Cytokinin regulates chloroplast development during de-etiolation through the two-component signaling system by binding of B-type ARRs to the promotors of chloroplast-related genes.
Abstract
One of the classical functions of the plant hormone cytokinin is the regulation of plastid development, but the underlying molecular mechanisms remain elusive. In this study, we employed a genetic approach to evaluate the role of cytokinin and its signaling pathway in the light-induced development of chloroplasts from etioplasts in Arabidopsis (Arabidopsis thaliana). Cytokinin increases the rate of greening and stimulates ultrastructural changes characteristic for the etioplast-to-chloroplast transition. The steady-state levels of metabolites of the tetrapyrrole biosynthesis pathway leading to the production of chlorophyll are enhanced by cytokinin. This effect of cytokinin on metabolite levels arises due to the modulation of expression for chlorophyll biosynthesis genes such as HEMA1, GUN4, GUN5, and CHLM. Increased expression of HEMA1 is reflected in an enhanced level of the encoded glutamyl-tRNA reductase, which catalyzes one of the rate-limiting steps of chlorophyll biosynthesis. Mutant analysis indicates that the cytokinin receptors ARABIDOPSIS HIS KINASE2 (AHK2) and AHK3 play a central role in this process. Furthermore, the B-type ARABIDOPSIS RESPONSE REGULATOR1 (ARR1), ARR10, and ARR12 play an important role in mediating the transcriptional output during etioplast-chloroplast transition. B-type ARRs bind to the promotors of HEMA1 and LHCB6 genes, indicating that cytokinin-dependent transcription factors directly regulate genes of chlorophyll biosynthesis and the light harvesting complex. Together, these results demonstrate an important role for the cytokinin signaling pathway in chloroplast development, with the direct transcriptional regulation of chlorophyll biosynthesis genes as a key aspect for this hormonal control.
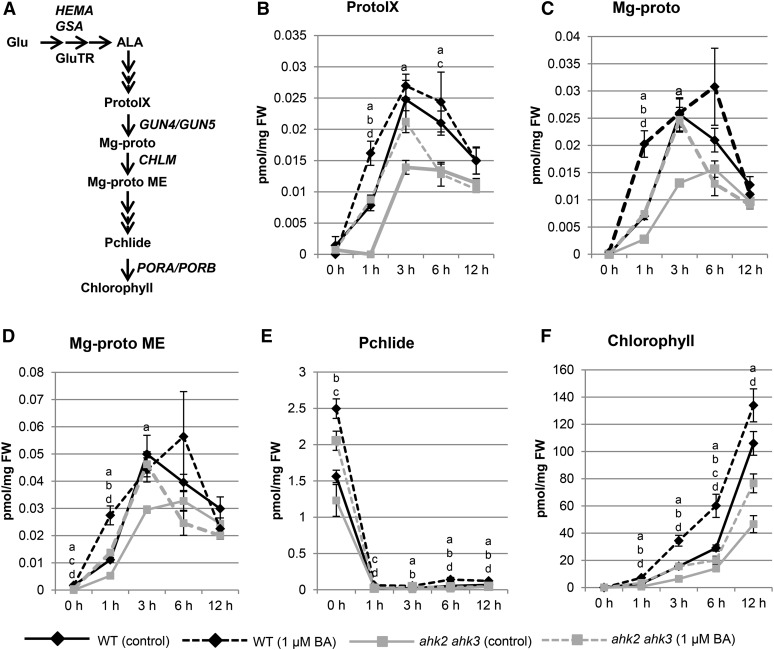
Chloroplasts are plant organelles that are vital for photosynthesis. In addition to this energy conversion process, they also play a role in primary and secondary metabolism (Lopez-Juez and Pyke, 2005). Chloroplasts develop from proplastids that are present in the immature cells of plant meristems. When plants are grown in the dark, proplastids develop into etioplasts that contain a semicrystalline structure, called the prolamellar body, which incorporates lipids and NADPH-dependent protochlorophyllide oxidoreductase (Armstrong et al., 1995). Upon exposure to light, the prolamellar body disperses, thylakoid membranes form, and a fully functional chloroplast develops. The transition of etioplasts to chloroplasts is part of the deetiolation process (for review, see von Arnim and Deng, 1996) and coincides with greening due to chlorophyll biosynthesis (for review, see Tanaka and Tanaka, 2007). Chlorophyll biosynthesis starts from Glu, which is converted to 5-aminolevulinic acid (ALA), and is then further processed to protochlorophyllide. Finally, after light activation of NADPH-dependent protochlorophyllide oxidoreductase, chlorophyll is formed (see Figure 3A for a simplified scheme).
Figure 3.
Effect of cytokinin on the steady-state levels of the metabolites of the tetrapyrrole biosynthesis pathway. A, Simplified scheme of the chlorophyll biosynthesis pathway. Names of genes and proteins analyzed in this study are given at the respective metabolic step. B to F, Levels of intermediate metabolites of chlorophyll biosynthesis at different time points following treatment with cytokinin. Wild-type (WT) and ahk2 ahk3 seedlings were grown for 3 d in dark on medium (MS, no Suc) with or without 1 µM BA prior to analysis. Letters indicate significant differences to the wild type (control conditions) (a, ahk2 ahk3, control conditions; b, wild type, BA; c, ahk2 ahk3, BA) and to the wild type (BA) (d, ahk2 ahk3, BA) (P < 0.05); error bars represent se (n = 5).
Light is the main environmental factor that regulates chloroplast development, but plant hormones are also of importance. Cytokinin, in particular, can partially replicate the effects of light on plant growth and development. Indeed, soon after the discovery of cytokinin as a plant hormone, it was found to regulate the development and function of chloroplasts in a variety of plant species (Miller et al., 1956; for a recent review, see Cortleven and Schmülling, 2015). In 1994, Chory and coworkers reported that cytokinin could induce a deetiolation response in plastids of Arabidopsis (Arabidopsis thaliana; Chory et al., 1994); in the absence of cytokinin, dark-grown seedlings had small etioplasts with a prolamellar body but, in presence of cytokinin, the seedlings had large lens-shaped plastids that lacked a prolamellar body and contained some bithylakoid membranes. Although similar reports on this cytokinin effect have been published for other species (e.g. Lupinus luteus [lupine], Hordeum vulgare [barley], and Nicotiana tabacum [tobacco]; Parthier, 1979; Cortleven and Schmülling, 2015), the molecular links between the core cytokinin signaling pathway and downstream elements involved in this process are largely missing.
Cytokinin is a versatile plant hormone that, in addition to its effect on chloroplast development, regulates numerous physiological and developmental processes such as the cell cycle, activity of root and shoot meristems, root and shoot branching, floral transition, and leaf senescence (for review, see Werner and Schmülling, 2009; Kieber and Schaller, 2014; Cortleven and Schmülling, 2015). Elements of the cytokinin signaling pathway have been revealed through genetic studies in Arabidopsis. Cytokinin is perceived by three sensor His kinases, named ARABIDOPSIS HIS KINASE2 (AHK2), AHK3, and CYTOKININ RESPONSE1 (CRE1)/AHK4 (Inoue et al., 2001; Suzuki et al., 2001), which have both overlapping and distinct functions (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006; reviewed by Heyl et al., 2012). A two-component signaling system transmits the signal from the receptors via His phosphotransfer proteins (AHPs; Hutchison et al., 2006) to type-B response regulators (type-B ARRs), which are transcription factors that regulate expression of early cytokinin-response genes (Sakai et al., 2001; Mason et al., 2005). The B-type response regulators ARR1 (ARABIDOPSIS RESPONSE REGULATOR1), ARR10, and ARR12 play the major role in mediating cytokinin signaling during the vegetative growth phase (Argyros et al., 2008; Ishida et al., 2008). Downstream of this core signaling pathway numerous early and late cytokinin response genes have been identified. These include some nuclear- and plastid-encoded genes involved in chloroplast function (e.g. Brenner et al., 2005; Zubo et al., 2008, 2009; Bhargava et al., 2013; Brenner and Schmülling, 2012; reviewed by Cortleven and Schmülling, 2015). Among the genes induced in response to cytokinin are two GATA-family transcription factors named GNC (GATA, NITRATE-INDUCIBLE, CARBON-METABOLISM INVOLVED) and CGA1/GNL (CYTOKININ-INDUCED GATA1/GNC-LIKE; Bi et al., 2005; Naito et al., 2007), which have been implicated in chloroplast development and chlorophyll biosynthesis (Richter et al., 2010; Hudson et al., 2011; Köllmer et al., 2011; Chiang et al., 2012). However, their transcriptional response to cytokinin during the chloroplast-to-etioplast transition is unknown.
In this study, we investigated the mechanistic basis by which cytokinin mediates the etioplast-to-chloroplast transition in Arabidopsis, focusing on how chlorophyll biosynthesis is regulated during this developmental process. By employing genetic analysis using mutants in the cytokinin signaling pathway, we reveal a central role for the cytokinin receptors AHK2 and AHK3, as well as the B-type response regulators ARR1, ARR10, and ARR12 in the etioplast-to-chloroplast transition. Our results demonstrate that cytokinin regulates expression of genes encoding multiple enzymes in the chlorophyll biosynthesis pathway, including HEMA1 (GLUTAMYL-tRNA REDUCTASE), which regulates an early rate-limiting step. These effects on gene expression are reflected in altered steady-state levels of metabolites in the chlorophyll biosynthetic pathway. Furthermore, we find that B-type ARRs bind to the promotors of HEMA1 and LHCB6 (LIGHT HARVESTING COMPLEX PHOTOSYSTEM II SUBUNIT6), indicating that cytokinin-dependent transcription factors directly regulate genes of chlorophyll biosynthesis and the light harvesting complex involved in the etioplast-to-chloroplast transition.
