SPIKE1 activates ROP GTPase proteins to regulate anisotropic cell expansion and petal anisotropic growth by affecting the organization of cortical microtubule arrays in Arabidopsis.
Abstract
Plant organ growth and final shape rely on cell proliferation and, particularly, on cell expansion that largely determines the visible growth of plant organs. Arabidopsis (Arabidopsis thaliana) petals serve as an excellent model for dissecting the coordinated regulation of patterns of cell expansion and organ growth, but the molecular signaling mechanisms underlying this regulation remain largely unknown. Here, we demonstrate that during the late petal development stages, SPIKE1 (SPK1), encoding a guanine nucleotide exchange factor, activates Rho of Plants (ROP) GTPase proteins (ROP2, ROP4, and ROP6) to affect anisotropic expansion of epidermal cells in both petal blades and claws, thereby affecting anisotropic growth of the petal and the final characteristic organ shape. The petals of SPK1 knockdown mutants were significantly longer but narrower than those of the wild type, associated with increased anisotropic expansion of epidermal cells at late development stages. In addition, ROP2, ROP4, and ROP6 are activated by SPK1 to promote the isotropic organization of cortical microtubule arrays and thus inhibit anisotropic growth in the petal. Both knockdown of SPK1 and multiple rop mutants caused highly ordered cortical microtubule arrays that were transversely oriented relative to the axis of cell elongation after development stage 11. Taken together, our results suggest a SPK1-ROP-dependent signaling module that influences anisotropic growth in the petal and defines the final organ shape.
A central question in plant organogenesis asks how gene activities contribute to organ growth and the final characteristic shape. Regulation of the growth and shape of plant organs is largely determined by the coordinated regulation of cell proliferation and cell expansion (Tsukaya, 2006; Barkoulas et al., 2007; Powell and Lenhard, 2012; Peaucelle et al., 2015). Petals are fascinating organs in flowering plants, with their fragrance and diverse color and shapes, which are important for attracting pollinators to ensure successful pollination (Willmer et al., 2009; Yuan et al., 2013). The Arabidopsis (Arabidopsis thaliana) petal has a laminar structure with epidermal cells overlying the mesophyll and vasculature; therefore, it has emerged as an excellent model for dissecting the mechanisms underlying organ growth and cell expansion (Irish, 2008; Huang and Irish, 2016). In Arabidopsis, petal organogenesis is divided into several stages and depends on cell proliferation and cell expansion (Smyth et al., 1990; Huang and Irish, 2016). During the past decade, large numbers of Arabidopsis genes that play crucial roles in regulating various aspects of cellular patterns and petal organogenesis have been identified (Dinneny et al., 2004; Takeda et al., 2004; Szécsi et al., 2006; Irish, 2008; Li et al., 2008b; Nag et al., 2009; Varaud et al., 2011; Sauret-Güeto et al., 2013; Fujikura et al., 2014; Schiessl et al., 2014; Huang and Irish, 2015, 2016). For example, Arabidopsis JAGGED, which encodes a zinc finger transcription factor expressed in the distal domain of the petal, functions in the regulation of anisotropic growth of the petal (Sauret-Güeto et al., 2013; Schiessl et al., 2014). However, regulators that function in coordinating cell expansion and anisotropic growth of the petal remain largely unknown.
Plant Rho-like small GTPase, usually termed Rho of Plants (ROP), belongs to a specific subfamily of the Rho GTPase family (Etienne-Manneville and Hall, 2002; Yang, 2002). In Arabidopsis, ROP proteins function as molecular signaling switches involved in a number of cellular processes, such as the regulation of cytoskeletal organization, cell wall patterning, the tip growth of pollen tubes, the interdigitated growth of leaf pavement cells, and the intracellular trafficking of PIN auxin efflux transporters (Lavy et al., 2007; Fu et al., 2009; Hazak et al., 2010; Qin and Yang, 2011; Wu et al., 2011; Craddock et al., 2012; Oda and Fukuda, 2012, 2013; Huang et al., 2014; Lin et al., 2015). As a molecular switch, ROP can shuttle between a GTP-bound active form and a GDP-bound inactive form, depending on its activating protein (ROPGAP) or guanine nucleotide exchange factor (ROPGEF). Once activated by upstream signals, active ROP interacts with effector proteins to relay signals into downstream cellular components and thus induce cellular responses (Yang, 2002).
ROPGEF proteins in the Arabidopsis genome have been classified as two types: one is named SPIKE1 (SPK1; Qiu et al., 2002; Basu et al., 2008; Zhang et al., 2010), a homolog of the animal single dock homology region; the other is a plant-specific ROPGEF family with 14 members that contain a plant-specific ROP nucleotide exchange (PRONE) domain (Berken et al., 2005; Gu et al., 2006). SPK1 loss-of-function mutation leads to dwarf plant organs and severe defective leaf epidermis, such as reduced cell-cell adhesion, loss of pavement cell interdigitation, and fewer trichome branches (Qiu et al., 2002; Basu et al., 2008; Zhang et al., 2010). SPK1 is involved in the activation of the ROP6-RIC1 pathway to regulate PIN2 internalization in root cells and, consequently, affects lateral root development (Lin et al., 2012). However, the molecular mechanisms by which SPK1 regulates organ growth remain to be determined.
Cortical microtubule (CMT) arrays orient the direction and deposition of cellulose microfibrils around the cell to build the cell wall and thus contribute to directional cell expansion (i.e. anisotropy; Baskin, 2001, 2005; Wasteneys and Galway, 2003; Wasteneys, 2004; Smith and Oppenheimer, 2005; Ehrhardt and Shaw, 2006; Paredez et al., 2006; Crowell et al., 2009; Bringmann et al., 2012; Wolf et al., 2012). Previous work has shown that, in the shoot apical meristem (SAM), CMT arrays in the peripheral SAM cells display ordered circumferential alignment to regulate growth anisotropy at both cellular and tissue levels (Hamant et al., 2008; Uyttewaal et al., 2012). Loss of function of the microtubule-severing protein katanin, a downstream component of the ROP6-RIC1 pathway (Lin et al., 2013), decreases the anisotropy of the CMT arrays in the SAM cells, which in turn affects the anisotropic growth of the SAM (Uyttewaal et al., 2012). A recent work has shown that the plant hormone auxin regulates growth anisotropy of the SAM by affecting the activity of ROP6 and its downstream components that control circumferential CMT alignment at the SAM (Sassi et al., 2014). In the absence of auxin accumulation, the ROP6-dependent signaling keeps CMT arrays at the SAM in an ordered circumferential state to inhibit spontaneous lateral outgrowth, which leads to the formation of a pin-like stem (Sassi et al., 2014).
To further consolidate our understanding of ROP-dependent signaling in the regulation of organ growth, we report here that SPK1 activates ROP proteins to inhibit anisotropic growth of petals at late development stages and thus affect the final characteristic shape. Consistent with this, petal blades and claws of SPK1 knockdown mutants have increased growth anisotropy, with significantly longer but narrower shape than those of the wild type, correlated with increased cell elongation and suppressed cell lateral expansion at late development stages. We also demonstrate that ROP proteins are activated by SPK1 to affect anisotropic growth of the petal by promoting isotropic expansion of epidermal cells in petal blades that is associated with the isotropic organization of CMT arrays. Thus, we suggest a ROP protein-dependent signaling module that contributes to anisotropic growth of the petal during late development stages.
