roles in (1) preventing aerobic fermentation, (2) regulating central metabolic processes, (3) modifying the levels of immune-regulatory metabolites, and (4) sustaining growth, development, and anoxic resistance of plants.
Oxygen Regulation of RAP2.12 Stabilization Is Indispensable to Prevent Aerobic Fermentation and the Associated Reduction in Plant Growth
The results presented in Figures 2–5 demonstrate that overexpression of an N-end-rule-insensitive form of RAP2.12 leads to induced expression of anoxic genes, increased activities of fermentative enzymes, and increased accumulation of fermentation products, such as ethanol, lactate, Ala, and GABA already under normoxic conditions, while these effects failed to appear after overexpression of an N-end-rule-sensitive form of RAP2.12. The induction of aerobic fermentation was accompanied by increased pyruvate levels (Fig. 5), decreased adenylate energy states (Fig. 6), decreased levels of starch (Fig. 5), and restricted plant growth (Fig. 1). These results provide genetic evidence that RAP2.12 oxygen sensing plays an important role to prevent aerobic fermentation and the attendant losses in energy and carbon reserves, which ultimately lead to a reduction of plant growth. This demonstrates that constitutive up-regulation of fermentation is detrimental for plant growth, while conditional regulation of fermentation is essential for plants. Fermentation is an ancient metabolic process and has been found to be regulated by oxygen availability in most organisms. There are a few exceptions where fermentation is regulated by sugars instead of oxygen to provide an additional metabolic capacity when sugar supply is increased. In baker’s yeast (Saccharomyces cerevisiae), high Glc levels lead to the induction of ethanol fermentation under aerobic conditions (Wills 1990), while in plants, specific cell types, such as pollen grains, show elevated expression of PDC and ADH and flux through the ethanol fermentation pathway in response to sugar supply (Bucher et al., 1995; Tadege et al., 1997, 1999). Although little is known on the regulatory processes leading to aerobic fermentation in these specific cell types, it will require oxygen-sensing mechanisms to be attenuated or to be modified by other signals, which may be linked to sugar sensing.
Deregulation of RAP2.12 Causes Impaired Anoxic Resistance Depending on the Availability of Carbohydrate Reserves
Overexpression of RAP2.12 has been found to lead to improved anoxic survival rates in previous studies (Licausi et al., 2011c; Gibbs et al., 2011). The data presented in Figure 7 show that overexpression of an N-end-rule-insensitive form of RAP2.12 leads to a decrease in anoxic resistance, which can be prevented by external Suc supply. This provides a possible explanation for the opposing results of previous studies. In Licausi et al. (2011c), 35S::Δ13RAP2.12 overexpressors as well as ate1ate2 and prt6 mutants of the N-end-rule pathway were grown on soil without sugars and revealed decreased anoxic resistance, while in Gibbs et al. (2011) ate1ate2 and prt6 mutants were grown on agar plates including sugars and revealed increased anoxic resistance. However, this does not rule out that also other reasons, such as light conditions and air humidity, can account for the contrasting effects in anoxic resistance of these two different studies (Riber et al., 2015). Taken together, these data support the suggestion that hypoxic tolerance depends to a great extent on the availability of carbohydrate reserves for fermentative metabolism. Constitutive up-regulation of fermentation upon normoxic conditions, such as in the 35S::Δ13RAP2.12 line, leads to depletion of starch, and concomitantly to a poor survival potential upon hypoxic stress.