RESULTS
Cytokinin Increases the Greening Rate and Accelerates the Etioplast-to-Chloroplast Transition in Arabidopsis
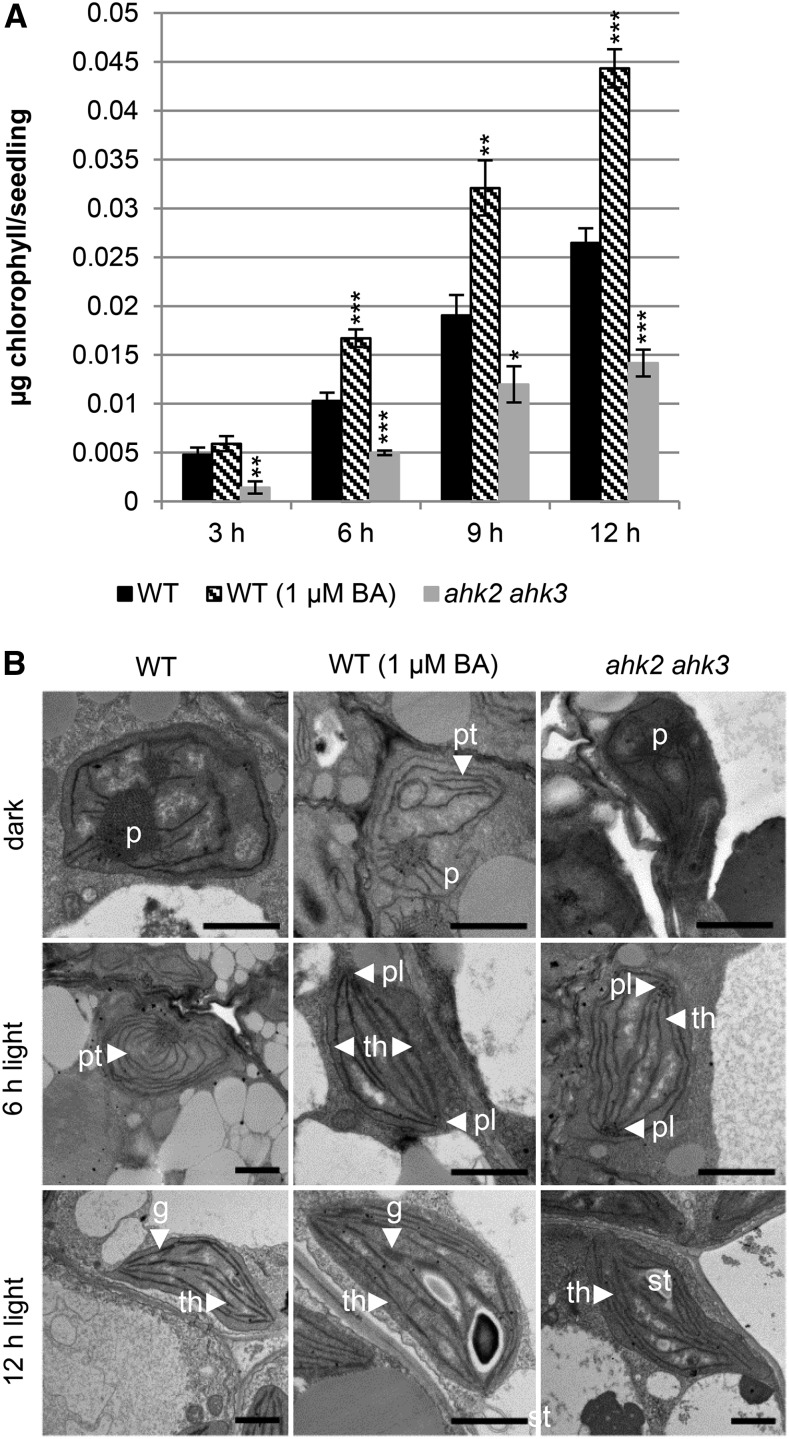
To test the influence of cytokinin on chlorophyll biosynthesis and the formation of chloroplasts, we first tested if under our experimental conditions similar results would be generated as in previous studies exploring the cytokinin effect during deetiolation (for review, see Cortleven and Schmülling, 2015). We germinated Arabidopsis seeds in the dark on medium with or without cytokinin (1 µM 6-benzylaminopurine [BA]). Three-day-old etiolated seedlings were transferred to light, and the formation of chlorophyll was thereafter analyzed in 3-h intervals up to 12 h (Fig. 1). Cytokinin treatment significantly accelerated the formation of chlorophyll in the light, which was apparent after 6 h. Twelve hours after the transfer to light, cytokinin-treated seedlings contained about 40% more chlorophyll than the untreated seedlings. For comparison to wild type, we first analyzed the cytokinin-hyposensitive ahk2 ahk3 mutant, which is affected in leaf development and has a reduced chlorophyll content (Riefler et al., 2006). Figure 1A shows that following dark-to-light transition, the ahk2 ahk3 mutant produced significantly less chlorophyll than wild type at all the time points examined (Fig. 1A). These results demonstrate that cytokinin acts as a positive regulator of the deetiolation process under our experimental conditions.
Figure 1.
Cytokinin increases the greening rate of etiolated Arabidopsis seedlings and alters chloroplast ultrastructure during etioplast-chloroplast transition. The wild type (WT) and ahk2 ahk3 cytokinin receptor mutants were grown for 3 d in dark on medium (MS, no Suc) before exposure to light. A, Chlorophyll content after exposure to light for 3 to 12 h. The average of three independent experiments each with three replicates per time point is shown. One replicate consisted of 30 etiolated seedlings grown on one plate. Error bars represent se (n = 3). Asterisks indicate significant differences to wild type control (* 0.05 > P > 0.01; ** 0.01 > P > 0.001; *** P < 0.001). B, Transmission electron microscope pictures illustrating the etioplast-chloroplast transition. Abbreviations: BA, 6-benzylaminopurine; (g), grana; (p), prolamellar body; (pl), prolamellar-like body; (pt), prethylakoids; (st), starch granules; (th), thylakoid membrane. Scale bars represent 1200 nm.
Besides its effect on the rate of greening, cytokinin also influences the ultrastructural alterations that characterize the etioplast-to-chloroplast transition (Fig. 1B). In dark, the cotyledons of wild-type plants contain etioplasts with the typical prolamellar body. Light treatment causes the differentiation from etioplasts to chloroplasts. After 6 h of light, prethylakoid membranes are formed starting from the center (prolamellar body). After 12 h of light, the prolamellar body has disappeared and chloroplasts are fully differentiated with grana and stroma thylakoids. Cytokinin treatment induces the formation of prethylakoid membranes without light exposure, indicating that the hormone mimics the mode of action of light (Fig. 1B), consistent with the previous observations of Chory et al. (1994). After 6 h of light, the prolamellar body is completely absent and the typical chloroplast ultrastructure with grana and stroma thylakoids is visible in wild-type seedlings growing on medium containing cytokinin. Finally, a more pronounced grana stacking and accumulation of starch is visible after 12 h of light in the presence of cytokinin. In dark-grown ahk2 ahk3 seedlings (Fig. 1B), the overall ultrastructure of the etioplast is comparable to wild type, although we observed fewer etioplasts. After 6 h of light, etioplasts in ahk2 ahk3 have largely lost their prolamellar body and have begun to form thylakoid-like membrane structures. However, we observed several paracrystalline structures or potential aggregations of small vesicles that resemble the prolamellar body. This could indicate that the differentiation of the prolamellar body into thylakoid membranes is disorganized and that these plants have compromised chlorophyll biosynthesis, because chlorophyll biosynthesis mutants exhibit an asymmetric organization of the prolamellar body (Solymosi and Schoefs, 2010). After 12 h of light, chloroplasts with grana and stroma thylakoids are also formed in the ahk2 ahk3 mutant; however, the overall structure is less organized than that found in wild type (Fig. 1B). Taken together, these results demonstrate that cytokinin can partially phenocopy and, in addition, accelerate the ultrastructural changes inherent to the etioplast-to-chloroplast transition during deetiolation.
Cytokinin-Signaling Mutants Exhibit Altered Greening Response
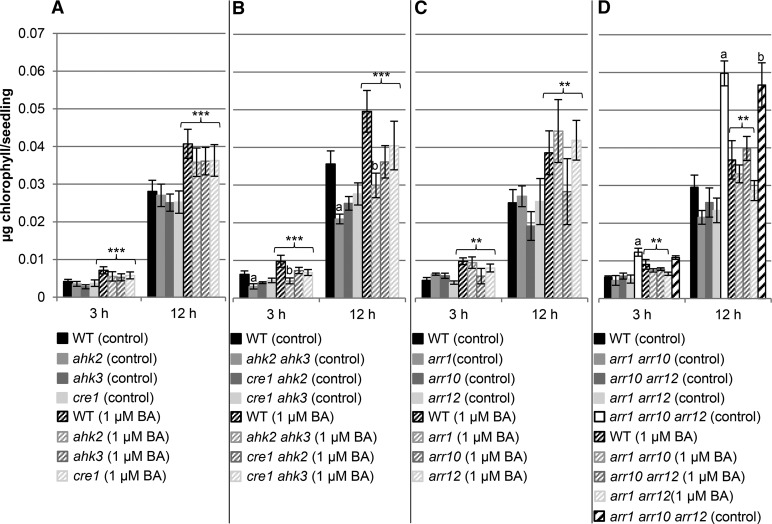
We took a genetic approach to investigate in more detail which elements of the cytokinin signaling pathway mediated the effects of cytokinin on chloroplast development. We analyzed the chlorophyll content in a set of cytokinin signaling mutants comprising all single and double receptor mutant combinations as well as all single and double mutant combinations of the three B-type response regulators, ARR1, ARR10, and ARR12. These three transcription factors functionally overlap and are the predominant regulators of cytokinin action during vegetative growth (Argyros et al., 2008; Ishida et al., 2008). More specifically, they were previously found to mediate cytokinin-based deetiolation of dark-grown seedlings (Argyros et al., 2008), a finding consistent with their expression level under this growth condition (normalized expression of ARR1 is 176, ARR12 186, and ARR10 97 in 4-d-old dark-grown seedlings based on microarray data analyzed through the Arabidopsis eFP browser; Winter et al., 2007). We therefore considered these three B-type ARRs to be good candidates to mediate the cytokinin-regulated transcriptional output during etioplast-chloroplast transition and chlorophyll biosynthesis (Fig. 2; additional time points can be found in Supplemental Fig. S1).
Figure 2.
Greening rate in mutants of the cytokinin signaling pathway. A, Single (ahk2, ahk3, and cre1) and B, double (ahk2 ahk3, cre1 ahk2, and cre1 ahk3) cytokinin receptor mutants, and C, single (arr1, arr10, and arr12) and D, double and triple arr (arr1 arr10, arr1 arr12, arr10 arr12, and arr1 arr10 arr12) mutants were grown for 3 d in dark on medium (MS, no Suc) with or without 1 µM BA. Control conditions are visualized by solid bars; treatment with BA with hatched bars. After exposure to light, chlorophyll content was measured at different time points. Diagrams shown are the average of at least four independent experiments each with three replicates per time point. One replicate consisted of 30 etiolated seedlings grown on one plate. Error bars represent se (n ≥ 3). Letters indicate significant differences between wild type (WT) and the different genotypes under control condition (a) and after BA treatment (b) (P < 0.05). Asterisks above brackets indicate a significant cytokinin effect in the respective genotype (** 0.01 > P > 0.001; *** P < 0.001). Additional time points can be found in Supplemental Figure S1.