RESULTS
SPK1 Regulates Petal Anisotropic Growth
To understand whether SPK1 functions in petal growth, we first investigated SPK1 expression patterns and found that SPK1 was highly expressed during petal development (Supplemental Fig. S1). Previous studies have shown that SPK1 loss of function caused seedling lethality when grown in soil at 30% to 50% humidity (Qiu et al., 2002; Lin et al., 2012). To better understand the role of SPK1 in petal growth, we used SPK1 knockdown mutants for the analysis of petal phenotype. In an earlier study, we isolated a SPK1 weak-allele mutant (spk1-4) that carries a mutation causing missplicing of the SPK1 pre-mRNA (Lin et al., 2012). We next examined the petal phenotype of the spk1-4 mutant. At development stage 14, the mature petal blades of the spk1-4 mutant had significantly longer but narrower shape and reduced petal blade areas compared with those of the wild type (Fig. 1), leading to an increased petal index (the ratio of length to width), a description of petal shape (Fig. 1D). Moreover, the spk1-4 mutant petal showed a significant increase in claw length and a decrease in claw width (Supplemental Fig. S2). Expressing SPK1 in the spk1-4 mutant by transforming pSPK1::SPK1 into the spk1-4 mutant rescued the mutant petal phenotype in six individual lines (two displayed in Supplemental Fig. S3). Thus, SPK1 knockdown mutants resulted in increased anisotropic growth of petals.
Figure 1.
Knockdown of SPK1 results in longer and narrower mature petals. A, Fully expanded flowers and petals at development stage 14 in the wild type, spk1-4, and SPK1 RNAi lines. Bar = 1 mm. B to E, Quantitative analyses of petal parameters for the indicated genotypes. Measurements are shown for petal blade length (B), petal blade width (C), petal blade index (i.e. length divided by width; D), and petal blade area (E). The spk1-4 mutant and the SPK1 RNAi lines had longer and narrower petals and reduced petal blade area than the wild type. Asterisks indicate significant differences from the wild type (**, P < 0.01, Student’s t test). Values are given as means ± sd of 16 petals.
To further confirm the role of SPK1 in the regulation of anisotropic growth in petals, we suppressed SPK1 expression by transforming an RNA interference (RNAi) construct, containing 309 bp of the SPK1 coding sequence designed to create a double-stranded RNA, into wild-type plants. Twenty SPK1 RNAi lines were isolated, and three lines with significantly reduced SPK1 transcriptional levels (Supplemental Fig. S4) were chosen for phenotype analysis. Compared with the wild type, at development stage 14, all three SPK1 RNAi lines had increased anisotropic growth with more elongated and narrower mature petals, as did the spk1-4 mutant (Fig. 1).
The morphological events of petal development are well described in Arabidopsis (Hill and Lord, 1989; Smyth et al., 1990). Before development stage 8, petals grow slowly depending on cell division, while petals undergo a rapid lengthening process from development stage 9 until full flower opening (Smyth et al., 1990; Pyke and Page, 1998; Dinneny et al., 2004). We next asked how SPK1 influenced petal growth during late development stages by comparing petal phenotype between the wild type and the spk1-4 mutant at development stages 10 to 14. Measuring petal blade length and width at development stages 10 and 11 showed that the spk1-4 mutant had similar petal sizes to the wild type (Fig. 2), while at development stage 12 and beyond, the spk1-4 mutant had increased anisotropic growth with significantly longer and narrower petals than those of the wild type (Fig. 2).
Figure 2.
Petal phenotypes and quantitative analysis of the wild type and spk1-4 at development stages 10 to 14. A, Flowers and petals at development stages 10 to 14 of the wild type and spk1-4. Bar = 1 mm. B to E, Quantitative analyses of petal blade parameters for the wild type and spk1-4 from development stages 10 to 14. The spk1-4 mutant displayed similar petal blade length, width, and area to those of the wild type at stages 10 and 11 (Student’s t test, P > 0.05), whereas the spk1-4 mutant had significantly longer and narrower petal blades and increased size in blade area than the wild type at development stage 12 and beyond (Student’s t test, P < 0.05). All data are represented as means ± sd of 16 petals.
Taken together, these results show that SPK1 participates in the inhibition of growth anisotropy of petals during late development stages, thus influencing the final characteristic shape.
SPK1 Regulates the Cell Expansion of Petal Epidermal Cells
To examine whether the petal phenotype observed in the spk1 mutant lines was associated with alterations in cell proliferation and/or cell expansion, we first analyzed cell numbers in mature spk1-4 petals (stage 14) and compared the results with those of the wild type. Our results demonstrated that cell numbers along the length and width directions of the petals and total cell numbers of petal blades in the spk1-4 mutant were similar to those of the wild type (Supplemental Fig. S5), which demonstrated that SPK1 might not participate in controlling cell proliferation. Previous reports have shown that loss-of-function SPK1 mutants exhibit severe defects in the interdigitated growth of leaf epidermal cells (Qiu et al., 2002). Thus, we expected that the petal phenotype observed in the spk1 mutant lines was correlated with changes in cell expansion patterns. We next analyzed the morphological phenotypes of both adaxial and abaxial epidermal cells in wild-type and mutant petals, respectively.
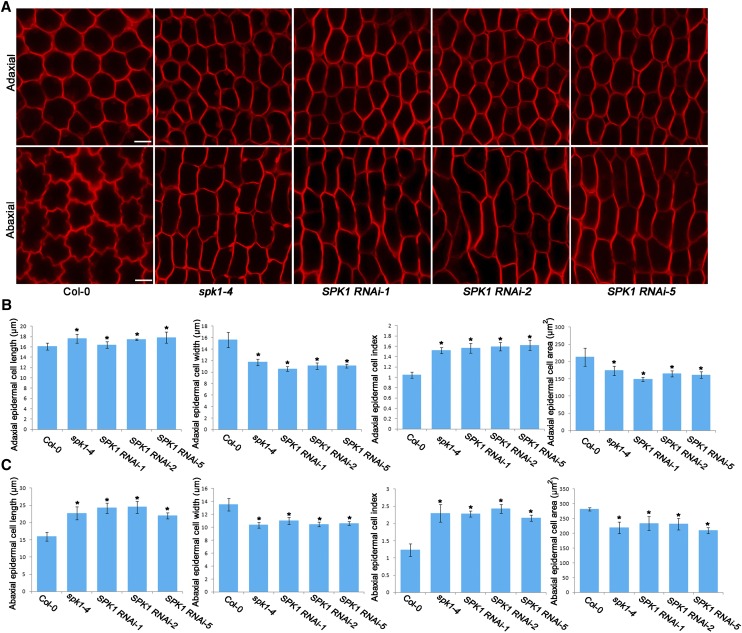
Epidermal cells of the adaxial and abaxial surfaces of Arabidopsis mature petal blades can be easily distinguished, as the adaxial epidermal cells have a conical shape with a pentagonal or hexagonal base, whereas the abaxial epidermal cells are flattened and shaped with interdigitated lobes (Irish, 2008). Analysis of the two-dimensional ground area of the cells in the middle part of the petal blades showed that, at development stage 14, adaxial epidermal cells in the spk1-4 mutant and the SPK1 RNAi lines showed increased anisotropic growth with increased cell elongation and suppressed cell lateral expansion compared with the wild type (Fig. 3), which was correlated with the morphological phenotype of petal blades in spk1-4 and the SPK1 RNAi lines (Fig. 1A). Similarly, abaxial epidermal cells in the spk1-4 mutant had increased growth anisotropy compared with the wild type (Fig. 3).
Figure 3.
Adaxial/abaxial epidermal cell shape of the wild type, spk1-4, and SPK1 RNAi lines at development stage 14. A, Epidermal cell shape in the middle part of petal blades from the wild type, spk1-4, and SPK1 RNAi lines at development stage 14. Epidermal cells of the spk1-4 mutant and the SPK1 RNAi lines had increased growth anisotropy with longer and narrower shape than the wild type. Bars = 10 µm. B and C, Quantitative analyses of adaxial/abaxial epidermal cell parameters for the indicated genotypes at development stage 14. The spk1-4 mutant and the SPK1 RNAi lines displayed significantly increased cell length, reduced cell width, and reduced cell area compared with the wild type. Asterisks indicate significant differences from the wild type at P < 0.05 (Student’s t test). Values are given as means ± sd of more than 500 cells from 12 petals.