Oxygen Regulation of RAP2.12 Stabilization Regulates Respiration, TCA Cycle, and Amino Acid Metabolism
The comparison of plants overexpressing N-end-rule-sensitive and -insensitive RAP2.12 proteins also points to a role of this oxygen-sensing mechanism in regulating central metabolic processes in plants (Fig. 8). Under normoxic conditions, N-end-rule-insensitive versus-sensitive RAP2.12 overexpression led to an inhibition of respiration (Fig. 4B); an accumulation of pyruvate, citrate, and malate; and an increase in the succinate/fumarate ratio (Fig. 5), indicating a down-regulation of the TCA cycle by inhibiting SDH. Moreover, the levels of GABA were strongly increased, indicating an activation of the GABA shunt. These alterations in respiratory metabolism resemble the metabolic changes that have been found to occur in response to a decrease in oxygen concentrations in previous work (Geigenberger et al., 2000; van Dongen et al., 2009; Narsai et al., 2011; António et al., 2016). As shown in previous studies, a decrease in internal oxygen concentration in roots, tubers, and other plant tissues leads to an adaptive inhibition of respiration as a proactive response to decrease oxygen consumption and prevent the tissue to drive into anoxia (Geigenberger et al., 2000; Zabalza et al., 2009; Geigenberger, 2014). Because inhibition of respiration occurs at oxygen concentrations well above the Km (oxygen) of cytochrome oxidase, an oxygen-sensing mechanism has been suggested to be involved (Geigenberger et al., 2000; Zabalza et al., 2009; Gupta et al., 2009). While the nature of this oxygen-sensing mechanism is unclear, decreased respiration rates and associated changes in the levels of TCA cycle intermediates in response to 35S::Δ13RAP2.12 overexpression implies a possible link between RAP2.12-mediated oxygen sensing and the regulation of respiration, although it cannot be excluded that this is due to indirect effects. Further metabolic studies will be necessary, such as stable isotope-labeling experiments to evaluate flux of pyruvate, Glu or other metabolites, to better understand the basis of the observed decrease in respiration.
Figure 8.
Summary model of RAP2.12 regulation of plant metabolism based on the results of this work. When oxygen falls to concentrations below 10% (v/v), the transcription factor RAP2.12 is stabilized and moves to the nucleus to activate the expression of specific genes (Kosmacz et al., 2015). First, mRNA levels and enzyme activities of AlaAT are increased, resulting in increased levels of Ala and γ-amino butyrate and associated changes in the flux modus of the TCA cycle, including an inhibition of succinate dehydrogenase, which finally leads to an inhibition of respiration. The precise mechanism of RAP2.12 regulation of AlaAT expression has not been clarified yet. Second, mRNA levels and enzyme activities of ADH, PDC, and LDH are increased, resulting in elevated rates of ethanol and lactate fermentation. In Arabidopsis plants overexpressing a deregulated (constitutively stabilized) RAP2.12, these metabolic changes are also observed under normoxic conditions, leading to decreased growth and anoxic stress resistance. Green = promoting effects; red = inhibiting effects; dotted arrow = precise mechanism is not known yet.
Under both normoxic and hypoxic conditions, the Δ13RAP2.12-induced increases in the succinate/fumarate ratio, Ala levels, and GABA shunt (Fig. 5) are indicative of a possible role of RAP2.12-mediated oxygen sensing in controlling the bifurcation of the TCA cycle by inhibiting SDH (Fig. 8). In previous studies it was already concluded from metabolome profiling analysis that the TCA cycle bifurcates in a parallel oxidative and reductive pathway during low oxygen stress (Rocha et al., 2010), while 13C-labeling experiments were able to show recently that this modification of the TCA modus is mediated by the inhibition of SDH and the activation the GABA shunt (António et al., 2016). In confirmation to this, characteristic low-oxygen-induced changes in the levels of succinate, fumarate, and GABA have been found in various studies with different plant tissues (Narsai et al., 2011), although it remained unclear how the regulation of the TCA cycle reactions is controlled. The results of this article indicate that oxygen-regulated RAP2.12 stabilization participates in the control of the flux-modus of the TCA cycle, probably by regulating the overall abundance of AlaAT and SDH activities. In confirmation to this, AlaAT1 and AlaAT2 mRNA levels (Fig. 2, C and D) as well as extractable overall AlaAT enzyme activities (Fig. 3D) were found to be increased in response to 35S::Δ13RAP2.12 overexpression.