Analysis of chlorophyll formation after transfer to light of dark-grown cytokinin receptor mutants showed a significant increase in the rate of chlorophyll production in all single mutants after cytokinin treatment (cytokinin effect), with a slight but not significant reduction compared to wild type (Fig. 2A; Supplemental Fig. S1A). Similar observations were made for the cytokinin-receptor double mutants (Fig. 2B; Supplemental Fig. S1B). A significant reduction in greening was observed in the ahk2 ahk3 mutant in comparison to wild type, both with and without BA treatment after 3 and 12 h of light (Fig. 2B). The effect of the ahk2 ahk3 mutant is consistent with what we observed in Figure 1A and demonstrates that AHK2 and AHK3 are key players in mediating the cytokinin signal during deetiolation.
In the single and double B-type arr mutants, the greening rate also increased significantly after cytokinin treatment, suggesting there might be functional redundancy between ARR1, ARR10, and ARR12 with respect to the cytokinin response (Fig. 2, C and D; Supplemental Fig. S1). In arr double mutants, chlorophyll accumulation was reduced under control conditions, although not significantly, compared to wild type by the 12-h time point for light treatment (Fig. 2D; Supplemental Fig. S1D). Interestingly, loss of all three ARR genes (arr1 arr10 arr12) resulted in a strong increase in greening rate under control conditions, although this effect on greening was not altered by cytokinin treatment consistent with the mutant being cytokinin insensitive (Fig. 2D). From these results, all three tested B-type ARRs appear to play redundant roles in the normal greening response to light both under control conditions and after CK treatment. Moreover, the results suggest that a compensatory mechanism may be engaged when cytokinin signaling is strongly hampered, the arr1 arr10 arr12 mutant exhibiting the strongest cytokinin insensitivity of all the mutants examined.
Cytokinin Affects the Steady-State Levels of the Metabolites in the Tetrapyrrole Biosynthesis Pathway
Since the largest differences of chlorophyll biosynthesis in response to cytokinin were found in the ahk2 ahk3 receptor mutant, we used this mutant to investigate which steps in the tetrapyrrole biosynthesis pathway leading to chlorophyll production were affected by cytokinin. For this purpose, the profiles of different chlorophyll metabolites and of chlorophyll were measured during deetiolation at multiple time points up to 12 h (Fig. 3) and expressed relative to fresh weight. The levels of the chlorophyll precursors protoporphyrinogen IX (ProtoIX), Mg-protoporphyrin IX (Mg-proto), and Mg-proto monomethylester (Mg-proto ME) follow a similar kinetic: metabolite levels increase after 1 h illumination, reach a maximum after 3 to 6 h, and then decrease. The protochlorophyllide (Pchlide) level has a different kinetic: it accumulates in the dark and decreases rapidly in response to light. This contrasts with chlorophyll, which accumulates steadily in response to light, an increase in chlorophyll levels being detected within 1 h after the start of the light treatment in wild type. These basic kinetic profiles are similar in the absence or presence of cytokinin, and in both wild type and the ahk2 ahk3 mutant (Fig. 3). However, ahk2 ahk3 mutants exhibit a delayed increase in protoporphyrinogen IX, Mg-proto, and Mg-proto monomethylester, and these reach lower levels in comparison to wild type. Cytokinin treatment results in a more rapid increase in the levels of these metabolites in both wild type and ahk2 ahk3 mutants, observable after 1 h illumination. The protochlorophyllide level is lower in the dark in ahk2 ahk3 mutants compared to wild type, and cytokinin increases the level in both genotypes (Fig. 3E). Thus, the levels of intermediates for chlorophyll synthesis (Fig. 3, B–E) correlate with the overall rate of formation of chlorophyll (Fig. 3F). After 12 h of light, ahk2 ahk3 mutant seedlings contain approximately half as much chlorophyll as the wild type, irrespective of being treated by cytokinin or not. In conclusion, treatment with exogenous cytokinin enhances the rate of chlorophyll formation, whereas cytokinin hyposensitivity as found in the ahk2 ahk3 mutant reduces the rate of chlorophyll formation. These effects found in the ahk2 ahk3 mutant indicate that cytokinin plays a normal physiological function in regulating chlorophyll levels. Moreover, cytokinin influences multiple steps of chlorophyll biosynthesis, as indicated by the changes in different metabolites.
Key Steps in the Chlorophyll Biosynthetic Pathway Are Transcriptionally Regulated by Cytokinin
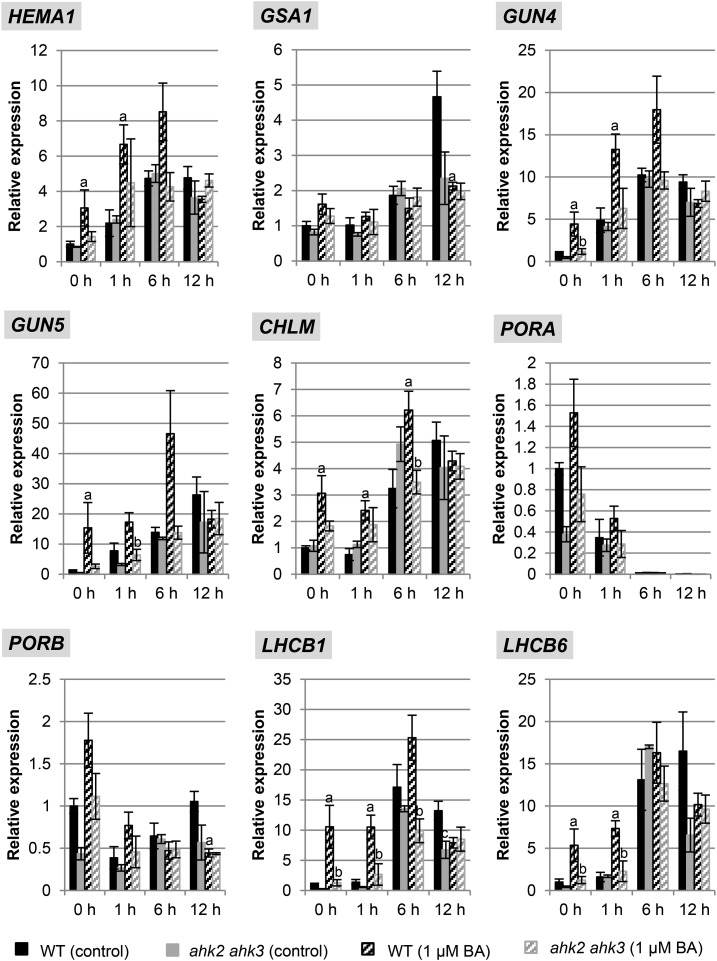
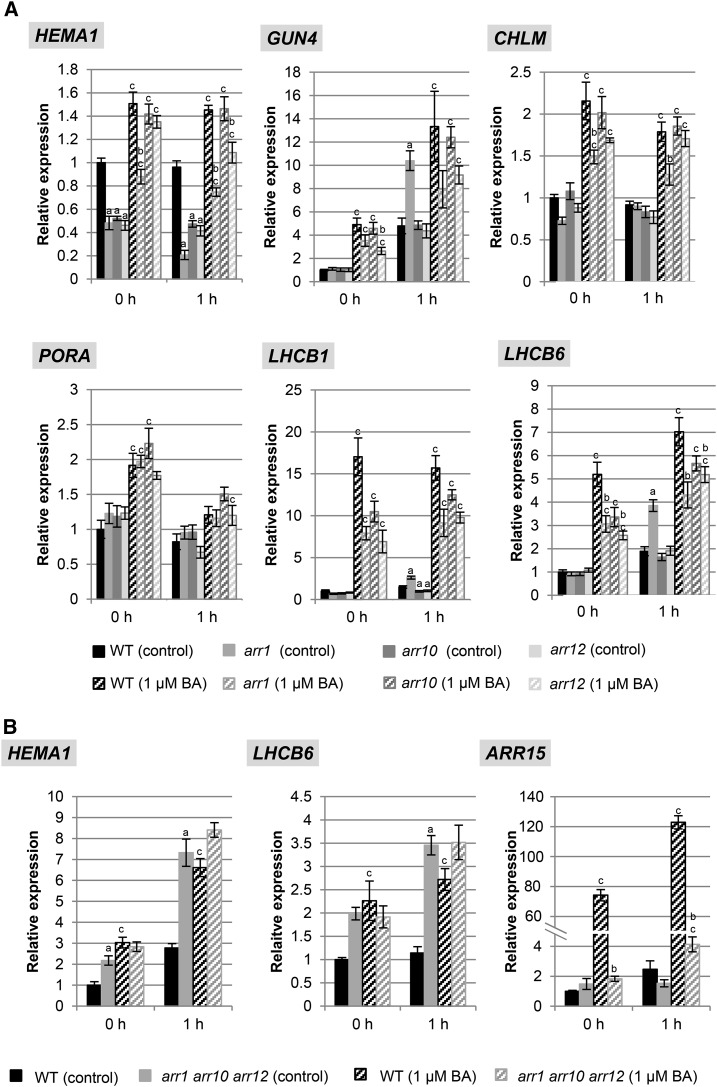
We hypothesized that the cytokinin-dependent changes in metabolites of the chlorophyll biosynthesis pathway could arise from differences in gene expression for biosynthetic enzymes of the pathway. We therefore characterized the expression of multiple genes involved in the pathway, doing so in the absence and presence of cytokinin and in both wild type and the hyposensitive ahk2 ahk3 mutant. These genes included HEMA1 and GSA1 encoding, respectively, glutamyl-tRNA reductase and Glu-1-semialdehyde aminotransferase, which catalyze the initial rate-limiting steps of ALA synthesis (Fig. 3A). We also analyzed several other genes involved in the subsequent steps following ALA biosynthesis; these were GUN4 and GUN5, involved in the conversion of protoporphyrin IX to Mg-proto; CHLM (MAGNESIUM-PROTOPORPHYRIN IX METHYL TRANSFERASE), encoding Mg-protoporphyrin IX methyltransferase; and PORA/PORB, encoding NADPH-protochlorophyllide oxidoreductase (Fig. 3A). In addition, because of the similar expression of chlorophyll biosynthesis and photosynthesis genes (McCormac and Terry, 2002), the transcript levels of LHCB1 and LHCB6, encoding light harvesting chlorophyll binding proteins, were also determined (Fig. 4).
Figure 4.