Interestingly, the spk1 mutant lines exhibited severe defects in the interdigitated growth of abaxial epidermal cells of petal blades (Fig. 3A), similar to the leaf pavement cell defects induced by SPK1 mutation (Qiu et al., 2002). In addition, epidermal cells in the middle part of petal claws of the spk1-4 mutant also had increased growth anisotropy (Supplemental Fig. S6), associated with the increased anisotropic shape of petal claws in the spk1-4 mutant (Supplemental Fig. S2). In the spk1-4 mutant, cell areas in both petal blades and claws were reduced in size (Fig. 3, B and C; Supplemental Fig. S6), correlated with changes in the petal size of the mutant.
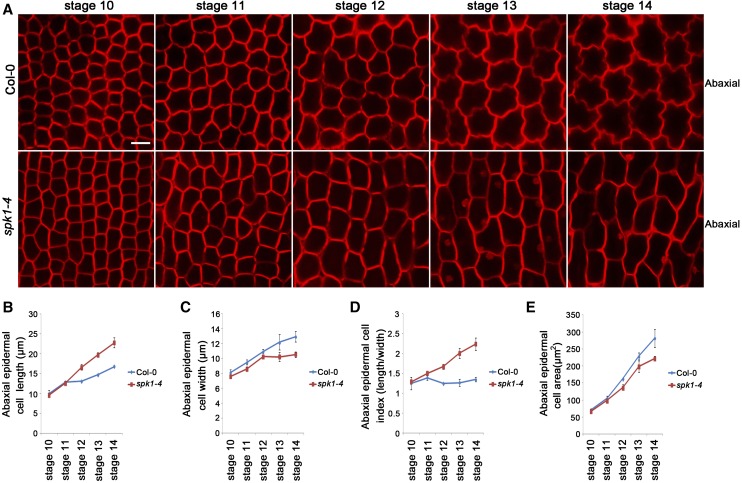
To determine whether the SPK1-regulated cell expansion is associated with anisotropic growth of petals during late development stages, we next performed detailed phenotype analyses in the spk1-4 mutant after development stage 10. We showed that, in wild-type petal blades, both adaxial and abaxial epidermal cells underwent both longitudinal and radial expansion at development stages 10 to 14 (Fig. 4; Supplemental Fig. S7). At development stages 10 and 11, the length and width of adaxial and abaxial epidermal cells in the spk1-4 mutant were similar to those of the wild type (Fig. 4; Supplemental Fig. S7). In striking contrast to the wild type, after development stage 12, abaxial epidermal cells in the spk1-4 mutant expanded less radially and more longitudinally than in the wild type throughout the late development stages 12 to 14, and mutant cells were marked by a cessation of radial expansion at stage 12 and a dramatic increase in longitudinal expansion after stage 12 (Fig. 4). In addition, adaxial epidermal cells in the spk1-4 mutant petal blades also showed increases in cell elongation after development stage 12 (Supplemental Fig. S7).
Figure 4.
Abaxial epidermal cell shape of the wild type and spk1-4 at development stages 10 to 14. A, Abaxial epidermal cell shape of wild-type and spk1-4 petal blades at development stages 10 to 14. Bar = 10 µm. B to E, Measurements for abaxial epidermal cell parameters of the wild type and spk1-4 at development stages 10 to 14. B, Cell length measurements at various stages showed that the spk1-4 mutant had similar cell length to the wild type at stages 10 and 11 (P > 0.05, Student’s t test) but displayed significantly increased cell length at stages 12 to 14 compared with the wild type (P < 0.05, Student’s t test). C, Cell width measurements showed that the spk1-4 mutant had similar cell width to the wild type at stages 10 and 11 (P > 0.05, Student’s t test) but displayed significantly increased cell width at stages 12 to 14 compared with the wild type (P < 0.05, Student’s t test). D, The spk1-4 mutant showed significantly increased cell index (cell length divided by cell width) at stage 11 and beyond. E, The spk1-4 mutant had similar size in cell area to the wild type before stage 11 (P > 0.05, Student’s t test) but had significantly reduced size in cell area at stages 11 to 14 (P < 0.05, Student’s t test). Values are given as means ± sd of more than 500 cells from 12 petals.
Taken together, these observations suggest that SPK1 function is required mainly in the later phase during late development stages to influence anisotropic expansion of petal epidermal cells, correlating with growth anisotropy in both the distal and basal parts of petals, which contributes to the final characteristic shape of the petal.
ROP2, ROP4, and ROP6 Act Redundantly to Regulate Anisotropic Growth in Petals
We next addressed the downstream mechanism by which SPK1 mediates the effect on growth anisotropy in petals. We speculated that, as a GEF, SPK1 must directly activate ROP GTPase proteins and transmit the developmental signals to downstream targets (Basu et al., 2008; Lin et al., 2012). We analyzed available rop knockout mutants for petal phenotype. However, no rop single mutant displayed an obvious defect in petal morphology (data not shown), suggesting that ROP genes may have overlapping functions during petal morphogenesis. Although neither rop2 nor rop6 single mutants showed significant alterations in petal blade length and width (Supplemental Fig. S8), the rop2 rop6 double mutant at development stage 14 displayed an increase in growth anisotropy with longer and narrower petal blades (Fig. 5), associated with increased epidermal cell length and reduced epidermal cell width (Fig. 6; Supplemental Fig. S9). Expressing ROP2 or ROP6 by transformation of pROP2::GFP-ROP2 or pROP6::GFP-ROP6 into the rop2 rop6 double mutant rescued the petal blade phenotype (Supplemental Fig. S10), which confirmed that the petal phenotype of the rop2 rop6 double mutant was caused by the loss of both ROP2 and ROP6 functions.
Figure 5.
ROP2, ROP4, and ROP6 redundantly regulate growth anisotropy of petal blades. A, Fully expanded flowers and petals at development stage 14 in the wild type, the rop2 rop6 double mutant, and the rop2 rop6 ROP4 RNAi lines. Bar = 1 mm. B to E, Quantitative analyses of petal parameters for the indicated genotypes. Measurements are shown for petal blade length (B), petal blade width (C), petal blade index (i.e. length divided by width; D), and petal blade area (E). The rop2 rop6 double mutant and the rop2 rop6 ROP4 RNAi lines had longer and narrower petals than the wild type (P < 0.05, Student’s t test) but displayed similar petal blade area to the wild type (P > 0.05, Student’s t test). Asterisks indicate significant differences from the wild type at P < 0.05 (Student’s t test). Values are given as means ± sd of 16 petals.
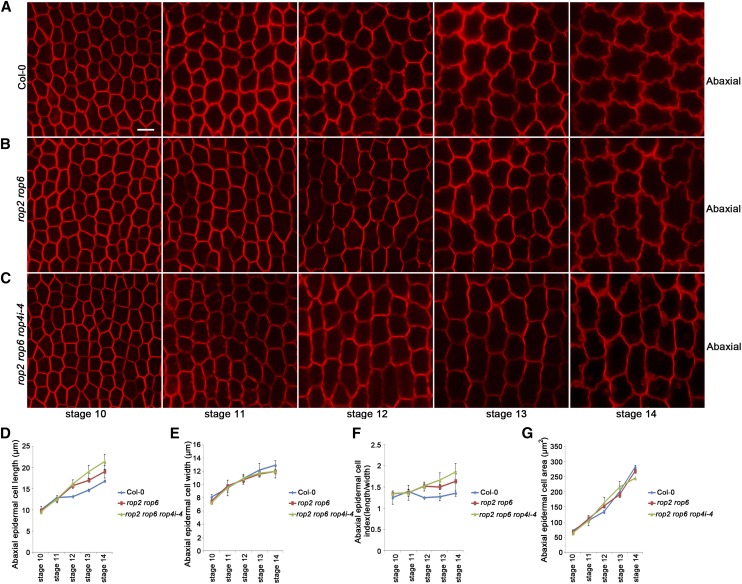
Figure 6.