Oxygen Regulation of RAP2.12 Stabilization Regulates the Levels of Immune-Regulatory Metabolites
Under both normoxia and hypoxia, overexpression of Δ13RAP2.12 led to strong increases in the levels of a large number of amino acids, including Phe, Trp, and Lys. This was accompanied by severe increases in the levels of immune-regulatory metabolites such as salicylic acid glucoside (derives from Phe), picolinic acid (derives from Trp), and pipecolic acid (derives from Lys), the latter showing a 10-fold increase (Fig. 5; Supplemental Table S1). Pipecolic acid has been found in previous studies as an endogenous mediator to orchestrate plant systemic required resistance and defense priming via salicylic acid-dependent and independent pathways in Arabidopsis (Návarová et al., 2012; Bernsdorff et al., 2016), while picolinic acid was found to stimulate defense responses and disease resistance in rice (Zhang et al., 2004). Interestingly, overexpression of RAP2.2—a member of ERF-VII transcription factors and a close homolog of RAP2.12 with similar functions (Gasch et al., 2016)—has been found to lead to increased disease resistance in Arabidopsis plants (Zhao et al., 2012). More studies are needed to resolve the role of immune regulatory metabolites in this context.
CONCLUSION
Recently, an oxygen-sensing system has been identified that leads to changes in the stability of the transcription factor RAP2.12, which regulates hypoxic gene expression and adaptation of Arabidopsis plants to low oxygen (Licausi et al., 2011c; Kosmacz et al., 2015). There is likely genetic redundancy in function of RAP2.2 and RAP2.12, as indicated in the study by Gasch et al. (2016). In this article, we show that operation of this oxygen-sensing system is indispensable to optimize metabolic performance and plant growth under normoxic as well as under hypoxic conditions. We provide direct evidence that conditional regulation of fermentation is necessary to prevent energy and carbon reserves to be wasted by aerobic fermentation. This is important to allow optimized growth under normoxic conditions as well as improved survival of anoxic stress conditions. Moreover, evidence is provided that oxygen sensing, mediated by RAP2.12 stabilization, regulates central metabolic processes, such as respiration, TCA cycle, and amino acid metabolism, as well as the levels of immune-regulatory metabolites.
MATERIALS AND METHODS
Plant Material
For most of the experiments, representative Arabidopsis (Arabidopsis thaliana) lines overexpressing RAP2.12 or Δ13RAP2.12 under control of the 35S CaMV promoter (characterized in Licausi et al., 2011c) and the respective Columbia-0 (Col-0) wild type were grown for 5 weeks under short-day conditions (8 h in 160 µmol photons m−2 s−1 at 20°C and 16 h in 0 µmol photons m−2 s−1 at 16°C). To do this, Arabidopsis seeds were surface-sterilized with 0.1% Tween 20 in 70% ethanol for 5 min, washed thoroughly with water one time, treated with 0.1% Tween 20 in 6% sodium hypochlorite for 5 min, and then washed thoroughly with water for five times again. For subsequent germination, seeds were grown vertically on plates containing 2% agar in 1/2 MS medium (pH 5.7) without sugars for 1 week, before they were transferred to potting soil (Stender) for another 4 weeks. For treatment with different oxygen concentrations, 5-week-old plants were exposed to a stream of air containing 21% (normoxia) or 1% (v/v) oxygen (hypoxia), supplemental with N and 350 ppm CO2, for 16 h in the dark, unless indicated otherwise in the figure legends. This duration coincided with the dark phase to which the plants were acclimated to in the growth chamber.
To analyze anoxic resistance, plants were grown for 1 week in horizontal agar plates containing 0.8% agar in 1/2 MS medium (pH 5.7) with 30 mm Suc or without Suc under long-day conditions (16 h in 100 µmol photons m−2 s−1 and 8 h in 0 µmol photons m−2 s−1), before they were subjected to anoxia for 8 or 6.5 h, respectively, by exposing to a stream of air containing zero oxygen.
Plants were harvested immediately after the respective treatment by freezing whole rosettes in liquid N. The deep-frozen rosette material was then powdered in a liquid-N cooled ball mill (MM400; Retsch) and subsequently stored at −80°C for further analysis of RNA, enzyme activities, and metabolite levels.
Developmental Analysis
Every 3 to 4 d, the developmental stage of plants growing under short-day conditions on soil (8-h d at 160 µmol photons m−2 s−1 and a constant temperature of 20°C) was determined according to the BBCH-scale (Hess et al., 1997). Additionally, the rosette fresh weight was recorded after 3, 4, and 5 weeks. All investigations referred to the vegetative state of plants exclusively.