Effect of cytokinin on transcript levels of genes encoding key steps in the chlorophyll biosynthesis pathway and of LHCB1 during dark-to-light transition. Wild-type (WT) and ahk2 ahk3 seedlings were grown for 3 d in dark on medium (MS, no Suc) with or without 1 µM BA prior to exposure to light. Analysis of transcript levels by quantitative real-time PCR (qPCR) of chloroplast-related genes. Transcript levels of the wild type (control conditions) in dark were set to 1. Statistical significant differences were found for the comparison of the wild type treated with cytokinin and the wild type under control conditions (a) and cytokinin-treated ahk2 ahk3 compared to cytokinin-treated wild type (b) (P < 0.05). Error bars represent se (n ≥ 3). Three independent experiments were performed with a similar outcome; results from one representative experiment are shown.
In wild type and ahk2 ahk3 mutants, similar basal transcript levels were noted in dark-grown seedlings for these genes (Fig. 4, time point 0 h). Growth on cytokinin-containing medium induced significant increases in transcript levels for most of the genes (HEMA1, GUN4, GUN5, CHLM, LHCB1, LHCB6) in wild type, while this response was absent in ahk2 ahk3 mutants. Light treatment also caused a strong increase of transcript abundance for most genes with different kinetics and leading to up to almost 30-fold increased steady-state transcript levels (GUN5). Cytokinin enhanced this effect of light in wild type, but not in the ahk2 ahk3 mutant, for all genes. Only the GSA1, PORA, and PORB transcripts responded differently. Expression of PORA transcripts dropped below detection levels during greening, and PORB mRNA abundance transiently decreased during light exposure. In this case no significant differences were observed in the expression behavior between wild type and the ahk2 ahk3 mutant. Taken together, these results demonstrate a role for cytokinin in the transcriptional regulation of chlorophyll biosynthesis and photosynthesis genes and the requirement of a fully functional cytokinin perception system for this regulation during greening.
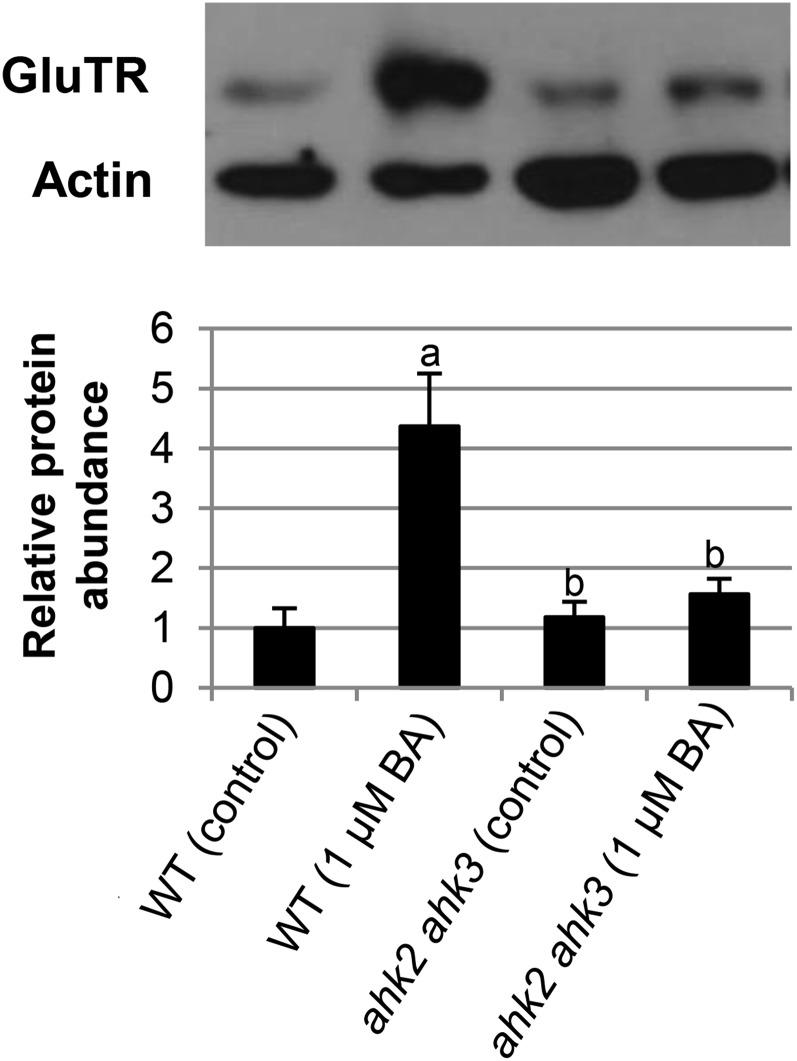
Because HEMA1 encodes an enzyme (glutamyl-tRNA-reductase [GluTR]) that catalyzes an early rate-limiting step in the pathway, we examined whether the changes in its transcript abundance (Fig. 4) were reflected at the protein level (Fig. 5). Levels of GluTR protein were strongly enhanced in response to exogenous cytokinin in wild type but not in the ahk2 ahk3 mutant, demonstrating that the transcriptional response to cytokinin is reflected at the protein level and confirming the relevance of AHK2 and AHK3 in mediating this cytokinin response.
Figure 5.
Effect of cytokinin on protein level of GluTR. Wild-type (WT) and ahk2 ahk3 seedlings were grown for 3 d in dark on medium (MS, no Suc) with or without 1 µM BA. Protein abundance is expressed relative to actin as loading control and wild type (control) was set to 1. One representative blot is shown. Bar graph shown is the average of five independent experiments. Error bars represent se. Letters indicate significant differences to the wild type under control condition (a) or to the wild type treated with BA (b) (P < 0.05).
B-Type ARRs Act in a Redundant Manner to Mediate Cytokinin Action in Etioplast-Chloroplast Transition
Next we investigated whether distinct B-type ARRs take part in the transcriptional regulation of chlorophyll biosynthesis genes, focusing on ARR1, ARR10, and ARR12. In single arr mutants, the strongest differences in expression were observed for HEMA1 and LHCB6 (Fig. 6A). HEMA1 expression was decreased in all three single arr mutants grown in the dark as well as after 1 h of light treatment. Cytokinin treatment increased the transcript level in the wild type and all arr mutants, but the response of arr1 in the dark and of arr1 and arr12 in the light was significantly dampened. The induction of LHCB6 by cytokinin following light treatment was significantly reduced in arr1 and arr12 single mutants. Effects on the expression of other chlorophyll biosynthesis genes were also observed in the arr single mutants, but these were less pronounced than those noted for HEMA1 and LHCB6 (Fig. 6). Taken together, these data demonstrate a role for the B-type ARRs to optimally regulate the expression of key photosynthesis-related genes in response to cytokinin. The altered sensitivity observed in the single mutants suggests that ARR1 plays the largest role in regulation, followed by ARR12, and then ARR10.
Figure 6.
Effect of cytokinin on transcript level of genes encoding key steps in the chlorophyll biosynthesis pathway and of LHCB genes in mutants of B-type ARR genes during dark-to-light transition. Seedlings were grown for 3 d in dark on medium (MS, no Suc) with or without 1 µM BA prior to exposure to light. Transcript levels of chloroplast-related genes in single arr1, arr10, and arr12 mutants (A) and the arr1 arr10 arr12 mutant (B) compared to the wild type. Transcript levels of additional genes can be found in Supplemental Figure S4. Transcript levels of the wild type (control conditions) in dark were set to 1 (n ≥ 3). Letters indicate significant differences between the wild type and the different genotypes under control condition (a) or after BA treatment (b) or between control and BA treatment for each genotype (c) (P < 0.05).
Interestingly, in the three double arr mutant combinations (arr1 arr10, arr1 arr12, arr10 arr12), the changes in gene expression found in the single arr mutants were not enhanced (Supplemental Fig. S2). Instead, the steady-state transcript levels of the genes in dark-grown double mutants were similar to or higher than that found in wild type. For example, the expression of HEMA1, LHCB1, and LHCB6 were higher in dark-grown arr10 arr12 and arr1 arr12 mutants than in the wild-type control. However, the transcript levels for many of these genes (e.g. HEMA1, CHLM, and LHCB1) exhibited decreased responsiveness to cytokinin in the double arr mutants compared to wild type, this response being most affected in arr1 arr12 (Supplemental Fig. S2). The tendency toward higher transcript levels of chlorophyll synthesis genes in the dark-grown higher-order mutants of the B-type ARR seedlings was further accentuated in the triple arr1 arr10 arr12 mutant (Fig. 6B). The basal transcript levels of the HEMA1 and LHCB6 genes (Fig. 6B), as well as those of GUN4, PORA, and LHCB1 (Supplemental Fig. S4), were increased in the triple arr mutant to a similar level as in wild type treated with cytokinin. The abundance of these chloroplast-related genes was not or only moderately increased further by cytokinin treatment. This behavior contrasts with that exhibited by the cytokinin primary response genes ARR15 and ARR5 (Fig. 6B, Supplemental Fig. S4), which have significantly reduced transcript levels in the triple arr mutant.
Taken together, the increase in chlorophyll synthesis gene expression and the corresponding increased greening rate (Fig. 2D) in mutants with strongly reduced cytokinin signaling activity indicate that a compensatory mechanism may be activated under conditions of cytokinin insensitivity.