Abaxial epidermal cell shape of the wild type, the rop2 rop6 mutant, and the rop2 rop6 RON4i-4 mutant at development stages 10 to 14. A to C, Abaxial epidermal cell shape of the indicated genotypes. The wild type (A), rop2 rop6 (B), and rop2 rop6 rop4i-4 (C) are shown at development stages 10 to 14. Bar = 10 µm. D to G, Quantitative analyses of parameters of abaxial epidermal cells for the wild type, the rop2 rop6 mutant, and the rop2 rop6 rop4i-4 mutant at development stages 10 to 14. D, Cell length measurements at various stages showed that both the rop2 rop6 mutant and rop2 rop6 rop4i-4 displayed similar cell length to the wild type at stages 10 and 11 (P > 0.05, Student’s t test). The rop2 rop6 mutant displayed similar cell length to the wild type at stage 12 (P > 0.05, Student’s t test) but had significantly increased cell length at stages 13 and 14 compared with the wild type (P < 0.05, Student’s t test), while the rop2 rop6 rop4i-4 mutant displayed significantly increased cell length at stage 12 and beyond compared with the wild type (P < 0.05, Student’s t test). E, Cell width measurements showed that both the rop2 rop6 mutant and rop2 rop6 rop4i-4 displayed similar cell width to the wild type at stages 10 to 12 (P > 0.05, Student’s t test) but displayed significantly increased cell width at stages 13 and 14 compared with the wild type (P < 0.05, Student’s t test). F, Quantitative analysis of cell index (cell length divided by cell width) for the indicated genotypes. G, Quantitative analysis of cell area for the indicated genotypes. The rop2 rop6 mutant and rop2 rop6 rop4i-4 displayed similar size in cell area to the wild type at stages 10 to 14 (P > 0.05, Student’s t test). All values are given as means ± sd of more than 500 cells from 12 petals.
The mature petal blade phenotype of the rop2 rop6 double mutant was milder than that of the spk1-4 mutant, which could be caused by the overlapping function of ROP2 and ROP4, which share 97% amino acid identity (Fu et al., 2005). Next, we suppressed ROP4 expression by transferring a ROP4 RNAi construct into rop2 rop6 double mutant plants. Multiple rop2 rop6 ROP4 RNAi lines (referred as rop2 rop6 ROP4i) were identified, and three lines with significantly reduced ROP4 transcriptional levels were chosen for phenotype analysis (Supplemental Fig. S11). Consistent with the overlapping roles of ROP4 and ROP2, the phenotypic defects (stage 14) of petal blades and epidermal cells in the rop2 rop6 ROP4i lines were more severe than those of the rop2 rop6 double mutant (Figs. 5 and 6; Supplemental Fig. S9). Moreover, a mutant line, referred to as rop2 rop6 ROP4i-4, which displayed the lowest expression level of ROP4 (Supplemental Fig. S11), had a similar phenotype to the spk1-4 mutant and the SPK1 RNAi lines (Figs. 5 and 6; Supplemental Fig. S9). Furthermore, cell numbers along the length and width directions of the petals in the rop2 rop6 double mutant and the rop2 rop6 ROP4i-4 mutant were similar to those of the wild type (Supplemental Fig. S5), which demonstrated that ROP protein-regulated growth anisotropy was correlated with cell expansion rather than cell proliferation.
Moreover, the lengths of petal claws and claw cells in both the rop2 rop6 mutant and the rop2 rop6 ROP4i-4 mutant showed significant increases compared with those of the wild type (Supplemental Figs. S12 and S13). The widths of petal claws and claw cells were normal in the rop2 rop6 mutant but were reduced significantly in the rop2 rop6 ROP4i-4 mutant (Supplemental Figs. S12 and S13). Therefore, we conclude that ROP2, ROP4, and ROP6 act redundantly to inhibit growth anisotropy at both the distal and basal parts of petals.
To determine whether ROP proteins function redundantly in the regulation of growth anisotropy at late development stages, we performed detailed phenotype analyses from development stages 10 to 14. Our results demonstrated that the rop2 rop6 mutant at development stages 13 and 14, and the rop2 rop6 ROP4i-4 mutant at development stage 12 and beyond, had significantly increased anisotropic growth with more elongated and narrower petals (Supplemental Fig. S14), which were associated with increased cell length and reduced cell width, than those of the wild type (Fig. 6; Supplemental Fig. S9).
SPK1 Is Required for the Activation of Both ROP2 and ROP6 in Floral Organs
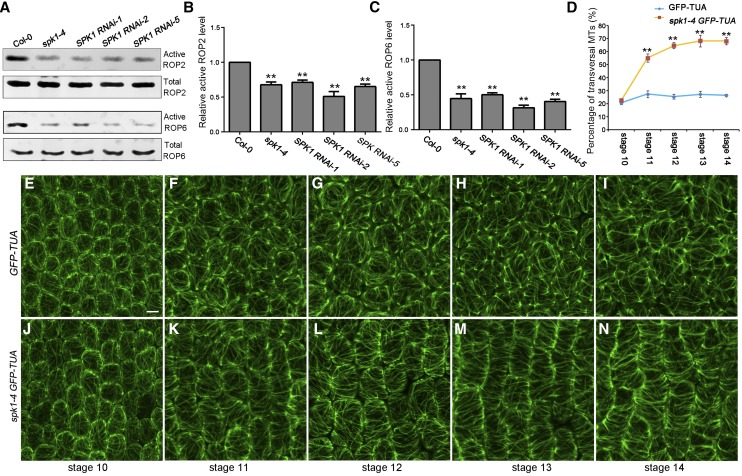
We next tested the hypothesis that SPK1 is the direct activator of ROP proteins in the regulation of anisotropic growth in petals. To determine whether SPK1 is required for the activation of ROP2 and ROP6 during petal growth, we examined ROP activities in the wild type and the spk1 mutant lines. The active form of a ROP protein (GTP-bound ROP) can bind its effector protein, such as RIC1 (Fu et al., 2009); therefore, bacterially expressed recombinant MBP-RIC1 proteins can be used for the effector binding-based pull-down assay (Tao et al., 2002; Duan et al., 2010; Lin et al., 2012) to measure the active form of ROP2 or ROP6 by western blotting with a ROP2- or ROP6-specific antibody, respectively. We then used this method for the quantitative analysis of ROP2 or ROP6 activity in the floral organs of the wild type, the spk1-4 mutant, and the SPK1 RNAi lines. If SPK1 activates both ROP2 and ROP6, we would expect both ROP2 and ROP6 activities to decrease in the spk1-4 mutant and the SPK1 RNAi lines. Indeed, the pull-down experiments based on the effector-binding assay demonstrated that, in spk1-4 and the SPK1 RNAi lines, both ROP2 and ROP6 activities were greatly decreased (Fig. 7, A–C), suggesting that SPK1 is required for the activation of both ROP2 and ROP6 in the floral organs.
Figure 7.