Analysis of mRNA Levels of Anoxic Marker Genes by Quantitative Real Time PCR
RNA was extracted from 50 mg of frozen plant powder using the RNeasy plant mini kit (Qiagen) according to the manufacturer’s instructions. cDNA synthesis was performed with 500 ng RNA using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer’s instructions, to a final volume of 30 μL. The cDNA was diluted 1:10 before 2 μL was added using the QuantiFast SYBR green kit (Bio-Rad) according to the manufacturer’s instructions. With the IQ5 Reader (Bio-Rad), quantitative real time PCR was carried out for the housekeeping gene RCE1 (AT4G36800) and the hypoxia-responsive genes ADH1 (AT1G77120), PDC1 (AT4G33070), HB1 (AT2G16060), SUS4 (AT3G43190), RAP2.12 (AT1G53910), AlaAT1 (AT1G17290), and AlaAT2 (AT1G72330) using the following primers: RCE1 FW (5′-CTGTTCACGGAACCCAATTC-3′), RCE1 RV (5′-GGAAAAAGGTCTGACCGACA-3′), ADH1 FW (5′-TATTCGATGCAAAGCTGCTGTG-3′), ADH1 RV (5′-CGAACTTCGTGTTTCTGCGGT-3′), PDC1 FW (5′-CGATTATGGCACTAACCGGATT-3′), PDC1 RV (5′-TGTTCACCACCGCCTGATAAC-3′), HB1 FW (5′-TTTGAGGTGGCCAAGTATGCA-3′), HB1 RV (5′-TGATCATAAGCCTGACCCCAA-3′), SUS4 FW (5′-CGCAGAACGTGTAATAACGCG-3′), SUS4 RV (5′-CAACCCTTGAGAGCAAAGCAAA-3′), RAP2.12 FW (5′-CGCTGCGGAAGGTTCAGTT-3′), RAP2.12 RV (5′-CAGCCCATTTTCCCCAAGGA-3′), AlaAT1 FW (5′-GTCAAATTCTTGCTAGCC-3′), AlaAT1 RV (5′-CCCTCTAGCTTGTTCAGA-3′), AlaAT2 FW (5′-GTCAAATTCTTGCTAGCC-3′), and AlaAT2 RV (5′-TAAACTGTTGAGAGCGTC-3′). Relative quantification of gene expression was calculated according to Pfaffl (2001) with data normalized to the wild type at normoxia.
Analysis of Enzyme Activities
For enzyme activity determination of ADH, PDC, and AlaAT, the protein extraction buffer consisted of 50 mm HEPES/KOH (pH 7.4), 5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 0.1% Triton X100, 10% glycerol, 2 mm benzamidine, 2 mm ε-aminocapronic acid, 5 mm DTT, 0.5% Protease Inhibitor Cocktail (Roche), and 1 mm PMSF. The extraction buffer for LDH activity determination had the same composition, except for benzamidine, ε-aminocapronic acid, and DTT, which were not included. The determination of ADH activity was done according to John and Greenway (1976). Therefore, 20 mg of frozen plant powder were extracted with 200 µL extraction buffer and 30 μL of the resulting supernatant were mixed with 170 µL reaction buffer consisting of 50 mm Bicine/KOH (pH 8.8), 5 mm MgCl2, and 1 mm NAD+. After addition of 1 µL 100% ethanol, the NADH production was measured photometrically at 340 nm. PDC and LDH activity measurements were performed according to Bouny and Saglio (1996), with extraction of 20 mg frozen plant powder in 500 µL extraction buffer for PDC activity determination, and in 300 µL extraction buffer for LDH activity quantification. In each case, 30 μL amounts of the supernatant were mixed with 170 μL reaction buffer for measurement. For PDC, the reaction mix contained 100 mm Tricine/KOH (pH 6.5), 2 mm MgCl2, 25 mm oxamate, 0.1 mm DTT, 0.1 mm thiamine pyrophosphate, 0.15 mm NADH, and 300 U mL−1 ADH. For LDH, the reaction buffer consisted of 100 mm Tricine/KOH (pH 7.5), 100 mm pyrazole, 10 mm potassium cyanide, and 0.15 mm NADH. After addition of 2 mm pyruvate the consumption of NADH was measured photometrically at 340 nm to measure PDC or LDH activity, respectively. The determination of AlaAT activity was done according to Hatch and Mau (1973). Therefore, 10 mg of frozen plant powder was extracted with 1 mL extraction buffer and 30 μL of the resulting supernatant were mixed with 170 μL reaction buffer consisting of 50 mm HEPES/KOH (pH 7.5), 2 mm EDTA, 10 mm l-Ala, 0.04 mm pyridoxal-5-P, 0.2 mm NADH, and 5.9 U mL−1 LDH. After addition of 0.5 mm α-ketoglutarate, the consumption of NADH was measured photometrically at 340 mm.