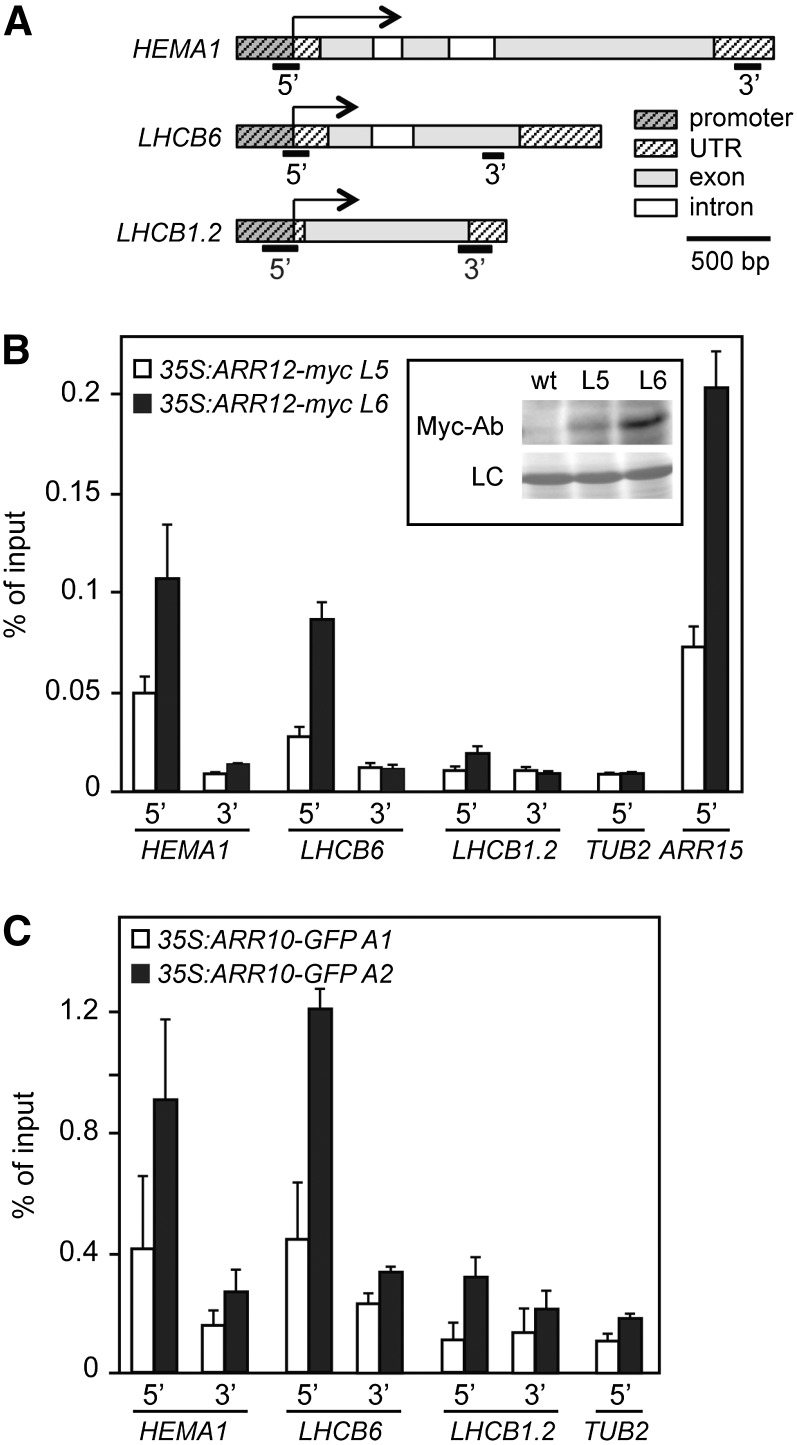
ARR12 and ARR10 Bind Specifically to the HEMA and LHCB6 Promotors
The analysis of transcript levels indicated the involvement of cytokinin in the regulation of the transcription of chlorophyll biosynthesis genes as well as several LHCB genes. To determine whether B-type ARRs could regulate the expression of these genes by directly interacting with their promoters, we performed chromatin immunoprecipitation (ChIP) assays (Fig. 7). For this purpose, we made use of ARR12 fused to an amino-terminal Myc epitope tag to allow for immunoprecipitation, with expression of the transgene driven by the CaMV 35S promoter (35S:myc-ARR12). Two independent lines expressing myc-ARR12 were used (L5 and L6), immunoblot analysis with the anti-Myc antibody confirming production of the full-length fusion protein (Fig. 7B, inset). ChIP-qPCR confirmed binding of myc-ARR12 to the promoter region of the type-A response regulator ARR15, a known primary response gene, as compared to TUB2 (TUBULIN BETA CHAIN2), which served as a negative control (Fig. 7B). We therefore examined myc-ARR12 binding to HEMA1, the gene product of which catalyzes an early rate-limiting step in chlorophyll biosynthesis, and LHCB6, the gene product of which functions in the light harvesting complex of chloroplasts; expression of both genes was responsive to cytokinin and partially dependent on ARR12 (Figs. 4 and 7; Supplemental Fig. S2). We observed binding of myc-ARR12 to the promoter regions of HEMA1 and LHCB6, based on primers designed to amplify the region around the transcriptional start site (TSS; Fig. 7, A and B). The differing levels of binding found in the two lines were consistent with the differing protein levels of myc-ARR12 found in the two lines (Fig. 7B, inset). In contrast, no binding was observed based on primers designed to amplify 3′ regions of the genes at least 1 kb distant from the promoter, which amplified similarly to the TUB2 negative control. Furthermore, we did not observe binding to the 5′-promoter or 3′-untranslated regions of LHCB1.2. Similar binding results were obtained when we examined target specificity of a second B-type response regulator, ARR10 (35S:ARR10-GFP; Fig. 7C). ChIP-qPCR performed with two independent lines demonstrated binding of ARR10-GFP to the promoter regions of HEMA1 and LHCB6, but not of LHCB1.2.
Figure 7.
ARR12 and ARR10 bind to the HEMA and LHCB6 promoters. ChIP analysis was performed on 3-week-old seedlings of 35S:ARR12-myc (lines L5 and L6) and 35S:ARR10-GFP (lines A1 and A2) treated with 5 µM BA for 30 min. Following cross-linking to target DNA, the Myc-tagged ARR12 transcription factor was immunoprecipitated by anti-Myc antibodies and the GFP-tagged ARR10 immunoprecipitated by anti-GFP antibodies ChIP-qPCR analysis was performed for HEMA, LHCB6, LHCB1.2, TUB2 (negative control), and ARR15 (positive control). Primers were designed to amplify 5′ promoter regions near the TSS as well as the 3′ ends of the genes. A, Positions for DNA regions amplified by qPCR are shown schematically by black bars below the gene scheme. B, ChIP-qPCR analysis for 35S:ARR12-myc based on three biological replicates (error bars = sd). The inset shows levels of ARR12-myc in lines L5 and L6 based on immunoblot analysis. C, ChIP-qPCR analysis for 35S:ARR10-GFP based on two biological replicates (error bars = sd).
We examined the promoter regions near the TSS for sequences implicated in the binding of type-B ARRs. For this purpose, we took advantage of recent analysis using protein binding microarrays, by which several high-affinity extended binding motifs have been identified that are longer than the minimal core binding motif originally determined (Hosoda et al., 2002; Sakai et al., 2001; Franco-Zorrilla et al., 2014; Weirauch et al., 2014). Consistent with an ability of ARR12 and ARR10 to bind near to the TSSs of HEMA1 and LHCB6, we identified an extended type-B ARR binding motif (AGATATG) on the reverse strand at position −78 relative to the TSS for HEMA1 along with multiple core binding motifs near the TSSs for both genes; for example, LHCB6 has seven core binding motifs (AGAT[T/C]; Sakai et al. (2001) between −110 and +80 of the TSS. Taken together, these data demonstrate that HEMA1 and LHCB6 are primary targets for cytokinin signaling and that cytokinin can directly regulate genes involved in the etioplast-chloroplast transition.
Interaction between Cytokinin and the GATA Transcription Factors GNL and GNC
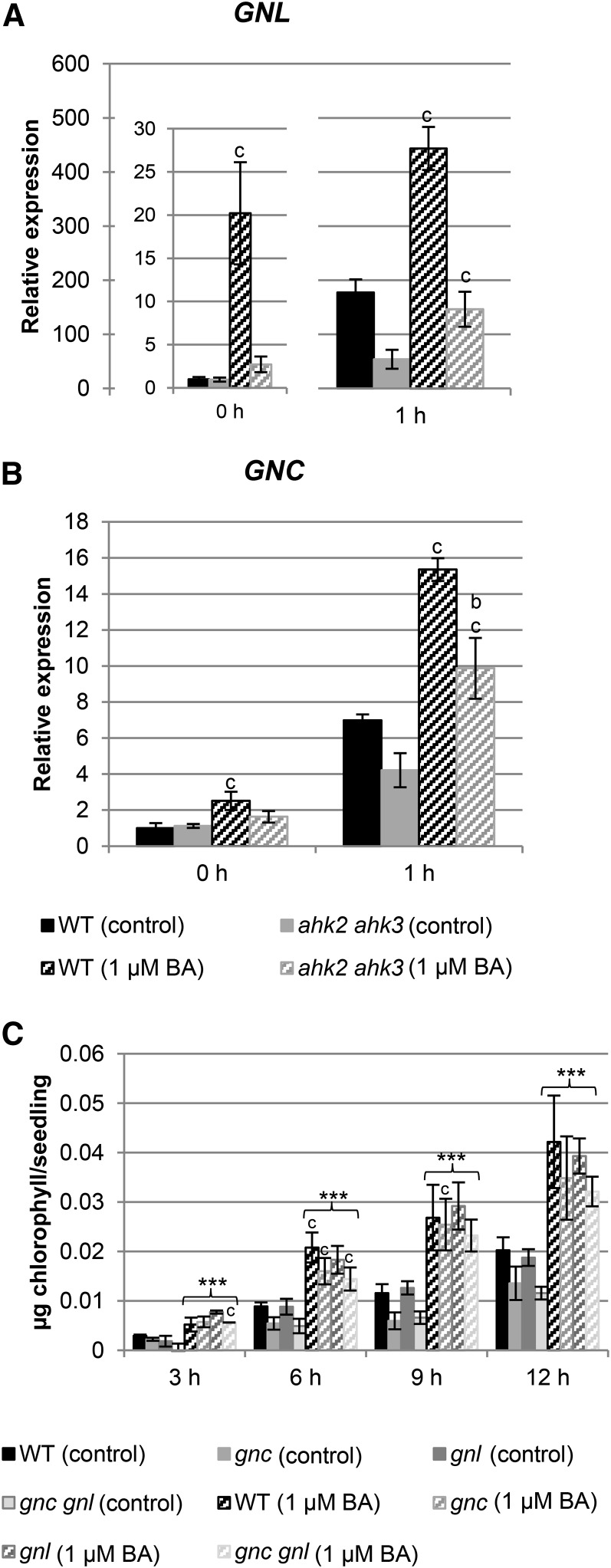
In recent years, several transcription factors that could be involved in mediating the cytokinin response during chloroplast development have been identified (for review, see Cortleven and Schmülling, 2015). Among these transcription factors, GNC and GNL/CGA1 are of particular interest. Belonging to the same clade, these paralogs have been implicated in chloroplast development and are inducible by cytokinin (Bi et al., 2005; Naito et al., 2007; Köllmer et al., 2011; Chiang et al., 2012). Therefore, they could be important factors connecting cytokinin with greening. Analysis of the cytokinin response of GNC and GNL transcript levels in wild type and in the ahk2 ahk3 mutant showed a strong increase of the GNL transcript level in wild type both in dark and after 1 h of light treatment (Fig. 8, A and B). GNC transcripts did not respond as strongly to cytokinin treatment as GNL, and the only approximately 2-fold induction was similar in wild type and the ahk2 ahk3 mutant. However, for both genes the amplitude of transcript level was lower in ahk2 ahk3 mutants.
Figure 8.