SPK1 activates both ROP2 and ROP6 and is required for the organization of CMTs in petals. A to C, Knockdown of SPK1 reduced both ROP2 and ROP6 activities by in vivo pull-down assays. A, Data shown at both top and bottom represent one of the three replicates. B and C, Quantification of active ROP level (amount of GTP-bound ROP divided by the amount of total ROP) relative to the wild-type control (as 1). Quantification data demonstrated that both ROP2 (B) and ROP6 activities (C) in SPK1 knockdown lines were reduced significantly in comparison with the wild type (**, P < 0.01, Student’s t test). Wild-type and mutant inflorescences were collected and used for protein extraction. Twenty micrograms of MBP-RIC1-conjugated agarose beads were added to the protein extracts for pull-down assays. Activated forms of GTP-bound ROP proteins that were associated with the MBP-RIC1 beads were boiled and used for analysis by western blotting with a ROP2- or ROP6-specific antibody (for details, see “Materials and Methods”). D, Quantitative analysis of the percentage of transverse CMTs. Asterisks indicate significant differences from the control (**, P < 0.01, Student’s t test). E to N, Knockdown of SPK1 resulted in well-ordered transverse CMT arrays in petal abaxial epidermal cells of the middle part of the petal blade. The organization of CMT arrays was analyzed at various development stages in the wild type and spk1-4 using GFP-tagged Tubulin6 (TUA) as described in the text. At stage 10, CMT arrays were oriented randomly in wild-type petal abaxial epidermal cells (E), and mutation of SPK1 did not affect the organization of CMT arrays at this stage (J). At stages 11 to 14, wild-type cells retained many randomly oriented CMT arrays and a few transverse CMTs that were only associated with the indentation regions of the cells (F–I), whereas the spk1-4 mutant generally contained more transversely arranged CMT arrays throughout stages 11 to 14 (K–N). Bar = 5 µm.
SPK1 Regulates the Organization of CMT Arrays in Petal Blades
Previous reports have shown that the isotropy or anisotropy of cells in the SAM depend on the presence of cellulose microfibrils in the extracellular matrix that are arranged in a highly ordered manner (Hamant et al., 2008; Corson et al., 2009; Uyttewaal et al., 2012). The orientation of these cellulose fibrils depends in turn on the organization of CMT arrays (Baskin, 2001; Ehrhardt and Shaw, 2006). It is well established that ROP GTPase-dependent signaling functions in the patterning of the cell wall (Oda and Fukuda, 2012, 2013). Well-ordered CMT arrays arranged transversely relative to the cell axis are associated with increased cell elongation but suppressed radial cell expansion (Wasteneys and Galway, 2003; Smith and Oppenheimer, 2005). Therefore, we hypothesized that the SPK1-ROP protein pathway functions in the orientation of CMT arrays, which contributes to the anisotropic expansion of petal cells and to the growth anisotropy of petals during late development stages.
Because the adaxial epidermal cell displays a three-dimensional geometric shape with a conical tip and a pentagonal or hexagonal base (Irish, 2008), we chose the flattened abaxial epidermal cell to study the correlation between CMT arrangements and cell expansion patterns. We next analyzed the organization of CMT arrays in a petal blade’s abaxial epidermal cells during late stages of petal development. We used a transgenic line expressing GFP-tagged α-tubulin (Ueda et al., 1999) to observe CMT arrays and crossed the spk1-4 mutant to the GFP-Tubulin6 line. We then compared the organization of CMT arrays in petal abaxial cells throughout late development stages between the GFP-Tubulin6 control line and a spk1-4 GFP-Tubulin6 line. In both wild-type and spk1-4 mutant petals at stage 10, the young abaxial epidermal cells displaying a polygonal shape exhibited a network of CMT arrays that were randomly oriented (Fig. 7, D, E, and J). Starting at stage 11, wild-type abaxial epidermal cells with small lobes that were beginning to extend retained many randomly oriented CMTs and a few transverse CMTs, which were only associated with the indentation regions of the cells (Fig. 7, D and F–I). When spk1-4 mutant abaxial epidermal cells started to elongate after stage 11, they generally contained more transversely arranged CMT arrays throughout the fast cell elongation stages (stages 11–13; Fig. 7, D and K–M). In mature petals at stage 14, CMT arrays in wild-type abaxial epidermal cells were oriented randomly (Fig. 7I), whereas the spk1-4 mutant cells that were longer and narrower than the wild-type cells exhibited highly ordered CMT arrays oriented transversely relative to the axis of cell elongation (Fig. 7N). This finding, together with the petal cell phenotype in the spk1-4 mutant, is consistent with previous observations that transverse CMT arrays are associated with increased cell elongation but suppressed radial cell expansion (Wasteneys and Galway, 2003; Smith and Oppenheimer, 2005). Moreover, our immunofluorescence analysis further confirmed that mutation of SPK1 caused highly ordered CMT arrays in petal epidermal cells after stage 11 (Fig. 8).
Figure 8.
Immunostaining of CMTs in petal abaxial epidermal cells. A, Immunostaining of CMTs in petal abaxial epidermal cells from the wild type, the rop2 rop6 double mutant, the rop2 rop6 ROP4i-4 line, and the spk1-4 mutant at development stages 12 to 14. Immunofluorescence was used for visualization of CMT organization. Starting at development stage 12, wild-type petal cells formed randomly oriented CMT arrays, whereas cells in rop2 rop6, rop2 rop6 ROP4i-4, and spk1-4 generally displayed transversely arranged CMT arrays at late stages of petal development (stages 12–14). Bar = 10 µm. B, Quantitative analysis of the percentage of transverse CMTs. Asterisks indicate significant differences from the control (***, P < 0.001, Student’s t test).
Taken together, our results show that mutation of SPK1 promotes a transition of microtubule reorientation from random to transverse at stage 11, which is consistent with the inhibition of lateral expansion and the promotion of longitudinal expansion in petal epidermal cells of the spk1-4 mutant during late petal development stages. The SPK1-promoted isotropic organization of CMT arrays contributes to the growth isotropy of epidermal cells likely by affecting cellulose microfibril arrangements and cell wall patterns.
ROP2, ROP4, and ROP6 Act Redundantly to Regulate the Organization of CMT Arrays
We next investigated the contribution of downstream signaling components of SPK1, the ROP proteins, to the organization of CMT arrays in petal abaxial epidermal cells. We visualized CMT arrays in the rop2 rop6 and rop2 rop6 ROP4i-4 mutant petal cells using immunofluorescence. Starting at development stage 12, wild-type petal cells formed randomly oriented CMT arrays, whereas cells in the rop2 rop6 or rop2 rop6 ROP4i mutant generally displayed transversely arranged CMT arrays at late stages of petal development (stages 12–14; Fig. 8), as did the spk1-4 mutant. These well-ordered transverse CMT arrays induced by loss of function of ROP proteins were correlated with reduced lateral expansion and increased longitudinal expansion in petal epidermal cells of the rop2 rop6 mutant or the rop2 rop6 ROP4i-4 mutant (Fig. 6; Supplemental Fig. S9). Taken together, these results suggest that ROP2, ROP4, and ROP6 act redundantly at late development stages to promote the formation of randomly oriented CMT arrays in petal abaxial epidermal cells, which contributes to cell expansion and the final characteristic shape of petals.
DISCUSSION
Our findings presented here have established a SPK1-ROP GTPase-dependent signaling module that functions in petals to regulate growth anisotropy and thus defines the final characteristic shape. This signaling module regulates the organization of CMT arrays, which affects the expansion patterns of petal epidermal cells, possibly through the arrangement and deposition of cellulose microfibrils. Previous results have shown that most anisotropically growing cells undergo two distinct expansion phases: cells expand both longitudinally and radially in the first phase, while cells expand only longitudinally in the second phase (Beemster and Baskin, 1998). Interestingly, our results suggest that petal epidermal cells undergo a homogenous expansion that requires both longitudinal and radial expansion throughout late development stages, which is crucial for the growth isotropy of petals and, consequently, to generate the characteristic shape of petals with length and width in proportion. A previous study has shown that loss of SPK1 causes abnormal cell expansion in leaves (Qiu et al., 2002); therefore, it remains an open question whether SPIKE1 has a specific role in regulating anisotropic cell expansion in petals.