Analysis of Starch Content and Metabolite Levels
The determination of starch was done according to Hendriks et al. (2003). Twenty milligrams of frozen plant powder were extracted two times with 250 μL 80% ethanol and one time with 250 µL 50% ethanol. After each ethanol addition step, the samples were heated for 30 min at 90°C, cooled down to room temperature and the supernatant was discarded. After ethanol extraction, the pellets were vacuum-dried at 30°C for 40 min, resuspended in 400 μL 100 mm NaOH, and heated to 95°C for 1 h shaking at 1400 rpm, followed by neutralization with 500 mm HCl and 100 mm acetate/NaOH (pH 4.9). Forty microliters of the extract were digested for 16 h at 37°C with 110 µL starch degradation mix containing 50 mm acetate/NaOH (pH 4.9), 3 U mL−1 amyloglucosidase, and 4 U mL−1 α-amylase. Fifty microliters of the supernatant was added to 160 μL Glc determination mix consisting of 100 mm HEPES/KOH (pH 7), 3 mm MgCl2, 3 mm ATP, 1.4 mm NADP, and 3.4 U mL−1 Glc-6-P dehydrogenase (G6PDH), and NADPH production was measured photometrically at 340 nm after addition of 2.4 U mL−1 hexokinase.
The extraction of pyridine nucleotides was performed as described in Hajirezaei et al. (2002). Twenty milligrams of frozen plant powder were extracted with 250 μL 100 mm HClO4 (NADP+) or 100 mm KOH (NADPH). The supernatant was heated to 95°C for 2 min, followed by addition of an equal volume of 200 mm Tris (pH 8.4), 100 mm KOH, or 200 mm Tris (pH 8.4), 100 mm HCLO4 for neutralization. The determination of the pyridine nucleotides was done according to Gibon et al. (2004), with the detection mix for NAD(H) containing 300 mm Tricine/KOH (pH 9), 12 mm EDTA, 0.3 mm phenazine ethosulfate, 1.8 mm methylthiazolyldiphenyl-tetrazolium bromide, 1.5 m ethanol, and 18 U mL−1 ADH. The mix for NADP(H) detection contained 300 mm Tricine/KOH (pH 9), 12 mm EDTA, 0.3 mm phenazine ethosulfate, 1.8 mm methylthiazolyldiphenyl-tetrazolium bromide, 9 mm Glc-6-P, and 9 U mL−1 G6PDH. The absorption was measured photometrically at 570 nm.
Extraction of ATP and ADP was performed according to Trethewey et al. (1998). Fifty milligrams of frozen plant powder were extracted with 600 μL of 16% trichloroacetic acid in water containing 5 mm EGTA. After 1 h of shaking at 4°C and centrifugation, the supernatant was mixed with cold diethylether (saturated with water) and centrifuged again. The upper phase was discarded and the washing procedure was repeated to remove trichloroacetic acid from the extract. The pH was finally adjusted to 6–7 using a solution containing 5 m KOH and 1 m triethanolamine. For ATP detection, 85 μL extract were measured in 200 µL detection mix containing 50 mm HEPES/KOH (pH 7), 0.4 mm NADP, 13 mm Glc, 5 mm MgCl2, 7.5 U mL−1 phosphoglucose isomerase, and 1 U mL−1 G6PDH by determining NADPH production using fluorescence spectroscopy at 360 nm after addition of 1.5 U mL−1 hexokinase. ADP was detected by measuring production of NAD+ of 75 µL extract in 200 µL mix consisting of 50 mm HEPES/KOH (pH 7), 0.02 mm NADH, 0.9 mm PEP, 5 mm MgCl2, and 15 U mL−1 LDH using fluorescence spectroscopy at 360 nm after adding 10 U mL−1 pyruvate kinase.