Interaction between cytokinin and the GATA transcription factor genes GNL and GNC during the greening response. A and B, Analysis of transcript levels of GNL and GNC genes in 3-d-old etiolated seedlings grown on medium (MS, no Suc) with or without 1 µM BA in the dark and after 1 h exposure to light. Transcript levels of the wild type (WT) in dark were set to 1. Error bars represent se (n ≥ 3). C, Greening rate in gnl, gnc, and gnc gnl mutants grown for 3 d in dark on medium (MS, no Suc) with or without 1 µM BA. After exposure to light, chlorophyll content was measured in 3-h intervals. Diagrams shown are the average of at least three independent experiments each with three replicates per time point. One replicate consisted of 30 etiolated seedlings grown on one plate. Error bars represent se (n = 3). Letters indicate significant differences between the wild type and the different genotypes under control condition (a) or after BA treatment (b) or between control and BA treatment for each genotype (c) (P < 0.05). Asterisks above brackets indicate a significant cytokinin effect (*** P < 0.001).
GNC and GNL transcript levels were also investigated in arr single and double mutants (Supplemental Fig. S3), and similar observations as described above for the transcript levels of chlorophyll biosynthesis genes (Fig. 6) were made. In dark-grown single arr mutants, the transcript level of GNL was slightly but not significantly decreased, whereas in the arr10 arr12 and arr1 arr12 double mutants, an increased expression level was observed. In the dark, the arr1 arr12 mutant combination was, in contrast to the two other arr double mutants, not responsive to cytokinin. Importantly, the induction by cytokinin after 1 h of light treatment was dampened in the single and double arr mutants, as indicated by the lack of significant differences compared to the corresponding control conditions (Supplemental Fig. S3). No differences in GNC transcript level were observed in the single and double arr mutants in comparison to wild type in the dark and after 1 h of light treatment.
The greening rate with and without exogenous cytokinin was also investigated in gnc, gnl, and gnc gnl mutants (Fig. 8C). These mutants still responded to cytokinin, indicating that these transcription factors are not instrumental in mediating the response to cytokinin.
DISCUSSION
Results from our study demonstrate that cytokinin acts as a positive regulator on different processes of the etioplast-to-chloroplast transition in Arabidopsis and realizes this action by signaling through the two-component signaling system. Cytokinin accelerates the light-dependent transformation of etioplasts to fully functional chloroplasts and stimulates chlorophyll formation. More specifically, in the dark cytokinin induces the transcripts of chlorophyll biosynthesis genes (e.g. HEMA1, GUN4) and enhances the level of glutamyl-tRNA reductase. Both these deetiolation-related cytokinin responses are strongly reduced in ahk2 ahk3 receptor mutants, indicating a role for AHK2 and AHK3. Three B-type ARRs (ARR1, ARR10, and ARR12) mediate redundantly the transcriptional output, and the binding of ARR12 and ARR10 to the promotors of HEMA1 and LHCB6 indicates direct regulation of processes related to chloroplast development by the cytokinin system. Below we discuss these results in terms of the role of the receptors and transcriptional regulation in regulating the deetiolation response to cytokinin, as well as evolutionary significance of this regulatory mechanism for mediating chloroplast development and function.
Cytokinin Regulates Multiple Processes during Deetiolation through Action of the AHK2 and AHK3 Receptors
Cytokinin promotes the formation of structural components during chloroplast formation in conjunction with regulating chlorophyll biosynthesis. In the dark, cytokinin induces early events of plastid differentiation such as the formation of prethylakoid membranes, which otherwise are strictly dependent on light and suppressed in darkness. Furthermore, in the presence of light it accelerates the development of thylakoid membranes and the disappearance of the prolamellar body, confirming earlier observations in Arabidopsis (Chory et al., 1994) and other species such as cucumber (Masuda et al., 1992), lupine (Kusnetsov et al., 1998), and barley (Yaronskaya et al., 2006). The molecular targets by which cytokinin influences such profound and complex changes are unknown, but one might suspect that they are at least partially realized through the cytokinin-dependent regulation of >100 genes encoding plastid-located proteins other than those involved in chlorophyll biosynthesis (Brenner and Schmülling, 2012). As discussed in the next section, our finding that LHCB6 is a cytokinin primary-response gene is consistent with this hypothesis.
Better known is the more specific action of cytokinin on the regulation of chlorophyll synthesis, of which the first report dates back to the early years after discovery of cytokinin (Sugiura, 1963). We observed that, in the absence of light, cytokinin stimulated the synthesis and accumulation of protochlorophyllide in Arabidopsis, the concentration of which almost doubled following cytokinin treatment compared to untreated controls. The induction of the HEMA1 transcript level (Fig. 4) and the strong increase in GluTR level (Fig. 5) after cytokinin treatment in wild type confirms that this important early step in chlorophyll biosynthesis leading to ALA is positively controlled by cytokinin in Arabidopsis, similar to what is found in cucumber (Masuda et al., 1992, 1994, 1995) and barley (Yaronskaya et al., 2006). The regulation of HEMA1 by cytokinin as well as of additional chlorophyll synthesis genes (GUN4, GUN5, CHLM) is reduced in the ahk2 ahk3 mutant, indicating a predominant role of AHK2 and AHK3 in mediating this cytokinin activity. The involvement of AHK2 and AHK3 is consistent with their activity in leaf mesophyll cells (Stolz et al., 2011) and their cooperative functions in regulating other developmental processes of the leaf and other organs (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006).
Transcriptional Regulation Is an Important Mechanism Mediating the Cytokinin-Induced Greening Response
Downstream of the AHK2 and AHK3 receptors, the transcription factors ARR1, ARR10, and ARR12 act redundantly to mediate cytokinin activity during the deetiolation process. A role of these transcription factors in regulating the chlorophyll level is also reflected by a pale green phenotype of arr1 arr10 arr12 mutants (Argyros et al., 2008). The tested chlorophyll biosynthesis genes still respond to the cytokinin treatment in all single and double arr mutants, but the responsiveness was reduced in the double mutants, indicating a functional role for the corresponding transcription factors. However, in contrast to the ahk2 ahk3 mutants, which showed both a reduced induction of chlorophyll biosynthesis genes and a reduced greening during deetiolation, we observed in the arr single and double mutants only a reduced transcriptional response but no reduced greening. This could be explained by the high functional redundancy of B-type ARRs and the fact that the lowered gene induction is still sufficient to allow for a full physiological response.
Previously it was not known whether cytokinin-regulated nuclear-encoded plastid-related genes are a direct target of B-type ARRs or whether they are regulated by transcription factors acting further downstream of the cytokinin signaling system. The result of the ChIP analysis revealed that at least HEMA1 and LHCB6 are under direct control of B-type ARRs, since both the ARR12 and ARR10 transcription factors interact with their promotors near the TSS. Thus, the cytokinin system directly regulates genes of the chlorophyll biosynthesis pathway as well as genes encoding structural components of the light harvesting complex. The extent of direct regulation of other chloroplast-related genes remains to be determined and is difficult to evaluate bioinformatically, since the core cis-acting sequence sufficient for B-type ARR binding is too short to be diagnostic for all cytokinin-regulated genes (Sakai et al., 2001; Brenner and Schmülling, 2015).
Our study also sheds light on the GNL/GNC family of transcription factors involved in chloroplast development (Richter et al., 2010; Köllmer et al., 2011; Hudson et al., 2011; Chiang et al., 2012). We confirm a role for cytokinin and the B-type response regulators in their induction (Chiang et al., 2012; Behringer and Schwechheimer, 2015) and extend on these earlier studies through our use of single and higher order mutant combinations. We find that ARR1, ARR12, and ARR10 each play a role in the induction of GNL, their relative contribution being in the order given. Interestingly, the higher order mutant combinations involving these family members do not accentuate the effects of the single mutants on GNL expression. This regulatory phenomenon may involve homeostatic mechanisms that control abundance of GNL and GNC, loss-of-function mutations in one family member having been found to result in increased expression of the other family member (Richter et al., 2010). We find that both single and double gnc gnl mutants still respond strongly to cytokinin during deetiolation, indicating that GNC and GNL are not obligatory players in mediating cytokinin action in this context, unless a compensatory mechanism masks their relevance in the respective loss-of-function mutants. This contrasts with a previously uncovered role for GNC and GNL in modulating the ability of cytokinin to induce chloroplast division and an increased greening rate in plants overexpressing members of the GNC family. It was shown that these transcription factors enhance the conversion of proplastids to etioplasts in the dark (Chiang et al., 2012). Additional studies including related members of the GATA transcription factor family are required to clarify their role in chlorophyll biosynthesis and the greening response. Recently, Ranftl et al. (2016) showed that other members of the B-GATA family are required for the full chlorophyll content in 7-d-old light-grown seedlings. However, whether these genes also have a function in the greening response during deetiolation has not yet been analyzed.
Cytokinin Is an Evolutionary Conserved Modulator of Chloroplast Development and Function
The arr1 arr10 arr12 mutant, just like the cre1 ahk2 ahk3 triple receptor mutant, lacks a response to cytokinin but still forms green leaves, indicating that chloroplasts and chlorophyll are formed independent of cytokinin (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006; Argyros et al., 2008; Ishida et al., 2008). However, the lower concentrations of chlorophyll in the mutant leaves suggest that although cytokinin is not absolutely required for chlorophyll biosynthesis per se, it is probably needed to obtain physiologically optimal concentrations. It could be that cytokinin acts as a modulator of chlorophyll content, mediating presumably information obtained from environmental cues and functions redundantly with other systems impinging on this all-important plant energy capturing system.
The existence of other systems acting redundantly with cytokinin is supported by the surprising finding that complete loss of all three B-type ARRs (ARR1, ARR10, ARR12) resulted in a strong up-regulation of the transcript levels of chlorophyll biosynthesis genes and a strongly increased greening rate under control conditions in comparison to wild type, although the cytokinin-regulated chloroplast genes were nonresponsive to the hormone in this mutant. This stronger greening response appears to be contradictory to the pale green phenotype of 2-week-old arr1 arr10 arr12 light-grown mutant seedlings (Argyros et al., 2008). However, this phenotype could be the consequence of oxidative stress arising from growth in continuous light in combination with hampered chloroplast functioning due to the lowered cytokinin status. It thus appears that lack of a functional cytokinin signal transduction pathway causes the activation of an as yet unknown compensatory mechanism to overcome this loss and results eventually in overcompensation visible as increased greening response already without additionally applied cytokinin.