Our results suggest that increased well-ordered CMT arrays result in the greater anisotropic growth of petals in the spk1 and rop mutants. However, the connection between microtubule orientation and anisotropic growth needs to be explored further. We cannot rule out the possibility that this connection is indirect. For example, perhaps the inability to form lobes in abaxial epidermal cells of the spk1 and rop mutants, which may be regulated by actin filaments, leads to a default anisotropic elongation mode and transverse microtubules.
Our previous study has demonstrated that SPK1 is required in roots for the auxin-activated ROP6-RIC1 pathway to inhibit PIN2 internalization by stabilizing actin microfilaments and, consequently, affect auxin distribution and lateral root development (Lin et al., 2012). In addition, SPK1 has been implicated in activating ROP signaling to regulate actin polymerization via WAVE and ARP2/3 complexes in leaves (Qiu et al., 2002; Basu et al., 2008; Zhang et al., 2010). We cannot rule out the possibility that SPK1 plays an additional role in other signaling mechanisms in the organization of actin microfilaments during cell expansion of petal adaxial or abaxial epidermis, especially in the formation of lobes of abaxial epidermal cells. Further research is required to investigate whether the SPK1-ROP pathway affects the organization of actin microfilaments to control the interdigitated growth of petal abaxial epidermal cells and the formation of the conical shape of petal adaxial epidermal cells.
It is well established that ROP2/4 and ROP6 function antagonistically in leaves (Fu et al., 2005, 2009; Xu et al., 2010); however, in this study, our results show that ROP2/4 and ROP6 act redundantly in petals to regulate cell shape and petal growth, which suggests that ROP proteins may have distinct roles in specific organs. The ROP GTPase proteins ROP2 and ROP6 function antagonistically in leaves to create the jigsaw puzzle piece shape of leaf pavement cells (Fu et al., 2005, 2009; Xu et al., 2010). ROP2 activates its effector protein RIC4 to promote the assembly of cortical actin microfilaments required for localized outgrowth, thus leading to lobe formation, while ROP6 activates its effector protein RIC1 to produce well-ordered transverse CMTs in neck regions, leading to the inhibition of neck outgrowth. In leaves, the rop2 rop4 RNAi double mutant had pavement cells with fewer lobes and narrow necks associated with transversely ordered CMT arrays (Fu et al., 2005), whereas loss of ROP6 function results in leaf pavement cells with wider necks but normal lobes associated with randomly oriented CMT arrays compared with the wild type (Fu et al., 2009). In this study, our results showed that, in petal blades at stage 12 and beyond, the rop2 rop6 and rop2 rop6 ROP4i-4 mutants had fewer lobed abaxial epidermal cells associated with transversely ordered CMT arrays (Fig. 6), which demonstrates that ROP2, ROP4, and ROP6 act redundantly to promote the isotropic organization of CMT arrays that are required for isotropic expansion of abaxial epidermal cells in petal blades. Previous studies have shown that the rop6 mutation alone decreases growth anisotropy in different organs, including hypocotyl and meristem (Fu et al., 2009; Sassi et al., 2014), whereas our study shows that the double rop2 rop6 mutation causes an increase in growth anisotropy in the petal. The detailed mechanisms underlying the regulation of growth anisotropy and CMT arrangements by ROP6 appear to differ between cell types and tissues (Fu et al., 2009; Chen et al., 2014), which may reflect the diverse developmental signals required for the activation of ROP signaling and the fact that ROP proteins may use diverse effectors (Lavy et al., 2007; Li et al., 2008a; Fu et al., 2009; Mucha et al., 2010).
Given that petal phenotypes of the rop mutants were not as strong as the spk1 mutants, this seems to suggest that other ROP proteins or regulators that function downstream of SPK1 also may be involved in the SPK1 pathway in the regulation of petal growth; therefore, future studies should identify novel components that function in the SPK1 pathway. In addition, future studies should determine the developmental signals required for the activation of SPK1-ROP signaling and identify the downstream targets of ROP proteins in the organization of CMT arrays during petal growth and should investigate the role of phytohormones, such as auxin and jasmonates, in the regulation of SPK1 activity. In addition, further studies should illustrate how the SPK1-ROP pathway influences the arrangement of cellulose microfibrils and cell wall patterning during petal growth and development.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Columbia-0 was used as the wild type. The spk1-4 mutant used in this study was described previously (Lin et al., 2012). The T-DNA insertion mutants of ROP2 and ROP6, named rop2 (SALK_055328C) and rop6 (SALK_091737C), respectively, were obtained from the Arabidopsis Biological Resource Center. The rop2 and rop6 homozygous mutants were identified with PCR amplification to confirm the presence of T-DNA insertions. The rop2 rop6 double mutant was generated by crossing parental single homozygous lines. The resulting F2 segregating progeny were genotyped to identify plants homozygous for each locus. Plants were grown in nutrition soil or in Murashige and Skoog agar petri dishes supplemented with 1% (w/v) Suc. Controlled environmental conditions were provided in the growth room at 22°C under a 16-h-light/8-h-dark cycle.
DNA Constructs and Plant Transformation
All constructs were made using the primers listed in Supplemental Table S1. For complementation experiments, full-length genomic DNAs (including its native promoter) of SPK1, ROP2, and ROP6 were amplified and cloned into vector PH35S-GW or PH35S-GFP-GW. The resulting pSPK1::SPK1 was introduced into the spk1-4 mutant by Agrobacterium tumefaciens-mediated transformation for the complementation experiments. The resulting pROP2::GFP-ROP2 and pROP6::GFP-ROP6 were introduced into the rop2 rop6 double mutant. To make the SPK1-RNAi and ROP4-RNAi constructs, cDNA fragments specific to SPK1 and ROP4 were amplified, respectively, using the PCR primers listed in Supplemental Table S1, and the fragments were further introduced into the modified binary vector pFGC5941. The SPK1-RNAi construct used the 35S promoter, but the ROP4-RNAi construct used the native promoter of ROP4 to replace the 35S promoter. The resulting construct, SPK1-RNAi, was then transformed into Columbia-0 to generate the SPK1-RNAi lines. And the resulting construct, ROP4-RNAi, was then transformed into the rop2 rop6 double mutant to generate the rop2 rop6 ROP4 RNAi lines.
Imaging of Petal Epidermal Cells and Measurements of Cell Phenotypes
We used young plants (wild-type and mutant plants with no mature siliques) grown under the same conditions. Petals from flowers at stages 10 to 14 were carefully dissected and then staining in a solution containing 10 μg mL−1 propidium iodide for 1 h. Then, petal adaxial epidermis or abaxial epidermis was imaged by confocal microscopy. For all mutants and the wild type, 12 to 16 petals from four individual plants were used to scan the epidermal cells. Petal blade cell length, cell width (distal region of the petal blade), and cell area were measured manually using ImageJ. More than 500 cells from 12 to 16 petals were measured. Student’s t test was used for statistical analysis.
Confocal Microscopy
Confocal images were collected using a Zeiss LSM 880 confocal microscope. To visualize petal epidermal cells, petal adaxial epidermis or abaxial epidermis was stained in propidium iodide solution and then imaged by confocal scanning using a 543-nm laser. To visualize CMT arrays, samples that express GFP-Tubulin6 were imaged by confocal scanning using a 488-nm laser, and a series of optical sections were taken at 0.6-µm increments with a 40× water lens and then three-dimensional projections were used.