GC-TOFMS Analysis of Polar Primary Metabolites
GC-TOFMS-based analysis of primary metabolites was performed exactly as described in Thormählen et al. (2013). To visualize the metabolite changes within an overview, the open-source software VANTED Ver. 2.1.0 (https://immersive-analytics.infotech.monash.edu/vanted/) was used.
Analysis of Respiration Rates
Five-week-old normoxic plants grown under short-day conditions were used to analyze respiration rates. At the end of the night, rosette material was introduced into vessels containing 50 mm MES/KOH buffer solution (pH 6.5) to measure oxygen consumption at different oxygen concentrations using the OXY-4 mini oxygen sensor (PreSens) in the absence of external sugars in the dark.
Analysis of Ethanol Levels
To analyze ethanol production, seedlings were grown in sterile liquid culture containing 1/2 MS medium (pH 5.7) and 30 mm Suc under long-day conditions (see above) for 1 week, while leaves were allowed to grow outside of the liquid medium. After 10 h into the photoperiod, the medium was exchanged to 1/2 MS medium (pH 5.7) containing 90 mm Suc and the seedlings were subjected to normoxia or anoxia by exposing to a stream of air containing 21% or zero oxygen, respectively, for further 16 h in the dark. After this treatment, 20 µL of the medium was sampled and mixed with 180 µL reaction buffer containing 72 mm Na4P2O7 (pH 8.7), 72 mm semicarbazide, 25 mm Gly, and 0.5 mm NAD. After addition of 7 U ADH the production of NADH was measured photometrically at 340 mm.
Analysis of Anoxic Resistance
After 2 h into the photoperiod, 1-week-old seedlings grown under long-day conditions (16 h light, 100 μmol photons, 21% v/v oxygen) on horizontal agar plates containing 0.8% agar in 1/2 MS medium (pH 5.7) with 30 mm Suc or no Suc were subjected to anoxic conditions in the dark for 8 h or 6.5 h, respectively, by exposing to a stream of air containing zero oxygen. After the anoxic treatment they were returned to normoxic conditions and grown for additional 7 d, before the number of surviving seedlings was determined.
Supplemental Data
The following supplemental material is available.
Supplemental Table S1. Changes in metabolite profiles in RAP2.12 and Δ13RAP2.12 overexpressing plants relative to wild type.
Supplementary Material
Acknowledgments
We are grateful to Jennifer Lindstadt (LMU Munich) for help with GC-MS analysis and to Benjamin Faix (LMU Munich) and Francesco Licausi (University of Pisa) for advice during experimental work.
Footnotes
Articles can be viewed without a subscription.