To the best of our knowledge, a similar compensatory mechanism acting during deetiolation has not been previously described. Nor have such effects on gene expression been previously reported when examining loss-of-function mutants of the B-type ARRs. The compensation would require that a different system, likely to operate otherwise redundantly with cytokinin to regulate chloroplast development and function, is able to sense the loss of the cytokinin system and respond accordingly. The altered response in the different B-type ARR mutant backgrounds suggests the presence of an incoherent feed-forward loop, in which the B-type ARRs control expression of the chlorophyll biosynthetic genes as well as of a repressor for those genes (i.e., an incoherent feed-forward loop). Under conditions of strong cytokinin insensitivity, such as in the arr1 arr10 arr12 mutant, the repressor is not induced and the biosynthetic genes express at higher levels due to the action of other transcription factors not directly involved in the cytokinin response. Such incoherent feed-forward loops and compensatory mechanisms for gene expression play important roles in regulating photosynthesis in the more tractable bacterial systems (Mank et al., 2013; Imam et al., 2014). Our results suggest that they may play similarly important roles in photosynthesis and mediating the effects of cytokinin plants.
Although the cytokinin system can apparently be replaced, at least in part, by another system to ensure the formation of chloroplasts and chlorophyll, cytokinin must have specific physiologically relevant functions in regulating chloroplast development and greening. This is strongly suggested by similar, evolutionary conserved responses in distantly related plant species, including monocots and dicots (Cortleven and Schmülling, 2015). This indicates that this function has been established early, before the separation of mono- and dicots, and has been ever since under selective constraint. Cytokinin functions related to chloroplasts could be more subtle than those observed during the deetiolation response. For example, Boonman et al. (2007, 2009) proposed that cytokinin is involved in the regulation of whole-plant photosynthetic acclimation to light gradients in canopies. Shaded leaves were shown to import less cytokinin than leaves exposed to light, and exogenous application of cytokinin or increased endogenous cytokinin levels rescued the effect of shading. Mutant analysis revealed that cytokinin is not absolutely required for the regulation of photosynthetic capacity, but rather acts as one of multiple, redundant operating mechanisms, as is probably also the case during deetiolation (Boonman et al., 2009). A recent survey on the role of cytokinin in photosynthesis concluded that the main function is the optimization of photosynthetic activity (Cortleven and Schmülling, 2015), which is achieved at least in part through enhanced protection against photodamage (Procházková et al., 2008; Cortleven et al., 2014).
CONCLUSION
A fully functional cytokinin signaling pathway is necessary for a normal greening response during deetiolation, and AHK2 and AHK3 are key mediators of cytokinin action in this process. We showed that direct transcriptional regulation by B-type ARRs of at least some genes relevant for chlorophyll biosynthesis and chloroplast function is likely to be part of the molecular mechanism mediating cytokinin action downstream of these receptors. A completely lacking cytokinin signaling pathway results in an as yet unknown compensatory mechanism causing a strong increase in greening rate and activation of chlorophyll biosynthesis genes. Importantly, evolutionary conservation of the functions of cytokinin in regulating chloroplast development and functions underpin physiologically important functions that need further investigations.
MATERIALS AND METHODS
Plant Material, Growth, and Treatment Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was used as the wild type. The cytokinin receptor mutants (ahk2-5, ahk3-7, cre1-2, ahk2-5 ahk3-7, cre1-2 ahk2-5, and cre1-2 ahk3-7; Riefler et al., 2006), B-type arr mutants (arr1-4, arr10-5, arr12-1, arr1-3 arr10-5, arr1-3 arr12-1, arr10-5 arr12-1, and arr1-3 arr10-5 arr12-1; Mason et al., 2005; Ishida et al., 2008), and GATA mutants (gnc, gnl, and gnc gnl; Chiang et al., 2012) were obtained from the Nottingham Arabidopsis Stock Centre. Seeds were germinated in the dark on full Murashige and Skoog (MS) medium without Suc at 22°C, and seedlings were transferred 3 d after stratification to light in a growth cabinet (Percival AR66L; Percival Scientific) at 22°C and a light intensity of 100 µmol m−2 s−1.
Pigment Analysis
Chlorophyll content was determined according to Porra et al. (1989) using methanol as solvent. Determination of chlorophyll intermediates by HPLC was done according to the method of Thayer and Björkman (1990). Pigment content was expressing per seedling or per fresh weight, which yielded similar results.
Transmission Electron Microscopy
Three-day-old seedlings were fixated for 3 d at 4°C using vacuum infiltration in 2% (v/v) paraformaldehyde, 2% (v/v) glutaraldehyde buffered in 50 mm cacodylate buffer with 50 mm NaCl. Samples were washed with 50 mm cacodylate buffer containing 50 mm NaCl and with 50 mm glycylglycin buffer containing 100 mm NaCl. Postfixation was performed in 1% (w/v) osmium tetroxide, buffered in 50 mm cacodylate buffer containing 50 mm NaCl for 3 h. After washing with distilled water, leaf tissues were incubated for 1 hour in 0.1% (w/v) tannic acid in 100 mm HEPES buffer, rinsed with water, and incubated overnight at 4°C in water. After staining in 2% (w/v) uranylacetate for 1.5 h, fixed tissues were dehydrated and embedded in Spurr’s epoxy resin. Ultra-thin sections (65 nm) were obtained using a Leica Ultracut UCT ultramicrotome and mounted on 0.7% (w/v) formvar-coated copper grids 200 mesh. The sections were contrasted with uranyl acetate (2% [w/v] in 50% ethanol) followed by lead citrate (4% [w/v] solution) and examined in a FEI Tecnai Spirit transmission electron microscope operated at 120 kV (Cortleven et al., 2014).
Protein Isolation and Protein Gel-Blot Analysis
Three-d-old seedlings were frozen, and the material was homogenized with a Retsch Mixer Mill MM2000 with two stainless steel beads (2-mm diameter). Total proteins were extracted from the homogenized leaf material in a TBS-T buffer containing 150 mm NaCl, 100 mm Tris, and 1% Triton X-100 (pH 7). Protein concentration was determined according to Bradford (1976). Proteins (4 µg) were separated by SDS-PAGE (10%) and blotted to a polyvinylidene fluoride membrane overnight at 4°C. The blots were blocked for 1.5 to 2 h in 1% skim milk (Sigma-Aldrich) in TBS-T buffer at room temperature. Afterward, the blots were incubated overnight at 4°C with the primary antibody (polyclonal GluTR-specific; kindly provided by Dr. Boris Hedtke, HU Berlin) or for 1 h (actin, ACT, polyclonal rabbit antiserum; Agisera) followed by the secondary antibody (bovine antirabbit IgG peroxidase conjugate; Calbiochem). The primary antibody was diluted 1:1,000 (GluTR) or 1:2,500 (ACT) in the blocking solution, and the secondary antibody was diluted 1:6,000 in 1% skim milk (in TBS-T). After each antibody treatment, blots were washed three times for 5 min in TBS-T. Afterward the GluTR and ACT protein was immuno-detected using the SuperSignal West Pico Chemiluminescent Substrate Kit (Fisher Scientific) according to the manufacturer’s instructions. Quantification of independent experiments (n = 5) was performed with ImageJ 1.46r (National Institutes of Health).
Analysis of Transcript Levels by qRT-PCR
Total RNA was extracted from entire seedlings using the NucleoSpin RNA plant kit (Machery and Nagel) as described in the user’s manual. The purified RNA concentration was determined spectrophotometrically at 260 nm using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies). The RNA purity was evaluated by means of the 260/280 ratio. After a second DNAse step (Fermentas, Life Technologies), equal amounts of starting material (1 µg RNA) were used in a 20-µL SuperScript III Reverse Transcriptase reaction. First-strand cDNA synthesis was primed with a combination of oligo(dT)-primers and random hexamers. Primer pairs were designed, using Primer 3 Software (http://www.genome.wi.mit.edu/cgibin/primer/primer3.cgi) under the following conditions: optimum Tm at 60°C, GC content between 20% and 80%, 150-bp maximum length. Primers used for reference genes and genes of interest are listed in Supplemental Table S1. Real-time PCR using FAST SYBR Green I technology was performed on an ABI PRISM 7500 sequence detection system (Applied Biosystems) and the following cycling conditions (15 min 95°C, 40 cycles of 15 s at 95°C, and 15 s at 55°C and 10s at 72°C), followed by the generation of a dissociation curve to check for specificity of the amplification. Reactions contained SYBR Green Master Mix (Applied Biosystems, Life Technologies), 300 nm of a gene specific forward and reverse primer and 2 µL of a 1:10 diluted cDNA in a 20-µL reaction. Gene expression data were normalized against two or three different nuclear-encoded reference genes (ACT2, UBC21, and/or TAFII15) according to Vandesompele et al. (2002) and presented relative to the control treatment.
ChIP Analysis
ChIP-qPCR was performed with the indicated numbers of biological replicates with two independent lines of 35S:Myc-ARR12 expressed in wild-type background and two independent lines of 35S:ARR10-GFP expressed in the arr1 arr10 arr12 background. The ARR12 construct was generated by cloning a genomic DNA fragment of ARR12 into the vector pEarlyGate 203 (Early et al., 2006), using primers ATGACTGTTGAACAAAATTTAGAA and TATGCATGTTCTGAGTGAACT for initial amplification of ARR12. The ARR10 construct has been described (Shanks et al., 2016). Seedlings were grown on vertical plates with 1× MS media with vitamins and 1% (w/v) Suc for 3 weeks under 50-µE light intensity, and then treated with 5 µM BA in liquid MS media for 30 min. Tissue cross-linking, chromatin isolation, and immunoprecipitation were performed as described (Zhang et al., 2013). Immunoprecipitation was performed using anti-Myc (ab9132, Abcam) or anti-GFP (ab290, Abcam) antibodies and MagnaChIP protein A+G Magnetic beads (EMD Millipore). Input and ChIP DNA were examined by qRT-PCR using the primers listed in Supplemental Table S1. Promoter (5′) and 3′ regions for HEMA1, LHCB6, and LHCB1.2 were amplified as indicated; the TUB2 promoter region was used as a negative control and the ARR15 promoter region as a positive control.
Statistical Analysis
Statistical analyses were performed using SAS v.9.2 (SAS Institute). Greening rate data (Figs. 2 and 8C; Supplemental Fig. S1) were first analyzed by 2-way ANOVA to define a significant cytokinin or genotype effect. Afterward, all datasets were analyzed for each time point by 1-way ANOVA, followed by Tukey’s post hoc test. Pairwise comparisons were made between wild type and the different genotypes in control conditions (labeled a), between wild type and the different genotypes after cytokinin treatment (b), and between control and cytokinin treatment for each genotype (c). Normality and homogeneity of variance were tested using the Shapiro-Wilk and Levene tests (Neter et al., 1996). To meet the assumptions, datasets were transformed using log or square root transformation. When assumptions were not met, a nonparametric Kruskall-Wallis test was performed followed by a Mann-Whitney test to perform a pairwise comparison (e.g. Fig. 3).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Greening rate in mutants of the cytokinin signaling pathway.