ROP2 and ROP6 Activity Assays
For ROP2 and ROP6 activity assays in the spk1-4 mutant and the SPK1 RNAi lines, the assay was conducted as described previously (Lin et al., 2012). Briefly, 0.1 g of wild-type and mutant inflorescences was collected and frozen in liquid nitrogen. Total proteins were extracted using extraction buffer (25 mm HEPES, pH 7.4, 10 mm MgCl2, 10 mm KCl, 5 mm dithiothreitol, 5 mm Na3VO4, 5 mm NaF, 1 mm phenylmethylsulfonyl fluoride, 1% Triton X-100, and protease inhibitor cocktail). Twenty micrograms (20 µL) of MBP-RIC1-conjugated agarose beads was added to the protein extracts and incubated at 4°C for 2 h on a rocker. The beads were washed four times in wash buffer (25 mm HEPES, pH 7.4, 1 mm EDTA, 5 mm MgCl2, 1 mm dithiothreitol, and 0.5% Triton X-100) at 4°C. GTP-bound ROP proteins that were associated with the MBP-RIC1 beads were boiled and used for analysis by western blotting with a ROP2- or ROP6-specific antibody (Xu et al., 2014). ROP2 and ROP6 polyclonal antibodies were generated against the peptides QFFIDHPGAVPITTNQG and LIGAPAYIECSAKTQQ, respectively (Abicode). Prior to the pull-down assay, a fraction of total proteins was analyzed by immunoblot assay to determine total ROP2 or ROP6 (GDP bound and GTP bound). The amount of the GTP-bound active form of ROPs was normalized to that of total ROPs.
Immunofluorescence
Immunostaining experiments were performed according to the procedure described previously (Wasteneys et al., 1997). Briefly, petals were fixed, frozen, shattered, and permeabilized. The fixed tissues were incubated with monoclonal anti-α-tubulin-fluorescein isothiocyanate antibody produced in mouse (1:300; Sigma F2168) The samples were analyzed using the Zeiss LSM 880 confocal microscopy system.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SPK1 (AT4G16340), ROP2 (AT1G20090), ROP4 (AT1G75840), and ROP6 (AT4G35020).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression analysis of SPK1 in floral organs.
Supplemental Figure S2. Phenotype and quantification of petal claws of the spk1-4 mutant.
Supplemental Figure S3. Complementation of the spk1-4 mutant.
Supplemental Figure S4. RT-PCR and qRT-PCR analysis of SPK1 RNAi lines.
Supplemental Figure S5. Quantitative analysis of cell numbers.
Supplemental Figure S6. Phenotype and quantification of epidermal cells in petal claws of the spk1-4 mutant.
Supplemental Figure S7. Adaxial epidermal cell shapes of the wild type and spk1-4 at development stages 10 to 13.
Supplemental Figure S8. Analysis of petal phenotypes of the rop2 and rop6 single mutants.
Supplemental Figure S9. Adaxial epidermal cell shapes of the wild type, the rop2 rop6 mutant, and the rop2 rop6 ROP4 RNAi lines.
Supplemental Figure S10. Complementation of the rop2 rop6 mutant.
Supplemental Figure S11. RT-PCR analysis of the rop2 rop6 mutant and the rop2 rop6 ROP4 RNAi lines.
Supplemental Figure S12. Phenotypes and quantification of petal claws in the rop2 rop6 mutant and the rop2 rop6 ROP4i-4 mutant.
Supplemental Figure S13. Phenotypes and quantification of epidermal cells in petal claws of the rop2 rop6 mutant and the rop2 rop6 ROP4i-4 mutant.
Supplemental Figure S14. Quantitative analysis of petal phenotypes for the wild type, the rop2 rop6 double mutant, and the rop2 rop6 ROP4i-4 line from development stages 10 to 14.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Zhenbiao Yang (University of California, Riverside) for helpful discussions and generously sharing seeds of Arabidopsis mutants, Lei Shi (Cell Biology Core Facility, Horticultural Plant Biology and Metabolomics Center, Fujian Agriculture and Forestry University) for technical assistance, and the Arabidopsis Biological Resource Center at Ohio State University.
Glossary
- CMT
cortical microtubule
- SAM
shoot apical meristem
- RNAi
RNA interference
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31570278 and 31500160), the Natural Science Foundation of Fujian Province (grant no. 2015J01093), and the Fujian-Taiwan Joint Innovative Center for Germplasm Resources and Cultivation of Crops (FJ 2011 Program, grant no. 2015–75).
References
- Barkoulas M, Galinha C, Grigg SP, Tsiantis M (2007) From genes to shape: regulatory interactions in leaf development. Curr Opin Plant Biol 10: 660–666 [DOI] [PubMed] [Google Scholar]
- Baskin TI. (2001) On the alignment of cellulose microfibrils by cortical microtubules: a review and a model. Protoplasma 215: 150–171 [DOI] [PubMed] [Google Scholar]
- Baskin TI. (2005) Anisotropic expansion of the plant cell wall. Annu Rev Cell Dev Biol 21: 203–222 [DOI] [PubMed] [Google Scholar]
- Basu D, Le J, Zakharova T, Mallery EL, Szymanski DB (2008) A SPIKE1 signaling complex controls actin-dependent cell morphogenesis through the heteromeric WAVE and ARP2/3 complexes. Proc Natl Acad Sci USA 105: 4044–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GT, Baskin TI (1998) Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol 116: 1515–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berken A, Thomas C, Wittinghofer A (2005) A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 436: 1176–1180 [DOI] [PubMed] [Google Scholar]
- Bringmann M, Li E, Sampathkumar A, Kocabek T, Hauser MT, Persson S (2012) POM-POM2/cellulose synthase interacting1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell 24: 163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Grandont L, Li H, Hauschild R, Paque S, Abuzeineh A, Rakusová H, Benkova E, Perrot-Rechenmann C, Friml J (2014) Inhibition of cell expansion by rapid ABP1-mediated auxin effect on microtubules. Nature 516: 90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corson F, Hamant O, Bohn S, Traas J, Boudaoud A, Couder Y (2009) Turning a plant tissue into a living cell froth through isotropic growth. Proc Natl Acad Sci USA 106: 8453–8458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock C, Lavagi I, Yang Z (2012) New insights into Rho signaling from plant ROP/Rac GTPases. Trends Cell Biol 22: 492–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Höfte H, Vernhettes S (2009) Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Yadegari R, Fischer RL, Yanofsky MF, Weigel D (2004) The role of JAGGED in shaping lateral organs. Development 131: 1101–1110 [DOI] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM (2010) FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA 107: 17821–17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt DW, Shaw SL (2006) Microtubule dynamics and organization in the plant cortical array. Annu Rev Plant Biol 57: 859–875 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A (2002) Rho GTPases in cell biology. Nature 420: 629–635 [DOI] [PubMed] [Google Scholar]
- Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z (2005) Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700 [DOI] [PubMed] [Google Scholar]
- Fu Y, Xu T, Zhu L, Wen M, Yang Z (2009) A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Curr Biol 19: 1827–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura U, Elsaesser L, Breuninger H, Sánchez-Rodríguez C, Ivakov A, Laux T, Findlay K, Persson S, Lenhard M (2014) Atkinesin-13A modulates cell-wall synthesis and cell expansion in Arabidopsis thaliana via the THESEUS1 pathway. PLoS Genet 10: e1004627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Li S, Lord EM, Yang Z (2006) Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth. Plant Cell 18: 366–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O, Heisler MG, Jönsson H, Krupinski P, Uyttewaal M, Bokov P, Corson F, Sahlin P, Boudaoud A, Meyerowitz EM, et al. (2008) Developmental patterning by mechanical signals in Arabidopsis. Science 322: 1650–1655 [DOI] [PubMed] [Google Scholar]