References
- António C, Päpke C, Rocha M, Diab H, Limami AM, Obata T, Fernie AR, van Dongen JT (2016) Regulation of primary metabolism in response to low oxygen availability as revealed by carbon and nitrogen isotope redistribution. Plant Physiol 170: 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Fukao T, Gibbs DJ, Holdsworth MJ, Lee SC, Licausi F, Perata P, Voesenek LA, van Dongen JT (2012) Making sense of low oxygen sensing. Trends Plant Sci 17: 129–138 [DOI] [PubMed] [Google Scholar]
- Bernsdorff F, Döring A-C, Gruner K, Schuck S, Bräutigam A, Zeier J (2016) Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid-dependent and -independent pathways. Plant Cell 28: 102–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouny JM, Saglio PH (1996) Glycolytic flux and hexokinase activities in anoxic maize root tips acclimated by hypoxic pretreatment. Plant Physiol 111: 187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher M, Brander KA, Sbicego S, Mandel T, Kuhlemeier C (1995) Aerobic fermentation in tobacco pollen. Plant Mol Biol 28: 739–750 [DOI] [PubMed] [Google Scholar]
- Gasch P, Fundinger M, Müller JT, Lee T, Bailey-Serres J, Mustroph A (2016) Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 28: 160–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P. (2014) Adaptation of storage metabolism to oxygen deprivation. In van Dongen JT and Licausi F, editors, Low-Oxygen Stress in Plants, Plant Cell Monographs Vol. 21, 223–244, Springer, Berlin, Germany [Google Scholar]
- Geigenberger P. (2003) Response of plant metabolism to too little oxygen. Curr Opin Plant Biol 6: 247–256 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Fernie AR, Gibon Y, Christ M, Stitt M (2000) Metabolic activity decreases as an adaptive response to low internal oxygen in growing potato tubers. Biol Chem 381: 723–740 [DOI] [PubMed] [Google Scholar]
- Gibbs DJ, Lee SC, Isa NM, Gramuglia S, Fukao T, Bassel GW, Correia CS, Corbineau F, Theodoulou FL, Bailey-Serres J, Holdsworth MJ (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479: 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JH, Palacios N, Cross J, Selbig J, Stitt M (2004) A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuntoli B, Lee SC, Licausi F, Kosmacz M, Oosumi T, van Dongen JT, Bailey-Serres J, Perata P (2014) A trihelix DNA binding protein counterbalances hypoxia-responsive transcriptional activation in Arabidopsis. PLoS Biol 12: e1001950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta KJ, Zabalza A, van Dongen JT (2009) Regulation of respiration when the oxygen availability changes. Physiol Plant 137: 383–391 [DOI] [PubMed] [Google Scholar]
- Hajirezaei MR, Peisker M, Tschiersch H, Palatnik JF, Valle EM, Carrillo N, Sonnewald U (2002) Small changes in the activity of chloroplastic NADP+-dependent ferredoxin oxidoreductase lead to impaired plant growth and restrict photosynthetic activity of transgenic tobacco plants. Plant J 29: 281–293 [DOI] [PubMed] [Google Scholar]
- Hatch MD, Mau SL (1973) Activity, location, and role of asparate aminotransferase and alanine aminotransferase isoenzymes in leaves with C4 pathway photosynthesis. Arch Biochem Biophys 156: 195–206 [DOI] [PubMed] [Google Scholar]
- Hendriks JH, Kolbe A, Gibon Y, Stitt M, Geigenberger P (2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess M, Barrali G, Bleiholder H, Buhr L, Eggers T, Hack H, Stauss R (1997) Use of the extended BBCH scale—general for the descriptions of the growth stages of mono- and dicotyledonous weed species. Weed Res 37: 433–441 [Google Scholar]
- Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, Dolferus R (2010) Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol 153: 757–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John CD, Greenway H (1976) Alcoholic fermentation and activity of some enzymes in rice roots under anaerobiosis. Aust J Plant Physiol 3: 325–336 [Google Scholar]
- Kosmacz M, Parlanti S, Schwarzländer M, Kragler F, Licausi F, Van Dongen JT (2015) The stability and nuclear localization of the transcription factor RAP2.12 are dynamically regulated by oxygen concentration. Plant Cell Environ 38: 1094–1103 [DOI] [PubMed] [Google Scholar]
- Licausi F, Giorgi FM, Schmälzlin E, Usadel B, Perata P, van Dongen JT, Geigenberger P (2011a) HRE-type genes are regulated by growth-related changes in internal oxygen concentrations during the normal development of potato (Solanum tuberosum) tubers. Plant Cell Physiol 52: 1957–1972 [DOI] [PubMed] [Google Scholar]
- Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LACJ, Perata P, van Dongen JT (2011c) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479: 419–422 [DOI] [PubMed] [Google Scholar]
- Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P (2010) HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J 62: 302–315 [DOI] [PubMed] [Google Scholar]
- Licausi F, Weits DA, Pant BD, Scheible WR, Geigenberger P, van Dongen JT (2011b) Hypoxia responsive gene expression is mediated by various subsets of transcription factors and miRNAs that are determined by the actual oxygen availability. New Phytol 190: 442–456 [DOI] [PubMed] [Google Scholar]
- Narsai R, Rocha M, Geigenberger P, Whelan J, van Dongen JT (2011) Comparative analysis between plant species of transcriptional and metabolic responses to hypoxia. New Phytol 190: 472–487 [DOI] [PubMed] [Google Scholar]
- Návarová H, Bernsdorff F, Döring A-C, Zeier J (2012) Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24: 5123–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papdi C, Abrahám E, Joseph MP, Popescu C, Koncz C, Szabados L (2008) Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol 147: 528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riber W, Müller JT, Visser EJW, Sasidharan R, Voesenek LACJ, Mustroph A (2015) The greening after extended darkness1 is an N-end rule pathway mutant with high tolerance to submergence and starvation. Plant Physiol 167: 1616–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha M, Licausi F, Araújo WL, Nunes-Nesi A, Sodek L, Fernie AR, van Dongen JT (2010) Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol 152: 1501–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R, Mustroph A (2011) Plant oxygen sensing is mediated by the N-end rule pathway: a milestone in plant anaerobiosis. Plant Cell 23: 4173–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Dupuis I, Kuhlemeier C (1999) Ethanolic fermentation: new functions for an old pathway. Trends Plant Sci 4: 320–325 [DOI] [PubMed] [Google Scholar]
- Tadege M, Kuhlemeier C (1997) Aerobic fermentation during tobacco pollen development. Plant Mol Biol 35: 343–354 [DOI] [PubMed] [Google Scholar]
- Thormählen I, Ruber J, von Roepenack-Lahaye E, Ehrlich SM, Massot V, Hümmer C, Tezycka J, Issakidis-Bourguet E, Geigenberger P (2013) Inactivation of thioredoxin f1 leads to decreased light activation of ADP-glucose pyrophosphorylase and altered diurnal starch turnover in leaves of Arabidopsis plants. Plant Cell Environ 36: 16–29 [DOI] [PubMed] [Google Scholar]
- Trethewey RN, Geigenberger P, Riedel K, Hajirezaei MR, Sonnewald U, Stitt M, Riesmeier JW, Willmitzer L (1998) Combined expression of glucokinase and invertase in potato tubers leads to a dramatic reduction in starch accumulation and a stimulation of glycolysis. Plant J 15: 109–118 [DOI] [PubMed] [Google Scholar]
- van Dongen JT, Fröhlich A, Ramírez-Aguilar SJ, Schauer N, Fernie AR, Erban A, Kopka J, Clark J, Langer A, Geigenberger P (2009) Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Ann Bot (Lond) 103: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JT, Licausi F (2015) Oxygen sensing and signaling. Annu Rev Plant Biol 66: 345–367 [DOI] [PubMed] [Google Scholar]
- Voesenek LACJ, Bailey-Serres J (2015) Flood adaptive traits and processes: an overview. New Phytol 206: 57–73 [DOI] [PubMed] [Google Scholar]
- Weits DA, Giuntoli B, Kosmacz M, Parlanti S, Hubberten H-M, Riegler H, Hoefgen R, Perata P, van Dongen JT, Licausi F (2014) Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat Commun 5: 3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills C. (1990) Regulation of sugar and ethanol metabolism in Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol 25: 245–280 [DOI] [PubMed] [Google Scholar]
- Zabalza A, van Dongen JT, Froehlich A, Oliver SN, Faix B, Gupta KJ, Schmälzlin E, Igal M, Orcaray L, Royuela M, Geigenberger P (2009) Regulation of respiration and fermentation to control the plant internal oxygen concentration. Plant Physiol 149: 1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HK, Zhang X, Mao BZ, Li Q, He ZH (2004) Alpha-picolinic acid, a fungal toxin and mammal apoptosis-inducing agent, elicits hypersensitive-like response and enhances disease resistance in rice. Cell Res 14: 27–33 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Wei T, Yin K-Q, Chen Z, Gu H, Qu L-J, Qin G (2012) Arabidopsis RAP2.2 plays an important role in plant resistance to Botrytis cinerea and ethylene responses. New Phytol 195: 450–460 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.