Supplemental Figure S2. Effect of cytokinin on transcript level of genes encoding key steps in the chlorophyll biosynthesis pathway and of LHCB in arr double mutants.
Supplemental Figure S3. Interaction between cytokinin and the GATA transcription factor GNL during the greening response.
Supplemental Figure S4. Effect of cytokinin on transcript level of genes encoding key steps in the chlorophyll biosynthesis pathway and of ARR5 and LHCB1 in the arr1 arr10 arr12 mutant during dark-to-light transition.
Supplemental Table S1. Sequences of primers used in this study.
Supplementary Material
Acknowledgments
We thank Thorsten Mielke and Beatrix Fauler (MPI for Molecular Genetics, Berlin) for technical support with the electron microscopy and Boris Hedtke (Humboldt University, Berlin) for kindly providing the GluTR antibody.
Glossary
- ALA
5-aminolevulinic acid
- ChIP
chromatin immunoprecipitation
- Mg-proto
Mg-protoporphyrin IX
- MS
Murashige and Skoog
- TSS
transcriptional start site
References
- Argyros RD, Mathews DE, Chiang Y-H, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong GA, Runge S, Frick G, Sperling U, Apel K (1995) Identification of NADPH:protochlorophyllide oxidoreductases A and B: a branched pathway for light-dependent chlorophyll biosynthesis in Arabidopsis thaliana. Plant Physiol 108: 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer C, Schwechheimer C (2015) B-GATA transcription factors - insights into their structure, regulation, and role in plant development. Front Plant Sci 6: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava A, Clabaugh I, To JP, Maxwell BB, Chiang Y-H, Schaller GE, Loraine A, Kieber JJ (2013) Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiol 162: 272–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y-M, Zhang Y, Signorelli T, Zhao R, Zhu T, Rothstein S (2005) Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J 44: 680–692 [DOI] [PubMed] [Google Scholar]
- Boonman A, Prinsen E, Gilmer F, Schurr U, Peeters AJ, Voesenek LA, Pons TL (2007) Cytokinin import rate as a signal for photosynthetic acclimation to canopy light gradients. Plant Physiol 143: 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonman A, Prinsen E, Voesenek LA, Pons TL (2009) Redundant roles of photoreceptors and cytokinins in regulating photosynthetic acclimation to canopy density. J Exp Bot 60: 1179–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T (2005) Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44: 314–333 [DOI] [PubMed] [Google Scholar]
- Brenner WG, Schmülling T (2012) Transcript profiling of cytokinin action in Arabidopsis roots and shoots discovers largely similar but also organ-specific responses. BMC Plant Biol 12: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Schmülling T (2015) Summarizing and exploring data of a decade of cytokinin-related transcriptomics. Front Plant Sci 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang Y-H, Zubo YO, Tapken W, Kim HJ, Lavanway AM, Howard L, Pilon M, Kieber JJ, Schaller GE (2012) Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol 160: 332–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Reinecke D, Sim S, Washburn T, Brenner M (1994) A role for cytokinins in de-etiolation in Arabidopsis (det mutants have an altered response to cytokinins). Plant Physiol 104: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortleven A, Nitschke S, Klaumünzer M, Abdelgawad H, Asard H, Grimm B, Riefler M, Schmülling T (2014) A novel protective function for cytokinin in the light stress response is mediated by the Arabidopsis histidine kinase2 and Arabidopsis histidine kinase3 receptors. Plant Physiol 164: 1470–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortleven A, Schmülling T (2015) Regulation of chloroplast development and function by cytokinin. J Exp Bot 66: 4999–5013 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R (2014) DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA 111: 2367–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A, Riefler M, Romanov GA, Schmülling T (2012) Properties, functions and evolution of cytokinin receptors. Eur J Cell Biol 91: 246–256 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al. (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda K, Imamura A, Katoh E, Hatta T, Tachiki M, Yamada H, Mizuno T, Yamazaki T (2002) Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14: 2015–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson D, Guevara D, Yaish MW, Hannam C, Long N, Clarke JD, Bi Y-M, Rothstein SJ (2011) GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis. PLoS One 6: e26765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, et al. (2006) The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18: 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam S, Noguera DR, Donohue TJ (2014) Global analysis of photosynthesis transcriptional regulatory networks. PLoS Genet 10: e1004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Yokoyama A, Mizuno T (2008) Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol 49: 47–57 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE (2014) Cytokinins. Arabidopsis Book 12: e0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllmer I, Werner T, Schmülling T (2011) Ectopic expression of different cytokinin-regulated transcription factor genes of Arabidopsis thaliana alters plant growth and development. J Plant Physiol 168: 1320–1327 [DOI] [PubMed] [Google Scholar]
- Kusnetsov V, Herrmann RG, Kulaeva ON, Oelmüller R (1998) Cytokinin stimulates and abscisic acid inhibits greening of etiolated Lupinus luteus cotyledons by affecting the expression of the light-sensitive protochlorophyllide oxidoreductase. Mol Gen Genet 259: 21–28 [DOI] [PubMed] [Google Scholar]
- Lopez-Juez E, Pyke KA (2005) Plastids unleashed: their development and their integration in plant development. Int J Dev Biol 49: 557–577 [DOI] [PubMed] [Google Scholar]
- Mank NN, Berghoff BA, Klug G (2013) A mixed incoherent feed-forward loop contributes to the regulation of bacterial photosynthesis genes. RNA Biol 10: 347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Komine Y, Inokuchi H, Kannangara CG, Tsuji H (1992) Sequence and expression of tRNA glu gene of cucumber chloroplast genome. Plant Physiol Biochem 30: 235–243 [Google Scholar]
- Masuda T, Ohta H, Shioi Y, Tsuji H, Takamiya K (1995) Stimulation of glutamyl-transfer RNA reductase activity by benzyladenine in greening cucumber cotyledons. Plant Cell Physiol 36: 1237–1243 [Google Scholar]
- Masuda T, Tanaka R, Shioi Y, Takamiya K, Kannangara CG, Tsuji H (1994) Mechanism of benzyladenine-induced stimulation of the synthesis of 5-aminolevulinic acid in greening cucumber cotyledons: benzyladenine increases levels of plastid tRNA-glu. Plant Cell Physiol 35: 183–188 [Google Scholar]
- McCormac AC, Terry MJ (2002) Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J 32: 549–559 [DOI] [PubMed] [Google Scholar]
- Miller CO, Skoog F, Okomura FS, von Salta MH, Strong FM (1956) Isolation, structure and synthesis of kinetin, a substance promoting cell division. J Am Chem Soc 78: 1345–1956 [Google Scholar]
- Naito T, Kiba T, Koizumi N, Yamashino T, Mizuno T (2007) Characterization of a unique GATA family gene that responds to both light and cytokinin in Arabidopsis thaliana. Biosci Biotechnol Biochem 71: 1557–1560 [DOI] [PubMed] [Google Scholar]
- Neter J, Kutner MH, Nachtsheim CJ, Wasserman W (1996) Applied Linear Statistic Models. McGraw-Hill, New York [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthier B. (1979) Role of phytohormones cytokinins in chloroplast development. Biochem Physiol Pflanz 174: 173–214 [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. BBA Bioenerg 975: 384–394 [Google Scholar]
- Procházková D, Haisel D, Wilhelmová N (2008) Antioxidant protection during ageing and senescence in chloroplasts of tobacco with modulated life span. Cell Biochem Funct 26: 582–590 [DOI] [PubMed] [Google Scholar]
- Ranftl QL, Bastakis E, Klermund C, Schwechheimer C (2016) LLM-domain containing B-GATA factors control different aspects cytokinin-regulated development in Arabidopsis thaliana. Plant Physiol 170: 2295–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Müller IK, Schwechheimer C (2010) The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev 24: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Shanks CM, Rice JH, Zubo Y, Schaller GE, Hewezi T, Kieber JJ (2016) The role of cytokinin during infection of Arabidopsis thaliana by the cyst nematode Heterodera schachtii. Mol Plant Microbe Interact 29: 57–68 [DOI] [PubMed] [Google Scholar]
- Solymosi K, Schoefs B (2010) Etioplast and etio-chloroplast formation under natural conditions: the dark side of chlorophyll biosynthesis in angiosperms. Photosynth Res 105: 143–166 [DOI] [PubMed] [Google Scholar]
- Stolz A, Riefler M, Lomin SN, Achazi K, Romanov GA, Schmülling T (2011) The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. Plant J 67: 157–168 [DOI] [PubMed] [Google Scholar]
- Sugiura M. (1963) Promotion of chlorophyll synthesis by kinetin. Botanical Magazin Tokyo 76: 309–310 [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T (2001) The Arabidopsis sensor His-kinase, AHk4, can respond to cytokinins. Plant Cell Physiol 42: 107–113 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Tanaka A (2007) Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol 58: 321–346 [DOI] [PubMed] [Google Scholar]
- Thayer SS, Björkman O (1990) Leaf Xanthophyll content and composition in sun and shade determined by HPLC. Photosynth Res 23: 331–343 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: 0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim A, Deng XW (1996) A role for transcriptional repression during light control of plant development. BioEssays 18: 905–910 [DOI] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. (2014) Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Schmülling T (2009) Cytokinin action in plant development. Curr Opin Plant Biol 12: 527–538 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaronskaya E, Vershilovskaya I, Poers Y, Alawady AE, Averina N, Grimm B (2006) Cytokinin effects on tetrapyrrole biosynthesis and photosynthetic activity in barley seedlings. Planta 224: 700–709 [DOI] [PubMed] [Google Scholar]
- Zhang W, Swarup R, Bennett M, Schaller GE, Kieber JJ (2013) Cytokinin induces cell division in the quiescent center of the Arabidopsis root apical meristem. Curr Biol 23: 1979–1989 [DOI] [PubMed] [Google Scholar]
- Zubo YO, Yamburenko MV, Kravtsov AK, Kulaeva ON, Kusnetsov VV (2009) Detached barley leaves as a model for studying cytokinin control of plastid gene regulation. Russ J Plant Physiol 56: 551–559 [Google Scholar]
- Zubo YO, Yamburenko MV, Selivankina SY, Shakirova FM, Avalbaev AM, Kudryakova NV, Zubkova NK, Liere K, Kulaeva ON, Kusnetsov VV, et al. (2008) Cytokinin stimulates chloroplast transcription in detached barley leaves. Plant Physiol 148: 1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.