- Hazak O, Bloch D, Poraty L, Sternberg H, Zhang J, Friml J, Yalovsky S (2010) A rho scaffold integrates the secretory system with feedback mechanisms in regulation of auxin distribution. PLoS Biol 8: e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JP, Lord EM (1989) Floral development in Arabidopsis thaliana: a comparison of the wild type and the homeotic pistillata mutant. Can J Bot 67: 2922–2936 [Google Scholar]
- Huang JB, Liu H, Chen M, Li X, Wang M, Yang Y, Wang C, Huang J, Liu G, Liu Y, et al. (2014) ROP3 GTPase contributes to polar auxin transport and auxin responses and is important for embryogenesis and seedling growth in Arabidopsis. Plant Cell 26: 3501–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Irish VF (2015) Temporal control of plant organ growth by TCP transcription factors. Curr Biol 25: 1765–1770 [DOI] [PubMed] [Google Scholar]
- Huang T, Irish VF (2016) Gene networks controlling petal organogenesis. J Exp Bot 67: 61–68 [DOI] [PubMed] [Google Scholar]
- Irish VF. (2008) The Arabidopsis petal: a model for plant organogenesis. Trends Plant Sci 13: 430–436 [DOI] [PubMed] [Google Scholar]
- Lavy M, Bloch D, Hazak O, Gutman I, Poraty L, Sorek N, Sternberg H, Yalovsky S (2007) A novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr Biol 17: 947–952 [DOI] [PubMed] [Google Scholar]
- Li S, Gu Y, Yan A, Lord E, Yang ZB (2008a) RIP1 (ROP Interactive Partner 1)/ICR1 marks pollen germination sites and may act in the ROP1 pathway in the control of polarized pollen growth. Mol Plant 1: 1021–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zheng L, Corke F, Smith C, Bevan MW (2008b) Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev 22: 1331–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Cao L, Zhou Z, Zhu L, Ehrhardt D, Yang Z, Fu Y (2013) Rho GTPase signaling activates microtubule severing to promote microtubule ordering in Arabidopsis. Curr Biol 23: 290–297 [DOI] [PubMed] [Google Scholar]
- Lin D, Nagawa S, Chen J, Cao L, Chen X, Xu T, Li H, Dhonukshe P, Yamamuro C, Friml J, et al. (2012) A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr Biol 22: 1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Ren H, Fu Y (2015) ROP GTPase-mediated auxin signaling regulates pavement cell interdigitation in Arabidopsis thaliana. J Integr Plant Biol 57: 31–39 [DOI] [PubMed] [Google Scholar]
- Mucha E, Hoefle C, Hückelhoven R, Berken A (2010) RIP3 and AtKinesin-13A: a novel interaction linking Rho proteins of plants to microtubules. Eur J Cell Biol 89: 906–916 [DOI] [PubMed] [Google Scholar]
- Nag A, King S, Jack T (2009) miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 106: 22534–22539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y, Fukuda H (2012) Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science 337: 1333–1336 [DOI] [PubMed] [Google Scholar]
- Oda Y, Fukuda H (2013) Rho of plant GTPase signaling regulates the behavior of Arabidopsis kinesin-13A to establish secondary cell wall patterns. Plant Cell 25: 4439–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Wightman R, Höfte H (2015) The control of growth symmetry breaking in the Arabidopsis hypocotyl. Curr Biol 25: 1746–1752 [DOI] [PubMed] [Google Scholar]
- Powell AE, Lenhard M (2012) Control of organ size in plants. Curr Biol 22: R360–R367 [DOI] [PubMed] [Google Scholar]
- Pyke KA, Page AM (1998) Plastid ontogeny during petal development in Arabidopsis. Plant Physiol 116: 797–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Yang Z (2011) Rapid tip growth: insights from pollen tubes. Semin Cell Dev Biol 22: 816–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Jilk R, Marks MD, Szymanski DB (2002) The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14: 101–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi M, Ali O, Boudon F, Cloarec G, Abad U, Cellier C, Chen X, Gilles B, Milani P, Friml J, et al. (2014) An auxin-mediated shift toward growth isotropy promotes organ formation at the shoot meristem in Arabidopsis. Curr Biol 24: 2335–2342 [DOI] [PubMed] [Google Scholar]
- Sauret-Güeto S, Schiessl K, Bangham A, Sablowski R, Coen E (2013) JAGGED controls Arabidopsis petal growth and shape by interacting with a divergent polarity field. PLoS Biol 11: e1001550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl K, Muiño JM, Sablowski R (2014) Arabidopsis JAGGED links floral organ patterning to tissue growth by repressing Kip-related cell cycle inhibitors. Proc Natl Acad Sci USA 111: 2830–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG, Oppenheimer DG (2005) Spatial control of cell expansion by the plant cytoskeleton. Annu Rev Cell Dev Biol 21: 271–295 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szécsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, Bendahmane M (2006) BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J 25: 3912–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Matsumoto N, Okada K (2004) RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 131: 425–434 [DOI] [PubMed] [Google Scholar]
- Tao LZ, Cheung AY, Wu HM (2002) Plant Rac-like GTPases are activated by auxin and mediate auxin-responsive gene expression. Plant Cell 14: 2745–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H. (2006) Mechanism of leaf-shape determination. Annu Rev Plant Biol 57: 477–496 [DOI] [PubMed] [Google Scholar]
- Ueda K, Matsuyama T, Hashimoto T (1999) Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma 206: 201–206 [Google Scholar]
- Uyttewaal M, Burian A, Alim K, Landrein B, Borowska-Wykręt D, Dedieu A, Peaucelle A, Ludynia M, Traas J, Boudaoud A, et al. (2012) Mechanical stress acts via katanin to amplify differences in growth rate between adjacent cells in Arabidopsis. Cell 149: 439–451 [DOI] [PubMed] [Google Scholar]
- Varaud E, Brioudes F, Szécsi J, Leroux J, Brown S, Perrot-Rechenmann C, Bendahmane M (2011) AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 23: 973–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys GO. (2004) Progress in understanding the role of microtubules in plant cells. Curr Opin Plant Biol 7: 651–660 [DOI] [PubMed] [Google Scholar]
- Wasteneys GO, Galway ME (2003) Remodeling the cytoskeleton for growth and form: an overview with some new views. Annu Rev Plant Biol 54: 691–722 [DOI] [PubMed] [Google Scholar]
- Wasteneys GO, Willingale-Theune J, Menzel D (1997) Freeze shattering: a simple and effective method for permeabilizing higher plant cell walls. J Microsc 188: 51–61 [DOI] [PubMed] [Google Scholar]
- Willmer P, Stanley DA, Steijven K, Matthews IM, Nuttman CV (2009) Bidirectional flower color and shape changes allow a second opportunity for pollination. Curr Biol 19: 919–923 [DOI] [PubMed] [Google Scholar]
- Wolf S, Hématy K, Höfte H (2012) Growth control and cell wall signaling in plants. Annu Rev Plant Biol 63: 381–407 [DOI] [PubMed] [Google Scholar]
- Wu HM, Hazak O, Cheung AY, Yalovsky S (2011) RAC/ROP GTPases and auxin signaling. Plant Cell 23: 1208–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Dai N, Chen J, Nagawa S, Cao M, Li H, Zhou Z, Chen X, De Rycke R, Rakusová H, et al. (2014) Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science 343: 1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z (2010) Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2002) Small GTPases: versatile signaling switches in plants. Plant Cell (Suppl) 14: S375–S388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YW, Byers KJ, Bradshaw HD Jr (2013) The genetic control of flower-pollinator specificity. Curr Opin Plant Biol 16: 422–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Kotchoni SO, Samuels AL, Szymanski DB (2010) SPIKE1 signals originate from and assemble specialized domains of the endoplasmic reticulum. Curr Biol 20: 2144–2149 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.